Abstract
Objectives
A novel beta coronavirus has been identified as responsible for the 2019 coronavirus infection (Covid-19). Clinical presentations range from asymptomatic cases to acute respiratory distress syndrome with fatal outcome. Such a broad spectrum of disease expression calls for an investigation of immune response characteristics.
Methods
We identified subjects admitted for Covid-19 in whom a large panel of immunological markers were measured, including B- and T- and NK-lymphocyte phenotypes, T-lymphocyte subpopulation cells and plasma cytokines. Patients were divided according to symptom severity during hospitalisation, in those with uncomplicated and complicated infection. Differences between groups were analyzed.
Results
Seventeen patients were included (mean age: 83 years; 9 women; mean delay of symptoms onset: 4 days). Six had uncomplicated infection, while 11 developed complicated forms during hospitalization. CD10 + B lymphocyte levels were inversely correlated with clinical severity (5.8% vs 2.0%, p = 0.04) and CD10+ levels above 3% were independently associated with uncomplicated forms [Odds Ratio 0.04 (CI 0.002-0.795, p = 0.034)]. TNF-alpha, IL-1, Il-6 and Il-8 measurements upon admission differed between patients who died and those who survived (p < 0.01 for all comparisons).
Conclusions
In a population of elderly patients recently infected with Covid-19, CD10 + B cell levels were inversely correlated with clinical severity. Cytokine values upon admission were highly predictive of fatal outcome during hospitalisation. These findings could explain differences in the clinical presentation and allow rapid identification of patients at risk for complications.
Keywords: Covid-19, Immune response, Clinical severity
Introduction
A cluster of pneumonia cases of unknown origin occurred in Wuhan Hubei Province, China, in December 2019 (Huang et al., 2020). The pathogen was then identified as a novel beta coronavirus now named severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) and responsible for the so-called 2019 novel coronavirus disease (Covid-19) (Gorbalenya et al., 2020). Human-to-human transmission of Covid-19 has been established and the virus has rapidly spread worldwide (Wang et al., 2020, Chang et al., 2020, Carlos et al., 2020).
The typical clinical presentation described was initially that of an acute respiratory syndrome, but several other clinical signs have since been reported, such as diarrhea, anosmia, confusion and vasculitis (Pinzon et al., 2020, D’Amico et al., 2020, Wai-Hin Chung et al., 2020, Meng et al., 2020, Buja et al., 2020, Boukhris et al., 2020, Román et al., 2020, Ng Kee Kwong et al., 2020).
However, clinical expression of Covid-19 is extremely variable, with, in some cases, asymptomatic or mild forms (Nikolai et al., 2020), while in other subjects severe forms culminating in acute respiratory distress syndrome (ARDS) have been described, requiring patients to be transferred to Intensive Care Units. Consequently, very high mortality rates have been reported worldwide (Alhazzani et al., 2020, Shang et al., 2020).
Among clinical predictive factors for severe forms of Covid-19, older age, male gender and obesity are the most described (Chen et al., 2020), while correlations between abnormal coagulation and hematological parameters with poor prognosis have been observed (Sun et al., 2020, Velavan and Meyer, 2020).
Immunity is probably one of the major determinants of clinical outcome, as ARDS has been associated with a so-called cytokine storm, characterized by an exaggerated and uncontrolled inflammatory response to the virus, potentially resulting in lung injury (Qin et al., 2020). This immune response to Covid-19 calls for in-depth investigation. Indeed, lymphocytes subsets and immune response during Covid-19 have only been partially studied. Qin et al. found lower levels of helper T cells and suppressor T cells in severe patients, but demographic characteristics of severe and non-severe patients were significantly different (Qin et al., 2020). Moreover, while the induction of a robust cellular immunity is likely essential for efficient virus control, dysregulated T cell and NK cell response may contribute to disease severity (Vabret et al., 2020). Therefore, it is urgent to better elucidate how different immune responses across patients affect the clinical presentation.
The aim of this study was to retrospectively analyze the immunological profile of patients at early stages of Covid-19 in order to identify potential risk factors for subsequent clinical deterioration.
Methods
Study design and participants
We conducted an observational, retrospective cohort study on patients admitted to the Internal Medicine and Infectious Diseases Department in Cannes General Hospital, from March to May 2020, with confirmed Covid-19 infection. Inclusion criteria were positive SARS-CoV-2 RT-PCR detected in nasal swabs and immunological investigation of blood samples, generally performed as a consequence of lymphopenia, upon admission. The immunological panel included B- and T- and NK-lymphocyte phenotyping, T-lymphocyte subpopulation cell counts and plasma cytokine measurements.
All patients enrolled in this study were diagnosed and treated according to national guidelines for Covid-19. Blood samples were collected in accordance with the clinical guidelines for inpatients with Covid-19. The study was submitted to the French National Institute for Health Data (Institut National de Données de Santé, reference CNIL MR004) and patients gave informed consent for retrospectively collecting data.
Procedures
Demographic characteristics, underlying comorbidities, duration of symptoms, clinical signs prior to admission, upon admission and during hospitalisation, laboratory findings during hospital stay and clinical outcome were collected from patients’ medical records.
Among laboratory findings, we focused particularly on immunological markers, which were mainly prescribed as a consequence of initial lymphopenia by the attending physicians and which included plasma cytokines (IL-1, Il-6, Il-8 and TNF-alpha), T- and B-lymphocytes and their sub-populations. Physicians who had been directly involved in patients’ management also actively took part in this retrospective analysis.
In line with WHO criteria for Covid-19 clinical severity (World Health Organization), patients were divided into two groups according to the clinical outcome during hospitalisation:
-
-
Subjects with either uncomplicated illness, defined either by a complete lack of symptoms and/or by the presence of symptoms such as fatigue or upper respiratory tract infection, with no clinical signs of pneumonia, hypoxia or clinical worsening during the course of the infection.
-
-
Subjects with complicated illness, in case patients experienced severe forms, either already at admission, or during hospitalization. Severity was defined by signs of pneumonia associated with at least one of the following additional criteria: respiratory rate >30 breaths/minute, severe respiratory distress, or spO2 < 90% on ambient air.
In case of patient transfer to another clinical setting, files were reviewed and physicians interviewed in order to confirm that no significant changes in clinical presentation had been observed for each patient.
The main reason for admitting uncomplicated Covid-19-positive patients was the identification of virus outbreaks in several local nursing homes, which required systematic testing of residents and their admission to our Department in order to reduce risks of spread in such high risk settings. In case of completely asymptomatic patients, duration of symptoms was calculated starting from the first RT-PCR positive nasal swab.
Multicolour Flow Cytometry (MFC) analysis of circulating T and B cells and their sub-populations
Flow cytometric analysis was conducted on blood collected on EDTA and labelled within 48 h. Briefly, immunophenotypical analysis was performed using 8-color flow cytometry (FACSCanto II, BD Biosciences, San Jose, CA). The following antibodies were used in this study: fluorescein isothiocyanate (FITC)-conjugated –CD27, -CD38, -CD8, -anti-kappa; phycoerythrin (PE)-conjugated -CD56, -CD25, -anti-Lambda, -anti-IgD; Peridinin chlorophyll protein-Cy5.5 (PerCP-Cy5.5)-conjugated –CD8, -CD24, -CD4, -CD5, PE-Cy7-conjugated -CD127; allophycocyanin (APC)-conjugated -CD45RA, -HLA-DR, -CD10, -anti-IgM; APC-H7-conjugated-CD3, -CD14; V450-conjugated-CD197, -CD4, -CD38; V500-conjugated-CD45; BV421-conjugated –CD8, all purchased from BD Biosciences. PE-Cy7-conjugated -CD19 was obtained from Beckman-Coulter.
Instrument set-up was performed according to the FranceFlow Standard Operating Procedures (Solly et al., 2019). Lymphocytes were identified on the basis of SSC/FSC properties and CD45 expression, and their sub-populations of naïve, memory, effector and effector memory lymphocytes according to expression of CD45RA and CD197 (Maecker et al., 2012). The Treg gating strategy was based on the gating of CD3+CD4+ and CD3+CD8+ lymphocytes, the CD127wkCD25+ gate being set on CD3+CD4 + T cells using the CD3+CD8+ T cells as a negative control (Liu et al., 2006). In addition to expression of CD10, CD20 kappa and lambda on B cells, at least 6 different B cell populations were identified in each sample, namely naive B cells, switched memory B cells, marginal zone-like memory B cells and CD27-negative memory B cells, transitional B cells and plasmablasts.
Plasma cytokines analysis
In plasma, cytokine measurement included Il-6, Il-1, Il-8 and TNF-alpha, measured with custom-designed cartridges Ella (ProteinSimple), following the manufacturers’ instructions.
Statistical analysis
Categorical variables were described as frequency rates and percentages, while continuous variables were detailed with mean, median and inter-quantile range (IQR).
Patients with asymptomatic or mild forms were compared to those with moderate-to-severe forms.
χ2-tests and Student's t-tests were performed to compare variables, and independent risk factors were identified by multivariate analysis.
Variables with P ≤ 0.20 in univariate analysis were initially selected for the multivariate model and only those with P ≤ 0.05 were retained in the final model. All analyses were performed using Statview© software.
Results
Study population
From February to May 2020, 101 patients were admitted to our Department for Covid-19.
Among them, 17 underwent immunological analysis upon admission and were included in this study (mean age 83 years, 9 women, mean time since symptoms onset 4 days, mean length of hospitalisation 10 days). Demographic characteristics of patients studied were quite similar to the total population admitted in the Department for Covid-19, with a slight older age and higher rate of comorbid conditions (data not shown). Among included patients, 16 out of 17 had at least one comorbid condition, i.e. hypertension (10/17, 59%), diabetes (6/17, 35%) and cognitive disorders (5/17, 29%). All patients had positive SARS-CoV-2 RT-PCR in nasal swabs at the admission.
According to their clinical severity at the admission, 15 out of 17 subjects were either asymptomatic or had mild symptoms, while just 2 patients already had criteria for severity.
Patients’ outcomes and risk factors associated with complicated illness
Six patients had uncomplicated illness during the hospitalization and completely recovered. The other 11 patients experienced a complicated Covid-19; 2 of them already had signs of severity at the admission and the other 9 developed clinical worsening during the hospitalization.
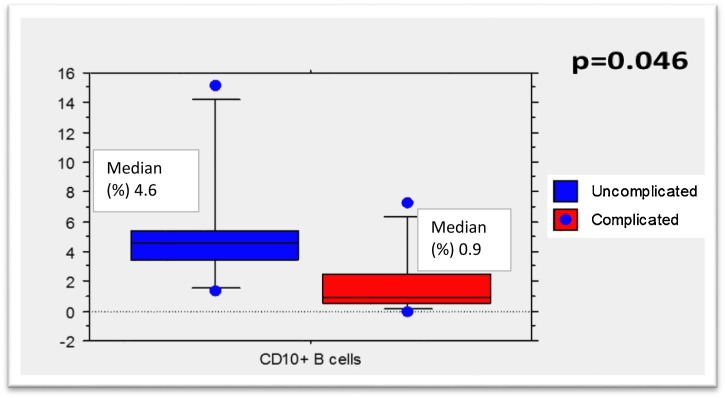
Patients with uncomplicated illness had higher levels of B CD19+CD10+ lymphocytes upon admission than patients with complicated infection, and multivariate analysis confirmed that levels above 3% were associated with uncomplicated illness (Table 1 , Figure 1 ). Moreover, as the presence of CD10+ cells has been associated with lymphopenia in HIV-infected patients (Ho et al., 2006), we measured these and found no correlation between lymphocyte count and CD10+ levels (data not shown). Even excluding the 2 patients who already had clinical signs of severity at admission, levels of B CD19+CD10+ levels were higher in patients with uncomplicated illness (data not shown). Neither lymphopenia at the admission nor during the hospitalization was associated with clinical severity or fatal outcome (data not shown).
Table 1.
Risk factors associated with complicated forms (N = 17).
| Clinical presentation |
||||||||
|---|---|---|---|---|---|---|---|---|
| Uncomplicated (n = 6) |
Complicated (n = 11) |
|||||||
| mean | [SD] | mean | [SD] | p-value | ORAdj | [IC 95%] | p | |
| Age | 84.8 | [16] | 82.7 | [10.7] | 0.748 | |||
| Symptoms onset (days) | 4.2 | [5.51] | 4.4 | [3.72] | 0.930 | |||
| Lymphocyte T CD4+ (%) | 44.9 | [14.21] | 38.4 | [16.09] | 0.419 | |||
| L. T CD8+ (%) | 22.2 | [15.15] | 20.1 | [12.5] | 0.767 | |||
| L. T CD4+CD8+ (%) | 3.4 | [2.87] | 2.1 | [1.14] | 0.197 | |||
| L. CD4-CD8- (%) | 1.4 | [1.35] | 3.6 | [2.93] | 0.108 | |||
| L. NK (%) | 23.0 | [13.36] | 26.97 | [17.73] | 0.641 | |||
| L. T CD56+ (%) | 3.2 | [1.63] | 7.8 | [5.78] | 0.314 | |||
| L. T CD5+ (%) | 14.0 | [11.77] | 5.4 | [4.74] | 0.786 | |||
| L. T CD4CD38HLADR (%) | 3.0 | [1.97] | 5.4 | [8.67] | 0.506 | |||
| L. T CD8CD38HLADR (%) | 13.2 | [13.14] | 13.3 | [15.51] | 0.993 | |||
| L. T regulators (%) | 3.0 | [1.15] | 2.6 | [0.80] | 0.465 | |||
| L. B CD19+ kappa (%) | 61.7 | [3.21] | 58.0 | [7.20] | 0.300 | |||
| L. B CD19+ lambda (%) | 37.7 | [3.54] | 41.7 | [7.14] | 0.266 | |||
| L. B CD19+ CD20+ (%) | 97.3 | [2.82] | 97.5 | [4.26] | 0.922 | |||
| L. B CD19+ CD10+ (%) | 5.8 | [4.85] | 2.0 | [2.34] | 0.046 | |||
| L. T CD4 naive (%) | 34.6 | [17.07] | 28.3 | [14.75] | 0.439 | |||
| L. T CD4 central memory (%) | 36.0 | [18.67] | 39.0 | [18.71] | 0.756 | |||
| L. T CD4 effector memory (%] | 26.3 | [20.33] | 30.2 | [22.59] | 0.726 | |||
| L. T CD4 EMRA* (%) | 3.1 | [3.37] | 5.4 | [10.97] | 0.627 | |||
| L. T CD8 naive (%) | 12.4 | [10.70] | 16.7 | [21.36] | 0.653 | |||
| L. T CD8 CM (%) | 10.3 | [19.79] | 5.9 | [3.75] | 0.473 | |||
| L. T CD8 EM (%) | 25.8 | [13.31] | 37.2 | [16.55] | 0.168 | |||
| L. T CD8 EMRA* (%) | 51.5 | [27.97] | 40.2 | [16.62] | 0.308 | |||
| n | % | n | % | p-value | ORAdj | [IC 95%] | p | |
|---|---|---|---|---|---|---|---|---|
| Gender | 0.399 | |||||||
| Men | 2 | (33.3) | 6 | (54.5) | ||||
| Women | 4 | (66.7) | 5 | (45.5) | ||||
| Hypertension | 0.113 | |||||||
| Yes | 2 | (33.3) | 8 | (72.7) | ||||
| No | 4 | (66.7) | 3 | (27.3) | ||||
| Diabetes | 0.219 | |||||||
| Yes | 1 | (16.7) | 5 | (45.5) | ||||
| No | 5 | (83.3) | 6 | (54.5) | ||||
| Cognitive disorders | 0.174 | |||||||
| Yes | 3 | (50.0) | 2 | (18.2) | ||||
| No | 3 | (50.0) | 9 | (81.8) | ||||
| Obesity | 0.515 | |||||||
| Yes | 0 | (0) | 2 | (18.2) | ||||
| No | 6 | (100.0) | 9 | (81.8) | ||||
| Total lymphocyte count >1000 cc/mm3 | ||||||||
| Yes | 5 | (83.3) | 5 | (45.5) | 0.116 | |||
| No | 5 | (16.7) | 6 | (54.5) | ||||
| L. B CD19+ CD10+ > 3% | 0.007 | |||||||
| No | 1 | (16.7) | 9 | (81.8) | 1 | |||
| Yes | 5 | (83.3) | 2 | (18.2) | 0.04 | [0.002;0.795] | 0.034 |
EMRA: effector memory CD45 + RA cells.
Figure 1.
CD10+ B cells according to the clinical outcome.
Values of TNF-alpha and IL-1, but not IL-6 nor IL-8, differed significantly between subjects with uncomplicated and complicated illness: TNF-alpha: 34.8 vs 13.0, p = 0.04, respectively; IL-1:0.8 vs 0.2, p = 0.04, respectively.
All patients in the complicated illness group received hydroxychloroquine, frequently associated with a macrolide. The dosage of hydroxychloroquine was a regimen of 200 mg three times daily for a 10 days, while the macrolide was co-administered the first 5 days. A close monitoring of potential side effects to such drugs was performed. In particular, patients had electrocardiograms every two days during their hospital stay and daily control of electrolyte count. No criteria for stopping these drugs earlier than scheduled were found in any patient.
Four patients died, all of them in the complicated group; the cause of death was either directly related to Covid or to associated complications. Values of TNF-alpha, IL-1, Il-6 and Il-8 were significantly different between subjects who died and those who survived (50 vs 15, 1.4 vs 0.2, 219.1 vs 13.0 and 52.1 vs 11.89, respectively, p < 0.01 for all comparisons). Among patients who died, under no circumstances was hydroxychloroquine suspected as related to death.
Corticosteroids were prescribed in 5 patients who had clinical worsening and 3 of them unfortunately died despite this treatment. Antibiotic therapy was prescribed in 10 out of 17 subjects. Main regimens included beta-lactams and macrolides. All four patients who died received antibiotics during their hospitalization.
Trends in immunological markers according to clinical severity
A second immunological analysis was conducted in a subgroup of 10 patients 10 days later. Levels of CD19+CD10 + B lymphocytes did not differ between clinical groups (4.7% in uncomplicated vs 2.8% in complicated illness, p = 0.361). A trend was observed in the slope of CD4CD38HLADR (-1.4% in uncomplicated vs +0.840 in complicated illness, p = 0.08) and CD8CD38HLADR cells (−10.7% vs −1.9%, p = 0.08).
Discussion
In a population of elderly patients at a very early stage of Covid-19, we found that levels of CD10 + B lymphocyte cells were predictive of the clinical severity.
In humans, CD10 is expressed on B cell progenitors in the bone marrow and subsequently disappears gradually with maturation, except on activated germinal center founder B cells, co-expressed with CD27 (Malaspina et al., 2006). These activated B cells are also detected in peripheral blood, the majority of them consisting of immature/transitional B cells (Sims et al., 2005). As CD10 is a marker known to be associated with immature/transitional cells, we compared transitional B cells between the two groups, which only revealed a trend (data not shown). Our study, which was not intended to specifically analyze this cell population, does not allow a conclusion on the origin of these cells and their significance in Covid-19 infection, as the difference in levels of transitional cells was not statistically significant between the two groups. However, several hypotheses can be put forward: if they are not immature/transitional cells, they could be activated precursor B cells with early Ig rearrangement in order to produce a broad repertoire of antibodies capable of responding to the viral infection (Vabret et al., 2020, van Zelm et al., 2005). Indeed, a robust B-cell response and plasmablasts expansion is generally detected early during SARS-CoV-2 infection (Stephens and McElrath, 2020). For reasons that still require investigation, only subjects who produce higher levels of precursor B-cells appear to develop less severe forms of Covid-19. Besides, Ho et al. showed that in HIV-infected patients with advanced disease there is an increased production of CD10+ cells, which are highly susceptible to intrinsic apoptosis, due to overexpression of the anti-apoptotic BCL-2 and BCL-xL proteins (Ho et al., 2006). Thus, increased levels of CD10+ in patients with mild forms of infection could also be explained by their better ability to eliminate the virus as a consequence of a pro-apoptotic profile of B lymphocytes and accelerated cell turnover. Unfortunately, the analysis did not include Il-7 which has been associated with the homeostatic compensation of overrepresented precursor B cells (Rosenberg et al., 2006). Finally, another explanation for lower levels of CD10+ cells in complicated forms could be a vigorous virally-induced immunosuppression, responsible for a blunted lymphocyte response (McGonagle et al., 2020).
If confirmed by larger studies, this work could furnish new insights about the immunological mechanisms of response to the virus and the broad spectrum of clinical presentation in a population at high risk of complication.
Interestingly, no difference between groups was found upon admission for values of the main plasma cytokines involved in the so-called “cytokine storm”. However, although the majority of patients had a mild clinical presentation upon admission, initial cytokine values were highly predictive of fatal outcome during hospitalisation. These results could have another important consequence for future management of Covid-19, suggesting that cytokine levels at entry could allow rapid identification of subjects who, despite moderate clinical symptoms, could be at risk for developing severe complications, and could thus be anticipated to benefit from anti-inflammatory strategies (Zhang et al., 2020).
To our knowledge, this is the first study to evaluate the immunological profile of patients preceding their clinical deterioration. As previously cited, lymphocyte subsets during Covid-19 have been already studied by Qin et al. (2020), but demographic characteristics of severe and non-severe patients, in contrast with ours, were significantly different between groups. Moreover, although time since symptoms onset is not described, we presume that patients were in a later stage of infection than in our cohort, as the immunological analysis was performed when patients already had signs of severity.
The study was not designed to determine the role of hydroxychloroquine regarding clinical response, as only subjects with severe forms received this compound. Only randomized prospective studies on recently infected patients can be expected to establish its role.
This study is limited by its retrospective character. However, files were systematically reviewed in order to confirm all described clinical data. Duration of infection might be another limitation, as it cannot be coincident with symptoms onset. We therefore cannot exclude a difference in duration of viral replication between the two groups. Only through systematic and prospective nasal swabs in asymptomatic subjects would it be possible to exactly determine the time of onset of infection.
In conclusion, our work shows that levels of CD10 + B cells are higher in patients with uncomplicated Covid-19 and that levels of cytokines at the early phase of infection are predictive of a poor outcome, arguing in favor of rapid interventional strategies for patients at risk.
Authors’ contributions
Conceived, designed the study and collected data: M.V., S.M., P.P. and E.B.
Analyzed the data: M.V.
Wrote the manuscript: M.V.
Edited the manuscript: M.T., B.S.P., A.P., L.L., A.S., P.F. and J.D.
Declaration of interest
There are no conflicts of interest to declare.
Funding
This work did not require any funding.
Ethical approval
The study was submitted to the French National Institute for Health Data (Institut National de Données de Santé, reference CNIL MR004) and patients gave informed consent for retrospectively collecting data.
Acknowledgments
We wish to thank all the nurses of the Department of Internal Medicine/Infectious Diseases, without whom this work would not have been possible.
Special thanks also to Nathalie Doux and to Aurelie Leguillermic for the work organization and to Brigitte Dunais for reviewing this paper.
References
- Alhazzani W., Hylander Møller M., Arabi Y.M., Loeb M., Ng Gong M., Fan E. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19) Intensive Care Med. 2020;28:1–34. doi: 10.1007/s00134-020-06022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhris M., Hillani A., Moroni F., Annabi M.S., Addad F., Harada Ribeiro M. Cardiovascular implications of the COVID-19 pandemic: a global perspective. Can J Cardiol. 2020;36(July (7)):1068–1080. doi: 10.1016/j.cjca.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja L.M., Wolf D.A., Zhao B., Akkanti B., McDonald M., Lelenwa L. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc Pathol. 2020;48 doi: 10.1016/j.carpath.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos W.G., Dela Cruz C.S., Cao B., Pasnick S., Jamil S. Novel Wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med. 2020;201:7–8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- Chang D., Lin M., Wei L., Xie L., Zhu G., Dela Cruz C.S. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA. 2020;323(March (11)):1092–1093. doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zheng Z., Zhang C., Xijiang Zhang X., Wu H., Wang J. Clinical characteristics of 145 patients with corona virus disease 2019 (COVID-19) in Taizhou, Zhejiang, China. Infection. 2020;28:1–9. doi: 10.1007/s15010-020-01432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico F., Baumgart D.C., Danese S., Peyrin-Biroulet L. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention, and management. Clin Gastroenterol Hepatol. 2020;18:1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A. Severe acute respiratory syndrome-related coronavirus: the species and its viruses—a statement of the Coronavirus Study Group. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J., Moir S., Malaspina A., Howell M.L., Wang W., DiPoto A.C. Two overrepresented B cell populations in HIV-infected individuals undergo apoptosis by different mechanisms. Proc Natl Acad Sci U S A. 2006;103:19436–19441. doi: 10.1073/pnas.0609515103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Putnam A.L., Xu-yu Z., Szot G.L., Lee M.R., Zhu S. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H.T., McCoy J.P., Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspina A., Moir S., Ho J., Wang W., Howell M.L., O’Shea M.A. Appearance of immature/transitional B cells in HIV-infected individuals with advanced disease: correlation with increased IL-7. Proc Natl Acad Sci U S A. 2006;103:2262–2267. doi: 10.1073/pnas.0511094103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonagle D., Sharif K., O’Regan A., Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev. 2020;19:102537. doi: 10.1016/j.autrev.2020.102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Deng Y., Dai Z., Meng Z. COVID-19 and anosmia: a review based on up-to-date knowledge. Am J Otolaryngol. 2020;41 doi: 10.1016/j.amjoto.2020.102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Kee Kwong K.C., Mehta P.R., Shukla G., Mehta A.R. COVID-19, SARS and MERS: a neurological perspective. J Clin Neurosci. 2020;77:13–16. doi: 10.1016/j.jocn.2020.04.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolai L.A., Meyer C.G., Kremsner P.G., Velavan T.P. Asymptomatic SARS Coronavirus 2 infection: invisible yet invincible. Int J Infect Dis. 2020;20 doi: 10.1016/j.ijid.2020.08.076. 30706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzon R.T., Wijaya V.O., Bijak Buana R., Al Jody A., Nalla Nunsio P. Neurologic characteristics in coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. Front Neurol. 2020;11:565. doi: 10.3389/fneur.2020.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020:ciaa248. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román G.C., Spencer P.S., Reis J., Buguet A., El Alaoui Faris M., Katrak S.M. The neurology of COVID-19 revisited: a proposal from the Environmental Neurology Specialty Group of the World Federation of Neurology to implement international neurological registries. J Neurol Sci. 2020;414 doi: 10.1016/j.jns.2020.116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S.A., Sportès C., Ahmadzadeh M., Fry T.J., Ngo L.T., Schwarz S.L. IL-7 administration to humans leads to expansion of CD8+ and CD4+ Cells but a relative decrease of CD4+ T-Regulatory cells. J Immunother. 2006;29:313–319. doi: 10.1097/01.cji.0000210386.55951.c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Pan C., Yang X., Zhong M., Shang X., Wu Z. Management of critically ill patients with COVID-19 in ICU: statement from front-line intensive care experts in Wuhan, China. Ann Intensive Care. 2020;10:73. doi: 10.1186/s13613-020-00689-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims G.P., Ettinger R., Shirota Y., Yarboro C.H., Illei G.G., Lipsky P.E. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solly F., Angelot-Delettre F., Ticchioni M., Geneviève F., Rambaud H., Baseggio L. Standardization of flow cytometric immunophenotyping for hematological malignancies: the franceflow group experience. Cytometry A. 2019;95:1008–1018. doi: 10.1002/cyto.a.23844. [DOI] [PubMed] [Google Scholar]
- Stephens D.S., McElrath M.J. COVID-19 and the path to immunity. JAMA. 2020 doi: 10.1001/jama.2020.16656. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Koh V., Marimuthu K., Tek Ng O., Young B., Vasoo S. Epidemiological and clinical predictors of COVID-19. Clin Infect Dis. 2020;25:ciaa322. doi: 10.1093/cid/ciaa322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M. Immunology of COVID-19: current state of the science. Immunity. 2020;16(52):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zelm M.C., van der Burg M., de Ridder D., Barendregt B.H., de Haas E.F.E., Marcel J.T. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol. 2005;175:5912–5922. doi: 10.4049/jimmunol.175.9.5912. [DOI] [PubMed] [Google Scholar]
- Velavan T.P., Meyer C.G. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai-Hin Chung T., Sridhar S., Zhang A.J., Chan K.H., Li H.L., Kai-Chuen Wong F. Olfactory dysfunction in coronavirus disease 2019 patients: observational Cohort Study and Systematic Review. Open Forum Infect Dis. 2020;7:ofaa199. doi: 10.1093/ofid/ofaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;23(March (11)):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Clinical management of Covid-19. Available at: https://www.who.int/publications/i/item/clinical-management-of-covid-19. [PubMed]
- Zhang J., Xie B., Hashimoto K. Current status of potential therapeutic candidates for the COVID-19 crisis. Brain Behav Immun. 2020;87(July):59–73. doi: 10.1016/j.bbi.2020.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]