Abstract
While DNA's perpetual role in biology and life science is well documented, its burgeoning digital applications are beginning to garner significant interest. As the development of novel technologies requires continuous research, product development, startup creation, and financing, this work provides an overview of each respective area and highlights current trends, challenges, and opportunities. These are supported by numerous interviews with key opinion leaders from across academia, government agencies and the commercial sector, as well as investment data analysis. Our findings illustrate the societal and economic need for technological innovation and disruption in data storage, paving the way for nature's own time-tested, advantageous, and unrivaled solution. We anticipate a significant increase in available investment capital and continuous scientific progress, creating a ripe environment on which DNA data storage-enabling startups can capitalize to bring DNA data storage into daily life.
Keywords: Biotechnology, Commercialization, Data, DNA computing, DNA data storage, DNA sequencing, DNA synthesis, Entrepreneurship, Investment landscape, Venture capital
Abbreviations: A, adenine; C, cytosine; CAGR, compound annual growth rate; CMOS, Complementary Metal Oxide Semiconductor; DARPA, Defense Advanced Research Projects Agency; DNA, deoxyribonucleic acid; G, guanine; HEDGES, Hash Encoded, Decoded by Greedy Exhaustive Search; IARPA, Intelligence Advanced Research Projects Activity; MIST, Molecular Information Storage Technology; NSF, National Science Foundation; PCR, polymerase chain reaction; T, thymine; WORN, Write Once Read Never
Highlights
-
•
Overview on current DNA data storage technologies and commercialization hurdles
-
•
Insights from leading DNA data storage experts and investment financing data
-
•
DNA synthesis remains the biggest challenge in the industry
-
•
Archiving cold data is the low-hanging fruit in DNA data storage
-
•
Upwards trend in investment landscape suggests optimal startup fundraising period
1. Introduction
The Digital Revolution triggered a paradigm shift in how we generate and store information, resulting in an unprecedented exponential increase in the amount of data that we produce and marking the beginning of the Information Age. Until now, data storage media including magnetic tapes and silicon chips have kept up with this demand, but they are fast approaching a critical limit in their physical storage capacities. In addition, the demand for data storage is expected to exceed the supply of silicon within the next 20 years (Zhirnov et al., 2016).
Currently, cloud-based systems are widely used for remote storage of data that does not need to be frequently accessed (Schadt et al., 2010). Whilst it might conjure up an ethereal image of how data are stored, the reality of cloud storage is much starker. Storage services use large warehouses stacked with constantly active servers that require a continuous supply of power and cooling systems to prevent overheating. Another major cost driver is archive replication. To attain redundancy, users can require multiple copies of data to be stored in geographically distinct locations. For petabyte to exabyte scale archives, this is non-trivial as the cost of each archive replicate is an integer multiple of the original archive cost. Consequently, cloud storage services are associated with substantial costs in terms of associated materials, storage space and electricity (Trelles et al., 2011). Therefore, how we currently meet our data storage needs is unsustainable environmentally, physically and financially. The urgent unmet need to develop truly disruptive technologies for the future of data storage has been widely recognized by organizations within both the public and private sectors. This has triggered a sharp increase of activity in pursuit of this goal.
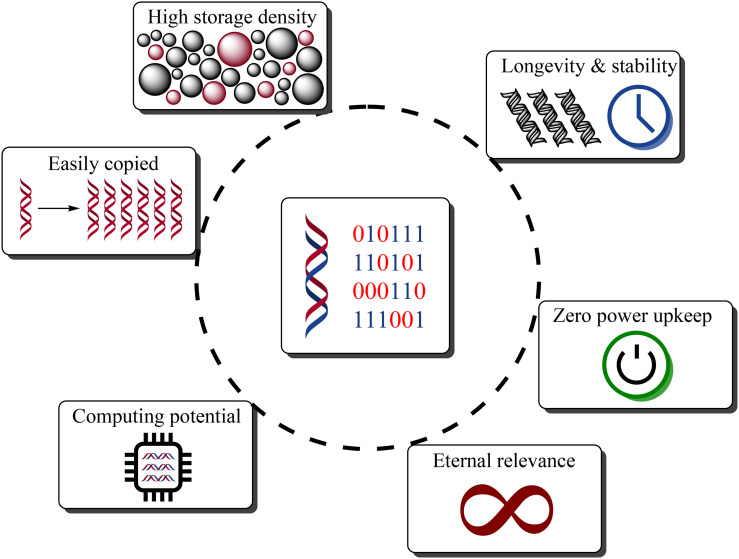
The solution may lie within nature's method of data storage. Deoxyribonucleic acid (DNA) carries the genetic information that is required for the development and maintenance of living organisms. The inherent properties that enable DNA to store biological data make it incredibly well-suited to store digital data (Fig. 1 ). This is a testament to an exciting new era where biology is providing novel solutions to engineering problems.
Fig. 1.
Distinct benefits and advantages of using DNA data storage as opposed to conventional technologies. DNA-based approaches promise to provide orders of magnitude higher storage density with outstanding long-term stability, while maintaining integrity without a power supply. Additionally, the intrinsic nature of DNA guarantees eternal relevance and provides exciting new opportunities for biocomputing. Finally, storage redundancies are easily achievable through the well-researched DNA amplification process.
2. Advantages of DNA data storage
A significant advantage of DNA over conventional data storage approaches is its longevity and stability. DNA has a half-life of approximately 500 years and can endure over millennia under appropriate storage conditions (Allentoft et al., 2012; Branzei and Foiani, 2008; Zhirnov et al., 2016). In contrast, current storage media including magnetic tapes and optical disks have a lifespan in the order of decades, requiring data to be regularly copied to new media for preservation (Ceze et al., 2019). Notably, DNA can be archived at room temperature without any power input (Grass et al., 2015).
Equally remarkable is the improvement in density that DNA can provide. One cubic millimeter of DNA can store up to 1018 bytes, which would give DNA an approximately six orders of magnitude higher theoretical storage density than the densest storage medium currently available (Ceze et al., 2019). In practice, a sample of DNA the size of a few dice would store the equivalent of an entire data center's worth of data (Bornholt et al., 2016).
Even though new copies of data stored in DNA do not need to be frequently produced for preservation, it can be done with ease. DNA can be copied exponentially using the same polymerase chain reaction (PCR) that is frequently used in laboratories for life science and medical purposes. This markedly improves the efficiency of producing data backups compared to current storage technologies. However, it should be noted that amplification is also a source of error, as small variations are compounded during amplification, leading to molecular bias (Chen et al., 2020).
There are no data storage media that share the eternal relevance of DNA, with its prominence in nature over billions of years of evolution. Inevitably, there will always be the desire to read and write DNA. Further, the storage medium DNA has the right characteristics to conduct computations, so-called DNA- or bio-computing (Adleman, 1994; Braich et al., 2002; Thachuk and Liu, 2019). Looking further into the future, one can imagine a holistic, novel solution for data handling completely run on DNA.
3. Technological strategies towards implementation
Norbert Wiener and Mikhail Neiman are considered the founding fathers of data storage in nucleic acid (U.S. News and World Report, 1964). In 1964, Norbert Wiener proposed that a memory system could be built from genetic material outside of a living organism in the future. Since then, many researchers from academia and industry have worked on developing this initial idea towards a viable product. Moving forward, the concerted efforts of academia, large corporates, innovative startup companies together with venture capital investment will be required to propel DNA data storage to commercial scale.
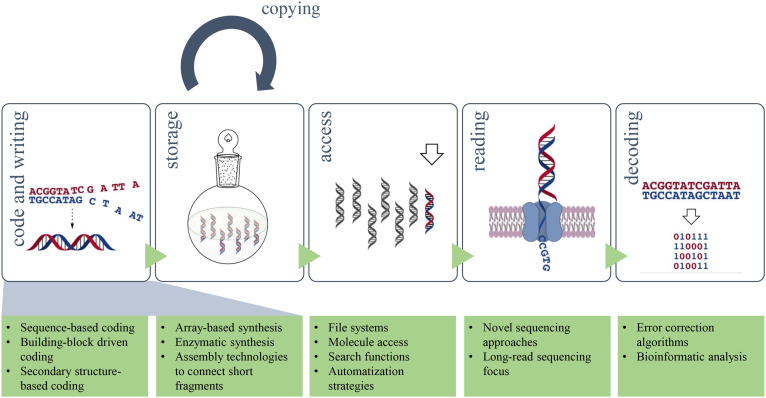
In general, storing information in a biological material follows the same principle as for common silicon-based hard drives or recording tapes (Fig. 2 ). The data needs to be transferred into a code (3.1.) that is written (3.2.) into DNA suitable for storage (3.3.), where the information can be accessed (3.4.) again to read (3.5.) the data. The ability to copy (3.6.) data from one device to another is also beneficial (Ceze et al., 2019). Interdisciplinary efforts spanning molecular biology, computer science, and information technology are required to reach a complete DNA data storage workflow and in the following paragraphs, several of these approaches are discussed in more depth.
Fig. 2.
Workflow of DNA data storage. Top panels show a simplified way of how coded information can be written into DNA, where it can be accessed in the storage device to read the code and retrieve the information. Additionally, the stored material can be copied. Each step is described in detail in the text. Bottom panels highlight development efforts in each area.
3.1. Code
DNA naturally carries an intrinsic code composed of its four nucleobases: adenine (A), thymine (T), cytosine (C) and guanine (G). Therefore, the most obvious approach is to translate binary (digital) code directly into this four-base alphabet which researchers have succeeded in. Nevertheless, programming code for storage is completely flexible and not universal across different approaches in the field. It has been recently shown that rational system design can yield higher bit densities, e.g. 1.55 bits per nucleotide through employing the four canonical nucleotides in order to approach the maximum theoretical number of two bits per base for a quaternary system like DNA (Erlich and Zielinski, 2017).
Even several nucleotide-long, pre-made DNA oligonucleotides can be utilized to encode information when they are assembled in a combinatorial manner. In addition, the four-letter alphabet code can be extended by chemical modifications of the nucleobases or the phosphate backbone. A striking example of an extended DNA alphabet is “Hachimoji” DNA, which uses eight non-canonical bases with Watson-Crick hydrogen binding (Hoshika et al., 2019).
Completely different approaches, however, use secondary structure elements that can be constructed into DNA. Naturally, DNA is a double helix of two antiparallel strands of nucleotides. These are connected via their sugar phosphates that build the backbone, with the four different nucleobases facing inwards.
The formation of hairpins of different lengths, in which one of the strands loops out, is an example of a nanostructure that has been shown to represent digital code (long hairpin = ‘1’, short hairpin = ‘0’). This nanostructure can be sensed and decoded through nanopores (see 3.5.) (Chen et al., 2019).
Introducing cuts into the phosphate backbone of one of the strands, called nicking, can serve as a code, too (Tabatabaei et al., 2020). The presence or absence of a nick at a certain position resembles a ‘1’ or a ‘0’, respectively, in digital binary code. For this approach, an existing DNA sequence is extracted from a native source, e.g. bacterial DNA, to select registers with desirable sequence elements for restrictions. The nicks are then enzymatically introduced in a parallel fashion. Denaturing the DNA separates the two strands and alignment of the fragments onto a known sequence retrieves the position of the nick, regenerating the code. The nicking approach leads to a 10-50-fold decrease in information density per base pair of DNA, compared to codes with a nucleotide resolution. Hairpin approaches also share this inherent density reduction. However, this could be a very low price to pay considering the ease of reading, and potential reductions in costs of DNA synthesis, providing adequate automation of these processes can be realized.
A further key aspect to consider in the encoding process is the choice of codec scheme before synthesis to facilitate accurate error correction during readout (see 3.5.). This can include using multiple copies, Reed Solomon block codes, repeat accumulate codes, and more (Blawat et al., 2016; Wang et al., 2019a). A descriptive example recently reported for storing quantized images in DNA uses signal processing and machine learning techniques to deal with error instead of redundant oligos or rewriting. This relies on decoupling and separating the color channels of images, performing specialized quantization and compression on the individual color channels, and using discoloration detection and image inpainting (Pan et al., 2020). It is worth noting that images are a medium the brain can self error correct to a certain degree, making it not necessary to recover every bit, nevertheless this is an example of rational system design and codec scheme choice.
3.2. Code writing and DNA synthesis
Depending on how information is encoded in DNA, the requirements for its synthesis vary. Producing long strands of DNA is currently the main challenge. While all synthetic oligonucleotides are prone to errors during synthesis (Hölz et al., 2018), oligonucleotides longer than 200 nucleotides are particularly difficult to obtain with high fidelity due to accumulated errors. During the reaction, costly reagents generate toxic byproducts. Most technologies still rely on a sequential one-by-one addition of nucleotides to the growing strand, where the speed of liquid handling in microfluidic devices limits production speed. This explains the industry push towards systems using shorter sequences, which has been demonstrated in academic research (Wang et al., 2019b). In array-based DNA synthesis, several strands that encode different DNA sequences are grown simultaneously while immobilized on a surface. This allows for higher parallelization, thereby increasing production speed (Kosuri and Church, 2014).
Novel approaches focus on enzymatic DNA synthesis (Lee et al., 2019; Lee et al., 2020a). While oligonucleotides produced with this methodology currently remain shorter, experts expect lower error rates, higher speed and longer fragments with this upcoming technology. To obtain larger sequence strings for ease of read, an assembly process connecting these 200 to 300 nucleotide-long fragments is needed. Currently, most efforts follow the same principle that is used in molecular biology for gene assembly.
Codes that are independent of the actual sequence but rely on secondary structure can be assembled from a pool of oligonucleotides, which can be produced by chemical synthesis in large amounts and at low costs. The same applies to code in which longer oligonucleotide sequences present one digital state (‘1’ or ‘0’). To promote the correct assembly of these in a fast and reliable fashion, researchers at CATALOG have developed an inkjet-printer like machine. In contrast to biological applications, the requirements for DNA synthesized for data storage are throughput, costs and few copies per unique sequence. Researchers across the field agree that DNA synthesis remains the biggest challenge and needs to become faster, more reliable, and significantly cheaper to advance data storage in DNA.
3.3. Storage
Whichever way encoded, the nucleic acid molecules themselves can adapt to any structure and could, in principle, be stored in any geometric shape or form. DNA molecules can be pooled for liquid storage in a suitable solvent. On the other hand, researchers have also developed storage devices where DNA molecules are immobilized on solid surfaces, or where DNA molecules are embedded in other materials such as glass or plastic (personal communication, unpublished data (Grass et al., 2015; Koch et al., 2020). So far, empirical values for the retention time (the time data can still be recovered reliably) of all storage forms still need to be confirmed. Initial experiments, in which DNA was encapsulated into an inorganic matrix, have shown promising results. In this form, information on DNA is predicted to be stably stored for 2000 years (Grass et al., 2015).
3.4. Accessing the information
To avoid reading a whole storage device, systems for organizing and accessing information are needed. A nested file address system has, for instance, been shown to increase the capacity of DNA storage further and provides progress towards a scalable DNA-based data storage system (Stewart et al., 2018; Tomek et al., 2019). Random access describes the reading of selected information in computer science, and it is a key feature that needs to be developed for DNA data storage to become viable. Indexing (adding a unique file identification sequence onto each DNA molecule) helps to identify the desired data; however, how to index data is not a unified system yet. In most approaches, the index will allow for targeted amplification of the requisite information by PCR, for example by having the same PCR primer target sequences form a unique file ID for each strand and including a one-of-a-kind, strand-specific address to order strands within a file (Organick et al., 2018). In other approaches, a complementary sequence of the index region is encoded on magnetic beads and hybridization allows for the physical separation of the desired DNA molecule from the pool (Tomek et al., 2019). These index regions must be designed carefully to access only the desired DNA molecule. Both approaches are derived from molecular biology techniques. Search functions have been designed in a similar way, generating query DNA strands to identify the searched information through hybridization. Recently, this content-based retrieval from a DNA database was scaled to include 1.6 million database images with a retrieval rate much greater than chance when prompted with new images (Bee et al., 2020).
In some use cases, data need to be rewritten, meaning only parts of the data change while other parts are retained. The first successful approach to create a rewritable DNA-based storage system was described in 2015. The technology is based on an elegant design of DNA blocks with recognition sequences that can be altered via PCR (Yazdi et al., 2015). More recent advances include a dynamic storage system based on a T7 promoter sequence with a single-stranded overhang, unlocking versatile editing and rewriting capabilities (Lin et al., 2020).
3.5. Reading the code by DNA sequencing and codecs
Retrieving information from DNA for storage can benefit from the sequencing ecosystem that is continuously being improved for life science and medical applications. As novel sequencing methods are developed, the cost per base sequenced decreases while the speed increases. Currently, the main sequencing approaches used by researchers in the DNA data storage field are sequencing by synthesis, promoted by Illumina, or nanopore sequencing. The novel nanopore sequencing technology, designed for longer molecules, can in principle also decode secondary structure elements and base modifications. However, the workflow to prepare DNA for sequencing is still currently laborious and additional steps are required for nanopore sequencers. Despite improvements in the workflow, experts agree that the entire process speed needs to be improved.
Despite inherent error rates, DNA data storage can in principle tolerate high error rates in both write and read channels through sufficient redundancy, appropriate codecs (coder-decoder), error correction codes and algorithm design (Erlich and Zielinski, 2017; Organick et al., 2018; Press et al., 2020).
Researchers describe that on average ten copies of a DNA molecule encoding the same sequence is sufficient to reliably retrieve the stored information with the current technologies (Organick et al., 2020). For biological applications, this so-called coverage is typically required to exceed 30 reads per nucleobase.
The required error correction algorithms are different and often more complex than those needed for biological purposes or algorithms used in conventional data storage. Beside the substitution errors that are found in the latter, nucleotide insertions or deletions are additional common error types that occur during DNA synthesis and sequencing. One such algorithm has recently been developed to repair all three error types, where insertions or deletions can be corrected directly within a single DNA strand, unlike previous codes that correct substitution errors (Press et al., 2020). A part of this algorithm, known as HEDGES (Hash Encoded, Decoded by Greedy Exhaustive Search), translates between a string of the four nucleobases and a binary string of the digital binary code (see 3.1.), without changing the number of bits. It can do this all while tackling practical challenges of storing information in DNA. In addition to correcting the three error types, HEDGES can convert unresolved insertions or deletions into substitutions, and it can also adapt to sequence constraints (e.g. having a balanced G-C content). HEDGES, therefore, has the potential to enable error-free recovery of data on a large scale. Another coding algorithm developed in 2018 can tolerate high error rates during reading, while also reducing the level of sequencing redundancy required for error-free decoding (Organick et al., 2018). This limits the number of required copies of DNA to recover stored data, which becomes increasingly important as throughput increases. To reduce the need for error correction, codes that avoid long stretches of the same nucleobase, which are difficult to synthesize and sequence, can be applied (see 3.1.).
3.6. Transferring information and amplifying DNA
The intrinsic property of DNA to make copies of itself for data transfer in living organisms is highly beneficial for data storage, as information is more valuable when it can be multiplied and distributed. Currently, a minimum of two copies of the data are required as a pre-sequencing step because modern sequencers are constructed to discard the DNA as part of the reading process. Standard PCR protocols can easily be transferred from molecular biology to DNA data storage to generate additional copies in a fast and parallel way. Together with the use of PCR-based random access, DNA sequencing combined with PCR-based copying are the primary reasons why most experts predict DNA will become the next generation storage medium instead of other organic or biological polymers. Most recent developments for the latter still rely on low-throughput mass spectrometry-based sequencing techniques (Lee et al., 2020b), in stark contrast to methods to rapidly sequence large quantities of DNA as outlined previously (see 3.2.).
3.7. Integration and automation
For DNA storage to be widely adopted as a commercial product, the whole process, including transfer steps between synthesis, storage and sequencing, needs to eventually be automated. The first end-to-end storage device handling 5 bytes of data encoding the word ‘hello’ (published in March 2019 by the University of Washington and Microsoft) sets the stage for further fully-integrated solutions (Takahashi et al., 2019). This approach is based on liquid DNA storage, where the main limiting factor is considered to be liquid handling. Progress in the field of nano- and micro-fluidics will help advance automation strategies, such as the novel runtime system, “Puddle”, a high-level dynamic, error-correcting, full-stack microfluidics platform (Willsey et al., 2019) and dehydrated DNA spots on glass (Newman et al., 2019).
Another approach towards fully integrated solutions is building on established complementary metal oxide semiconductor (CMOS) technology to increase throughput via bespoke, microfabricated, and highly-parallel synthesis and sequencing devices, such as those in development by Twist Biosciences and Roswell Biotechnologies.
Beside the technical hurdles in this interdisciplinary field, researchers have also realized that a more extensive network with effective communication is needed to advance data storage in DNA. Recently, for instance, a glossary and controlled vocabulary was introduced to increase accessibility (Hesketh et al., 2018).
4. Commercial hurdles
While DNA data storage technologies are immensely intriguing from a scientific point of view, companies are still facing key challenges towards achieving large-scale commercial success. Widely recognized as the central bottleneck, DNA synthesis is costly, time consuming and prone to errors. The synthesis price per base has seen a rapid decline over the past decades, with companies like Twist Bioscience or DNAScript continuously pushing the boundaries of what is possible. Twist Bioscience provides large quantities of error-free DNA fragments up to 300 nucleotides using their silicon-based writing technology. In early 2019, DNAScript announced the successful production of the first 200 nucleotide-long DNA fragment by enzymatic synthesis. However, we speculate that initial go-to-market technologies will still need to circumvent codes that depend on long-strand, error-free DNA synthesis.
On the flip-side, hurdles in DNA sequencing are often overlooked as sequencing is currently much cheaper and faster than synthesis. Nevertheless, for DNA data storage to become a widely-implemented technology, further decreases in costs are essential. Illumina has succeeded in lowering prices by roughly five orders of magnitude since the early 2000s, but this rate is now slowing down. In addition, it is worth highlighting that the technical requirements of sequencing for data storage are orthogonal to traditional life sciences applications – the latter cannot tolerate errors. This is one of the factors that may provide additional leverage for emerging technologies in the field.
When considering the nature of DNA data storage, it becomes clear that instantaneous and random access presents a substantial problem. Especially for large quantities of data, full sequencing and decoding will not always be practical and entails higher latency. Commercially, this will push development towards the low-hanging fruit of archiving “cold data”, often referred to as “Write Once, Read Never” (“WORN”). An extremely promising entry point into the archiving market is image storage: the developed error correction codes and the eye's fault tolerance level mean that image fidelity does not have to be 100%, thereby compensating for error rates (personal communication, unpublished data). We therefore expect first commercial adopters to be sourced in market segments such as image backup or streaming services.
These described challenges represent intrinsic hurdles for DNA data storage from a molecular perspective. More issues arise when considering the required operational infrastructure. In structural terms, DNA molecules cannot just be applied to existing chip architectures. Thus, the silicon-to-DNA interface will have to be optimized and accounted for by software and physical interconnects. Moving forward from current prototypes (see 3.7.) to larger setups will entail fluidic difficulties, as a liquid-based data storage system will ideally operate under zero-human-contact conditions. An end-to-end solution will require a standardization of data formats when stored in DNA, processable by all data-hardware interfaces, and subsequently streamlined workflow steps to enable cross-platform storage and seamless embedding into existing data architectures (see 3.). Currently, the individual approaches consume considerable resources and efforts – DNA data storage will require strategic investments in holistic frameworks to reach its full potential.
In terms of DNA data storage reaching mass markets and becoming a large-scale commercial success, the aforementioned hurdles seem quite daunting and obtrusive. However, this should not deter research, investments and public interest in this highly promising field. Several indicators show that this area is already gaining substantial traction. The general public is being introduced to the concept and technology by recent articles in prominent mainstream outlets, such as Forbes and Wired ( Forbes, 2019; Wired, 2018 ). Meanwhile, significant funding opportunities are arising from the US government's Defense Advanced Research Projects Agency (DARPA), Intelligence Advanced Research Projects Activity (IARPA), and National Science Foundation (NSF). IARPA most recently launched its Molecular Information Storage Technology (MIST) program, currently involving DNA Script and Illumina, which aims to develop technologies that can write 1 TB and read 10 TB of data per day with DNA. The considerable interest from large corporations and prominent universities, like Microsoft and the University of Washington, is fostering fruitful collaborations.
The key opinion leaders interviewed for this article - spanning from academics to business leaders - consider a timeline of 4–10 years, depending on the application, level of automation and scale size, to be a realistic estimate for market entry and success. This technology maturity horizon places DNA data storage into the scope of venture capital funds, particularly for strategic investments from corporate venture capital branches and financial funds with a longer return-on-investment mandate or an evergreen fund structure. Tangible benefits will arise for investments in robotics for automation and scaling solutions, as these will require a high capital expenditure to develop but do not incur an intrinsic chemical problem. Additionally, focusing cash flows into areas with key differences to DNA's biomedical applications will aid the rise of DNA data storage, ultimately leading to the different technological approaches branching off into respective best-in-class applications. A predominant example for such an area is fast-throughput DNA synthesis and sequencing.
5. Venture capital landscape
Increased public and corporate interest in a novel technology often garners an uptake in investments and startup funding. Additionally, in this case, the venture capital perspective on the overarching industrial landscape should be considered. Looking at the current general data storage market, it is a harsh business for hardware manufacturers and providers, who operate on low margins and rely on technological improvements to drive new products. Typically, venture capitalists avoid investing in such a commoditized industry as it is not attractive for exit potentials and high returns. However, the imminent crisis upon reaching physical limits in classical data storage will also drastically change the investment landscape. Not only will succeeding technologies be highly contested acquisition targets by numerous large corporations, they will simultaneously generate very substantial capital returns for investors. Furthermore, novel methods and scientific breakthroughs achieved in pursuit of molecular data storage will generate complimentary use cases and applications in other industries, like bio-computing and life sciences. From a business model and scaling perspective, such a broad range of markets is appealing. This paradigm shift in the DNA data storage investment landscape has already begun and can be quantified by tracking the number and size of venture capital deals made in the relevant market segments and technologies. Thus, we have performed a database search and analysis of investment activity in the DNA data storage space over the past 10 years. Using appropriate key words and input criteria, a list of companies that have been - or are still - venture capital backed was generated (Fig. 3 ).
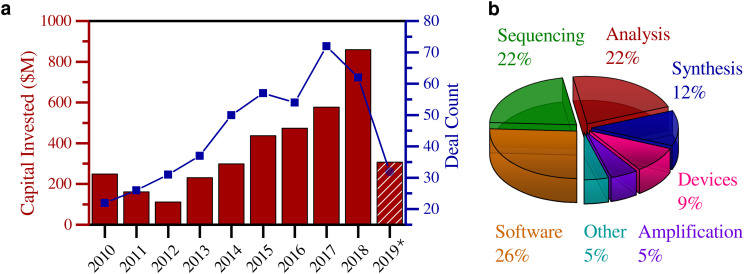
Fig. 3.
Analysis of the venture capital landscape for DNA data storage according to custom search results extracted from PitchBook, private capital market data provider. Venture capital funding was only included into the analysis if the respective company disclosed to work on a technology that has a potential to advance DNA data storage (e.g. modifications of sequencing technologies for specific biologic set-ups were not included). For startups active in the development of various technologies and/or areas, it is not possible to attribute the percentage of funding specifically supporting DNA data storage technologies. a. Column graph, left axis: Total capital invested per year in companies in the DNA data storage space, specifically including novel nucleic acid synthesis and sequencing technologies, software for sequencing data analysis, nucleic acid analysis methods (excluding single nucleotide sequencing), DNA amplification and manipulation techniques, biological computing, and storage devices. Line graph, right axis: Corresponding total deal count per year. *Note that the 2019 values are assessed with fully completed and public deals by the 26th of September. b. Segmentation of all the companies found in the database search into their respective technology and market areas as described under a.
Several key takeaways are apparent from this data. First, the past seven years have seen a rapid increase in total capital invested in companies developing DNA data storage-enabling technologies (Fig. 3a), averaging a 44% compound annual growth rate (CAGR). The number of concluded deals per year follows a similar trend. This significant upswing corresponds with the previously noted uptake in academic activity and accomplishments in this space. Second, the predominant sectors that the invested companies are active in are novel nucleic acid analysis, sequencing and software solutions (Fig. 3b). While this is not surprising, it does stress the need for innovation and technological progress, fueled by increased funding, in the nucleic acid synthesis area. As this is widely regarded as the current key bottleneck and pain point, this space provides significant opportunities for large value generation. We anticipate that the funding focus, which currently stems from life science-oriented venture capital companies, will shift to include multidisciplinary and deep tech-oriented funds. As a result, additional capital sources will become available. Third, the data confirm DNA data storage to be an attractive and fast-growing investment sector, and we expect significant funding in the coming years.
One major, and likely defining, event of 2020 – the COVID-19 pandemic – has disrupted almost every industry and economy around the world, introducing a level of economic uncertainty not seen in generations. Increased timelines on development and commercialization due to the pandemic have led to an increase in capital needed to achieve milestones. This has, in turn, decreased the total number of deals a fund can conduct to support new and existing portfolio companies. Despite this, it is important to note that venture capital deals have continued (Milkove, 2020). Even during the pandemic, venture capital firms likely need to deploy the capital they have already raised in order to reach their target return on investments. Given DNA data storage's technology maturity time-scale, venture capital firms companies investing in this space (see 5.) would already be operating with a long-term view and longer timelines than are typical for other fields of investment. COVID-19 has actually lead to increased investment in biotech companies listed on the stock exchanges with the Nasdaq Biotechnology Index up 11% since the start of the year (Senior, 2020), as well as highlighted the importance of investing in life sciences technologies like sequencing. The increased worldwide interest in such technologies sparked by this global health crisis could result in a boost in investments in technologies that will ultimately advance DNA data storage. In addition, COVID-19 has accelerated digitalization across industries, further increasing the demand for data storage services (Tilley, 2020). Big tech- and data-oriented companies, like Microsoft, have already benefited from the pandemic and will likely emerge from COVID-19 in an even stronger financial position to invest in and secure their stake in the DNA data storage space. With global economies beginning to recover, potential vaccine candidates looking promising, and markets looking forward, the venture capital landscape is also likely to recover. Though there is a risk of a potential prolonged market shock, we deduce that it is still a favorable time for startups in the DNA data storage space to seek funding from venture capital firms.
6. Conclusions
The dawn of the modern Information Age has drastically accelerated the rate of data generation to the point where almost overwhelming quantities threaten our current storage capacities. Paradoxically, one of the most promising long-term prospects towards this challenge is storing data in DNA – the very substance that defines and holds the code to human life. Nature's time-tested and durable solution to complex data encoding and storage guarantees its longevity and eternal relevance. However, large-scale commercialization of non-genetic DNA data storage is faced with considerable technical, engineering and financial barriers. We believe that the latter will be increasingly addressed in the future by investment companies such as venture capital firms, given the substantial increase in invested capital in the field over of the last seven years (Fig. 3a). Considering the growing public and corporate traction, DNA data storage-enabling startups are firmly on investment radars for strategic and long-term funding. As the overarching storage industry, currently a commoditized industry, is rapidly approaching disruption, this paves the path for technological innovations and will offer high returns. We anticipate that a significant increase in available capital, together with continuing scientific progress, will enable the rise and mainstream implementation of DNA data storage into daily life.
Author contributions
P.M.S., L.M.S., A.M.V. and K.C.K.L. conceived the project scope and conducted the expert interviews. P.M.S. and L.M.S. performed the custom Pitchbook search and A.M.V. assisted with the data analysis. P.M.S. and L.M.S. designed the figures. P.M.S., L.M.S. and A.M.V. drafted the manuscript, with all authors contributing to the discussion, editing and completion.
Declaration of Competing Interest
The authors of the article were employees of M Ventures, the strategic corporate venture capital arm of Merck KGaA, Darmstadt, Germany. Correspondingly, this work was supported by M Ventures.
Acknowledgements
This work was supported by numerous interviews with key opinion leaders on DNA data storage – in research and industry – as well as industry specialists for traditional CMOS-based storage. We are especially grateful to the many experts who gave their time freely for providing insight and feedback on the topic, including the following individuals: Albert Keung (NC State University), Anne Fischer (DARPA), David Markowitz (MIST, IARPA), Emily Leproust (Twist Bioscience), Fahim Farzadfard (Schmidt Science Fellow at Massachusetts Institute of Technology), James Gagnon (University of Utah), Karin Strauss (Microsoft), Luis Ceze (University of Washington), Michael Anderson Burley (Merck KGaA, Darmstadt, Germany), Monika Brenner-Rieck (Merck KGaA, Darmstadt, Germany), Olgica Milenkovic (University of Illinois), Ulrich Keyser (University of Cambridge), Xavier Gordon (DNAScript), and Yaniv Erlich (MyHeritage, Columbia University). We thank Daniel Franke, Jasper Bos, Joey Mason, Owen Lozman, W.L.E. Wallace (all at M Ventures), and Isabel De Paoli (Merck KGaA, Darmstadt, Germany) for the helpful discussions and support.
The views, opinions, and findings contained in this article are those of the authors and do not represent those of the institutions or organizations listed in the acknowledgements. Furthermore, they should not be interpreted as representing the official views or policies, either expressed or implied, of the Defense Advanced Research Projects Agency, the Intelligence Advanced Research Projects Activity, or the Department of Defense.
References
- Adleman L.M. Molecular computation of solutions to combinatorial problems. Science (New York, N.Y.) 1994;266(5187):1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- Allentoft M.E., Collins M., Harker D., Haile J., Oskam C.L., Hale M.L. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. Biol. Sci. 2012;279(1748):4724–4733. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bee C., Chen Y.-J., Ward D., Liu X., Seelig G., Strauss K. Content-based similarity search in large-scale DNA data storage systems. bioRxiv. 2020 doi: 10.1101/2020.05.25.115477. [DOI] [Google Scholar]
- Blawat M., Gaedke K., Hütter I., Chen X.-M., Turczyk B., Inverso S. Forward error correction for DNA data storage. Procedia Comput. Sci. 2016;80:1011–1022. [Google Scholar]
- Bornholt J., Lopez R., Carmean D.M., Ceze L., Seelig G., Strauss K. Conte T, Zhou Y. Proceedings of the Twenty-First International Conference on Architectural Support for Programming Languages and Operating Systems - ASPLOS ’16 02/04/2016–06/04/2016: Atlanta, Georgia, USA. New York, New York. ACM Press; USA: 2016. A DNA-based archival storage system. [Google Scholar]
- Braich R.S., Chelyapov N., Johnson C., Rothemund P.W.K., Adleman L. Solution of a 20-variable 3-SAT problem on a DNA computer. Science (New York, N.Y.) 2002;296(5567):499–502. doi: 10.1126/science.1069528. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9(4):297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Ceze L., Nivala J., Strauss K. Molecular digital data storage using DNA. Nat. Rev. Genet. 2019;20(8):456–466. doi: 10.1038/s41576-019-0125-3. [DOI] [PubMed] [Google Scholar]
- Chen K., Kong J., Zhu J., Ermann N., Predki P., Keyser U.F. Digital data storage using DNA nanostructures and solid-state Nanopores. Nano Lett. 2019;19(2):1210–1215. doi: 10.1021/acs.nanolett.8b04715. [DOI] [PubMed] [Google Scholar]
- Chen Y.-J., Takahashi C.N., Organick L., Bee C., Ang S.D., Weiss P. Quantifying molecular bias in DNA data storage. Nat. Commun. 2020;11(1):3264. doi: 10.1038/s41467-020-16958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich Y., Zielinski D. DNA fountain enables a robust and efficient storage architecture. Science (New York, N.Y.) 2017;355(6328):950–954. doi: 10.1126/science.aaj2038. [DOI] [PubMed] [Google Scholar]
- Forbes [Internet] DNA Data Storage Is about to Go Viral. 2019. https://www.forbes.com/sites/johncumbers/2019/08/03/dna-data-storage-is-about-to-go-viral/#22389c7f7721 [cited 2020 Jul 9]. Available from:
- Grass R.N., Heckel R., Puddu M., Paunescu D., Stark W.J. Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. Eng. 2015;54(8):2552–2555. doi: 10.1002/anie.201411378. [DOI] [PubMed] [Google Scholar]
- Hesketh E.E., Sayir J., Goldman N. Improving communication for interdisciplinary teams working on storage of digital information in DNA. F1000Res. 2018;7:39. doi: 10.12688/f1000research.13482.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölz K., Hoi J.K., Schaudy E., Somoza V., Lietard J., Somoza M.M. High-efficiency reverse (5′→3′) synthesis of complex DNA microarrays. Sci. Rep. 2018;8(1):15099. doi: 10.1038/s41598-018-33311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshika S., Leal N.A., Kim M.-J., Kim M.-S., Karalkar N.B., Kim H.-J. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science (New York, N.Y.) 2019;363(6429):884–887. doi: 10.1126/science.aat0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J., Gantenbein S., Masania K., Stark W.J., Erlich Y., Grass R.N. A DNA-of-things storage architecture to create materials with embedded memory. Nat. Biotechnol. 2020;38(1):39–43. doi: 10.1038/s41587-019-0356-z. [DOI] [PubMed] [Google Scholar]
- Kosuri S., Church G.M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods. 2014;11(5):499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.H., Kalhor R., Goela N., Bolot J., Church G.M. Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat. Commun. 2019;10(1):2383. doi: 10.1038/s41467-019-10258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Wiegand D.J., Griswold K., Punthambaker S., Chun H., Kohman R.E. Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage. bioRxiv. 2020 doi: 10.1101/2020.02.19.956888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Koo M.B., Lee S.W., Lee H., Kwon J., Shim Y.H. High-density information storage in an absolutely defined aperiodic sequence of monodisperse copolyester. Nat. Commun. 2020;11(1):56. doi: 10.1038/s41467-019-13952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.N., Volkel K., Tuck J.M., Keung A.J. Dynamic and scalable DNA-based information storage. Nat. Commun. 2020;11(1):2981. doi: 10.1038/s41467-020-16797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkove H [Internet] COVID-19's Impact on Early Stage Venture Capital. Medium. 2020. https://medium.com/swlh/covid-19s-impact-on-early-stage-venture-capital-2851230c0c64 [cited 2020 Aug 31]. Available from.
- Newman S., Stephenson A.P., Willsey M., Nguyen B.H., Takahashi C.N., Strauss K. High density DNA data storage library via dehydration with digital microfluidic retrieval. Nat. Commun. 2019;10(1):1706. doi: 10.1038/s41467-019-09517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organick L., Ang S.D., Chen Y.-J., Lopez R., Yekhanin S., Makarychev K. Random access in large-scale DNA data storage. Nat. Biotechnol. 2018;36(3):242–248. doi: 10.1038/nbt.4079. [DOI] [PubMed] [Google Scholar]
- Organick L., Chen Y.-J., Dumas Ang S., Lopez R., Liu X., Strauss K. Probing the physical limits of reliable DNA data retrieval. Nat. Commun. 2020;11(1):616. doi: 10.1038/s41467-020-14319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Hossein Tabatabaei Yazdi S.M., Kasra Tabatabaei S., Hernandez A.G., Schroeder C., Milenkovic O. 2020. Image Processing in DNA. ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) [Google Scholar]
- Press W.H., Hawkins J.A., Jones S.K., Schaub J.M., Finkelstein I.J. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints. Proc. Natl. Acad. Sci. U. S. A. 2020;117(31):18489–18496. doi: 10.1073/pnas.2004821117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Linderman M.D., Sorenson J., Lee L., Nolan G.P. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 2010;11(9):647–657. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior M. The biopharmaceutical anomaly. Nat. Biotechnol. 2020;38(7):798–805. doi: 10.1038/s41587-020-0593-1. [DOI] [PubMed] [Google Scholar]
- Stewart K., Chen Y.-J., Ward D., Liu X., Seelig G., Strauss K. A content-addressable DNA database with learned sequence encodings. In: Doty D., Dietz H., editors. DNA Computing and Molecular Programming. Springer International Publishing; Cham: 2018. [Google Scholar]
- Tabatabaei S.K., Wang B., Athreya N.B.M., Enghiad B., Hernandez A.G., Fields C.J. DNA punch cards: storing data on native DNA sequences via nicking. bioRxiv. 2020:672394. doi: 10.1038/s41467-020-15588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi C.N., Nguyen B.H., Strauss K., Ceze L. Demonstration of end-to-end automation of DNA data storage. Sci. Rep. 2019;9(1):4998. doi: 10.1038/s41598-019-41228-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thachuk C., Liu Y. Springer International Publishing; Cham: 2019. DNA Computing and Molecular Programming. [Google Scholar]
- Tilley A [Internet] One business winner amid coronavirus lockdowns: the cloud. Wall Street J. 2020 https://www.wsj.com/articles/one-business-winner-amid-coronavirus-lockdowns-the-cloud-11585327905 [cited 2020 Aug 31]. Available from. [Google Scholar]
- Tomek K.J., Volkel K., Simpson A., Hass A.G., Indermaur E.W., Tuck J.M. Driving the scalability of DNA-based information storage systems. ACS Synth. Biol. 2019;8(6):1241–1248. doi: 10.1021/acssynbio.9b00100. [DOI] [PubMed] [Google Scholar]
- Trelles O., Prins P., Snir M., Jansen R.C. Big data, but are we ready? Nat. Rev. Genet. 2011;12(3):224. doi: 10.1038/nrg2857-c1. [DOI] [PubMed] [Google Scholar]
- U.S. News and World Report [Internet] U.S. News & World Report, Inc; 1964. Machines Smarter Than Men? Interview with Dr. Norbert Wiener, Noted Scientist; pp. 84–87.https://profiles.nlm.nih.gov/spotlight/bb/catalog/nlm:nlmuid-101584906X7699-doc [cited 2020 Jul 9]. Available from: [Google Scholar]
- Wang Y., Noor-A-Rahim M., Zhang J., Gunawan E., Guan Y.L., Poh C.L. High capacity DNA data storage with variable-length oligonucleotides using repeat accumulate code and hybrid mapping. J. Biol. Eng. 2019;13:89. doi: 10.1186/s13036-019-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Noor-A-Rahim M., Zhang J., Gunawan E., Guan Y.L., Poh C.L. Oligo design with single primer binding site for high capacity DNA-based data storage. IEEE/ACM Trans. Comput. Biol. Bioinf. 2019 doi: 10.1109/TCBB.2019.2940177. [DOI] [PubMed] [Google Scholar]
- Willsey M., Stephenson A.P., Takahashi C., Vaid P., Nguyen B.H., Piszczek M. Puddle: A dynamic, error-correcting, full-stack microfluidics platform. ASPLOS. 2019;19:183–197. [Google Scholar]
- Wired [Internet] The Rise of DNA Data Storage. 2018. https://www.wired.com/story/the-rise-of-dna-data-storage/ [cited 2020 Jul 9]. Available from:
- Yazdi S.M.H.T., Yuan Y., Ma J., Zhao H., Milenkovic O. A rewritable, random-access DNA-based storage system. Sci. Rep. 2015;5:14138. doi: 10.1038/srep14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirnov V., Zadegan R.M., Sandhu G.S., Church G.M., Hughes W.L. Nucleic acid memory. Nat. Mater. 2016;15(4):366–370. doi: 10.1038/nmat4594. [DOI] [PMC free article] [PubMed] [Google Scholar]