Abstract
Background
Fetal nucleated red blood cells (NRBC) from maternal circulation are rare events but can be enriched and used to evaluate the genetics of the fetus. We compared two simplified selection methods of the fetal cells from the maternal blood.
Methods
We isolated fetal cells from maternal blood through double‐density gradient centrifugation followed either by magnetic cell selection, based on the paramagnetic proprieties of the NRBC hemoglobin, converted to methemoglobin, or by a positive magnetic‐activated cell sorting (MACS) enrichment, using anti‐CD71 monoclonal antibodies. Finally, the cells were identified through fluorescence in situ hybridization (FISH) with specific chromosome X and Y probes.
Results
We processed 10 mL of peripheral blood samples from 27 pregnant women with singleton normal male fetuses. Hemoglobin‐based enrichment isolated significantly more NRBCs: 29.7 × 104 cells than anti‐CD71 MACS: 10.1 × 104 cells (P < .001). The FISH analysis found at least one XY cell in 81.5% and 61.5% of cases, respectively, for paramagnetic and anti‐CD71 selection. Also, the average number of XY cells identified through paramagnetic selection was 5.09 ± 2.5, significantly higher than those observed through CD71 sorting: 3.38 ± 1.7 cells (average ± SE) (P = .03).
Conclusion
The combination of density gradient centrifugation with paramagnetic selection has the advantage of simplicity and achieves a minimal manipulation and treatment of cells. It yields an increased number of NRBCs and FISH confirmed fetal cells, compared to the anti‐CD71 sorting.
Keywords: CD71, fluorescent in situ hybridization, magnetic‐activated cell sorting, nucleated red blood cells, prenatal diagnosis
1. INTRODUCTION
Prenatal diagnosis has become critical for the management of high‐risk patients and fetal ultrasound defects. Currently, a definitive prenatal diagnosis needs an invasive procedure, most frequent a chorionic villous biopsy or an amniocentesis. These procedures are associated with a low, measurable risk for the fetus and the mother. 1 Therefore, significant efforts have been made to develop non‐invasive diagnosis methods with similar performance.
Currently, non‐invasive prenatal testing (NIPT) techniques are based mainly on cell‐free fetal DNA analysis in the maternal blood. However, these methods can accurately identify common aneuploidies (trisomy 21, 18, and 13), 2 but they are, at this moment, ineffective in detecting copy‐number variations, which are another important group of prenatal abnormalities.
Many studies proved the presence of fetal cells in maternal blood. 3 Intact fetal cells from the maternal circulation can overcome the cell‐free NIPT's limitations because they could be a source of pure fetal genomes. They are a rare event, with a frequency of one in 104 to 109, which represents a challenge for actual technologies. 4 However, these cells can be enriched from the maternal circulation and used in different tests to assess the fetus genetics. That leads to interesting opportunities for prenatal evaluation.
After years of oblivion, the interest in the analysis of the fetal cells non‐invasively isolated from maternal circulation reborn. 5 That is due to recent developments in single‐cell analysis technologies, which opens opportunities for prenatal screening. 6 , 7
Fetal nucleated red blood cells (NRBC) are a reliable candidate target because they have a limited lifespan, 8 can be differentiated morphologically from maternal cells, 9 contain a representation of whole fetal genome, 10 have specific markers (eg, embryonic and fetal globin) and allow analysis with chromosomal fluorescence in situ hybridization (FISH). 11
Because the number of fetal NRBC in maternal blood is small, many attempts have been made to enrich these cells: density gradient centrifugation, magnetic‐activated cell sorting (MACS), fluorescence‐activated cell sorting (FACS), lectin‐binding method, micro‐beads sedimentation, 12 and micromanipulation. However, the procedures are complex that limits extensive studies. In recent years, many approaches based on microfluidics chips have been developed to capture circulating fetal cells. 13 , 14 , 15 , 16 , 17
Despite considerable progress, reproducibility, and reliability of isolation and detection of fetal cells from maternal blood remain poor. 18 That was attributed to the rarity and variability of fetal cells among pregnancies. 19 Also, current methods require a great deal of work and specific cellular localization or manipulation technologies that loose or affect the cells. Thus, the selection methods should be relatively simple, efficient, and reproducible.
This study aimed to compare two simplified selection methods of the fetal cells in the blood of normal pregnant women. First was a magnetic cell selection based on the paramagnetic proprieties of the hemoglobin, converted to methemoglobin. Second was a positive MACS enrichment, using anti‐CD71 monoclonal antibodies. The fetal cells were recognized through the visualization of the Y chromosome by FISH.
2. MATERIAL AND METHODS
2.1. Patients
We included women with singleton pregnancies with a male fetus, attending for routine antenatal care or prenatal diagnosis. After an informed consent, approved by the University Ethics Committee, we collected 10‐16 mL of maternal peripheral venous blood into K2 EDTA vacutainers and processed within 1‐4 hours. In all cases, there was a normal fetal development without structural or chromosomal abnormalities. The fetal sex was determined by ultrasound, confirmed by amniocentesis or at birth. We harvest the maternal blood samples before any invasive fetal procedure.
2.2. Separation and enrichment of nucleated cells from the maternal blood
In the laboratory, maternal samples were mixed for 15 minutes on a roller, diluted 1:1 with phosphate‐buffered saline (PBS) Dulbecco without Ca and Mg (Biochrom AG) and carefully layered on a double‐density gradient prepared with 10 mL of 1.119 g/mL and 10 mL of 1.077 g/mL Percoll (Fluka). Then, at room temperature (20°C), samples were centrifuged at 500 g for 30 minutes (2 acceleration, no brake). After that, the cell ring at the interface of the two density gradients was collected with a Pasteur pipette and washed twice with PBS Dulbecco's solution (1:1 dilution). Then, another by centrifugation at 500 g for 10 minutes followed. The cell layer of interest was localized with density marker beads (green 1.102 g/mL and orange 1.035 g/mL, Sigma). After removal of the supernatant, the washed cells were re‐suspended in 3 mL buffer prepared by a 1:10 dilution of the MACS BSA Stock Solution with autoMACS™ Rinsing Solution (named Buffer B). The viable cells thus selected were counted using a Bürker‐Türk chamber, after which the sample was divided into two aliquots. These were processed as paired samples using the hemoglobin enrichment and magnetic‐activated cell sorting (MACS) techniques.
Magnetic cell selection with hemoglobin was based on the paramagnetic properties of the nucleated cells when the contained hemoglobin is converted to methemoglobin. For this, one aliquot of the suspension was incubated with 1.5 mL of 50 mmol/L NaNO2 solution for 10 minutes at room temperature. The probe was then passed through a MiniMACS MS+ magnetic column under a magnetic field of 1.4T provided by a VarioMACS™ magnet (Miltenyi Biotech). We used a low flow rate of 270 s/mL controlled with a NE‐500 programmable syringe pump (World Precision Instruments Inc). 14 The column was then washed with 500 µL buffer B at the same flow rate. After rinsing, the column, removed from the magnetic field, was eluted with 2.5 mL Hanks’ balanced salt solution (HBSS) at high flow, according to the manufacturer's protocol.
Cells from the other aliquot of the suspension were enriched by CD71 magnetic positive sorting technique, following a slightly modified manufacturer protocol (Miltenyi Biotech), previously described. 20
Viable cells from the paramagnetic and CD71 positive fractions were counted using a Burker‐Turk chamber. The suspension was gently mixed with a fresh Carnoy solution (1:1 dilution) and centrifuged at 300 g for 10 minutes. The pellet was re‐suspended in 500 µL Carnoy fixative for a minimum of 2 hours. We placed the enriched NRBCs on increased adherence slides (Superfrost®Plus Gold, Menzel‐Glaser) by cytocentrifugation at 270 g for 5 minutes (Rotofix 32, Hettich) at medium acceleration and brake, using a mean size cyto chamber (60 mm2 area, Hettich Cyto System 2). We obtained, for each case, a single‐cell spot, with 0.3‐3 × 105 total cells/slide. Then, the slides were cytocentrifuged at 1100 g for 1 minute and air‐dried. Finally, the slides were successively fixed with 100% methanol for 5‐7 minutes and formaldehyde (2% in PBS) for 10 minutes at room temperature.
To assess the protocol results and to check the presence of hemoglobin‐rich cells, slides from some patients were stained with benzidine and May‐Grunwald/Giemsa. In practice, the slides were exposed to a solution of 0.25% benzidine (Sigma‐Aldrich, #3503‐1G) in methanol for 3 minutes, then developed in a solution of hydrogen peroxide and methanol for 1.5 minutes in the dark (1.25 mL 30% hydrogen peroxide in 50 mL 50% ethanol). After two rapid rinses in dH2O, the slides were fixed for 10‐15 minutes in absolute methanol. We performed the May Grunwald Giemsa staining according to the standard methods, the slides being finally washed with dH2O, and then with tap water for color enhancement. 14 However, we found that these treatments affected the quality of FISH, so this step was removed from the study protocol.
2.3. Fluorescence in situ hybridization analysis (FISH)
The FISH analysis was performed using specific satellite enumeration probes for chromosomes X and Y (Kreatech Poseidon™, Leica Biosystems), and the rapid protocol was presented elsewhere. 11 The fluorescent signals were analyzed using a Axioscop 1.0 microscope with ×100 objective and triple band‐pass filters (Zeiss). The images were captured and processed using a digital camera and TissueFAXS software (TissueGnostics). The system automatically recorded the cells from the whole cytocentrifugation spot, followed by manual analysis of images. We choose for analysis only intact cells that were not overlapping. When a Y chromosome was suspected, the field was rechecked manually. Generally, all identified spots were counterchecked for non‐specificity in all other filters at a magnification of ×1000.
The enrichment steps and analysis of fetal cells were performed without knowledge of clinical details or fetal sex.
2.4. Statistical analysis
Data were analyzed using the SPSS program, version 23.0 (SPSS). The Gaussian distribution of data was evaluated with the Shapiro‐Wilk test. Non‐parametric tests (Wilcoxon Signed Ranks Test) were used for comparison between paramagnetic and anti‐CD71 sorting of nucleated cells as well as for differentiation of XY cell numbers in the two groups, as all these data had a non‐gaussian distribution. The purity of cell isolation was expressed as 1‐NRBC/total cells for each enrichment method. The efficiency of the two methods in detecting fetal sex (at least one XY cell) was compared using the McNemar test. A P‐value of <.05 was considered significant.
3. RESULTS
We tested peripheral blood samples from 27 normal pregnancies with unique, healthy fetuses. Seventy‐seven percent of them were at their first pregnancy, and the others have had no history of a male fetus. The gestational age had a mean of 21 weeks (range 12‐30). Maternal age has a median of 26 years (range 16‐38). From 10 mL of maternal blood, we isolated a mean of 9.6 ± 1.2 × 106 mononuclear cells (Figure 1) (mean ± SE).
FIGURE 1.
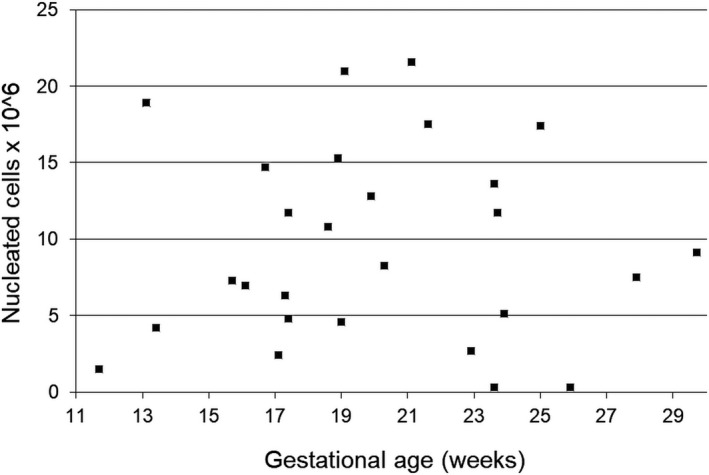
Number of NRBC isolated by centrifugation in the density gradient, depending on gestational age
The median number of isolated NRBC, corrected for 10 mL blood, was 29.7 × 104 cells (range: 0.9‐76.8 × 104) after hemoglobin enrichment and 10.1 × 104 cells (range: 1‐28.8 × 104) after anti‐CD71 magnetic‐activated sorting. There is a significant statistical difference between the numbers of total NRBC isolated by the two techniques (Wilcoxon Signed Ranks Test; P < .001) (Figure 2). The median depletion rate (1‐NRBC/total cells) was significantly lower for hemoglobin selection (96.7%) than for the anti‐CD71 sorting (99.1%) (range: 89.2%‐99.8% vs 95.7%‐99.8%; Wilcoxon Signed Ranks Test; P < .001).
FIGURE 2.
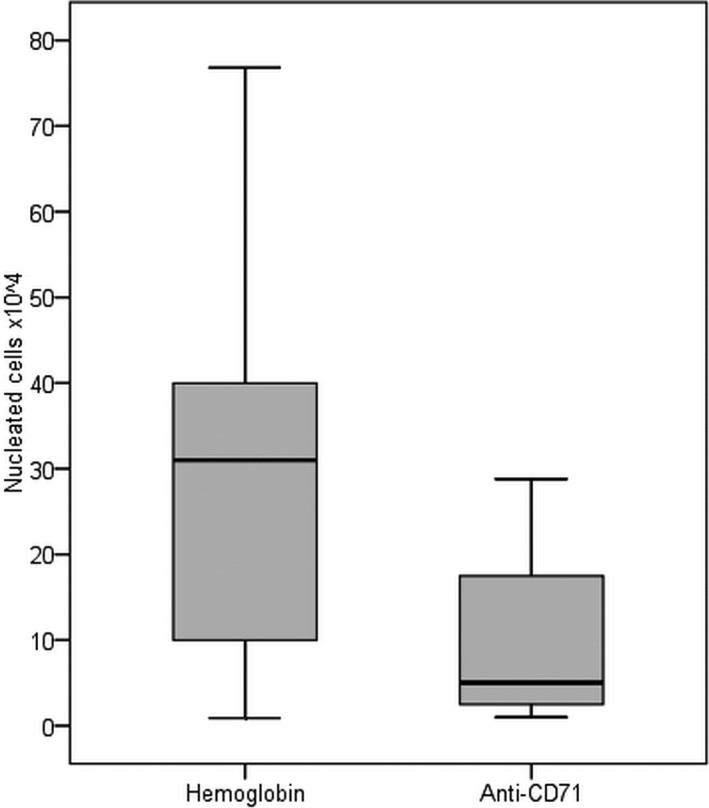
NRBC isolated by paramagnetic hemoglobin and anti‐CD71 techniques. The horizontal line represents the median. The bottom and top of each box shows 25% and 75% of all values, respectively. The upper and lower bars represent the 90% and 10% limits, respectively
The total number of NRBC harvested from double‐density gradient centrifugation, or the total number of cells from hemoglobin or MACS enrichment were not influenced by parity or gestational age.
In the case of paramagnetic selection, the FISH analysis found at least one male cell in 22 of the 27 (81.5%) cases. The average number of XY cells viewed was 5.09 ± 2.5 (mean ± SE) cells, with a range between 2 and 10. For anti‐CD71 selection, the FISH analysis found at least one male cell 16 from the 27 women (61.5%). The average number of XY cells, thus visualized, was 3.38 ± 1.7 (average ± SE), with a range between 1 and 6. That represents about one cell per 1 mL of maternal blood. There was no statistically significant difference between the percentage of nuclei with an XY signal in hemoglobin and anti‐CD71 sorted samples (McNemar test, N = 26, P = .109). However, the number of NRBC was significantly higher in hemoglobin selected samples (P = .03, Wilcoxon Signed Ranks Test) (Figure 3).
FIGURE 3.
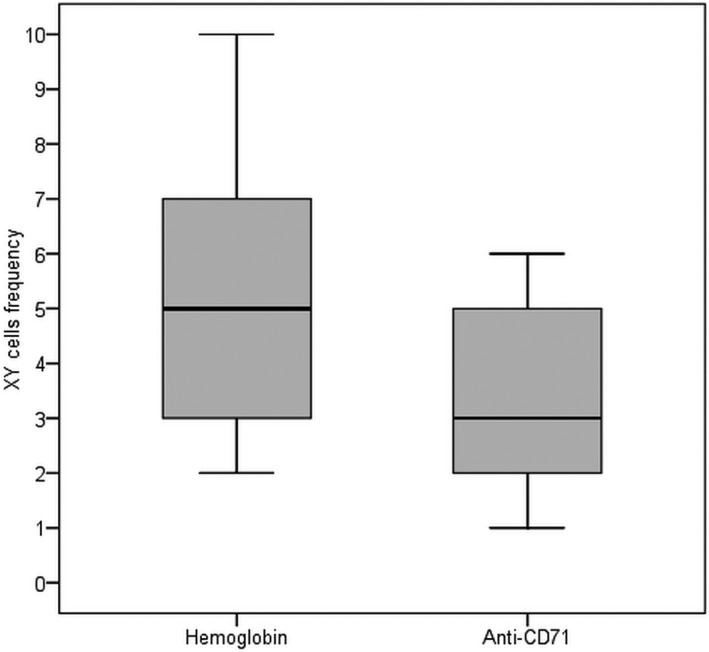
The number of XY nuclei identified by FISH for hemoglobin enriched and anti‐CD71 sorted samples. The horizontal line represents the median, the inferior and superior bars represent the limits of 10%, respectively, 90%. The bottom and top of each box shows 25% and 75% of all values, respectively
All control slides, with male cord blood samples, achieved easily identifiable spots for all fluorochromes. Our FISH protocol achieves an efficiency over 90% for the X and Y chromosome samples. All probes were processed by female assistants only, to limit the contamination.
The final enriched fractions were assessed using benzidine/May‐Grunwald/Giemsa staining. That found the isolation of several cell types, including erythrocytes, white blood cells, and NRBCs. The NRBCs were recognized as cells with a low nucleus to cytoplasm ratio, a small dense nucleus, and an orthochromatic nongranular cytoplasm. 21 The hemoglobin in the erythroid cells was specifically stained using a pseudo‐peroxidase reaction with benzidine. Thus, the cytoplasm of the hemoglobin containing NRBCs appears golden‐brown, improving their detection (Figure 4).
FIGURE 4.
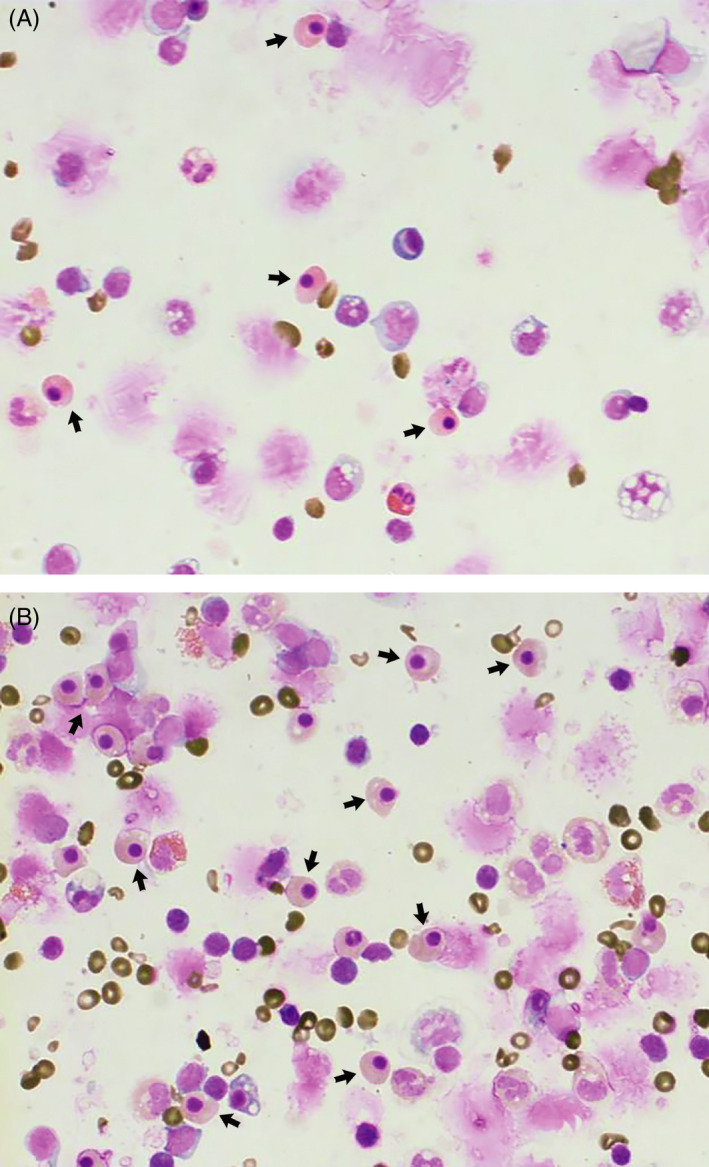
Cytospin preparation of enriched cells from anti‐CD71 positive magnetic sorting (A) and paramagnetic selection (B), stained with benzidine/May Grunwald/Giemsa (×400). The NRBCs (arrows) were recognized as cells with a low nucleus to cytoplasm ratio, a small dense nucleus, and an orthochromatic nongranular cytoplasm. Also, by benzidine staining, the cells with a golden‐brown cytoplasm contain hemoglobin (NRBC, RBC), and cells without hemoglobin (WBC) have a blue cytoplasm
4. DISCUSSION
Our study assessed the ability to isolate the fetal nucleated erythroblasts by a simplified method. After a common double‐density gradient enrichment, we compared the efficacy of the magnetic selection using the paramagnetic properties of hemoglobin with the magnetic sorting using anti‐CD71 antibodies. In the first case, the method selects erythroblasts containing magnetic active hemoglobin, converted to methemoglobin, while in the second, the fetal erythroblasts are screened according to the presence of CD71 antigens. The assessment of the two methods was performed by highlighting the Y chromosome using FISH.
Double‐density gradient centrifugation provided mononuclear concentrations with a wide range of variations. Similar values also acquired Lim and col. 22 (mean of 11.8 × 106 cells/ 10 mL blood) and Reading and col. 23 (mean of 17.47 × 106 cells/ 10 mL whole blood), using a 1.077 g/mL density gradient centrifugation with Percoll or Histopaque, respectively. Kwon and col., using the same double‐density gradient as in our study, but with other osmolarities, found similar results: a mean of 7, respective 4.9 × 106 NRBC cells per 10 mL blood, for gestational ages of 17 and 28 weeks. 24 Our value (950 NRBC/µL) also fits in the range described in neonates. 25 The number of cells recovered by the double‐density gradient had a considerable variation. That may directly influence the number of cells from magnetic sorting and probably the number of identified fetal cells.
The stationary capture of fetal nucleated erythroblasts in the magnetic column through a single separation cycle allowed the elimination of non‐specific cells with a 96% efficiency (median 96.7%, percentiles 20 and 80, 94.9% and 98.7%, respectively). However, these values are lower than those obtained by Huang et al in a similar experiment (median separation efficiency 98.6%, percentiles 20 and 80, 98.1% and 99%, respectively). This difference could appear by the higher number of cells initially introduced in the experiment because the separation was carried out by a microfluidic device. 14
The second method was a positive magnetic sorting using CD71, the transferrin receptor present on erythroid cells such as nucleated erythrocytes. We achieved a mean depletion of 98.6%, similar to that obtained by others. 23 However, other groups, using a similar method, have obtained significantly more CD71 positive cells (mean of 34 to 149 × 104 cells/10 mL blood), with a lower depletion rate (94%‐97%). 24 , 26 Variations in the initial cell number and the different measurement methods (automatic vs manual) can explain these differences.
We did not find the variation of isolated total NRBC with gestational age like others, using a microfluidic or microbead selection. 12 , 27
In this study, we found through FISH at least one XY cell in 81.5% of the pregnant women bearing a male fetus, in the case of the paramagnetic selection and in 59% for that with anti‐CD71 antibodies, respectively.
The rate at which the CD71 positive selection followed by FISH found a male sex fetus varies widely in the literature from 24% to 100%. 24 , 28 Most studies report values ranging from 50% to 60%. 29 , 30 This phenomenon could appear because: (a) only a small part of the fetal cells are nucleate, 31 (b) we can select only a reduced proportion of the fetal erythroblasts, (c) the fetal erythroblasts have a large number variation in the maternal circulation, (d) the fetal erythroblasts do not survive or are strongly affected through the enrichment process, with numerous cell treatments. From this point of view, some studies have found that most fetal erythroblasts are not suitable for FISH analysis, about 43% of them being apoptotic. 32 Erythroblasts could find in the maternal blood a higher oxygen concentration, which promotes apoptosis 33 and reduces nucleus size. 34 Thus, probably most fetal erythroblasts are not suitable for the FISH analysis. Other authors suggest contamination, especially with persistent fetal cells from previous pregnancies. 24 In our study, the percentage of women at their first pregnancy was high (80%), and the others did not had male children. These could partially explain the reduced frequency of identification of XY fetal cells.
Finally, we found a low number of confirmed XY cells in all samples, an average of five cells in 10 mL blood for paramagnetic selection, and three cells in 10 mL blood for anti‐CD71 selection. New technologies based on microfluidic devices achieved a slightly improved number, in normal pregnancies: 2.38‐7.25 fetal cells per mL of maternal blood in Huang et al 17 study, or at least 1‐11 fetal cells per mL in Ma et al 15 study.
However, there are some limitations to the study. First, the CD71 and hemoglobin are not specific to fetal cells, being expressed as well in maternal erythroid precursors. Therefore, we expect many maternal cells selected by the two enrichment methods, which must be differentiated by FISH. Second, the number of available fetal cells is inevitably reduced by the numerous procedures like centrifugation, re‐suspension. Third, the cytospin itself could be a limiting factor, leading to an uneven cell distribution, not fixing all cells, and even degrading some of them. At this moment we cannot estimate the weight of these procedures in the result. Fourth, the automatic microscopy has allowed the study of hundreds of thousands of cells on a single slide, the time required to scan a single centrifugation spot requiring about 12 hours. The scan is limited by the fact that the fluorescent signals are on different levels in the same nucleus. We needed a special algorithm to focus and combine multiple acquired planes. Fluorescence visualization signaled a lot of candidate cells, some with red, as well as green signals. We do not know the cause of these false‐positive red signals. They may appear by non‐specific binding of the sample or maybe small particles present in the wash solution. 35
Our methods were based on a single phase of detection and confirmation of the fetal cells, applying the FISH detection to all cells resulting from the positive selection process. Thus, we excluded many centrifugations, various cell staining and discoloration treatments, which could make the fetal cells unsuitable for FISH analysis. This approach, however, required automatic slide assessment on the fluorescence microscope.
From a technical point of view, the combination of density gradient centrifugation with paramagnetic selection represents a unique, original approach. It has the advantage of simplicity and achieves a minimal manipulation and treatment of cells. The technology is much easier to be applied compared to most approaches commonly used to enrich and screen fetal nucleated erythroblasts.
The isolation and identification of fetal cells in the maternal blood is a considerable challenge due to the low number of these cells. To be used in prenatal screening, new methods of cell enrichment or other specific antibodies are required.
ACKNOWLEDGMENTS
Part of this work was founded initially through National Program for Research Development and Innovation II, grant 1196.
Nemescu D, Constantinescu D, Gorduza V, Carauleanu A, Caba L, Navolan DB. Comparison between paramagnetic and CD71 magnetic activated cell sorting of fetal nucleated red blood cells from the maternal blood. J Clin Lab Anal. 2020;34:e23420 10.1002/jcla.23420
REFERENCES
- 1. Salomon LJ, Sotiriadis A, Wulff CB, Odibo A, Akolekar R. Risk of miscarriage following amniocentesis or chorionic villus sampling: systematic review of literature and updated meta‐analysis. Ultrasound Obstet Gynecol. 2019;54(4):442‐451. [DOI] [PubMed] [Google Scholar]
- 2. Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell‐free DNA in maternal blood in screening for aneuploidies: updated meta‐analysis. Ultrasound Obstet Gynecol. 2017;50(3):302‐314. [DOI] [PubMed] [Google Scholar]
- 3. Sekizawa A, Purwosunu Y, Matsuoka R, et al. Recent advances in non‐invasive prenatal DNA diagnosis through analysis of maternal blood. J Obstet Gynaecol Res. 2007;33(6):747‐764. [DOI] [PubMed] [Google Scholar]
- 4. Hamada H, Arinami T, Kubo T, Hamaguchi H, Iwasaki H. Fetal nucleated cells in maternal peripheral blood: frequency and relationship to gestational age. Hum Genet. 1993;91(5):427‐432. [DOI] [PubMed] [Google Scholar]
- 5. Pin‐Jung C, Pai‐Chi T, Zhu Y, et al. Noninvasive prenatal diagnostics: recent developments using circulating fetal nucleated cells. Curr Obstet Gynecol Rep. 2019;8(1):1‐8. [PMC free article] [PubMed] [Google Scholar]
- 6. Vossaert L, Wang Q, Salman R, et al. Reliable detection of subchromosomal deletions and duplications using cell‐based noninvasive prenatal testing. Prenat Diagn. 2018;38(13):1069‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hua R, Barrett AN, Tan TZ, et al. Detection of aneuploidy from single fetal nucleated red blood cells using whole genome sequencing. Prenat Diagn. 2015;35(7):637‐644. [DOI] [PubMed] [Google Scholar]
- 8. Pearson HA. Life‐span of the fetal red blood cell. J Pediatr. 1967;70(2):166‐171. [DOI] [PubMed] [Google Scholar]
- 9. Huang Z, Fong CY, Gauthaman K, Sukumar P, Choolani M, Bongso A. Novel approaches to manipulating foetal cells in the maternal circulation for non‐invasive prenatal diagnosis of the unborn child. J Cell Biochem. 2011;112(6):1475‐1485. [DOI] [PubMed] [Google Scholar]
- 10. Liou JD, Pao CC, Hor JJ, Kao SM. Fetal cells in the maternal circulation during first trimester in pregnancies. Hum Genet. 1993;92(3):309‐311. [DOI] [PubMed] [Google Scholar]
- 11. Nemescu D, Martiniuc V, Gorduza V, Onofriescu M. Fetal aneuploidy diagnosis through rapid fluorescence in situ hybridization (FISH) on uncultured amniocytes. Rev Romana Med Lab. 2011;19(2):161‐167. [Google Scholar]
- 12. Wei X, Ao Z, Cheng L, et al. Highly sensitive and rapid isolation of fetal nucleated red blood cells with microbead‐based selective sedimentation for non‐invasive prenatal diagnostics. Nanotechnology. 2018;29(43):434001. [DOI] [PubMed] [Google Scholar]
- 13. Easley CJ, Karlinsey JM, Bienvenue JM, et al. A fully integrated microfluidic genetic analysis system with sample‐in‐answer‐out capability. Proc Natl Acad Sci USA. 2006;103(51):19272‐19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang R, Barber TA, Schmidt MA, et al. A microfluidics approach for the isolation of nucleated red blood cells (NRBCs) from the peripheral blood of pregnant women. Prenat Diagn. 2008;28(10):892‐899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma GC, Lin WH, Huang CE, et al. A silicon‐based coral‐like nanostructured microfluidics to isolate rare cells in human circulation: validation by SK‐BR‐3 cancer cell line and its utility in circulating fetal nucleated red blood cells. Micromachines. 2019;10(2):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng C, He Z, Cai B, et al. Non‐invasive prenatal diagnosis of chromosomal aneuploidies and microdeletion syndrome using fetal nucleated red blood cells isolated by nanostructure microchips. Theranostics. 2018;8(5):1301‐1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang CE, Ma GC, Jou HJ, et al. Noninvasive prenatal diagnosis of fetal aneuploidy by circulating fetal nucleated red blood cells and extravillous trophoblasts using silicon‐based nanostructured microfluidics. Mol Cytogenet. 2017;10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bianchi DW, Simpson JL, Jackson LG, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. National Institute of Child Health and Development Fetal Cell Isolation Study. Prenat Diagn. 2002;22(7):609‐615. [DOI] [PubMed] [Google Scholar]
- 19. Choolani M, Mahyuddin AP, Hahn S. The promise of fetal cells in maternal blood. Best Pract Res Clin Obstet Gynaecol. 2012;26(5):655‐667. [DOI] [PubMed] [Google Scholar]
- 20. Nemescu D, Constantinescu D, Martiniuc V, Onofriescu M, Carasevici E. Detection of nucleated red blood cells in maternal circulation by magnetic sorting and in situ hybridization. Gineco Ro. 2011;7(1):15‐18. [Google Scholar]
- 21. Troeger C, Zhong XY, Burgemeister R, et al. Approximately half of the erythroblasts in maternal blood are of fetal origin. Mol Hum Reprod. 1999;5(12):1162‐1165. [DOI] [PubMed] [Google Scholar]
- 22. Lim TH, Tan A, Goh VH. Enrichment of fetal trophoblasts and nucleated erythrocytes from maternal blood by an immunomagnetic colloid system. Hum Genet. 1999;104(5):399‐404. [DOI] [PubMed] [Google Scholar]
- 23. Reading JP, Huffman JL, Wu JC, et al. Nucleated erythrocytes in maternal blood: quantity and quality of fetal cells in enriched populations. Hum Reprod. 1995;10(9):2510‐2515. [DOI] [PubMed] [Google Scholar]
- 24. Kwon KH, Jeon YJ, Hwang HS, et al. A high yield of fetal nucleated red blood cells isolated using optimal osmolality and a double‐density gradient system. Prenat Diagn. 2007;27(13):1245‐1250. [DOI] [PubMed] [Google Scholar]
- 25. Christensen RD, Henry E, Andres RL, Bennett ST. Reference ranges for blood concentrations of nucleated red blood cells in neonates. Neonatology. 2011;99(4):289‐294. [DOI] [PubMed] [Google Scholar]
- 26. Prieto B, Candenas M, Ladenson JH, Alvarez FV. Comparison of different CD71 monoclonal antibodies for enrichment of fetal cells from maternal blood. Clin Chem Lab Med. 2002;40(2):126‐131. [DOI] [PubMed] [Google Scholar]
- 27. He Z, Guo F, Feng C, et al. Fetal nucleated red blood cell analysis for non‐invasive prenatal diagnostics using a nanostructure microchip. J Mater Chem B. 2017;5(2):226‐235. [DOI] [PubMed] [Google Scholar]
- 28. Jansen MW, Brandenburg H, Wildschut HI, et al. The effect of chorionic villus sampling on the number of fetal cells isolated from maternal blood and on maternal serum alpha‐fetoprotein levels. Prenat Diagn. 1997;17(10):953‐959. [PubMed] [Google Scholar]
- 29. Fernandez A, Prieto B, Escudero A, Ladenson JH, Alvarez FV. A monoclonal antibody with potential for aiding non‐invasive prenatal diagnosis: utility in screening of pregnant women at risk of preeclampsia. J Histochem Cytochem. 2005;53(3):345‐350. [DOI] [PubMed] [Google Scholar]
- 30. Zhao XX, Suzumori N, Ozaki Y, Sato T, Suzumori K. Examination of fetal cells and cell‐free fetal DNA in maternal blood for fetal gender determination. Gynecol Obstet Invest. 2004;58(1):57‐60. [DOI] [PubMed] [Google Scholar]
- 31. Seppo A, Frisova V, Ichetovkin I, et al. Detection of circulating fetal cells utilizing automated microscopy: potential for noninvasive prenatal diagnosis of chromosomal aneuploidies. Prenat Diagn. 2008;28(9):815‐821. [DOI] [PubMed] [Google Scholar]
- 32. Sekizawa A, Samura O, Zhen DK, Falco V, Farina A, Bianchi DW. Apoptosis in fetal nucleated erythrocytes circulating in maternal blood. Prenat Diagn. 2000;20(11):886‐889. [DOI] [PubMed] [Google Scholar]
- 33. Kondo T, Sekizawa A, Saito H, Jimbo M, Sugito Y, Okai T. Fate of fetal nucleated erythrocytes circulating in maternal blood: apoptosis is induced by maternal oxygen concentration. Clin Chem. 2002;48(9):1618‐1620. [PubMed] [Google Scholar]
- 34. Babochkina T, Mergenthaler S, De Napoli G, et al. Numerous erythroblasts in maternal blood are impervious to fluorescent in situ hybridization analysis, a feature related to a dense compact nucleus with apoptotic character. Haematologica. 2005;90(6):740‐745. [PubMed] [Google Scholar]
- 35. Kolvraa S, Christensen B, Lykke‐Hansen L, Philip J. The fetal erythroblast is not the optimal target for non‐invasive prenatal diagnosis: preliminary results. J Histochem Cytochem. 2005;53(3):331‐336. [DOI] [PubMed] [Google Scholar]


