Abstract
Background
Long non‐coding RNAs (lncRNAs) perform pivotal regulatory roles in tumor development. Our previous work revealed that the lncRNA gastric cancer‐associated transcript 3 (GACAT3) was significantly overexpressed and associated with tumor size and metastasis in gastric cancer.
Methods
Total RNAs were extracted from colorectal cancer (CRC) and reverse transcribed, and then quantitative real‐time PCR (qRT‐PCR) was conducted. Cell counting was performed to assess the effect of GACAT3 on CRC cell line proliferation. Bioinformatics prediction, dual luciferase assay, miRNA mimics, siRNAs, and transfection experiments were applied to determine whether GACAT3 and LINC00152 are reciprocally regulated by miR‐103. The relationship between their expression levels and clinicopathological factors of patients was explored. A receiver operating characteristic (ROC) curve was used to assess the potential diagnostic value of GACAT3 and LINC00152.
Results
GACAT3 was identified to be highly expressed in CRC tissues and associated with cell proliferation. Furthermore, we demonstrated that GACAT3 acted as a competing endogenous RNA of LINC00152 and they were both regulated by miR‐103. Moreover, analysis of clinicopathological characteristics revealed that GACAT3 and LINC00152 were positively correlated with the depth of invasion, TNM stage, lymph node metastasis, and CA19‐9 level. Importantly, a combination of GACAT3 and LINC00152 showed a superior diagnostic capacity compared with the use of the two molecules alone.
Conclusion
Our work shows that GACAT3 and LINC00152 are both overexpressed in CRC and they act as a ceRNA network. Therefore, our data suggest that GACAT3 and LINC00152 may be a promising potential diagnostic biomarker for CRC.
Keywords: colorectal cancer, competing endogenous RNA, GACAT3, LINC00152, LncRNA
1. INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide and the third leading cause of cancer death in both sexes, 1 generally due to the late diagnosis and recurrence or metastasis of tumor cells. 2 In China, CRC is the fifth leading cause of cancer‐related morbidity and mortality with a 5‐year survival rate of only 31%. 3 , 4 Therefore, exploring the key molecules involved in colon cancer progression and demonstrating their functional mechanisms would provide potential diagnostic biomarkers and have a significant impact for CRC therapy.
Long non‐coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without protein‐coding ability. LncRNAs are reported to play crucial roles in fundamental biological processes through regulating gene expression. 5 Recently, growing evidence indicates that aberrantly expressed lncRNAs not only have roles in tumorigenesis but also could be potential diagnostic biomarkers, 6 such as H19 in non‐small cell lung cancer, 7 metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) in osteosarcoma cells, 8 and prostate cancer antigen 3 (PCA3) in prostate cancer. 9 One representative function of lncRNA is that it acts as a microRNA (miRNA) sponge to regulate the stability and/or translation efficiency of other RNAs, and such a lncRNA is called a competing endogenous RNA (ceRNA). 10 Increasing evidence has revealed that lncRNAs act as ceRNAs and possess significant regulatory effects on carcinogenesis and tumor development. 11 For example, LINC00152 eliminates miR‐139‐5p‐mediated inhibition of NOTCH1 by competitively binding to miR‐139‐5p, promoting colon cancer cell proliferation and metastasis. 12 MALAT1 regulates HDAC4‐mediated proliferation and apoptosis by acting as a decoy of miR‐140‐5p in osteosarcoma cells. 8 In previous study, we have identified GACAT3 as a novel lncRNA in gastric cancer tissues based on a lncRNA microarray 13 , 14 and found it to be positively correlated with tumor size, distant metastasis, and tumor‐node‐metastasis (TNM) stage. Recent research identified GACAT3 as a downstream target of the IL‐6/STAT3 signaling pathway and a ceRNA of high mobility group protein A1 (HMGA1) that promotes gastric cancer cell proliferation. 15 , 16 Moreover, further studies have shown that GACAT3 participates in the development of breast cancer, non‐small cell lung cancer, and glioma. 17 , 18 , 19 Thus, GACAT3 might have fundamental roles and serve as a potential biomarker in various types of cancer. However, the role of GACAT3 in colon cancer remains unclear.
In the present study, we demonstrate that GACAT3 is overexpressed in CRC and plays a role in regulating colon cancer cell growth. Furthermore, GACAT3 and LINC00152 are positively correlated and act as ceRNAs, and their expression is associated with clinicopathological characteristics of CRC.
2. MATERIALS AND METHODS
2.1. Cell culture
HCT‐116 human CRC cells were cultured in RPMI 1640 medium (Corning) supplemented with 10% (v/v) fetal bovine serum and 100 U/mL penicillin‐streptomycin (Sigma‐Aldrich).
2.2. RNA extraction and reverse transcription
Total RNAs were extracted from cultured cells or tissues using TRIzol reagent (Invitrogen). For RNA extraction from formalin‐fixed and paraffin‐embedded CRC tissues, EZNA® FFPE RNA Kit (OMEGA) was used. The extracted RNAs were reverse transcribed into cDNAs using HiFiScript 1st strand cDNA Synthesis Kit (CWBIO).
2.3. Quantitative real‐time PCR
QRT‐PCR was performed using the Light Cycler 480 SYBR Green I Master (Roche). The PCR conditions were 95°C for 5 minutes, 95°C for 10 seconds, 56°C for 20 seconds, and 72°C for 30 seconds with the latter three steps repeated for 45 cycles. The primer sequences of LINC00152 were 5′‐CACCAGCATCTTTTCCAACC‐3′ (forward primer) and 5′‐AAGGCCGACTCTCCTACACA‐3′ (reverse primer). The primer sequences of GACAT3 and β‐actin were as described in a previous publication. 16 The relative expression was shown as ∆C t value by subtracting the β‐actin C t value from the lncRNA C t value. 20 Higher gene expression was indicated by a smaller ∆C t value.
2.4. miRNA mimics, siRNAs, and transfection experiments
The mimics of miR‐103, miR‐128, and miR‐138, as well as the non‐targeting negative control were purchased from GenePharma. Lipofectamine 2000 (Invitrogen) was used for transfection. Two LINC00152 siRNAs with sequences of siLINC00152‐1:5‐CAUUUGGUCUUCAUUGAACATT‐3 and siLINC00152‐2:5‐GCUCUAUGACACACUUGAUTT‐3 were used for the knockdown experiment. The GACAT3 siRNAs were previously described 16 and knockdown experiments were performed using transfection reagent RNAiMAX (Invitrogen).
2.5. Dual luciferase assay
The LINC00152 cDNA sequence (1198 bp, ENSG00000222041) was synthesized and inserted into pGL3 to construct pGL3‐LINC00152 by Novobio Biotechnology. pGL3‐GACAT3 was described previously. 15 Dural luciferase assay was conducted according to the protocol from the manufacturer (Promega). In brief, a constructed pGL3 reporter vector was co‐transfected into 293T cells with the reference vector pRL‐SV40 by X‐tremeGENETM HP DNA (Roche). Then, the miRNA mimics was applied 48 hours post‐transfection. The activities of firefly luciferase were normalized to that of Renilla luciferase.
2.6. Subject recruitment and data collection
Sample from two clinical patient cohorts was collected at the Changhai Hospital (Shanghai, China) from 2011 to 2015 as described previously. 20 In brief, a total of 30 fresh CRC tissues and the adjacent colorectal mucosa were obtained to be as cohort 1. Cohort 2 consisted of 406 formalin‐fixed and paraffin‐embedded CRC biopsies. The clinicopathological information was collected, including sex, age, body mass index (BMI), morphological classification (ulcerative type and mass type), depth of invasion (Tis, T1, T2, T3, and T4), differentiation (poor and intermediate/well), intestinal lymph nodes (0, 1‐3, and ≥4), lymph node metastasis (negative and positive), TNM (I–II and III–IV), carcinoembryonic antigen (CEA), and carbohydrate antigen 19‐9 (CA19‐9). The above research was approved by the Human Ethics Committee of Chancel Hospital, Second Military Medical University and performed according to Declaration of Helsinki principles. Each patient had been informed with the written informed consent.
2.7. Statistical analysis
Continuous variables (age, BMI, CEA, CA19‐9, GACAT3 ∆C t, and LINC00152 ∆C t) were described as mean ± standard deviation (SD) or median values and quartiles depending on whether the data had a normal distribution. Student's t tests were used to analyze the continuous variables between two groups. Categorical variables were presented as proportions, and chi‐square test or Fisher’ exact test was used for the comparisons between groups.
The correlation between the expression of lncRNAs and the clinicopathologic data was analyzed using the Pearson or Spearman rank correlation methods. Receiver operating characteristic (ROC) curves were performed to evaluate the diagnostic value of lncRNAs. The graphs were created using GraphPad Prism 6. Statistical analyses were performed using SPSS 18.0 statistical software. A statistically significant difference in all comparisons was defined as P‐value < .05.
3. RESULTS
3.1. GACAT3 promotes cell growth and is overexpressed in CRC
To determine the expression of GACAT3, qRT‐PCR was performed using samples from CRC tissues of cohort 1 as described in Materials and Methods. As shown in Figure 1A, GACAT3 expression was significant (P < .001) higher in CRC tissues than in the adjacent normal tissues based on the statistical analysis. To uncover the role of GACAT3 overexpression, knockdown experiment was performed in the human CRC HCT‐116 cell line, where the expression of GACAT3 was decreased approximately 2.2‐fold by two independent sets of specific siRNAs (Figure 1B). As a result, both GACAT3 siRNAs inhibited the growth of CRC cells (Figure 1C).
Figure 1.
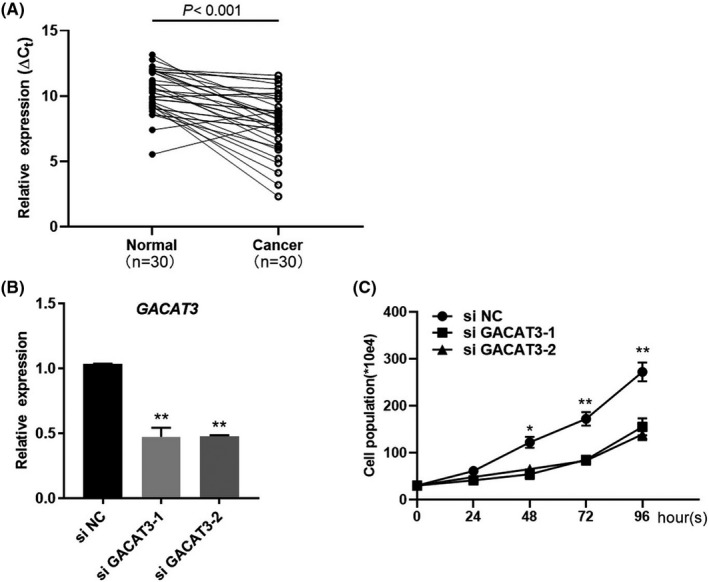
GACAT3 is overexpressed in CRC and promotes cell growth. A, GACAT3 expression in CRC tissues of cohort 1. The ∆C t value by subtracting the β‐actin C t value from the GACAT3 C t value was used. Higher gene expression was indicated by a smaller ∆C t value. B, Knockdown of GACAT3 by gene‐specific siRNAs was validated in human colon cancer cell line HCT116. GACAT3 expression was determined by qRT‐PCR. C, Effects of GACAT3 on cell proliferation. Cell numbers were counted on days 0, 1, 2, 3, and 4 after GACAT3 knockdown. Data represent the mean ± SD of three independent experiments. *P < .05; **P < .01
3.2. GACAT3 and LINC00152 are reciprocally regulated by miR‐103
LncRNAs could regulate each other as ceRNA through the same miRNAs. 10 We therefore asked whether GACAT3 and LINC00152 act as ceRNAs. The entire mRNA sequences of GACAT3 and LINC00152 were obtained from the NCBI database, and common putative miRNA binding sites were scanned using microT and miRcode. As shown in Figure 2A, miR‐103, miR‐128, and miR‐138 had putative binding sites in both the GACAT3 and LINC00152 genes. Next, to verify the miRNA binding sites, miRNA mimics were synthesized and applied to the human CRC cells. The mimics of miR‐103 significantly decreased expression of both GACAT3 (1.4‐fold) and LINC00152 (1.6‐fold) (Figure 2B). The downregulation of LINC00152 by miR‐103 was also reported by a previous study. 21 Although miR‐128 decreased GACAT3 expression, it did not show any effects on the expression of LINC00152, while miR138 had no effects on either lncRNA. To further identify the direct regulatory role of miR‐103 on both lncRNAs, a dual luciferase activity assay was performed and miR‐103 decreased the luciferase expression driven by either GACAT3 or LINC00152 (Figure 2C) suggesting that GACAT3 and LINC00152 are both regulated by miR‐103.
Figure 2.
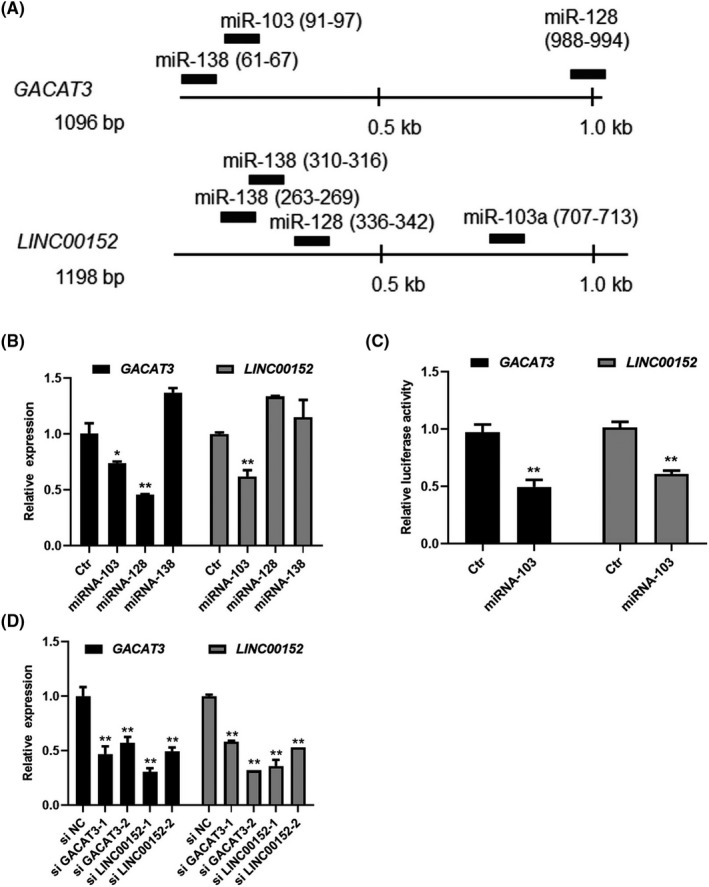
Reciprocal regulation between GACAT3 and LINC00152 through miR‐103. A, GACAT3 and LINC00152 share common binding sites of three miRNAs. Common miRNA binding sites were predicted by microT and miRcode. B, GACAT3 and LINC00152 regulation by the miRNAs. The miRNA mimics of miR‐103, miR‐128, and miR‐138 were applied to HCT116, and then GACAT3 and LINC00152 expression was determined by qRT‐PCR. C, Direct regulation of GACAT3 and LINC00152 by miR‐103 shown by a dual luciferase reporter assay. Mimics of miR‐103 were applied after the transfection of plasmid pGL3‐GACAT3 or pGL3‐LINC00152 into 293T cells, and luciferase assay was performed. D, GACAT3 and LINC00152 have the reciprocal regulatory effects. Knockdown was performed by specific siRNAs of GACAT3 and LINC00152, and then their expression was determined by qRT‐PCR. Data represent the mean ± SD of three independent experiments. *P < .05; **P < .01
We next evaluated whether GACAT3 and LINC00152 had the reciprocal regulatory effects. First, the expression of GACAT3 or LINC00152 was knocked down by specific siRNAs and identified through the qRT‐PCR (Figure 2D). Under GACAT3 knockdown, LINC00152 expression was decreased 1.7‐fold (siGACAT3‐1) and 2.2‐fold (siGACAT3‐2) (Figure 2D). Similarly, GACAT3 expression was downregulated 2.3‐fold (siLINC00152‐1) and 2‐fold (siLINC00152‐2) in cells treated with LINC00152 siRNAs (Figure 2D). These results strongly indicated that GACAT3 and LINC00152 had a reciprocal regulatory association and acted as ceRNAs of each other in CRC cells.
3.3. GACAT3 and LINC00152 are positively correlated in CRC
To evaluate the relationship between GACAT3 and LINC00152 in clinical CRC, two cohorts of samples were used. We first assessed their expression in 30 fresh CRC samples (cohort 1) and observed that GACAT3 expression was positively correlated with LINC00152 expression (r = 0.5707, P < .001) (Figure 3A). Moreover, a positive correlation between the expression of GACAT3 and LINC00152 was observed in the 406 CRC samples embedded in paraffin in cohort 2 (r = 0.9348, P < .001) (Figure 3B).
Figure 3.
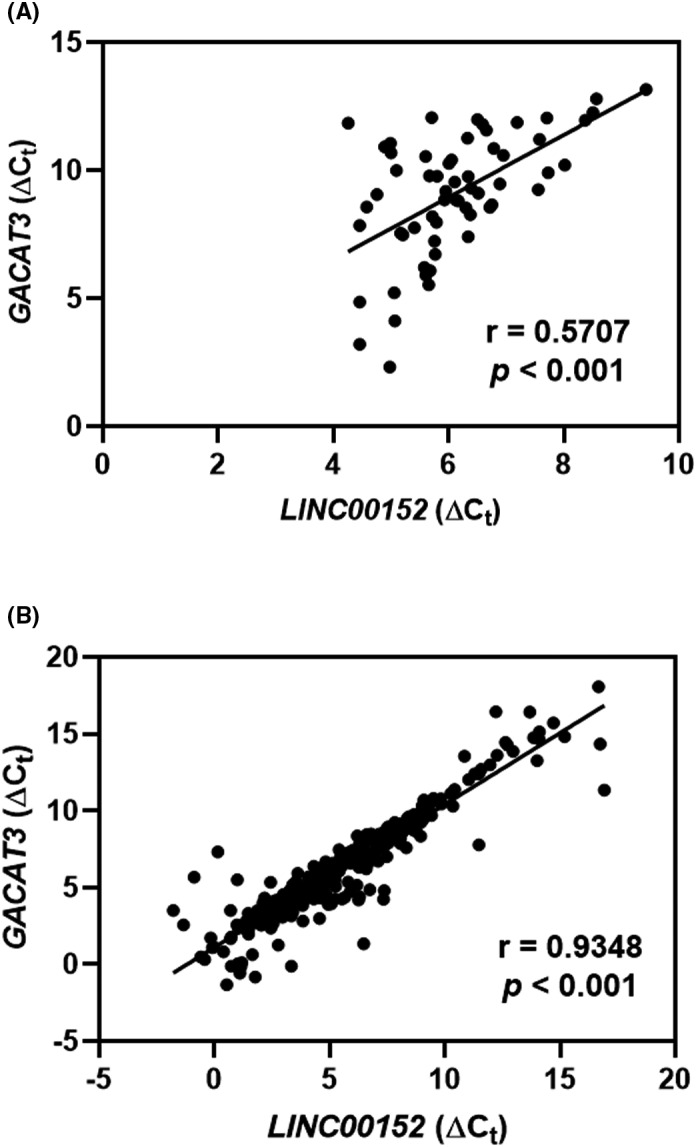
GACAT3 and LINC00152 are positively correlated in CRC. A, Correlation between GACAT3 expression and LINC00152 expression in cohort 1. B, Correlation between GACAT3 expression and LINC00152 expression in cohort 2. The ∆C t value was calculated by subtracting the β‐actin C t value from the GACAT3 or LINC00152 C t value
3.4. GACAT3 and LINC00152 are associated with depth of invasion, lymph node metastasis, and TNM stage
To explore the relationship of GACAT3 and/or LINC00152 expression with the clinicopathological characteristics, cohort 2 was divided into GACAT3‐low and ‐high or LINC00152‐low and ‐high groups. Both GACAT3 and LINC00152 were found to be associated with the depth of invasion, lymph node metastasis, TNM, and CA19‐9 level (Table 1). Furthermore, significantly higher expression of GACAT3 and LINC00152 was observed in the T3‐T4 group than in the Tis & T1‐T2 group (P = .0029 and P = .0458, respectively; Table 2). Moreover, the levels of GACAT3 and LINC00152 were higher in TNM stage III–IV tissues than in stage I–II tissues (P < .001 for both; Table 2). Similarly, higher levels of GACAT3 and LINC00152 were positively related to lymph node metastasis (P < .001 for both; Table 2).
Table 1.
Associations between clinicopathological features and levels of GACAT3 and LNIC00152 in 406 CRC patients
| Variables | GACAT3 | P‐value | LINC00152 | P‐value | ||
|---|---|---|---|---|---|---|
|
Low (n = 203) |
High (n = 203) |
Low (n = 203) |
High (n = 203) |
|||
| Gender | ||||||
| Male | 121 | 113 | .325 | 131 | 106 | .012* |
| Female | 79 | 90 | 72 | 97 | ||
| Age (y) | 59.5 ± 13.9 | 59.15 ± 12.5 | .742 | 59.9 ± 13.5 | 58.6 ± 12.9 | .302 |
| BMI (kg/m2) | 22.9 ± 3.2 | 22.8 ± 3.3 | .733 | 22.8 ± 3.2 | 23.0 ± 3.2 | .604 |
| Morphological classification | ||||||
| Ulcerative type | 125 | 134 | .405 | 127 | 132 | .586 |
| Mass type | 66 | 59 | 65 | 60 | ||
| Depth of invasion | ||||||
| Tis | 18 | 5 | .016** | 19 | 4 | .018** |
| T1 | 10 | 7 | 7 | 10 | ||
| T2 | 38 | 43 | 35 | 46 | ||
| T3 | 60 | 81 | 70 | 71 | ||
| T4 | 73 | 62 | 69 | 66 | ||
| Differentiation | ||||||
| Poor | 12 | 14 | .782 | 13 | 13 | .92 |
| Intermediate/Well | 167 | 174 | 167 | 174 | ||
| Intestinal lymph nodes | ||||||
| 0 | 106 | 112 | .916 | 107 | 113 | .989 |
| 1‐3 | 46 | 51 | 48 | 49 | ||
| ≥4 | 34 | 33 | 33 | 34 | ||
| Lymph node metastasis | ||||||
| Negative | 120 | 99 | <.001*** | 118 | 101 | .0038** |
| Positive | 58 | 104 | 63 | 104 | ||
| TNM | ||||||
| I‐II | 134 | 100 | <.001*** | 135 | 99 | <.001*** |
| III‐IV | 69 | 103 | 68 | 104 | ||
| CEA | 10.6 (6.7‐14.4) | 15.9 (6.0‐25.9) | .324 | 10.8 (6.6‐15.0) | 16.9 (6.8‐3.9) | .27 |
| CA19‐9 | 16.6 (14.2‐19.0) | 33.7 (18.1‐43.3) | .0143* | 20.2 (15.6‐24.9) | 37.0 (21.2‐52.8) | .047* |
Abbreviations: BMI, Body mass index; CA19‐9, Carbohydrate antigen 19‐9; CEA, Carcinoembryonic antigen; TNM, Tumor‐node‐metastasis.
P < .05.
P < .01.
P < .001.
Table 2.
Comparisons of GACAT3 and LNIC00152 expression (∆C t) in groups stratified by depth of invasion, lymph node metastasis, and TNM stage
| Variables | GACAT3 (∆Ct) | P‐value | LINC00152 (∆Ct) | P‐value |
|---|---|---|---|---|
| Depth of invasion | ||||
| Tis & T1‐T3 (n = 128) | 5.92 ± 3.09 | .0029** | 5.15 ± 3.14 | .0458* |
| T3‐T4 (n = 271) | 5.03 ± 2.50 | 4.54 ± 2.50 | ||
| TNM | ||||
| I‐II (n = 234) | 6.17 ± 3.25 | <.001*** | 5.42 ± 3.27 | <.001*** |
| III‐IV (n = 172) | 4.95 ± 2.74 | 4.15 ± 2.73 | ||
| Lymph node metastasis | ||||
| Negative (n = 219) | 5.85 ± 3.02 | <.001*** | 5.07 ± 3.05 | <.001*** |
| Positive (n = 162) | 4.77 ± 2.52 | 4.17 ± 2.69 | ||
P < .05.
P < .01.
P < .001.
3.5. Potential diagnostic values of GACAT3 and LINC00152
We next explored the potential diagnostic value of GACAT3 and LINC00152. The ROC curve analysis was conducted, and the area under ROC curve was 0.8183, 0.7533, and 0.8411 for GACAT3, LINC00152, and their combination, respectively, in cohort 1 (Figure 4A). Moreover, the area under ROC curve was 0.6238, 0.6284, and 0.6675 for GACAT3, LINC00152, and their combination, respectively, in cohort 2 (Figure 4B). The combined use of GACAT3 and LINC00152 slightly increased the diagnostic value.
Figure 4.
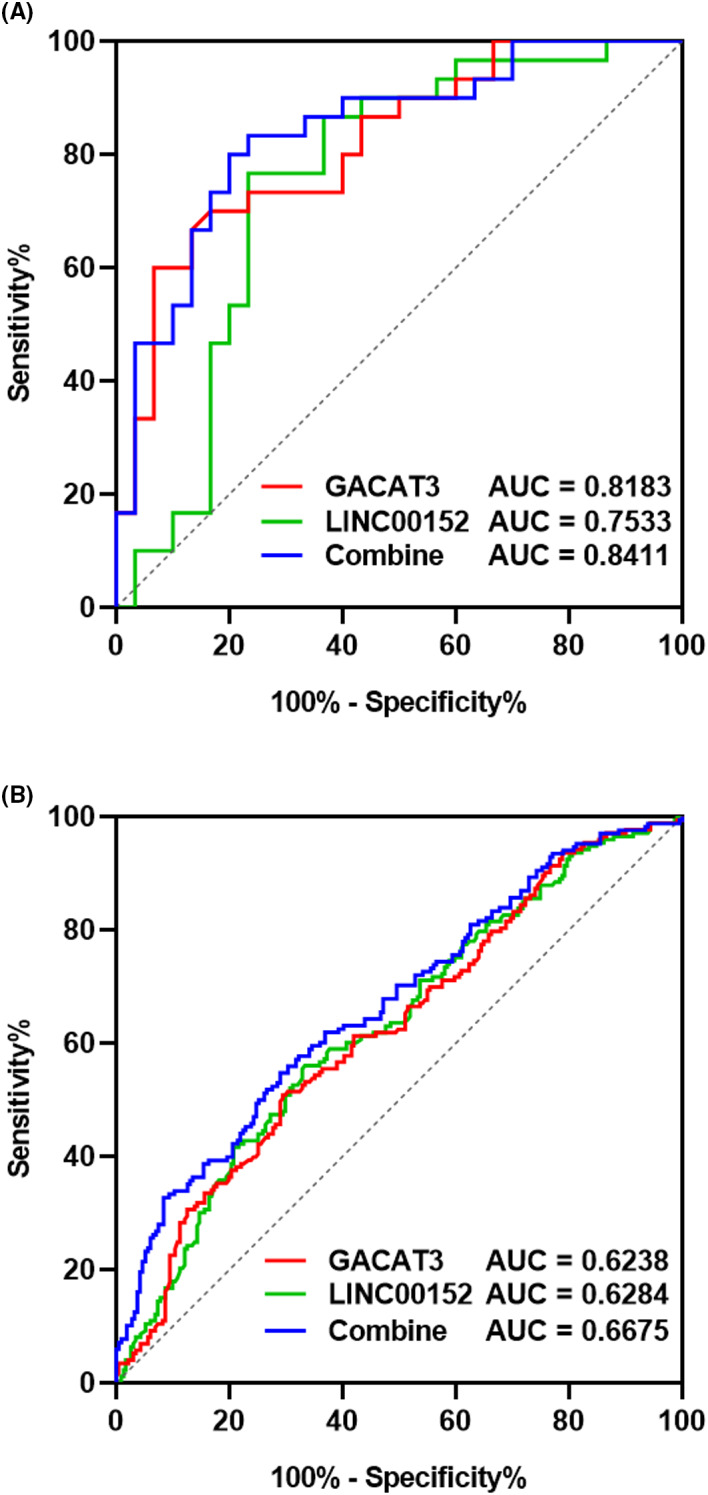
ROC analysis of the expression of GACAT3, LINC00152, and their combination for diagnosis and prognosis of CRC. A, ROC analysis for CRC diagnosis in cohort 1. B, ROC analysis of the colon cancer prognostic value in TNM staging in cohort 2. ROC, receive operating curve; AUC, area under the curve
4. DISCUSSION
Increasing evidence has revealed that lncRNAs perform important regulatory roles in gene expression and contribute to human diseases, including tumor development. 22 , 23 , 24 One of the key mechanisms is that lncRNAs act as miRNA sponges, building a ceRNA network to regulate key signaling pathways in oncogenesis. For example, HOX transcript antisense RNA (HOTAIR) promotes renal cell carcinoma proliferation via miR‐217/HIF‐1α/AXL signaling, 25 and long non‐coding RNA XIST promotes colon cancer development through regulating Wnt/β‐catenin signaling pathway by competitively binding to miR‐34a. 26 Our previous work revealed that GACAT3 is significantly overexpressed in gastric cancer and associated with tumor size and metastasis. 16 Moreover, GACAT3 promotes cell proliferation through inhibiting the expression of cell cycle‐related genes. 15 Furthermore, GACAT3 was reported to be regulated by several miRNAs including miR‐128. 15 Recently, high expression level of LINC00152 has been detected in human malignant tumors of the digestive system. 27 LINC00152 can specifically recognize the EGFR‐binding site and activate the PI3K/AKT signaling pathway to promote the proliferation of gastric cancer cells. 28 In the present study, although several common miRNA binding sites were discovered in both GACAT3 and LINC00152 sequences, only miR‐103 was found to inhibit the expression of both lncRNAs suggesting that GACAT3 might have an oncogenic role by acting as the ceRNA of LINC00152 via competitive binding to miR‐103. A previous study found that miR‐138 could downregulate GACAT3 in gastric cancer cells 15 ; however, this inhibition was not observed CRC cells in the present study suggesting a cancer type‐specific regulatory role of miR‐138.
GACAT3 is a lncRNA recently identified in gastric cancer, and studies have suggested that it plays oncogenic roles and is overexpressed in many types of human tumor tissues. 18 , 29 Here, we report that GACAT3 was highly expressed in CRC tissues and associated with cell proliferation. Importantly, analysis of clinicopathological characteristics revealed that GACAT3 expression was positively associated with the depth of invasion, TNM stage, lymph node metastasis, and CA19‐9 level, which is in line with previous discoveries that GACAT3 is a potential diagnostic biomarker of gastric cancer, 14 , 30 non‐small cell lung cancer, 17 and glioma, 29 suggesting a common fundamental regulatory role of GACAT3 in tumor development.
CRC currently ranks third in tumor incidence rates, which may bring a huge economic burden on patients. Although great advancement in CRC treatment has been seen, the survival rate of CRC patients remains low, which may be related to the lack of a specific and efficient screening method for early stage disease. Some molecular biomarkers, such as CEA and CA19‐9, are widely used in clinical medicine. Nonetheless, the specificity and sensitivity of these conventional molecular biomarkers are dissatisfactory for clinical diagnosis and prognosis. Increasing evidence shows that lncRNAs are potential biomarkers in many types of human malignant tumors, including H19, 20 HOTAIR, 25 GACAT3, 16 and LINC00152. 27 In the present study, we demonstrated that the levels of GACAT3 and LINC00152 were significantly correlated to the depth of invasion, lymph node metastasis, TNM stage, and CA19‐9 level, which are known prognostic features of colon cancer. 31 , 32 , 33 Furthermore, the diagnostic accuracy of GACAT3 and LINC00152 combined is higher than that of either one alone.
In summary, our work shows that GACAT3 and LINC00152 are both overexpressed in CRC and they act as a ceRNA network. The levels of both lncRNAs are correlated with the depth of invasion, lymph node metastasis, TNM stage, and CA19‐9 level. A combination of GACAT3 and LINC00152 showed a superior diagnostic capacity compared with the use of the two molecules alone. Therefore, our data suggest that GACAT3 and LINC00152 may be a promising biomarker for CRC.
ACKNOWLEDGMENTS
This work was supported by the Zhejiang Provincial Natural Science Foundation of China [grant number LY17C060002]; the Natural Science Foundation of Ningbo [grant number 2019A610325]; the Scientific Innovation Team Project of Ningbo [grant number 2014B82002]; and the KC Wong Magna Fund in Ningbo University.
Ye S, Lu Y, Ru Y, et al. LncRNAs GACAT3 and LINC00152 regulate each other through miR‐103 and are associated with clinicopathological characteristics in colorectal cancer. J Clin Lab Anal. 2020;34:e23378 10.1002/jcla.23378
Contributor Information
Shizhong Bu, Email: bushizhongbu@nbu.edu.cn.
Yang Xi, Email: xiyang@nbu.edu.cn.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7‐34. [DOI] [PubMed] [Google Scholar]
- 2. Fedewa SA, Flanders WD, Ward KC, et al. Racial and ethnic disparities in interval colorectal cancer incidence: a population‐based cohort study. Ann Intern Med. 2017;166:857‐866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015. Ca‐Cancer J Clin. 2016;66:115‐132. [DOI] [PubMed] [Google Scholar]
- 4. Jiang Z, Li Y, Han G, et al. Association of serum albumin level with clinicopathologic features and prognosis in colon cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2016;19:80‐83. [PubMed] [Google Scholar]
- 5. Kunej T, Obsteter J, Pogacar Z, et al. The decalog of long non‐coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344‐357. [DOI] [PubMed] [Google Scholar]
- 6. Xiong X‐D, Ren X, Cai M‐Y, et al. Long non‐coding RNAs: an emerging powerhouse in the battle between life and death of tumor cells. Drug Resist Updat. 2016;26:28‐42. [DOI] [PubMed] [Google Scholar]
- 7. Qian B, Wang D‐M, Gu X‐S, et al. LncRNA H19 serves as a ceRNA and participates in non‐small cell lung cancer development by regulating microRNA‐107. Eur Rev Med Pharmacol Sci. 2018;22:5946‐5953. [DOI] [PubMed] [Google Scholar]
- 8. Sun Y, Qin B. Long noncoding RNA MALAT1 regulates HDAC4‐mediated proliferation and apoptosis via decoying of miR‐140‐5p in osteosarcoma cells. Cancer Med. 2018;7:4584‐4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016;70:45‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5:317‐333. [DOI] [PubMed] [Google Scholar]
- 11. Lin P, Wen D‐Y, Li Q, et al. Genome‐wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell Physiol Biochem. 2018;48:1953‐1967. [DOI] [PubMed] [Google Scholar]
- 12. Bian Z, Zhang J, Li M, et al. Long non‐coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5‐FU resistance in colorectal cancer by inhibiting miR‐139‐5p. Oncogenesis. 2017;6(11):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song H, Sun W, Ye G, et al. Long non‐coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu C, Shao Y, Xia T, et al. lncRNA‐AC130710 targeting by miR‐129‐5p is upregulated in gastric cancer and associates with poor prognosis. Tumor Biol. 2014;35:9701‐9706. [DOI] [PubMed] [Google Scholar]
- 15. Lin Y, Li J, Ye S, et al. LncRNA GACAT3 acts as a competing endogenous RNA of HMGA1 and alleviates cucurbitacin B‐induced apoptosis of gastric cancer cells. Gene. 2018;678:164‐171. [DOI] [PubMed] [Google Scholar]
- 16. Shen W, Yuan Y, Zhao M, et al. Novel long non‐coding RNA GACAT3 promotes gastric cancer cell proliferation through the IL‐6/STAT3 signaling pathway. Tumour Biol. 2016;37:14895‐14902. [DOI] [PubMed] [Google Scholar]
- 17. Yang X, Zhang W, Cheng S‐Q, et al. High expression of lncRNA GACAT3 inhibits invasion and metastasis of non‐small cell lung cancer to enhance the effect of radiotherapy. Eur Rev Med Pharmacol Sci. 2018;22:1315‐1322. [DOI] [PubMed] [Google Scholar]
- 18. Zhong H, Yang J, Zhang B, et al. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation in breast cancer through regulation of miR‐497/CCND2. Cancer Biomark. 2018;22:787‐797. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, Zhang M, Lu W. Long noncoding RNA GACAT3 promotes glioma progression by sponging miR‐135a. J Cell Physiol. 2019;234:10877‐10887. [DOI] [PubMed] [Google Scholar]
- 20. Zhao M, Wang H, Chen J, et al. Expression of long non‐coding RNA H19 in colorectal cancer patients with type 2 diabetes. Arch Physiol Biochem. 2019:1‐7. [DOI] [PubMed] [Google Scholar]
- 21. Yu M, Xue Y, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR‐103a‐3p/FEZF1/CDC25A pathway. Mol Cancer. 2017;16:110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Liu B, Ye B, Yang L, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499‐508. [DOI] [PubMed] [Google Scholar]
- 23. Guo Z, Zhou C, Zhong X, et al. The long noncoding RNA CTA‐941F9.9 is frequently downregulated and may serve as a biomarker for carcinogenesis in colorectal cancer. J Clin Lab Anal. 2019;33:e22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu X, Wang D, Lin Q, et al. Screening key lncRNAs for human rectal adenocarcinoma based on lncRNA‐mRNA functional synergistic network. Cancer Med. 2019;8:3875‐3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong Q, Li O, Zheng W, et al. LncRNA HOTAIR regulates HIF‐1α/AXL signaling through inhibition of miR‐217 in renal cell carcinoma. Cell Death Dis. 2017;8:e2772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Sun N, Zhang G, Liu Y. Long non‐coding RNA XIST sponges miR‐34a to promotes colon cancer progression via Wnt/β‐catenin signaling pathway. Gene. 2018;665:141‐148. [DOI] [PubMed] [Google Scholar]
- 27. Xu J, Guo J, Jiang Y, et al. Improved characterization of the relationship between long intergenic non‐coding RNA Linc00152 and the occurrence and development of malignancies. Cancer Med. 2019;8(10):4722‐4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007‐2012. [DOI] [PubMed] [Google Scholar]
- 29. Pan B, Zhao M, Xu L. Long noncoding RNA gastric cancer‐associated transcript 3 plays oncogenic roles in glioma through sponging miR‐3127‐5p. J Cell Physiol. 2019;234:8825‐8833. [DOI] [PubMed] [Google Scholar]
- 30. Xian H‐P, Zhuo Z‐L, Sun Y‐J, et al. Circulating long non‐coding RNAs HULC and ZNFX1‐AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689‐4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Al Natour RH, Saund MS, Sanchez VM, et al. Tumor size and depth predict rate of lymph node metastasis in colon carcinoids and can be used to select patients for endoscopic resection. J Gastrointest Surg. 2012;16:595‐602. [DOI] [PubMed] [Google Scholar]
- 32. Otani T, Yoshimatsu K, Yokomizo H, et al. Clinicopathological differences between proximal and distal pT3 colon cancer and the clinical significance of the depth of cancer invasion beyond the muscularis propria. Hepatogastroenterology. 2012;59:395‐399. [DOI] [PubMed] [Google Scholar]
- 33. Zhou W, Yang F, Peng J, et al. High pretreatment serum CA19‐9 level predicts a poor prognosis for patients with stage III colon cancer after curative resection and adjuvant chemotherapy. Journal of Cancer. 2019;10:3810. [DOI] [PMC free article] [PubMed] [Google Scholar]


