Abstract
Purpose
To explore the effects of PAK4/LIMK1/Cofilin‐1 signaling pathway on the proliferation, invasion, and migration of human osteosarcoma cells.
Methods
The expression of PAK4/LIMK1/Cofilin‐1 was detected by immunohistochemistry in osteosarcoma tissues. The osteosarcoma cell line MG63 was transfected and divided into Mock, Control siRNA, si‐PAK4, LIMK1, and si‐PAK4+LIMK1 groups. Then, the cellular biological features of MG63 cells were detected by CCK‐8, wound‐healing, Transwell, and flow cytometry methods. The relationship of PAK4 and LIMK1 was performed by co‐immunoprecipitation test, and the protein expression of PAK4/LIMK1/Cofilin‐1 was determined by Western blotting. Finally, the effect of PAK4 on the growth of osteosarcoma was verified by subcutaneous transplantation model of osteosarcoma in nude mice.
Results
The expression of PAK4/LIMK1/Cofilin‐1 in both osteosarcoma tissues and cells was up‐regulated. Positive PAK4, LIMK1, and Cofilin‐1 expressions in osteosarcoma were associated with the clinical stage, distant metastasis, and tumor grade. The MG63 cell viability, migration, and invasion, as well as the expression of PAK4, p‐LIMK/LIMK, and p‐Cofilin‐1/Cofilin‐1, were restrained by the knock down of PAK4 while it promoted apoptosis. PAK4 silencing also suppressed the growth of subcutaneous transplanted tumor in nude mice. Co‐immunocoprecipitation showed that LIMK and PAK4 protein can form complex in osteosarcoma cells. Besides, LIMK1 overexpression reversed the inhibition effect of PAK4 siRNA on the growth of osteosarcoma cells.
Conclusion
The expression of PAK4/LIMK1/Cofilin‐1 pathway in osteosarcoma tissues was up‐regulated. Thus, PAK4 inhibition may restrict the osteosarcoma cell proliferation, invasion, and migration but promote its apoptosis via decreasing the activity of LIMK1/Cofilin‐1 pathway.
Keywords: Cofilin‐1, LIMK1, osteosarcoma, PAK4
1. INTRODUCTION
As one of the most common primary malignant bone tumors, osteosarcoma is originated from the mesenchymal tissue. 1 In recent years, the five‐year survival rate of osteosarcoma surgical patients has increased to about 70% with the use of neoadjuvant chemotherapy drugs and the standard application of surgical system. 2 However, due to the high degree of malignancy of osteosarcoma, lung metastasis often occurs before the removal of the primary tumor, and a large number of patients would die owing to its metastasis even if the tumor is under control proportionally. 3 , 4 The pathogenesis of osteosarcoma is an extremely complex process involving many factors and many specific genes in various cellular processes, such as tumor migration, invasion, and proliferation. 5 , 6 , 7 Therefore, a new therapeutic, which aims to improve the prognosis of osteosarcoma patients, is urgently needed. 8
PAK (p‐21‐activated protein kinase), an evolutionarily conserved serine/threonine protein kinase, is thought to be one of the key factors in the signaling network of tumor cells. 9 As a representative member of type II PAK, PAK4 has been shown to be abnormally overexpression in numerous tumors, and the overactivation of PAK4 can improve the malignant transformation and invasion and metastasis of tumor cells via a number of different mechanisms. 10 , 11 In terms of LIMK1 gene, it is located on human chromosome 7q11.23, mainly participating in tumor cell invasion and metastasis. 12 Cofilin, a ubiquitous actin‐binding protein including two subtypes (Cofilin‐1 and Cofilin‐2), is not only essential for cell viability and for actin‐based motility, but also important for the metastasis and invasion of tumor cells. 13 , 14 As reported previously, LIMK1 is a downstream target protein of PAK4, which is involved in the tumor angiogenesis and cell migration by phosphorylating and inactivating Cofilin and regulating the reorganization of cytoskeleton of activator protein. 15 What coincide is that, Ahmed T et al demonstrated a similar finding in prostate cancer cells that PAK4 binds and phosphorylates LIMK1 in a HGF‐dependent manner, thus promoting the cell migration and proliferation. 16 In the study by Chen P et al, miR‐138 was shown to regulate the LIMK1/Cofilin/p‐Cofilin signaling pathway by targeting LIMK1, thus inhibiting the invasion and migration of ovarian cancer cells. 12 More importantly, overexpression of LIMK1 can promote the migration of multidrug resistant osteosarcoma cells in the work from Zhang H et al. 17 However, whether LIMK1/Cofilin‐1 signaling pathway in the growth of osteosarcoma can be mediated by has not been confirmed.
Therefore, the purpose of this study was aimed to explore the influence of PAK4/LIMK1/Cofilin‐1 signaling pathway on the proliferation, invasion, and migration ability of human osteosarcoma cells in vitro and in vivo, to find out an ideal therapeutic target for osteosarcoma.
2. MATERIALS AND METHODS
2.1. Ethical statement
All experiments in this study got permission from the Ethical Committee of our hospital, and consent obtained from all patients or patients’ legal guardian before the study protocol. The animal experiments strictly followed the guidelines for the management and use of laboratory animals published by the U.S. National Institutes of Health. 18
2.2. Tissue samples
The tumor samples were collected from 56 patients who were confirmed from the pathological examination with osteosarcoma during the operation in our hospital. These tissue samples were frozen rapidly and stored at −80°C. In addition, 56 cases of matched adjacent non‐tumor tissues form osteosarcoma patients were selected as the normal control. Demographic information of those patients and clinical features were collected.
2.3. Immunohistochemical staining
The 56 paired osteosarcoma tissues and adjacent non‐tumor tissues were embedded in paraffin, and each paraffin samples was sectioned continuously according to 4 μm. Paraffin section was dewaxed and dehydrated with xylene and gradient alcohol, respectively. After incubation with 3% hydrogen peroxide for 15 minutes, the activity of endogenous peroxidase was eliminated, and then, the antigen was repaired by immersion in 0.01 M citrate buffer. Then, a drop of 5% BSA blocking solution was added to incubate for about 20 minutes, with the appropriate diluted primary antibody (PAK4, LIMK1, Cofilin‐1, 1: 50 dilution; Abcam, Cambridge, MA, USA), followed by putting into a wet box at 4°C overnight and washing with PBS for 3 × 3 times/min. After that, the secondary antibody labeled with biotin was added to incubate at 37°C for 20 minutes and then added a drop of SABC reagent to incubate for 20 minutes. The color reaction was developed with diaminobenzidine (DAB) and then sealed by neutral gum for 30 seconds after hematoxylin re‐dyeing. The results were observed under the microscope.
2.4. Cultivation of osteosarcoma cells
The human osteosarcoma cell lines (MG63, U2OS, OS732, and Saos2) and human osteoblasts (hFOB1.19) cells were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). Cells were cultivated in DMEM solution, which contained 10% inactivated fetal bovine serum (FBS), streptomycin and penicillin, and incubated in a cell cultivator, which was at 37°C and 5% CO2.
2.5. Cell grouping and transfection
MG63 cells in logarithmic growth phase were counted under a microscope after trypsin digestion and inoculated into 24‐well cell culture plate with 1 × 105 cells/mL. Cells were transfected by using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) in accordance with the manufacturer's protocols. Then, osteosarcoma cells were grouped into Mock group (cells without transfected), si‐PAK4 group (cells were transfected with siRNA PAK4), Control siRNA group (cells were transfected with Negative Control siRNA), LIMK1 group (cells were transfected with LIMK1 plasmid), and si‐PAK4+LIMK1 (cells were transfected with siRNA PAK4 and LIMK1 plasmid simultaneously). Meanwhile, siRNA PAK4, Negative Control siRNA, and LIMK1 plasmid were synthesized by Shanghai Jima Biotechnology Co., Ltd.
2.6. CCK‐8 detection
Cells in the logarithmic phase were laid with the density of 1 × 104 cells/well evenly, and the final volume of each well was 100 μL with the peripheral wells filled with PBS. The 96‐well plates were placed incubator, which was at 37°C and 5% CO2. After cells attached to the wall, 10 μL of Cell Counting Kit‐8 (CCK‐8) reagent (Japan Tongren Chemical Research Institute, Dojindo, Japan) was added into each plate. After 2 hours of culture in the incubator, the absorbance value at 450 nm was measured in an enzyme, which labeled instrument at 24, 48, 72, and 96 hours, respectively, with the experimental results recorded.
2.7. Wound‐healing
The cells in the logarithmic growth phase were made into a single cell suspension and counted by a counting plate. 1 × 106 cells were inoculated in the 6‐well plate, respectively, and the cells were cultured to reach about 80%‐90% confluence. Next, the cells were washed with serum‐free medium for 3 times, with fresh serum‐free RPMI‐1640 added into them. Then, the cells were scraped by a 10 μL tip to generate wounds, took pictures (0 hour), and then cultured in incubator, which was at 37°C in 5% CO2. Finally, 6‐well plate was taken out after 48‐hour incubation, and the cells, which were washed with PBS and serum‐free RPMI‐1640 medium for 3 times. Five fields were randomly selected and their images were acquired to count the number of cells that migrated into the wounds.
2.8. Transwell invasion
Each Transwell cell was covered with 50 μL of Matrigel matrix protein, 5 × 104 cells were added with serum‐free Matrigel to invade cells. RPMI‐1640 medium with 10% FBS was added into cells. After 48 hours of cultivation, cells, which were in the upper layer, were wiped off with cotton swabs, and cells, which were in the lower layer, were fixed with 4% paraformaldehyde and stained with crystal violet. The results were observed in 5 fields under the microscope with 400‐times magnification.
2.9. Flow cytometry
The cells in the logarithmic growth phase were taken out and digested with trypsin. The cell concentration was adjusted to about 5 × 104 cells/mL and centrifuged at 1500 rpm for 5 minutes. The supernatant was discarded and 1 × binding buffer was added. Annexin V/FITC with a volume of 5 μL was added, mixed gently, and cultured at room temperature in dark for 10 minutes. Besides, the cells were then incubated in 5 μL of PI for 10 minutes. The results of apoptosis were measured by flow cytometer (BD Biosciences, San Jose, CA, USA).
2.10. Co‐immunoprecipitation method
Osteosarcoma cells were lysed in lysate for 10 minutes, centrifuged at 4°C and 14 000 g for 15 minutes, and swiftly transferred to another centrifuge tube. The supernatant was added with 4 μg LIMK1 antibody and cultivated at 60 μL protein A/G Plus Agarose 4°C for 3 hours. The solution was washed 3 times, each time for 10 minutes, and the precipitated compound was added into the same volume of 2 × sodium dodecyl sulfate (SDS) buffer, and the other steps were the same as the Western blotting test.
2.11. Western blotting
At the logarithmic growth phase, the total protein was extracted from cells by adding lysate, and then, the protein concentration was detected by a bicinchoninic acid (BCA) method. The protein was transferred to polyvinylidene fluoride (PVDF) membrane by 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) electrophoresis, and the membrane was sealed with 5% skimmed milk for 4 hours. Then, the protein was incubated at 4°C overnight with primary antibody PAK4, LIMK1, p‐LIMK1, Cofilin‐1, p‐Cofilin‐1, and β‐actin (Abcam); washed with trisbuffered saline tween‐20 (TBST) for 4 times with 10 minutes each time. The corresponding secondary antibody was added for 1 to 2 hours incubation, washed with TBST for 4 times, with 10 minutes each time. The chemiluminescence (ECL) reagent was developed with β‐actin as the internal control, and the gray value of the target protein was analyzed by ImageJ software.
2.12. Subcutaneous transplantation model of osteosarcoma in nude mice
The MG63 cells in the logarithmic growth phase from each transfection group were used to prepare the single cell suspension with a density of 1 × 107/mL (produced by PBS) and injected into the right forelimb of Balb/c nude mice (purchased from Shanghai SLAC Laboratory Animal Co., Ltd). The mice were divided into Mock, Control siRNA, si‐PAK4, LIMK1, and si‐PAK4+LIMK1 groups with 5 mice in each. The long axes (a) and the short axes (b) of the tumor were measured with the vernier caliper every week to calculate the volume of the transplanted tumor, and the growth curve of the tumor was drawn. At the end of the experiment, the nude mice were killed, whose tumor tissues were taken out to weigh and photograph.
2.13. Statistical analysis
All experiments were repeatedly operated for three times. All data were analyzed by SPSS21.0. The measurement data were expressed by mean ± SD. Comparison between two groups was tested by t test, and among multiple groups was analyzed by one‐way ANOVA. Chi‐squared test was used to make comparison among the enumeration data. Significant differences were represented by P < .05.
3. RESULTS
3.1. Expression of PAK4/LIMK1/Cofilin‐1 in osteosarcoma tissues and cells
In accordance with the immunohistochemistry which was shown in Figure 1A, the expression of PAK4, LIMK1, and Cofilin‐1 were brownish yellow or brown granules in cytoplasm, and the positive expression rates of PAK4, LIMK1, and Cofilin‐1 in osteosarcoma tissues were significantly higher than that in normal tissues (all P < .05). Table 1 showed the clinicopathological differences between positive and negative PAK4/LIMK1/Cofilin‐1 expression among groups. Positive PAK4, LIMK1, and Cofilin‐1 expressions in osteosarcoma were found to be significantly associated with the clinical stage, distant metastasis, and tumor grade (all P < .05). No significant difference was observed between PAK4, LIMK1, and Cofilin‐1 expressions and patients’ age, gender, tumor size, and anatomic location (all P > .05). Besides, the protein expression of PAK4, p‐LIMK1/LIMK1, and p‐Cofilin‐1/Cofilin‐1, which were obviously higher in osteosarcoma cell lines (MG63, U2OS, OS732, and Saos2) than that in osteoblasts (hFOB1.19) (all P < .05, Figure 1A). Among them, the expression of MG63 cells is the most significant, so this cell line was used for further study in the subsequent in vitro experiments.
FIGURE 1.
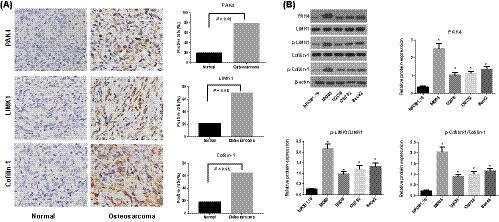
Expression of PAK4/LIMK1/Cofilin‐1 in osteosarcoma tissues and cells. A, The expression of PAK4/LIMK1/Cofilin‐1 in osteosarcoma tissues and adjacent non‐tumor tissues detected by immunohistochemistry; B, The protein expression of PAK4, LIMK1, and Cofilin‐1 in osteosarcoma cell lines and osteoblasts cells determined by Western blotting, *, Compared with osteoblasts hFOB1.19, P < .05
TABLE 1.
Correlation of PAK4, LIMK1 and Cofilin‐1 expressions with clinicopathological features of osteosarcoma
| Clinicopathological features | No. of cases | PAK4 expression | P value | LIMK1 expression | P value | Cofilin‐1 expression | P value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |||||
| Age (year) | .494 | .518 | .772 | |||||||
| >20 | 18 | 13 | 5 | 12 | 6 | 11 | 7 | |||
| ≤20 | 38 | 31 | 7 | 27 | 11 | 25 | 13 | |||
| Gender | .732 | .224 | .382 | |||||||
| Female | 19 | 16 | 3 | 11 | 8 | 14 | 5 | |||
| Male | 37 | 28 | 9 | 28 | 9 | 22 | 15 | |||
| Anatomic location | .744 | .562 | .413 | |||||||
| Tibia/femur | 32 | 26 | 6 | 21 | 11 | 19 | 13 | |||
| Elsewhere | 24 | 18 | 6 | 18 | 6 | 17 | 7 | |||
| Tumor size | .189 | .763 | .143 | |||||||
| ≥8 cm | 37 | 27 | 10 | 25 | 12 | 21 | 16 | |||
| <8 cm | 19 | 17 | 2 | 14 | 5 | 15 | 4 | |||
| Clinical stage | .025 | .040 | .025 | |||||||
| I‐IIA | 30 | 20 | 10 | 17 | 13 | 15 | 15 | |||
| IIB‐III | 26 | 24 | 2 | 22 | 4 | 21 | 5 | |||
| Metastasis | .039 | .016 | .021 | |||||||
| Yes | 20 | 19 | 1 | 18 | 2 | 17 | 3 | |||
| No | 36 | 25 | 11 | 21 | 15 | 19 | 17 | |||
| Tumor grade | .040 | .008 | .003 | |||||||
| Low | 21 | 13 | 8 | 10 | 11 | 8 | 13 | |||
| High | 35 | 31 | 4 | 29 | 6 | 28 | 7 | |||
3.2. Proliferation and apoptosis of osteosarcoma cells in each transfection group
According to Figure 2, the proliferation of osteosarcoma cells in si‐PAK4 group was greatly decreased, while the apoptosis rate was markedly increased compared with Mock group (all P < .05). On the contrary, the speed of cell proliferation in LIMK1 group was accelerated, but the rate of apoptosis was significantly reduced as compared with Mock group (all P < .05). In addition, as compared to si‐PAK4 group, the cell proliferation in si‐PAK4+LIMK1 group was also significantly accelerated, with the decreased rate of apoptosis (all P < .05).
FIGURE 2.
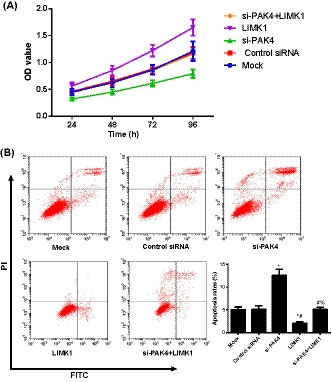
Comparison of the proliferation and apoptosis of transfected osteosarcoma cells in each group. A, The proliferation of MG63 cells in each transfected group detected by CCK‐8 method; B, The apoptosis of MG63 cells in each transfected group measured by flow cytometry; *, Compared with Mock group; P < .05; #, Compared with si‐PAK4 group, P < .05; %, Compared with LIMK1 group, P < .05
3.3. Invasion and migration of osteosarcoma cells in each transfected group
Compared with Mock group, the migration and invasion of osteosarcoma cells were reduced in si‐PAK4 group, while significantly increased in LIMK1 group (all P < .05, Figure 3). Furthermore, migration and invasion of the cells were higher in si‐PAK4+LIMK1 group than in si‐PAK4 group (all P < .05).
FIGURE 3.
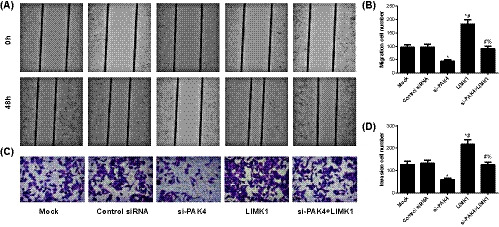
Comparison of invasion and migration of MG63 cells in each transfected group. A,B, Detection of the migration ability of MG63 cells in each transfected group with wound‐healing test; C,D, Measurement of the invasion ability of osteosarcoma cells in each transfected group with Transwell test. *, Compared with Mock group, P < .05; #, Compared with si‐PAK4 group, P < .05; %, Compared with LIMK1 group, P < .05
3.4. LIMK1 was a downstream protein of PAK4
On the basis of co‐immunoprecipitation method (Figure 4A), the protein of LIMK and PAK4 could be detected in the immunoprecipitated with the antibody of LIMK protein, but cannot be detected with IgG immunoprecipitation. The results showed that LIMK and PAK4 can form complex in osteosarcoma cells, demonstrating an interaction between LIMK and PAK4 proteins in osteosarcoma cells. Besides, the expression of PAK4 protein, p‐LIMK/LIMK, and p‐Cofilin‐1/Cofilin‐1 in osteosarcoma cells in si‐PAK4 group were significantly decreased compared with Mock group (all P < .05), whereas the expression of p‐LIMK/LIMK and p‐Cofilin‐1/Cofilin‐1 was increased (all P < .05) with no significant difference of PAK4 protein expression in LIMK1 group (all P > .05). In addition, compared with si‐PAK4 group, p‐LIMK/LIMK and p‐Cofilin‐1/Cofilin‐1 were increased in si‐PAK4+LIMK1 group (all P < .05, Figure 4B,C).
FIGURE 4.
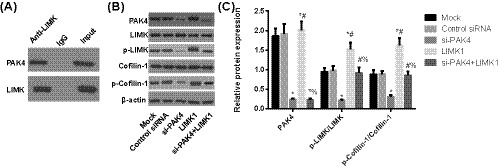
The protein expression of PAK4/LIMK1/Cofilin‐1 in MG63 cells in each group. A, The relationship between PAK4 and LIMK1 detected by co‐immunoprecipitation method; B,C, Determination of the protein expression of PAK4/LIMK1/Cofilin‐1 in MG63 cells in each group by using Western blotting, *, Compared with Mock group, P < .05; #, Compared with si‐PAK4 group, P < .05; %, Compared with LIMK1 group, P < .05
3.5. Subcutaneous growth of osteosarcoma in nude mice
By comparison with Mock group, the growth and weight of subcutaneous tumor in si‐PAK4 group were significantly decreased, but were significantly increased in LIMK1 group (all P < .05, Figure 5). However, compared with si‐PAK4 group, the growth and weight of subcutaneous tumor in si‐PAK4+LIMK1 group were increased (all P < .05).
FIGURE 5.
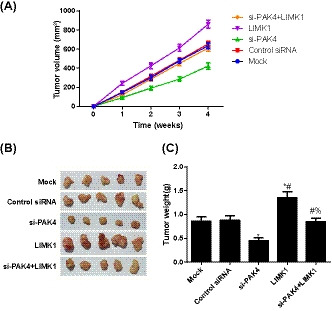
Comparison of subcutaneous tumor of nude mice in each group. A, Growth curve of nude mice in each group; B, Representative images of the tumor morphology in nude mice; C, Comparison of the weight of transplanted tumor of nude mice in each group, *, Compared with Mock group, P < .05; #, Compared with si‐PAK4 group, P < .05; %, Compared with LIMK1 group, P < .05
4. DISCUSSION
Firstly, it was found that the expression of PAK4, LIMK1, and Cofilin‐1 in osteosarcoma tissues and cells were all up‐regulated. Accordingly, other previous studies agree with our results in different types of cancers. For example, the expression of PAK4 was significantly increased in the breast cancer tissues, and gradually increased with the growth of breast cancer. 19 In pancreatic cancer, Cofilin‐1 was up‐regulated and a higher Cofilin‐1 expression predicted a poorer prognosis. 20 Similarly, in osteosarcoma, Jian‐Zeng Yang and his colleagues also noted the increased LIMK1 in both tissues and cells, 21 which suggested that PAK4, LIMK1, and Cofilin‐1 may play vital roles in the development of osteosarcoma. PAK, a protein which is a downstream target of the Rho subfamily proteins Rac and Cdc42, is activated by binding to the activated (GTP‐bound) Cdc42, while LIMK1 can be activated by upstream PAK4. 16 , 22 , 23 Besides, Cofilin is the specific substrate of LIMK1, which could increase the phosphorylation level of Cofilin at Ser3 site. 24 Therefore, the up‐regulation of PAK4, LIMK1, and Cofilin‐1 in osteosarcoma tissues may be due to the activation of Cdc42, which could further activate PAK4 and then activate LIMK1 and Cofilin‐1 considering with the above.
Next, the osteosarcoma cells were selected for transfection experiments, and consequently, PAK4 gene knockout can inhibit the proliferation, invasion and migration of osteosarcoma cells, whereas overexpression of LIMK1 can promote its growth. In agreement with our findings, the proliferation and migration of gastric cancer cells was promoted by miR‐224 via targeting PAK4. 25 The overexpression of LIMK1 was involved in osteosarcoma cell proliferation through regulation of insulin/PI3K/LIMK1 pathway, 26 which could also induce the proliferation and metastasis of gastric cancer cells. 27 In the present study, PAK4 was confirmed to specifically interact with LIMK1 by co‐immunocoprecipitation experiment. Mechanistically, inhibition of PAK4 can significantly reduce the level of p‐LIMK/LIMK and p‐Cofilin‐1/Cofilin‐1, and overexpression of LIMK can significantly reverse the inhibitory effect of PAK4 siRNA on the progress of osteosarcoma cells. In other cancers, like colorectal cancer, the phosphorylation expression levels of LIMK1 and Cofilin were down‐regulated owing to the deficiency of PAK4, with the inhibited migration and invasion. 28 In prostate cancer cells, PAK4 was found to bind and phosphorylate LIMK1, which directly influenced the expression level of LIMK1 to promote the migration rate of cancer cells. 16 The evidence, which was mentioned above, suggested that PAK4 siRNA may suppress the growth and metastasis of osteosarcoma by restraining LIMK1/Cofilin‐1 pathway.
Moreover, LMK1 was identified to be crucial in cell cycle, since it could regulate the recombination of actin, which was a key factor in cell mitosis. 29 Cofilin was the only known enzyme substrate of LMK1 and was phosphorylated by regulating actin recombination with LMK1 to accelerate the cell cycle progress and promote cell proliferation. 30 , 31 As indicated by Guo B et al, after inhibition of PAK4, the proliferation of lung adenocarcinoma cells was restricted, with the arrested cell cycle at the G1 phase and the promoted early apoptosis of lung adenocarcinoma cells. 32 Also, the inhibited PAK4/LIMK1/Cofilin pathway resulted in the cell cycle arrest at the G1 phase and the declined cell proliferation in accordance with the study of Jian Zhang et al. 33 Additionally, LIMK1/Cofilin pathway modulates cytoskeletal dynamics to affect the formation of microfilament actin stress fibers and adhesion plaque, eventually influencing the metastasis behavior of tumor cells. 34 Dan C et al indeed confirmed that PAK4 can act on LIMK1 and promote the phosphorylation of LIMK1 to further phosphorylate cofilin; however, the absence of LIMK1 and cofilin can affect the regulation of PAK4 on cytoskeletal and cell shape changes. 15 The inhibited phosphorylation of LIMK1 and Cofilin‐1 affect the reorganization of cytoskeleton, thus suppressing the invasion and migration of tumor cells, such as in colorectal cancer. 35 In this regard, PAK4 can influence cell mitosis, and indirectly affect the cytoskeleton shaping, ultimately regulating the cell proliferation, invasion, and migration of colorectal cancer by regulating LIMK1/Cofilin‐1 signaling pathway. Last but not least, PAK4 inhibition was further verified in vivo in our work that it may inhibit the progress of osteosarcoma via the subcutaneous transplantation model of osteosarcoma in nude mice.
In a word, our research found that the expression of PAK4, LIMK1, and Cofilin‐1 was up‐regulated in osteosarcoma tissues. Moreover, PAK4 gene knockout can inhibit the proliferation, invasion and migration of osteosarcoma cells by inhibiting LIMK1/Cofilin‐1 signaling pathway, and promote cell apoptosis, which would be a new insight for the targeted treatment of osteosarcoma.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interests regarding the publication of this paper.
Li Z‐F, Yao Y‐D, Zhao Y‐Y, et al. Effects of PAK4/LIMK1/Cofilin‐1 signaling pathway on proliferation, invasion, and migration of human osteosarcoma cells. J Clin Lab Anal. 2020;34:e23362 10.1002/jcla.23362
REFERENCES
- 1. Wu PK, Chen WM, Lee OK, et al. The prognosis for patients with osteosarcoma who have received prior manipulative therapy. J Bone Joint Surg Br. 2010;92(11):1580‐1585. [DOI] [PubMed] [Google Scholar]
- 2. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283‐292. [DOI] [PubMed] [Google Scholar]
- 3. Clark JC, Dass CR, Choong PF. A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol. 2008;134(3):281‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Janeway KA, Grier HE. Sequelae of osteosarcoma medical therapy: a review of rare acute toxicities and late effects. Lancet Oncol. 2010;11(7):670‐678. [DOI] [PubMed] [Google Scholar]
- 5. Kushlinskii NE, Fridman MV, Braga EA. Molecular mechanisms and microRNAs in osteosarcoma pathogenesis. Biochemistry (Moscow). 2016;81(4):315‐328. [DOI] [PubMed] [Google Scholar]
- 6. Broadhead ML, Clark JC, Myers DE, et al. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26(1):1‐18. [DOI] [PubMed] [Google Scholar]
- 8. Broadhead ML, Clark JCM, Choong PFM, et al. Making gene therapy for osteosarcoma a reality. Expert Rev Anticancer Ther. 2010;10(4):477‐480. [DOI] [PubMed] [Google Scholar]
- 9. Dart AE, Wells CM. P21‐activated kinase 4–not just one of the PAK. Eur J Cell Biol. 2013;92(4‐5):129‐138. [DOI] [PubMed] [Google Scholar]
- 10. Radu M, Semenova G, Kosoff R, et al. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14(1):13‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, Lock JG, Olofsson H, et al. Integrin‐mediated cell attachment induces a PAK4‐dependent feedback loop regulating cell adhesion through modified integrin alpha v beta 5 clustering and turnover. Mol Biol Cell. 2010;21(19):3317‐3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen P, Zeng M, Zhao Y, et al. Upregulation of Limk1 caused by microRNA‐138 loss aggravates the metastasis of ovarian cancer by activation of Limk1/cofilin signaling. Oncol Rep. 2014;32(5):2070‐2076. [DOI] [PubMed] [Google Scholar]
- 13. Pavlov D, Muhlrad A, Cooper J, et al. Actin filament severing by cofilin. J Mol Biol. 2007;365(5):1350‐1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Borensztajn K, Peppelenbosch MP, Spek CA. Coagulation factor Xa inhibits cancer cell migration via LIMK1‐mediated cofilin inactivation. Thromb Res. 2010;125(6):e323‐e328. [DOI] [PubMed] [Google Scholar]
- 15. Dan C, Kelly A, Bernard O, et al. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J Biol Chem. 2001;276(34):32115‐32121. [DOI] [PubMed] [Google Scholar]
- 16. Ahmed T, Shea K, Masters JR, et al. A PAK4‐LIMK1 pathway drives prostate cancer cell migration downstream of HGF. Cell Signal. 2008;20(7):1320‐1328. [DOI] [PubMed] [Google Scholar]
- 17. Zhang H, Wang Y, Xing F, et al. Overexpression of LIMK1 promotes migration ability of multidrug‐resistant osteosarcoma cells. Oncol Res. 2011;19(10‐11):501‐509. [DOI] [PubMed] [Google Scholar]
- 18. Mason TJ, Matthews M. Aquatic environment, housing, and management in the eighth edition of the Guide for the Care and Use of Laboratory Animals: additional considerations and recommendations. J Am Assoc Lab Anim Sci. 2012;51(3):329‐332. [PMC free article] [PubMed] [Google Scholar]
- 19. Bi Y, Tian M, Le J, et al. Study on the expression of PAK4 and P54 protein in breast cancer. World J Surg Oncol. 2016;14(1):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Satoh M, Takano S, Sogawa K, et al. Immune‐complex level of cofilin‐1 in sera is associated with cancer progression and poor prognosis in pancreatic cancer. Cancer Sci. 2017;108(4):795‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang JZ, Huang LH, Chen R, et al. LIM kinase 1 serves an important role in the multidrug resistance of osteosarcoma cells. Oncol Lett. 2018;15(1):250‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, Shao Y, Tong Y, et al. Nucleo‐cytoplasmic shuttling of PAK4 modulates beta‐catenin intracellular translocation and signaling. Biochim Biophys Acta. 2012;1823(2):465‐475. [DOI] [PubMed] [Google Scholar]
- 23. King H, Nicholas NS, Wells CM. Role of p‐21‐activated kinases in cancer progression. Int Rev Cell Mol Biol. 2014;309:347‐387. [DOI] [PubMed] [Google Scholar]
- 24. Fu Y‐M, Yu Z‐X, Lin H, et al. Selective amino acid restriction differentially affects the motility and directionality of DU145 and PC3 prostate cancer cells. J Cell Physiol. 2008;217(1):184‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xia M, Wei J, Tong K. MiR‐224 promotes proliferation and migration of gastric cancer cells through targeting PAK4. Pharmazie. 2016;71(8):460‐464. [DOI] [PubMed] [Google Scholar]
- 26. Zhang H‐S, Zhao J‐W, Wang H, et al. LIM kinase 1 is required for insulin‐dependent cell growth of osteosarcoma cell lines. Mol Med Rep. 2014;9(1):103‐108. [DOI] [PubMed] [Google Scholar]
- 27. You T, Gao W, Wei J, et al. Overexpression of LIMK1 promotes tumor growth and metastasis in gastric cancer. Biomed Pharmacother. 2015;69:96‐101. [DOI] [PubMed] [Google Scholar]
- 28. Sheng N, Tan G, You W, et al. MiR‐145 inhibits human colorectal cancer cell migration and invasion via PAK4‐dependent pathway. Cancer Med. 2017;6(6):1331‐1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rombouts K, Mello T, Liotta F, et al. MARCKS actin‐binding capacity mediates actin filament assembly during mitosis in human hepatic stellate cells. Am J Physiol Cell Physiol. 2012;303(4):C357‐C367. [DOI] [PubMed] [Google Scholar]
- 30. Ritchey L, Chakrabarti R. Aurora A kinase modulates actin cytoskeleton through phosphorylation of Cofilin: implication in the mitotic process. Biochim Biophys Acta. 2014;1843(11):2719‐2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nadella KS, Saji M, Jacob NK, et al. Regulation of actin function by protein kinase A‐mediated phosphorylation of Limk1. EMBO Rep. 2009;10(6):599‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo B, Li X, Song S, et al. (‐)‐beta‐hydrastine suppresses the proliferation and invasion of human lung adenocarcinoma cells by inhibiting PAK4 kinase activity. Oncol Rep. 2016;35(4):2246‐2256. [DOI] [PubMed] [Google Scholar]
- 33. Zhang J, Zhang H‐Y, Wang J, et al. GL‐1196 suppresses the proliferation and invasion of gastric cancer cells via targeting PAK4 and inhibiting PAK4‐mediated signaling pathways. Int J Mol Sci. 2016;17(4):470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Manetti F. Recent findings confirm LIM domain kinases as emerging target candidates for cancer therapy. Curr Cancer Drug Targets. 2012;12(5):543‐560. [DOI] [PubMed] [Google Scholar]
- 35. Zhou Y, Su J, Shi L, et al. DADS downregulates the Rac1‐ROCK1/PAK1‐LIMK1‐ADF/cofilin signaling pathway, inhibiting cell migration and invasion. Oncol Rep. 2013;29(2):605‐612. [DOI] [PubMed] [Google Scholar]


