Abstract
Background
Given the reliability of circRNAs in symbolizing cancer progression, this investigation was designed to expound the involvement of hsa_circ_0028007 in regulating chemosensitivity of nasopharyngeal carcinoma (NPC) cells.
Methods
Altogether, 241 pairs of NPC tissues and para‐cancerous normal tissues were collected to identify NPC‐symbolic circRNAs, which have been screened by circRNA microarray in advance. Expressions of the circRNAs were determined by means of real‐time polymerase chain reaction (PCR). Besides, human NPC cell lines (ie, CNE2 and HONE1) were transfected by si‐hsa_circ_0028007 and si‐NC. Scratch assay, transwell assay, and MTT assay were performed to assess migration, invasion, and paclitaxel/cisplatin‐resistance of NPC cell lines.
Results
Hsa_circ_0028007 expression was abnormally heightened within NPC tissues in comparison with matched non‐tumor tissues (P < .05). Over‐expressed hsa_circ_0028007 was strongly associated with advanced (III‐IV) tumor stage, aggressive infiltration, and metastatic lymph nodes of NPC patients (P < .05). Regarding in vitro experiments, hsa_circ_0028007 expression was elevated in CNE2 and HONE1 cell lines as compared with HENE cell line (P < .05). Silencing of hsa_circ_0028007 not merely sensitized CNE2 and HONE1 cells against paclitaxel and cisplatin (P < .05), but also significantly repressed migration and invasion of the cell lines (P < .05).
Conclusion
Hsa_circ_0028007 was involved in facilitating progression and chemo‐resistance of NPC, which might offer an alternative for NPC treatment.
Keywords: cell invasion, cell migration, chemosensitivity, hsa_circ_0028007, nasopharyngeal carcinoma
1. INTRODUCTION
Nasopharyngeal carcinoma (NPC), owing to dysfunction of nasopharyngeal epithelium cells (NPECs), was a prevalent head and neck malignancy in China, and its incidence (ie, around 0.7‰) reached a peak in Guangdong district. 1 , 2 Virtually, a majority of NPC patients have exacerbated to the advanced stage when they sought medical attentions, since that early‐stage NPC usually occurred in hidden spaces (eg, recess and top wall of pharyngeal) that were hard to perceive. 3 Although chemoradiotherapy has benefited vast numbers of NPC patients, 4 , 5 drug tolerance that arose during the treatment impeded noticeable improvement of NPC prognosis. 6 , 7 Hence, identifying biomarkers that modified NPC progression and chemo‐resistance was critical to improving outcome of NPC patients.
CircRNAs, stemming from back splicing of protein‐encoding genes, outperformed linear RNAs in indicating pathophysiological changes within organisms, owing to their high stability and long half‐life period. 8 On account of this, circRNAs were no longer ignored, and some of them were anticipated to participate in the etiology of head and neck malignancies. 9 , 10 For instance, ciRS‐7 was expected to implicate in pathogenesis of tongue squamous cell carcinoma (TSCC) due to its interaction with miR‐7, 11 which slowed down proliferation and induced cell cycle arrest of tumor cells by debilitating Akt signaling. 12 Besides, cir‐ITCH seemed as an anti‐oncogene in esophageal squamous cell carcinoma, and its sponging miR‐7 could facilitate ubiquitin‐mediated DLV2 degradation. 13 Furthermore, hsa_circ_0000543 (gene symbol: DAAM1) enhanced radioresistance of NPC cells by negative regulation of miR‐9‐led axis, 14 and hsa_circ_0000285 (gene symbol: HIPK3) level was markedly raised in radioresistant NPC patients as relative to radiosensitive NPC patients. 15 Interestingly, genes that engendered the above oncogenic circRNAs, such as HIPK3 and DAAM1, 16 were also powerful motivators of tumorigenesis, 17 , 18 which triggered an assumption that oncogene‐derived circRNAs might also perform tumor‐promoting/suppressing functions.
NUAK1, also termed as AMPK‐related protein kinase 5 (ARK5), revealed strong associations with NPC deterioration, 19 and only after phosphorylation by LKB1 or Akt kinase could it function biologically. 20 , 21 For decades, NUAK1‐centric signalings were successively confirmed to involve in cell growth, cell apoptosis, and angiogenesis, which were entwined with tumorigenesis. 22 Among them, one accepted theory held that NUAK1 strengthened tumor invasion through an IGF‐1/Akt‐dependent manner, 21 and others revealed that intentionally curbing NUAK1 expression in non–small‐cell lung cancer and pancreatic carcinoma could elevate sensitivity of these solid tumors in response to chemo‐drugs. 23 , 24 Despite the intimate relation of NUAK1 with neoplastic development and chemo‐tolerance, it remained ambiguous as for whether NUAK1‐derived circRNAs also played similar roles in tumors, especially NPC.
Hence, this investigation was intended to screen out circRNAs that were potentially relevant to NPC progression and chemo‐resistance, which might provide a novel direction for NPC treatment.
2. MATERIALS AND METHODS
2.1. Collection of NPC specimens
Back from January 2012 to February 2016, 241 patients histopathologically diagnosed as NPC were recruited from Ren Ji Hospital affiliated to Shanghai Jiaotong University School of Medicine. Both NPC tissues and para‐cancerous normal tissues were resected from the NPC patients during surgery. There were 4 requirements for NPC cases: (1) They hardly underwent any NPC‐aimed treatments before this project, (2) they were treated by intensity‐modulated radiation therapy (IMAP), and (3) their Karnofsky performance 25 scored higher than 70. The NPC patients were excluded if (1) they were complicated by severe injuries in liver or kidney; (2) they were intolerant of surgery; (3) they had tumor history; and (4) they were during gestation or pregnancy. The NPC patients were classified into stages I‐IV according to the grading system specifically designed for Chinese NPC patients. 26 All the enrollees have signed informed consents, and approvals were gained from Ren Ji Hospital affiliated to Shanghai Jiaotong University School of Medicine and the ethics committee of Ren Ji Hospital affiliated to Shanghai Jiaotong University School of Medicine.
2.2. Real‐time polymerase chain reaction (PCR)
Total RNAs, extracted from NPC tissues and cell lines using TRIzol kit (Invitrogen, USA), were quantified on a nucleic acid protein analyzer (model: NanoDrop ND‐2000/2000C, Thermo Fisher Scientific). Then, the RNAs were reversely transcribed into cDNAs (Takara), which were amplified on the real‐time PCR instrument (model: StepOnePlus™, Life Tech). The PCR reaction was accomplished following steps particularized in the SYBR Green qRT‐PCR kit (Takara, Japan), and primers were enlisted in Table S1. Expressions of circRNAs were calculated adhering to 2−ΔΔCt method, 27 and GAPDH was designated as the internal reference. All these experiments were repeated for ≥3 times.
2.3. Cell culture
Human NPC cell lines (ie, CNE2 and HONE1) and human embryonic nasopharyngeal epithelium (HENE) cell line were supplied by cell bank affiliated to Chinese Academy of Sciences (Shanghai, China). They were cultured in Dulbecco's modified Eagle's medium (DMEM, Hyclone) that was blended by 10% fetal calf serum (FCS, Gibco) and 1% penicillin‐streptomycin, and their culture atmosphere was maintained as 5% CO2 and 37°C.
2.4. Cell transfection
The siRNA against hsa_circ_0028007 (5′‐TCTTAAGTATTCCTGTGCACA‐3′) was designed and synthesized by GenePharma. In strict accordance with instruction of Lipofectamine™ 2000 kit (Invitrogen), si‐NC and si‐hsa_circ_0028007 were separately transfected into CNE2 and HONE1 cell lines, whose concentration was adjusted to 2 × 105/well. Around 48 hours later, the NPC cell lines were collected, and the experiments were repeated for ≥3 times.
2.5. MTT assay for appraising chemosensitivity of NPC cell lines
CNE2 and HONE1 cell lines of logarithmic growth phase were digested by 0.25% trypsin (Gibco), and they were inoculated into 96‐well plates at a density of 3 × 103/well. Then, the NPC cell lines were treated by cisplatin (Qilu Pharmaceutical corporation) and paclitaxel (Sihuan Pharmaceutical Technology corporation) for 48 hours. Afterward, 25 μL MTT solution (5 mg/mL, Sigma) was supplemented into each well to cultivate the NPC cells for 4 hours, and 100 μL DMSO (Sigma) was prepared to treat each cell sample for 10 minutes. The absorbance value of each well was measured at the wavelength of 490 nm using a microplate reader (Bio‐Tek), and the experiments were repeated for ≥3 times.
2.6. CCK‐8 assay for assessing proliferation of NPC cells
CNE2 and HONE1 cell lines were inoculated into 96‐well plates at a concentration of 2 × 103/well, and 10 μL CCK‐8 reagent (Dojindo) was rfemented into each well. After cultivation at 37°C for 2 hours, absorbance value of the NPC cells was monitored at the wavelength of 450 nm on a microplate spectrophotometer (Beckman). The experiments were repeated for ≥3 times.
2.7. Wound healing assay for estimating migratory potential of NPC cells
Every 5 × 104 NPC cells were inoculated into each well of 6‐well plates, and they were cultivated within 10% fetal bovine serum (FBS)‐containing RPMI‐1640 medium (Gibco) until 90% confluence. Scratches were drawn onto the back of cell plates using the tip of pipette (model: 100 μL), and scratch widths of each sample were photographed at the time points of 0 and 48 hours under an inverted microscope (Olympus). Difference of the scratch widths was recorded, and the experiments were repeated for ≥3 times.
2.8. Transwell assay for evaluating invasion of NPC cell lines
The upper Transwell chamber (Coming) was coated by man‐made basal membrane (BD), where 5 × 104 well‐grown NPC cells and 200 μL serum‐free RPMI‐1640 medium (Gibco) were inoculated. On the other hand, 600 μL RPMI‐1640 medium (Gibco) that consisted of 10% FBS was gently added to the lower Transwell chamber. After 24‐hour incubation, NPC cells that failed to pass through the filter membrane of upper chamber were removed with a swab, and the remaining NPC cells were fixated by methanol and dyed by crystal violet. Ultimately, 9 views randomly chosen from each sample were observed under a light microscope (×200), and the average cell number was counted. These experiments were repeated for ≥3 times.
2.9. Statistical analyses
All statistical analyses were accomplished by feat of SPSS (version 18.0) software. The measurement data [mean ± standard deviation (SD)] were compared via Student's t test or one‐way analysis of variance (ANOVA). Besides, logistic regression was carried out to screen out clinical items that were significantly associated with NPC prognosis, and Kaplan‐Meier curves were plotted to estimate the correlation between hsa_circ_0028007 expression and 3‐year survival of NPC patients. Furthermore, receiver operator characteristic (ROC) curves were devised to assess the potential of hsa_circ_0028007 expression in symbolizing NPC severity. Differences were statistically significant in the event of P < .05.
3. RESULTS
3.1. Association of hsa_circ_0028007 expression with clinical items of NPC patients
In the aggregate, 20 circRNAs, whose expressions were altered most remarkably between NPC tissues and adjacent normal tissues, were enlisted in Table S2, as concluded from the results of circRNA microarray. Then, expressions of the top 10 circRNAs were further confirmed in NPC patients (Figure S1A), which indicated that hsa_circ_0028007 expression was elevated pronouncedly within NPC tissues as relative to adjacent normal tissues (P < .05) (Figure 1A). With mean expression of hsa_circ_0028007 (ie, 8.515) as the cutoff value, the NPC patients were grouped into ones with over‐expressed hsa_circ_0028007 (n = 160) and ones carrying lowly expressed hsa_circ_0028007 (n = 81) (Table 1). It was demonstrated that over‐expressed hsa_circ_0028007 was associated with advanced (III‐IV) clinical stage (OR = 3.13, 95%CI: 1.72‐5.88, P < .001), aggressive (ie, T3 + T4) infiltration (OR = 1.49, 95%CI: 1.25‐3.57, P = .005), and lymph‐node metastasis (OR = 5.84, 95%CI: 2.99‐11.38, P < .001) of NPC patients.
Figure 1.
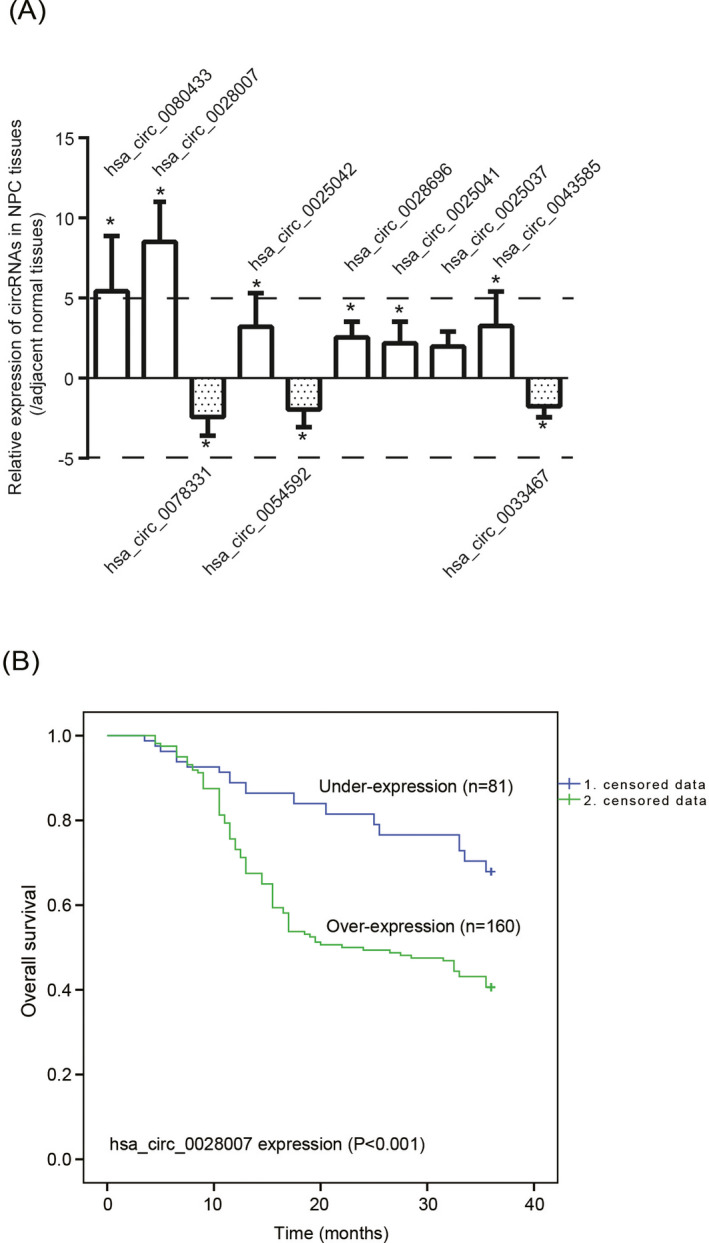
Clinical implication of hsa_circ_0028007 expression in nasopharyngeal carcinoma (NPC). A, Expressions of 10 circRNAs were compared between 241 pairs of NPC tissues and matched para‐cancerous normal tissues. *P < .05 in comparison with matched para‐cancerous normal tissues. B, Over‐expressed hsa_circ_0028007, in comparison with under‐expressed hsa_circ_0028007, was indicative of poor survival of NPC patients
Table 1.
Association of hsa_circ_0028007 expression with clinco‐pathological items of nasopharyngeal carcinoma patients
| Clinco‐pathological items | hsa_circ_0028007 | Chi‐square | P value | OR (95% CI) | |
|---|---|---|---|---|---|
| Over‐expression (n = 160) | Under‐expression (n = 81) | ||||
| Age (years old, n) | |||||
| ≥55 | 66 | 33 | |||
| <55 | 94 | 48 | 0.01 | .939 | 1.02 (0.59‐1.76) |
| Gender (n) | |||||
| Female | 81 | 41 | |||
| Male | 79 | 40 | 1.281e‐006.1 | .999 | 1.00 (0.59‐1.71) |
| Histological type (n) | |||||
| Differentiated non‐keratinizing carcinoma | 12 | 9 | |||
| Undifferentiated carcinoma | 148 | 72 | 0.88 | .348 | 1.54 (0.62‐3.85) |
| Clinical staging (n) | |||||
| I‐II | 38 | 21 | |||
| III‐IV | 122 | 60 | 14.49 | <.001* | 3.13 (1.72‐5.88) |
| Depth of infiltration (n) | |||||
| T1 + T2 | 75 | 34 | |||
| T3 + T4 | 85 | 47 | 8.01 | .005* | 1.49 (1.25‐3.57) |
| Lymph‐node metastasis (n) | |||||
| No | 35 | 21 | |||
| Yes | 125 | 60 | 30.24 | <.001* | 5.84 (2.99‐11.38) |
Abbreviations: CI, confidence interval; OR, odds ratio.
Statistical significance.
3.2. Possible role of hsa_circ_0028007 in reflecting NPC severity
As illustrated by Figure S1B, NPC patients of severer clinical symptoms (ie, stages III‐IV, T3‐T4 infiltration, and metastatic lymph nodes) were associated with slightly higher hsa_circ_0028007 expression than ones with relatively mild symptoms (ie, stages I‐II, T1‐T2 infiltration, and nonmetastatic lymph nodes). Moreover, ROC curves (Figure S1C) exhibited that hsa_circ_0028007 expression not merely could discriminate NPC patients at stages III‐IV from patients at stages I‐II (AUC = 0.625, cutoff level = 8.695, P = .005), but also could separate NPC population with deeper (T3 + T4) infiltration from patients with shallow (T1 + T2) depth (AUC = 0.621, cutoff level = 8.695, P = .001). In addition, expression of hsa_circ_0028007 was also able to identify NPC patients with metastatic lymph nodes from patients without lymph‐node metastasis (AUC = 0.705, cutoff level = 8.859, P < .001).
3.3. Implication of hsa_circ_0028007 in predicting NPC prognosis
The longevity of NPC patients in the over‐expressed hsa_circ_0028007 group was evidently shortened in comparison with lowly expressed hsa_circ_0028007 group (P = .004) (Figure 1B). Besides, clinical tumor staging (HR = 1.857, 95%CI: 1.14‐3.03, P = .013), infiltration depth (HR = 1.910, 95%CI: 1.31‐2.79, P = .001), lymph‐node metastasis (HR = 3.004, 95%CI: 1.64‐5.45, P < .001), and hsa_circ_0028007 level (HR = 2.550, 95%CI: 1.61‐4.05, P < .001) were outstanding parameters in estimating poor survival of recruited NPC patients, as displayed by the results of univariate analyses (Table 2). After eliminating impacts exerted by other parameters, the results of multivariate analysis implied that infiltration depth (HR = 1.760, 95%CI: 1.19‐2.59, P = .004), lymph‐node metastasis (HR = 2.256, 95%CI: 1.21‐4.20, P = .010) and hsa_circ_0028007 expression (HR = 1.738, 95%CI: 1.07‐2.83, P = .026) were independent predictors of NPC prognosis.
Table 2.
Implication of clinicopathological items in forecasting 3‐y survival of nasopharyngeal carcinoma patients
| Clinco‐pathological items | Number of cases (n) | Univariate | P | Multivariate | P | ||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | ||||
| Age (years old, n) | |||||||
| <55 | 99 | ||||||
| ≥55 | 142 | 1.023 | 0.710‐1.472 | .904 | 0.99 | 0.687‐1.439 | .976 |
| Gender (n) | |||||||
| Male | 122 | ||||||
| Female | 119 | 1.21 | 0.847‐1.729 | .296 | 1.21 | 0.841‐1.742 | .304 |
| Histological type (n) | |||||||
| Differentiated non‐keratinizing carcinoma | 21 | ||||||
| Undifferentiated carcinoma | 220 | 1.032 | 0.540‐1.971 | .924 | 1.089 | 0.563‐2.106 | .800 |
| Clinical staging (n) | |||||||
| I‐II | 59 | ||||||
| III‐IV | 182 | 1.857 | 1.137‐3.033 | .013 | 1.371 | 0.829‐2.269 | .219 |
| Depth of infiltration (n) | |||||||
| T1 + T2 | 109 | ||||||
| T3 + T4 | 132 | 1.91 | 1.31‐2.785 | .001 | 1.76 | 1.194‐2.594 | .004* |
| Lymph‐node metastasis (n) | |||||||
| Yes | 185 | ||||||
| No | 56 | 3.004 | 1.654‐5.456 | <.001 | 2.256 | 1.212‐4.201 | .010* |
| Relative expression of hsa_circ_0028007 (n) | |||||||
| Under‐expression | 160 | ||||||
| Over‐expression | 81 | 2.55 | 1.605‐4.053 | <.001 | 1.738 | 1.067‐2.831 | .026* |
Abbreviations: CI, confidence interval; HR, hazard ratio.
Statistical significance.
3.4. Silencing of hsa_circ_0028007 sensitized NPC cell lines against cisplatin and paclitaxel
Hsa_circ_0028007 expression was raised markedly in CNE2 and HONE1 cell lines, as compared with HENE cell line (P < .05) (Figure 2A). Besides, si‐hsa_circ_0028007 dramatically reduced hsa_circ_0028007 expression in CNE2 and HONE1 cell lines (P < .05) (Figure 2B). Proliferation of CNE2 (IC50 = 2.72 μg/mL) and HONE1 (IC50 = 1.82 μg/mL) cell lines was vastly restrained by cisplatin, and paclitaxel also diminished multiplication of CNE2 (IC50 = 446 nmol/L) and HONE1 (IC50 = 1621 nmol/L) cell lines (Figure 2C). Besides, silencing of hsa_circ_0028007 (ie, si‐hsa_circ_0028007 group) made CNE2 (cisplatin: IC50 = 1.65 μg/mL; paclitaxel: IC50 = 30.41 nmol/L) and HONE1 (cisplatin: IC50 = 0.99 μg/ml; paclitaxel: IC50 = 134.89 nmol/L) cell lines less tolerant against cisplatin and paclitaxel, when compared with NC group and si‐NC group (P < .05).
Figure 2.
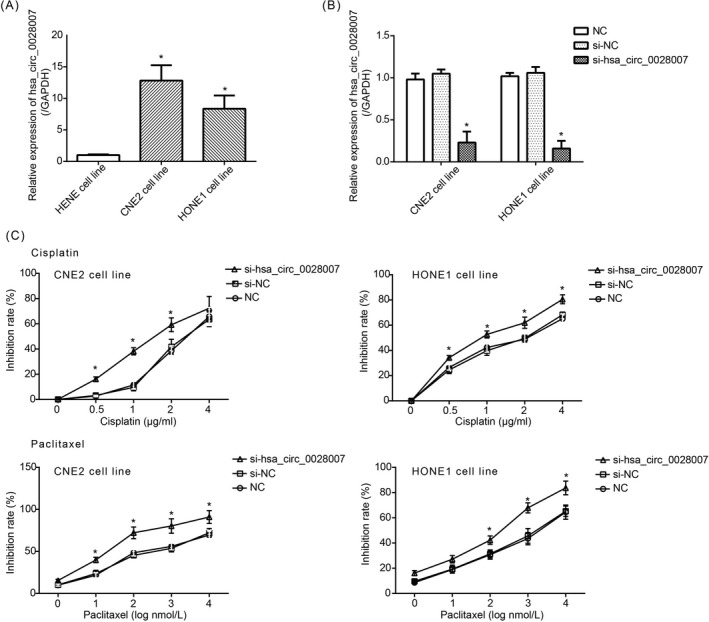
Impact of si‐hsa_circ_0028007 on chemosensitivity of nasopharyngeal carcinoma (NPC) cells. A, Hsa_circ_0028007 expression was upregulated in CNE2 and HONE1 cell lines as relative to HENE cell line. *P < .05 in comparison with HENE cell line. B, Hsa_circ_0028007 expression was lessened in CNE2 and HONE1 cell lines by silencing of hsa_circ_0028007. *P < .05 in comparison with NC group and si‐NC group. C, Silencing of hsa_circ_0028007 heightened sensitivity of CNE2 and HONE1 cell lines against cisplatin and paclitaxel. *P < .05 in comparison with NC group and si‐NC group
3.5. Silencing of hsa_circ_0028007 disabled proliferation, invasion and migration of NPC cell lines
Silencing of hsa_circ_0028007 significantly weakened proliferation of CNE2 and HONE1 cell lines, according to the results of CCK8 assay (P < .05) (Figure 3A). The metastatic trend of CNE2 and HONE1 cell lines was also undermined after intentional silencing of hsa_circ_0028007 (P < .05) (Figure 3B). To be specific, the scratch width of si‐hsa_circ_0028007 group was extended in comparison to NC group and si‐NC group at the time point of 48 hours (P < .05), yet there was no significant distinction among the three groups at the time point of 0 hours (P > .05). More than that, invasion of CNE2 and HONE1 cell lines in the si‐hsa_circ_0028007 group was depressed significantly as compared with NC group and si‐NC group (P < .05) (Figure 3C).
Figure 3.
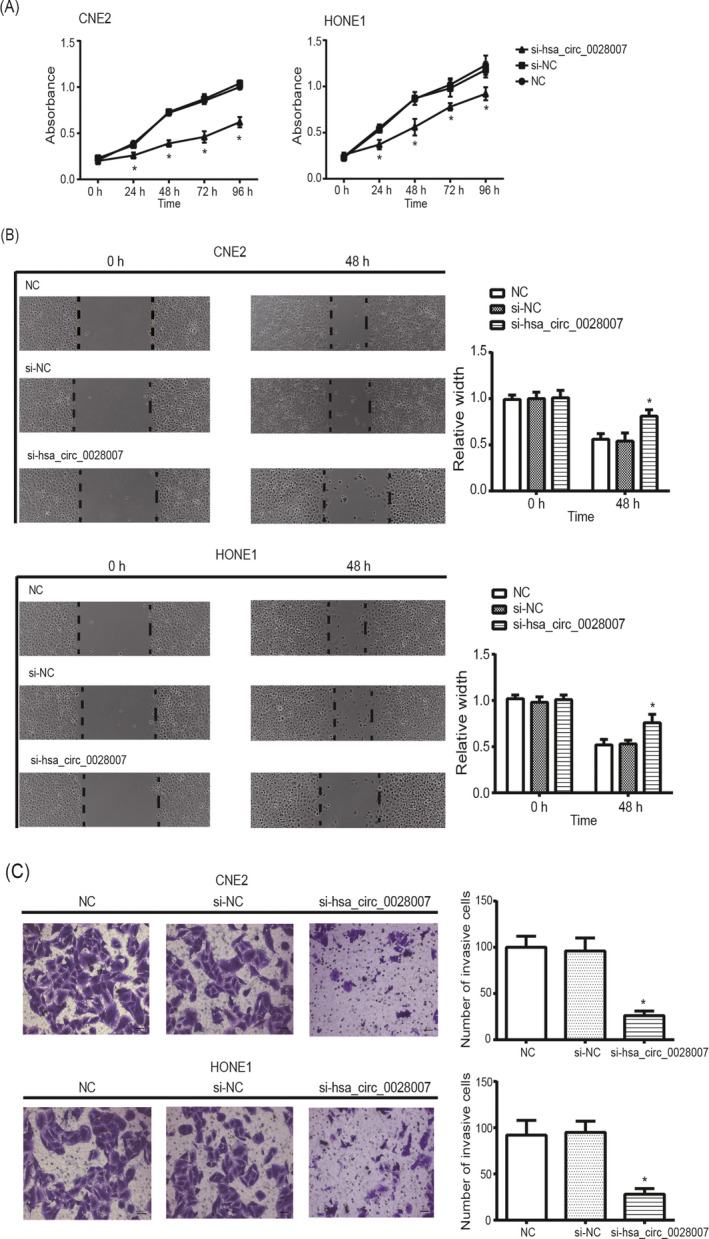
Silencing of hsa_circ_0028007 influenced activity of nasopharyngeal carcinoma (NPC) cells. A, Proliferation of CNE2 and HONE1 cell lines was evaluated among si‐hsa_circ_0028007, NC, and si‐NC groups. *P < .05 in comparison with NC group and si‐NC group. B, The migratory capability of CNE2 and HONE1 cell lines was monitored after transfection of si‐hsa_circ_0028007 and si‐NC. *P < .05 in comparison with NC group and si‐NC group at the time point of 48 h. C, The invasive ability of CNE2 and HONE1 cell lines was dampened after silencing of hsa_circ_0028007. *P < .05 in comparison with NC group and si‐NC group
4. DISCUSSION
Confronted by the killing effect of chemo‐drugs, tumor cells gradually upgraded their molecular network so that they could adapt to the extreme surroundings. In this fashion, chemo‐resistance was stimulated, suggesting that consciously regulating tumor‐regulatory genes might reverse drug tolerance of tumor cells. There have been several accounts for incremental drug tolerance of tumor cells, such as membrane‐pump protein (eg, p‐glycoprotein), drug‐target protein (eg, topoisomerase), and DNA repair capacity (DRC). 28 However, it remained challenging to improve chemotherapeutic effectiveness of tumor patients, which spurred discovery of other molecular explanations.
With the swift advancement of RNA sequencing, plentiful circRNAs have been documented to participate in tumorigenesis. 29 For instance, hsa_circ_0000096 and hsa_circ_002059 exhibited high diagnostic value for gastric cancer, 30 and low hsa_circ_0001649 expression was predictive of large tumor size and thrombus formation of gastric cancer patients. 31 In fact, there were several reasons why circRNAs could sensitively reflect disease progression. Firstly, circRNAs were tolerant to exonuclease‐induced degradation, so that their stability was maintained. Secondly, circRNAs were broadly present in eukaryotes, and their expressional change was observable among tissues of different features. 32 , 33 Last but not the least, circRNAs were available from blood, urine, and cerebrospinal fluid, 34 which stressed the convenience of applying circRNAs for disease diagnosis. This study also introduced a competitive circRNA (ie, hsa_circ_0028007) for signalizing NPC onset (Figure 1A), and its expressional change could reflect NPC progression and prognosis sensitively (Tables 1, 2, Figures 1B, S1B,C).
Furthermore, there was a speculation that circRNAs generated from oncogenes and anti‐oncogenes were strong propellers or blockers of neoplastic progression. For instance, circRNAs generated form Foxo3, a tumor‐suppressing gene, were able to undermine proliferation of tumor cells. 35 , 36 Besides, f‐circPR, derived from PML‐RARα, and f‐circM9, derived from MLL‐AF9, both promoted growth of acute promyelocytic leukemia cells. 37 Following an analogous logic, the hsa_circ_0028007 studied here was originated from NUAK1 gene (Table S1), whose excessive expression was correlated with poor disease‐free survival of NPC patients. 38 Besides NPC, aberrant over‐expression of NUAK1 was also associated with unfavorable survival of patients who were plagued by non–small‐cell lung cancer, primary hepatocellular carcinoma, glioma, and melanoma. 39 , 40 , 41 , 42 These clinical linkages might be ascribed to the role of NUAK1 to promoting multiplication and metastasis of tumor cells, including head and neck squamous cell carcinoma, 43 intrahepatic cholangiocarcinoma, 44 and non–small‐cell lung cancer. 45 Similar to NUAK1, silencing of hsa_circ_0028007 also suppressed proliferation, migration, and invasion of NPC cells (Figure 3A‐C), which could mechanically explain the clinical significance of hsa_circ_0028007 in NPC.
In addition, paclitaxel and cisplatin were commonly applied for treating patients with advanced NPC, and their anticancer mechanism differed from each other. 46 , 47 To be specific, paclitaxel, which belonged to tetracyclic diterpenoids, held up depolymerization of microtublins and interdicted replication of neoplasm cells, 48 whereas cisplatin, a complex compound of platinum, impeded DNA replication by forming intrachain links with DNAs. 49 However, drug tolerance reduced the treatment effect of paclitaxel and cisplatin on NPC patients. In this study, we observed that silencing of hsa_circ_0028007 elevated paclitaxel/cisplatin‐sensitivity of poorly differentiated NPC cell lines (ie, CNE2 and HONE1) (Figure 2C), which implied that targeting hsa_circ_0028007 could improve chemotherapeutic efficacy on NPC. Since that intensified cell migration and invasion could explain incremental chemo‐resistance of tumors, 50 silencing of hsa_circ_0028007 might heighten chemosensitivity of NPC cells through depressing these activities of NPC cells (Figure 3). However, detailed mechanisms that accounted for how hsa_circ_0028007 resisted against cisplatin and paclitaxel entailed more proofs.
Conclusively, silencing of hsa_circ_0028007 was expected to improve NPC chemosensitivity by curbing proliferation and metastasis of NPC cells, which might give a novel direction for NPC treatment. Nonetheless, there were a couple of defects about the experimental design. On one hand, cell models carrying high level of hsa_circ_0028007, owing to technical defects, failed to be established, which might reduce persuasiveness of study results. On the other hand, experiments relevant to animal models were not performed, so the effect of hsa_circ_0028007 on in vivo tumor growth could not be vividly presented. Shortcomings described as above demanded perfecting in future.
AUTHORS’ CONTRIBUTIONS
We declare that we have no conflict of interest, and all authors approved the final version of article.
Supporting information
Figure S1
Table S1
Table S2
ACKNOWLEDGMENT
We appreciate the helpful comments received from our reviewers.
Qiongna D, Jiafeng Z, Yalin H, et al. Implication of hsa_circ_0028007 in reinforcing migration, invasion, and chemo‐tolerance of nasopharyngeal carcinoma cells. J Clin Lab Anal. 2020;34:e23409 10.1002/jcla.23409
REFERENCES
- 1. He YQ, Xue WQ, Shen GP, et al. Household inhalants exposure and nasopharyngeal carcinoma risk: a large‐scale case‐control study in Guangdong, China. BMC Cancer. 2015;15:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu ZX, Lin ZX, Fang JY, et al. Mortality characteristic and prediction of nasopharyngeal carcinoma in China from 1991 to 2013. Asian Pac J Cancer Prev. 2015;16:6729‐6734. [DOI] [PubMed] [Google Scholar]
- 3. Yang XL, Wang Y, Liang SB, et al. Comparison of the seventh and eighth editions of the UICC/AJCC staging system for nasopharyngeal carcinoma: analysis of 1317 patients treated with intensity‐modulated radiotherapy at two centers. BMC Cancer. 2018;18:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li X, Zhou J, Chu C, et al. Home enteral nutrition may prevent myelosuppression of patients with nasopharyngeal carcinoma treated by concurrent chemoradiotherapy. Head Neck. 2019;41:3525‐3534. [DOI] [PubMed] [Google Scholar]
- 5. Liu F, Jin T, Liu L, et al. The role of concurrent chemotherapy for stage II nasopharyngeal carcinoma in the intensity‐modulated radiotherapy era: A systematic review and meta‐analysis. PLoS One. 2018;13:e0194733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu F, Tai Y, Ma J. LncRNA NEAT1/let‐7a‐5p axis regulates the cisplatin resistance in nasopharyngeal carcinoma by targeting Rsf‐1 and modulating the Ras‐MAPK pathway. Cancer Biol Ther. 2018;19:534‐542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y, Cheng N, Luo J. Downregulation of lncRNA ANRIL represses tumorigenicity and enhances cisplatin‐induced cytotoxicity via regulating microRNA let‐7a in nasopharyngeal carcinoma. J Biochem Mol Toxicol. 2017;31:e21904. [DOI] [PubMed] [Google Scholar]
- 8. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 11. Li RC, Ke S, Meng FK, et al. CiRS‐7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR‐7/HOXB13. Cell Death Dis. 2018;9:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang L, Liu X, Chen Z, et al. MicroRNA‐7 targets IGF1R (insulin‐like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432:199‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta‐catenin pathway. Oncotarget. 2015;6:6001‐6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen L, Zhou H, Guan Z. CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR‐9/platelet‐derived growth factor receptor B axis. Biochem Biophys Res Commun. 2019;512:786‐792. [DOI] [PubMed] [Google Scholar]
- 15. Shuai M, Hong J, Huang D, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16:6495‐6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao H, Xia D, Li ZL, et al. MiR‐382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci Rep. 2019;39:BSR20180441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu Y, Tian Y, Du J, et al. Dvl2‐dependent activation of Daam1 and RhoA regulates Wnt5a‐induced breast cancer cell migration. PLoS One. 2012;7:e37823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR‐145a‐5p on the down‐regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 2019;23:2351‐2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerjee S, Zagorska A, Deak M, et al. Interplay between Polo kinase, LKB1‐activated NUAK1 kinase, PP1betaMYPT1 phosphatase complex and the SCFbetaTrCP E3 ubiquitin ligase. Biochem J. 2014;461:233‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suzuki A, Lu J, Kusakai G, et al. ARK5 is a tumor invasion‐associated factor downstream of Akt signaling. Mol Cell Biol. 2004;24:3526‐3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang X, Lv W, Zhang JH, Lu DL. miR96 functions as a tumor suppressor gene by targeting NUAK1 in pancreatic cancer. Int J Mol Med. 2014;34:1599‐1605. [DOI] [PubMed] [Google Scholar]
- 23. Wang X, Song Z, Chen F, et al. AMPK‐related kinase 5 (ARK5) enhances gemcitabine resistance in pancreatic carcinoma by inducing epithelial‐mesenchymal transition. Am J Transl Res. 2018;10:4095‐4106. [PMC free article] [PubMed] [Google Scholar]
- 24. Li M, Zheng C, Xu H, et al. Inhibition of AMPK‐related kinase 5 (ARK5) enhances cisplatin cytotoxicity in non‐small cell lung cancer cells through regulation of epithelial‐mesenchymal transition. Am J Transl Res. 2017;9:1708‐1719. [PMC free article] [PubMed] [Google Scholar]
- 25. Modesto AP, Usvyat L, Calice‐Silva V, et al. Impact of the Karnofsky performance status on survival and its dynamics during the terminal year of peritoneal dialysis patients. Perit Dial Int. 2018;38:24‐29. [DOI] [PubMed] [Google Scholar]
- 26. Zong J, Huang Q, Guo Q, Pan J. Evolution of the Chinese staging system for nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5:19. [DOI] [PubMed] [Google Scholar]
- 27. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods. 2001;25:402‐408. [DOI] [PubMed] [Google Scholar]
- 28. Scheffer GL, Scheper RJ. Drug resistance molecules: lessons from oncology. Novartis Found Symp. 2002;243:19‐31; discussion 31–17, 180–185. [PubMed] [Google Scholar]
- 29. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116:626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161‐169. [DOI] [PubMed] [Google Scholar]
- 32. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417‐1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non‐coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919‐3931. [DOI] [PubMed] [Google Scholar]
- 37. Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion‐circRNAs derived from cancer‐associated chromosomal translocations. Cell. 2016;166:1055‐1056. [DOI] [PubMed] [Google Scholar]
- 38. Liu J, Tang G, Huang H, et al. Expression level of NUAK1 in human nasopharyngeal carcinoma and its prognostic significance. Eur Arch Otorhinolaryngol. 2018;275:2563‐2573. [DOI] [PubMed] [Google Scholar]
- 39. Cui J, Yu Y, Lu GF, et al. Overexpression of ARK5 is associated with poor prognosis in hepatocellular carcinoma. Tumour Biol. 2013;34:1913‐1918. [DOI] [PubMed] [Google Scholar]
- 40. Chen P, Li K, Liang Y, et al. High NUAK1 expression correlates with poor prognosis and involved in NSCLC cells migration and invasion. Exp Lung Res. 2013;39:9‐17. [DOI] [PubMed] [Google Scholar]
- 41. Lu S, Niu N, Guo H, et al. ARK5 promotes glioma cell invasion, and its elevated expression is correlated with poor clinical outcome. Eur J Cancer. 2013;49:752‐763. [DOI] [PubMed] [Google Scholar]
- 42. Namiki T, Tanemura A, Valencia JC, et al. AMP kinase‐related kinase NUAK2 affects tumor growth, migration, and clinical outcome of human melanoma. Proc Natl Acad Sci USA. 2011;108:6597‐6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Obayashi M, Yoshida M, Tsunematsu T, et al. microRNA‐203 suppresses invasion and epithelial‐mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget. 2016;7:8223‐8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiong X, Sun D, Chai H, et al. MiR‐145 functions as a tumor suppressor targeting NUAK1 in human intrahepatic cholangiocarcinoma. Biochem Biophys Res Commun. 2015;465:262‐269. [DOI] [PubMed] [Google Scholar]
- 45. Shi L, Zhang B, Sun X, et al. MiR‐204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111:2316‐2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen C, Wang FH, An X, et al. Triplet combination with paclitaxel, cisplatin and 5‐FU is effective in metastatic and/or recurrent nasopharyngeal carcinoma. Cancer Chemother Pharmacol. 2013;71:371‐378. [DOI] [PubMed] [Google Scholar]
- 47. He XY, Hu CS, Ying HM, et al. Paclitaxel with cisplatin in concurrent chemoradiotherapy for locally advanced nasopharyngeal carcinoma. Eur Arch Otorhinolaryngol. 2010;267:773‐778. [DOI] [PubMed] [Google Scholar]
- 48. Merchan JR, Jayaram DR, Supko JG, et al. Increased endothelial uptake of paclitaxel as a potential mechanism for its antiangiogenic effects: potentiation by Cox‐2 inhibition. Int J Cancer. 2005;113:490‐498. [DOI] [PubMed] [Google Scholar]
- 49. Wang X, Masters JR, Wong YC, et al. Mechanism of differential sensitivity to cisplatin in nasopharyngeal carcinoma cells. Anticancer Res. 2001;21:403‐408. [PubMed] [Google Scholar]
- 50. Wang J, Feng Y, Chen X, et al. SH3BP1‐induced Rac‐Wave2 pathway activation regulates cervical cancer cell migration, invasion, and chemoresistance to cisplatin. J Cell Biochem. 2018;119:1733‐1745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Table S2


