Abstract
Background
Numerous studies have assessed the association between xeroderma pigmentosum complementation group C (XPC) polymorphisms and susceptibility of prostate cancer (PCa); however, the findings remain inconsistent.
Methods
We performed an updated analysis utilizing data from electronic databases to obtain a more accurate estimation of the relationship between XPC rs2228001 A/C polymorphism and PCa risk. We further used in silico tools to investigate this correlation.
Results
Totally, 5,305 PCa cases and 6,499 control subjects were evaluated. When all studies pooled together, we detected no positive result (recessive genetic model: OR = 1.14, 95% CI = 0.93‐1.40, P heterogeneity = 0.001, P = .212); nevertheless, the XPC rs2228001 A/C variant was associated with PCa risk in Asian descendants in the subgroup analysis (OR = 1.21, 95% CI = 1.01‐1.43, P heterogeneity = 0.008, P = .034). In silico tools showed that more than 20 proteins can participate in the protein crosstalk with XPC. The expression of XPC was down‐regulated in all Gleason scores of prostate cancer.
Conclusions
The present study indicated that the XPC rs2228001 A/C variant may be associated with elevated PCa risk in Asian patients.
Keywords: analysis, prostate cancer, variant, XPC
1. INTRODUCTION
Prostate cancer (PCa) is the most common malignant tumor among males all over the world. Previous publications reported that PCa is the second and third leading cause of male death in the United States and Europe, respectively. 1 , 2 In the United States, about 174 650 new PCa cases were diagnosed and 31,620 patients died from this disease in 2019 estimated by the National Cancer Institute (https://seer.cancer.gov/csr/1975_2015). In Asian descendants, the PCa incidence and mortality rates were increasing extensively in recent years. 3 , 4 Up to now, the specific mechanisms and exact cause of PCa are not clear. 5 Due to the stage of this disease and the choice of patients, the prevention and treatment of PCa remains complicated. 6 Hence, it is necessary to demonstrate the molecular mechanism and explore new targeted therapies for PCa.
Studies have shown that genetic factors may play a crucial role in the development of PCa. Down‐regulation of DNA repair is a pivotal factor in the progression of PCa. 7 Nucleotide excision repair (NER), a main human DNA repair pathway, is one of the most significant defense mechanism against mutagenic exposure. 8 Xeroderma pigmentosum group C (XPC) is involved in the early damage initiation of NER. 9 The XPC gene located on c3p25 of homo sapiens. 10 Mutation of XPC can alter the capacity of NER and lead to carcinoma of human. 11 The substitution of A to C at position 939 is the most widely studied single nucleotide polymorphisms in XPC. 12
Previous publications demonstrated that the XPC rs2228001 A/C variant may be associated with increased risk for colorectal, bladder, breast, and lung cancer. 13 , 14 , 15 , 16 Association between this XPC variant and PCa likelihood was previously assessed. 17 , 18 , 19 , 20 However, there are vague conclusions about the relationship between XPC rs2228001 A/C polymorphism and PCa susceptibility in different case‐controlled studies. Hence, a systematic analysis based on all eligible studies was conducted to further investigate the correlation between the XPC rs2228001 A/C polymorphism and PCa risk. 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28
2. METHODS AND MATERIALS
2.1. Search strategy
We performed a comprehensive literature search on electronic databases including Web of Science, Cochrane Library, Google Scholar, PubMed, EMBASE, and Chinese Biomedical Database to retrieve all publications on the XPC rs2228001 A/C polymorphism and PCa susceptibility. The search terms were as follows: “XPC OR xeroderma pigmentosum group C,” “polymorphism OR single nucleotide polymorphism OR mutation OR variant,” and “carcinoma OR tumor OR adenocarcinoma OR cancer.” The last search was updated on April 10, 2020. We further screened the supplementary material of accept articles to maximize the search.
2.2. Study selection and inclusion criteria
Two investigators independently searched the studies according to inclusion criteria. The inclusion criteria were as follows: (a) evaluating the association between the XPC rs2228001 A/C polymorphism and PCa risk; (b) including available genotype frequencies to calculate odds ratio; and (c) using a case‐control design.
2.3. Exclusion criteria
The exclusion criteria were as follows: (a) Data of control were not available; (b) with no available genotype frequency for the XPC rs2228001 A/C polymorphism; (c) review articles; and (d) duplication with overlapping data from the same authors.
2.4. Data extraction
For every included study, the following information was extracted: name of author, year of publication, control source, ethnicity, genotyping method, PSA level (ng/mL), age range, sample size of case and control, genotyping data of the XPC rs2228001 A/C variant, and P‐value of Hardy‐Weinberg equilibrium (HWE) for case and control. Any disagreement should be addressed by discussion with a third investigator to achieve a final decision.
2.5. Methods for quantitative synthesis
Odds ratio and 95% confidence interval were adopted to evaluate the correlation between the XPC rs2228001 A/C polymorphism and PCa risk. Four genetic models were employed in the current analysis: allelic comparison (C‐allele vs A‐allele), heterozygous contrast (CA vs AA), dominant genetic model (CC + CA vs AA), and recessive model (CC vs CA + AA). P‐value of heterogeneity was calculated by the Q test. If P‐value for Q tests more than .005, a fixed‐effect model (Mantel‐Haenszel method) was applied. On the other hand, a random‐effect model (DerSimonian‐Laird method) was conducted. Publication bias was checked by Egger's test and Begg's plot. Moreover, sensitivity analysis was applied to examine the impact of each study on the combined OR P‐value of HWE was detected by chi‐square test. P‐value more than 0.05 indicates an HWE balance. The subgroup analysis included ethnic types and source of control. The above analyses were conducted utilizing Stata software (Stata Corporation, Lakeway, TX, v11.0).
2.6. Expression of XPC utilizing in silico analysis
Online gene expression database was applied to further investigate the expression of XPC in PCa tissues and control (http://ualcan.path.uab.edu/). A total of 497 PCa participants and 52 controls were included for investigating the XPC expression. The Cancer Genome Atlas (TCGA) samples were also employed to assess the effect of XPC expression in PCa based on patients’ Gleason score. Furthermore, we adopt the online String server (http://string‐db.org/) to explore the protein‐protein correlation regarding XPC. Protein Variation Effect Analyzer (PROVEAN, v1.1) was employed to evaluate the mutation of the XPC rs2228001 A/C variant in human (http://provean.jcvi.org/seq_submit.php). Gene‐gene interaction of XPC was also investigated by TCGA samples (http://ualcan.path.uab.edu/analysis.html).
3. RESULTS
3.1. Study Characteristics
As described in Table 1, a total of 12 publications based on 13 case‐controlled studies evaluating the XPC rs2228001 A/C polymorphism were retrieved in our analysis. Finally, 5305 PCa patients and 6,499 control subjects were included in the present study. Moreover, we checked the minor allele frequencies (MAF) of XPC reported in the genome aggregation database (gnomAD, https://www.ncbi.nlm.nih.gov/snp/rs2228001#frequency_tab): for global population, 0.367; Europeans, 0.391; Asians, 0.356; Americans, 0.294; Africans, 0.270; Ashkenazi Jewish, 0.472; and others, 0.396. Therefore, in the subgroup analysis by race, a total of seven studies were based on Asian populations, three studies were based on European populations, two analyzed African descendants, and the remaining was on Arabians. In the subgroup analysis by the source of control, there were six hospital‐based studies, and the rest seven studies focused on population‐based controls. The classic genotyping method, PCR‐restriction fragment length polymorphism (RFLP), was conducted in seven of the studies.
Table 1.
Basic information of included studies for XPC rs2228001 A/C variant and PCa risk
| Author | Year | Source | Ethnicity | Method | PSA level (ng/mL) | Age (years) | Case | Control | Case | Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | CC | CA | AA | P HWE | CC | CA | AA | P HWE | |||||||
| Perloy | 2018 | PB | European | iPLEX assay | NA | NA | 61.7 ± 4.1 | 61.2 ± 4.2 | 130 | 477 | 392 | 0.420 | 298 | 815 | 607 | 0.390 | 999 | 1720 |
| Said | 2018 | HB | African | PCR‐RFLP | mean 111.41 | 2.225 ± 1.5 | 71.8 ± 11.3 | 69.0 ± 8.51 | 16 | 55 | 39 | 0.632 | 26 | 158 | 82 | <0.001 | 110 | 266 |
| Wang | 2017 | PB | Asian | RT‐PCR | NA | NA | NA | NA | 131 | 459 | 414 | 0.831 | 125 | 495 | 435 | 0.379 | 1004 | 1055 |
| Kahnamouei | 2016 | HB | Asian | PCR‐RFLP | 9.95 (7.05‐16.5) | 2.80 (1.9‐9.1) | mean 61.7 | mean 69.2 | 47 | 59 | 47 | 0.005 | 62 | 88 | 55 | 0.044 | 153 | 205 |
| Zhang | 2014 | HB | Asian | MassARRAY | NA | NA | 66.7 ± 8.2 | 67.3 ± 7.5 | 33 | 38 | 158 | <0.001 | 31 | 37 | 170 | <0.001 | 229 | 238 |
| Mirecka | 2014 | PB | European | RT‐PCR | mean 12.0 | mean < 4.0 | mean 68.3 | mean 64.6 | 98 | 290 | 214 | 0.988 | 122 | 384 | 265 | 0.380 | 602 | 771 |
| Sorour | 2013 | HB | Arabian | PCR‐RFLP | mean 48.0 | mean < 4.0 | 65.4 ± 8.7 | NA | 9 | 25 | 16 | 0.888 | 5 | 27 | 18 | 0.263 | 50 | 50 |
| Mandal | 2012 | PB | Asian | PCR‐RFLP | 221 ± 57.4 | 2.3 ± 0.8 | 62.6 ± 8.9 | 59.1 ± 10.4 | 28 | 71 | 93 | 0.022 | 16 | 94 | 114 | 0.570 | 192 | 224 |
| Mittal | 2012 | PB | Asian | PCR‐RFLP | 221 ± 57.4 | 2.3 ± 0.8 | 66.0 ± 5.46 | 64.7 ± 5.71 | 28 | 73 | 94 | 0.031 | 19 | 104 | 127 | 0.727 | 195 | 250 |
| Liu | 2012 | HB | Asian | PCR‐RFLP | 161.45 ± 464.15 | 0.81 ± 0.90 | 70.7 ± 8.4 | 70.4 ± 10.0 | 31 | 85 | 86 | 0.196 | 19 | 100 | 102 | 0.426 | 202 | 221 |
| Agalliu | 2010 | PB | European | AB | NA | NA | NA | NA | 205 | 595 | 457 | 0.628 | 190 | 600 | 461 | 0.819 | 1257 | 1251 |
| Agalliu | 2010 | PB | African | AB | NA | NA | NA | NA | 16 | 61 | 70 | 0.623 | 9 | 38 | 36 | 0.827 | 147 | 83 |
| Hirata | 2007 | HB | Asian | PCR‐RFLP | NA | NA | 68 ± 5.0 | 67 ± 15 | 10 | 78 | 77 | 0.090 | 23 | 70 | 72 | 0.372 | 165 | 165 |
Abbreviations: AB, Applied Biosystems; HB, hospital‐based; HWE, Hardy‐Weinberg equilibrium; NA, not available; PB, population‐based; PCR‐RFLP, polymerase chain reaction and restrictive fragment length polymorphism; PSA, prostate‐specific antigen; RT, real‐time.
3.2. Quantitative synthesis
When all the studies pooled together (Table 2), no positive result was observed (C‐allele vs A‐allele, OR = 0.99, 95% CI = 0.94 ‐ 1.04, P heterogeneity = 0.058, P = .708; heterozygous contrast, OR = 0.95, 95% CI = 0.87 ‐ 1.03, P‐value for heterogeneity = 0.994, P = .194; dominant genetic model, OR = 0.96, 95% CI = 0.89 ‐ 1.04, P‐value for heterogeneity = 0.837, P = .343; recessive model, OR = 1.14, 95% CI = 0.93 ‐ 1.40, P heterogeneity = 0.001, P = .212). In a stratified analysis by ethnicity, a considerable increased risk was observed in Asian populations (OR = 1.21, 95% CI = 1.01 ‐ 1.43, P heterogeneity = 0.008, P = .034, I 2 = 65.2, Figure 1). However, we observed no obvious association between XPC rs2228001 A/C variant and PCa risk in European populations (allelic contrast: OR = 0.94, 95% CI = 0.88 ‐ 1.01, P heterogeneity = 0.033, P = .107; heterozygous contrast: OR = 0.95, 95% CI = 0.85 ‐ 1.06, P‐value for heterogeneity = 0.720, P = .333; dominant model: OR = 0.93, 95% CI = 0.84 ‐ 1.03, P heterogeneity = 0.260, P = .179; recessive model: OR = 0.91, 95% CI = 0.80 ‐ 1.05, P heterogeneity = 0.019, P = .199). Additionally, no positive association was identified in African individuals (allelic contrast: OR = 0.97, 95% CI = 0.75 ‐ 1.24, P heterogeneity = 0.709, P = .785; heterozygous comparison: OR = 0.77, 95% CI = 0.53 ‐ 1.12, P‐value for heterogeneity = 0.754, P = .169; dominant model: OR = 0.82, 95% CI = 0.58 ‐ 1.18, P heterogeneit = 0.918, P = .287; CC vs CA + AA: OR = 1.32, 95% CI = 0.77 ‐ 2.25 P heterogeneity = 0.422, P = .306). Moreover, in the subgroup analysis according to the source of control, no positive association of this XPC polymorphism was found in population‐based studies (recessive model: OR = 1.01, 95% CI = 0.90 ‐ 1.13, P heterogeneity = 0.002, P = .879, Figure 2). No positive correlation was detected in hospital‐based studies (allelic contrast: OR = 1.03, 95% CI = 0.90 ‐ 1.18, P heterogeneity = 0.305, P = .660; heterozygous comparison: OR = 0.94, 95% CI = 0.77 ‐ 1.15, P‐value for heterogeneity = 0.814, P = .544; CC + CA vs AA: OR = 0.98, 95% CI = 0.82 ‐ 1.18, P heterogeneity = 0.740, P = .858; CC vs CA + AA: OR = 1.14, 95% CI = 0.89 ‐ 1.46, P heterogeneity = 0.037, P = .289).
Table 2.
Stratified analysis of XPC rs2228001 A/C polymorphism on PCa risk
| Variables | N a | Case/Control | OR(95% CI) P heterogeneity P | OR(95% CI) P heterogeneity P | OR(95% CI) P heterogeneity P | OR(95% CI) P heterogeneity P |
|---|---|---|---|---|---|---|
| C‐allele vs A‐allele | CA vs AA | CC + CA vs AA | CC vs CA + AA | |||
| Total | 13 | 5305/6499 | 0.99 (0.94‐1.04) 0.058 0.708 | 0.95 (0.87‐1.03) 0.994 0.194 | 0.96 (0.89‐1.04) 0.837 0.343 | 1.14 (0.93‐1.40) 0.001 0.212 |
| Ethnicity | ||||||
| Asian | 7 | 2140/2358 | 1.06 (0.97‐1.16) 0.208 0.177 | 0.97 (0.85‐1.10) 0.981 0.643 | 1.02 (0.91‐1.15) 0.896 0.710 | 1.21 (1.01‐1.43) 0.008 0.034 |
| European | 3 | 2858/3742 | 0.94 (0.88‐1.01) 0.033 0.107 | 0.95 (0.85‐1.06) 0.720 0.333 | 0.93 (0.84‐1.03) 0.260 0.179 | 0.91 (0.80‐1.05) 0.019 0.199 |
| African | 2 | 257/349 | 0.97 (0.75‐1.24) 0.709 0.785 | 0.77 (0.53‐1.12) 0.754 0.169 | 0.82 (0.58‐1.18) 0.918 0.287 | 1.32 (0.77‐2.25) 0.422 0.306 |
| Arabian | 1 | 50/50 | 1.28 (0.73‐2.26)‐0.387 | 1.04 (0.44‐2.48)‐0.926 | 1.20 (0.52‐2.74)‐0.673 | 1.98 (0.61‐6.38)‐0.255 |
| Source of control | ||||||
| HB | 6 | 909/1145 | 1.03 (0.90‐1.18) 0.305 0.660 | 0.94 (0.77‐1.15) 0.814 0.544 | 0.98 (0.82‐1.18) 0.740 0.858 | 1.14 (0.89‐1.46) 0.037 0.289 |
| PB | 7 | 4396/5354 | 0.98 (0.93‐1.04) 0.029 0.547 | 0.95 (0.87‐1.04) 0.986 0.249 | 0.96 (0.88‐1.04) 0.608 0.338 | 1.01 (0.90‐1.13) 0.002 0.879 |
Abbreviations: HB, hospital‐based; PB, population‐based.
Number of case‐control studies.
Figure 1.
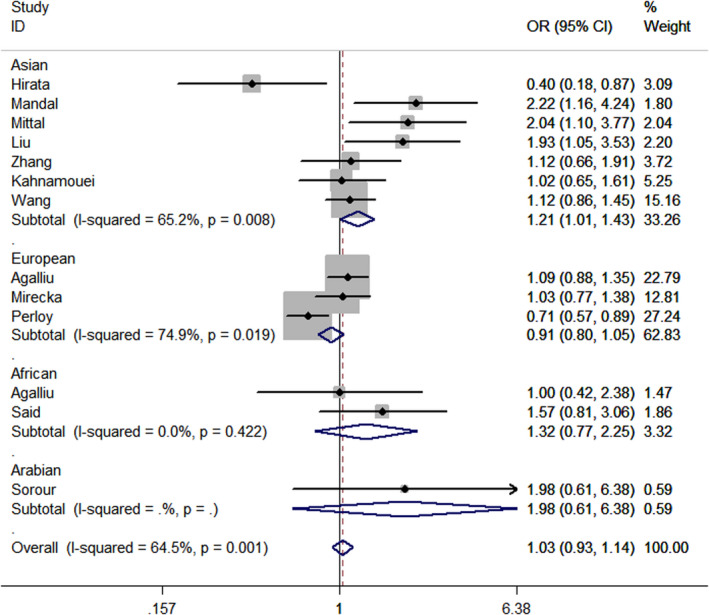
Forest plot of cancer risk correlated with XPC rs2228001 A/C variant (CC vs CA + AA) in stratified analysis by race
Figure 2.
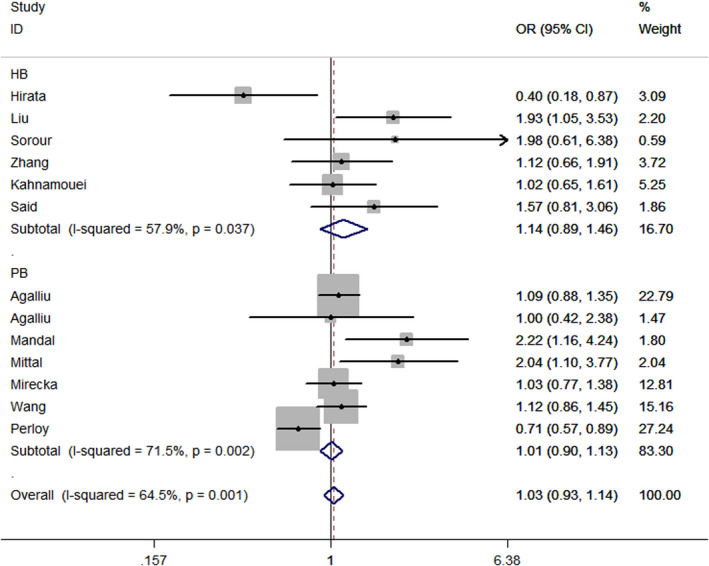
Forest plot of CC vs CA + AA model of XPC rs2228001 A/C polymorphism in the subgroup analysis by source of control
3.3. Expression of XPC utilizing in silico analysis
The in silico tool was used to evaluate the expression of XPC in 497 primary tumors and 52 normal tissues. XPC expression was lower in PCa tissues than in the control (P < .001, Figure 4A). The expression of XPC was down‐regulated in all Gleason scores of PCa. (P < .05, Figure 3). Furthermore, we investigated whether XPC expression influenced the overall survival and disease‐free survival rate in PCa cases. As shown in Figure 4B and Figure 4C, no significant correlation was observed between the high and low expression of XPC (P > .05). In order to investigate whether the rs2228001 A/C variant could have an impact on the expression of XPC, we adopted the Protein Variation Effect Analyzer (PROVEAN, v1.1) to predict the mutation of XPC. Sensitivity and specificity at different PROVEAN score cutoffs are shown in Figure 5A (default threshold is −2.5). PROVEAN score distribution for deleterious and neutral human protein variations is shown in Figure 5B. The PROVEAN score of the XPC rs2228001 A/C variant is 1.667, which indicates that this variant is neutral (Figure 5C). As shown in Figure 6A, at least 25 genes are involved in the interaction of XPC. The AKAP10 (A‐kinase anchoring protein 10), RBM9 (RBFOX2, RNA binding fox‐1 homolog 2), and BRPF1 (bromodomain and PHD finger containing 1) gene are the top three related genes. Results from TCGA samples indicated a significant correlation between XPC and AKAP10 in prostate cancer (Figure 6B). Similar findings were indicated for RBM9 (Figure 6C) and BRPF1 gene (Figure 6D). Nevertheless, there are few studies on their connections in present publications. We further used the online String server tools to explore the protein‐protein correlation regarding XPC. As described in Figure 8A, at least 20 proteins participate in the protein crosstalk with XPC. The top 10 proteins are as follows: CETN2: centrin‐2; RAD23A: UV excision repair protein RAD23 homolog A; RAD23B: UV excision repair protein RAD23 homolog B; GTF2H1: general transcription factor IIH subunit 1; XPA: DNA repair protein complementing XP‐A cells; ERCC4: DNA repair endonuclease XPF; ERCC1: DNA excision repair protein ERCC‐1; CHD1L: chromodomain‐helicase‐DNA‐binding protein 1‐like; RPA2: replication protein A 32 kDa subunit; DDB2: DNA damage‐binding protein 2 (Figure 8B).
Figure 4.
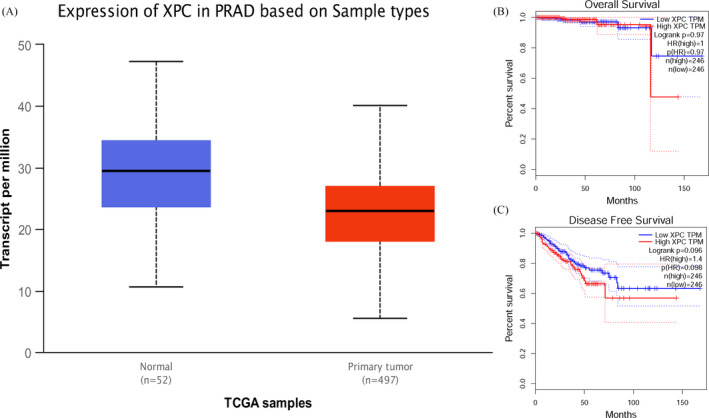
Association of the XPC expression in PCa based on sample types (Figure A) and the overall survival (Figure B) and disease‐free survival probability (Figure C)
Figure 3.
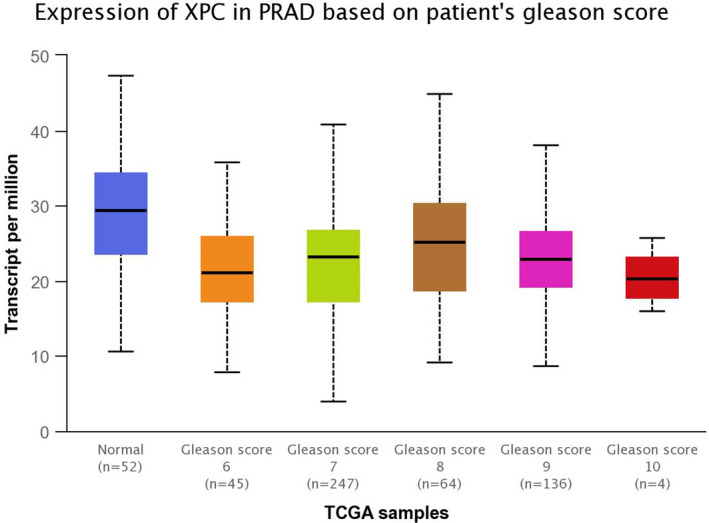
In silico analysis of XPC expression in PCa subjects based on patients’ Gleason score
Figure 5.
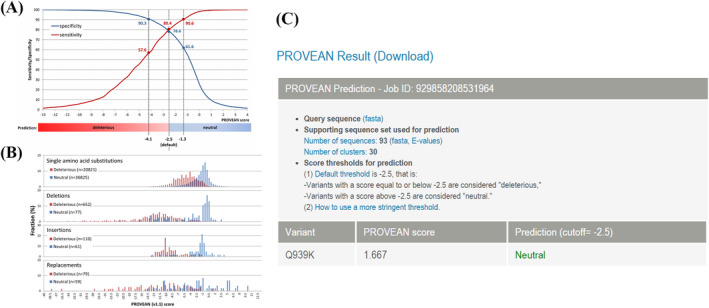
Evaluation of the XPC rs2228001 A/C variant by Protein Variation Effect Analyzer (PROVEAN, v1.1). Sensitivity and specificity at different PROVEAN score cutoffs are shown in Figure A (default threshold is −2.5). PROVEAN score distribution for deleterious and neutral UniProt human protein variations is shown in Figure B. The PROVEAN score of the XPC rs2228001 A/C variant is 1.667, which indicates that this variant is neutral (Figure C). Figure A and B is quoted from http://provean.jcvi.org/seq_submit.php
Figure 6.
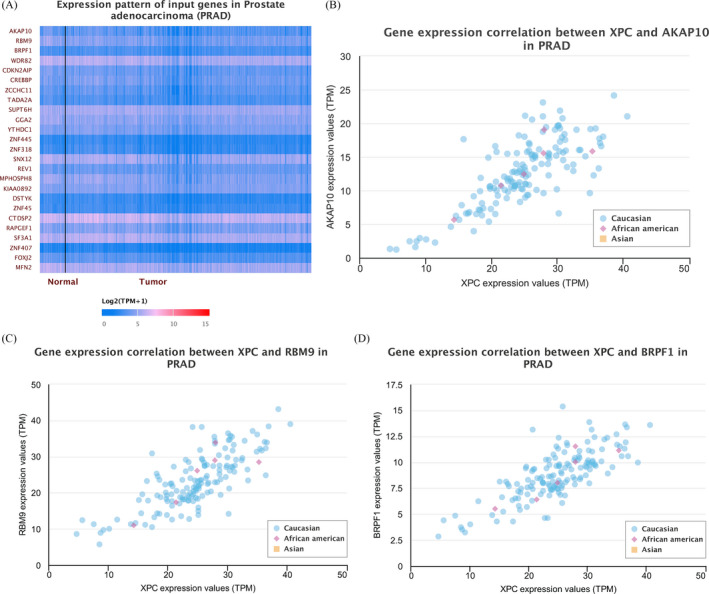
Gene‐gene crosstalk of XPC. As shown in Figure A, a total of 25 genes participate in the interaction of XPC. The AKAP10 gene is the most related gene. There was a significant correlation between XPC and AKAP10 in PCa (Figure B). Similar findings were indicated for RBM9 (Figure C) and BRPF1 gene (Figure D)
Figure 8.

XPC correlations with other proteins determined by String server (homo sapiens). At least 20 proteins participate in the protein crosstalk with XPC (Figure A). The top 10 proteins are as follows: CETN2: centrin‐2; RAD23A: UV excision repair protein RAD23 homolog A; RAD23B: UV excision repair protein RAD23 homolog B; GTF2H1: general transcription factor IIH subunit 1; XPA: DNA repair protein complementing XP‐A cells; ERCC4: DNA repair endonuclease XPF; ERCC1: DNA excision repair protein ERCC‐1; CHD1L: chromodomain‐helicase‐DNA‐binding protein 1‐like; RPA2: replication protein A 32 kDa subunit; DDB2: DNA damage‐binding protein 2 (Figure B)
3.4. Publication bias
We conducted Egger's test and Begg's funnel plot to detect the publication bias. Moreover, sensitivity analysis was applied to examine the impact of each study on the combined OR. No publication bias for the XPC rs2228001 A/C variant was observed from Egger's test (Figure 7A). For C‐allele vs A‐allele: t = 1.30, P = .219; heterozygous contrast: t = 1.22, P = .246; CC + CA vs AA: t = 1.22, P = .247; CC vs CA + AA: t = 1.36, P = .202. The symmetry of the funnel plot indicated no evidence of publication bias in our analysis as described in Figure 7B. The sensitivity analysis for the XPC variant is shown in Figure 7C. No individual study would influence the pooled OR.
Figure 7.
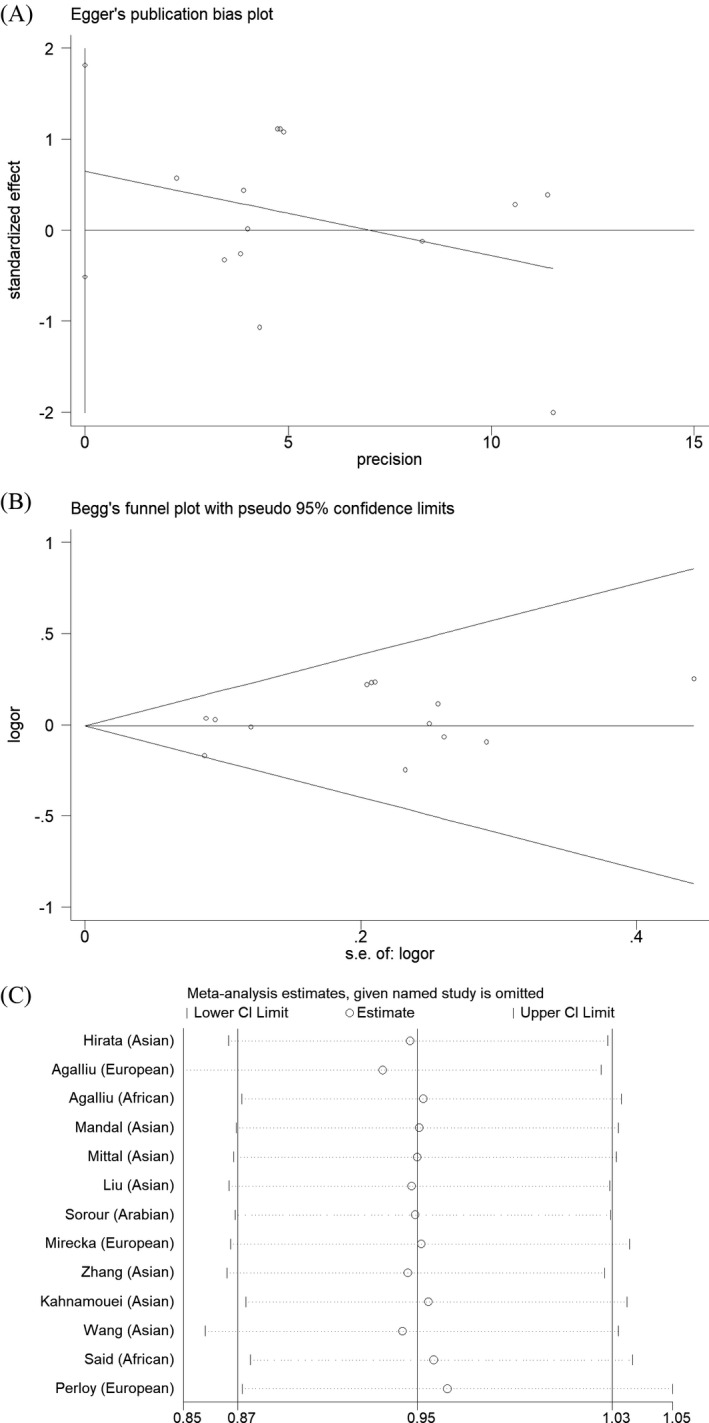
Publication bias analysis for the XPC rs2228001 A/C variant. No publication bias was observed from Egger's test (Figure A). The symmetry of Begg's funnel plot also indicated no evidence of publication bias (Figure B). The sensitivity analysis for the XPC variant is shown in Figure C. No individual study would influence the pooled OR
4. DISCUSSION
The pathogenesis of PCa remains complex. Previous research showed that genetic variants of XPC may be involved in down‐regulation of the DNA repair capacity (DRC). 29 , 30 Decreased DRC could cause genetic instability and contribute to susceptibility to PCa. 31 , 32 Previous case‐controlled studies were conducted to investigate whether the XPC rs2228001 A/C polymorphism confers the risk of PCa, but with controversial results. 17 , 18 , 19 , 20 , 21 , 22 , 23 A Japanese population‐based research showed that the XPC rs2228001 A/C polymorphism might be a risk factor for PCa. 17 However, another study based on Egyptian population suggested no significant difference between the XPC rs2228001 A/C variant and PCa susceptibility. 22 In 2013, a meta‐analysis conducted by He et al indicated elevated colorectal, lung, and bladder cancer susceptibility correlated with this polymorphism. 33 However, their conclusions cannot be confirmed by other researchers two years later. 34 Since then, new case‐control studies have emerged. The aim of the present study was to summarize all eligible data to draw more accurate conclusions.
In this study, 5305 cases and 6,499 control subjects were finally included to evaluate the effect of the XPC rs2228001 A/C variant in PCa susceptibility. When all studies pooled together, no positive result was observed (recessive genetic model: OR = 1.14, 95% CI = 0.93 ‐ 1.40, P heterogeneity = 0.001, P = .212). However, we found that the XPC rs2228001 A/C variant is associated with PCa risk in Asian populations (OR = 1.21, 95% CI = 1.01 ‐ 1.43, P = .034). Our finding is in line with the conclusions reported by He et al 33 Furthermore, in silico analysis was used to assess the expression of XPC in different grade of PCa. It showed evidence that XPC expression was down‐regulated in all Gleason scores of prostate cancer. We also evaluated whether the XPC expression influenced the overall survival probability of PCa cases; however, no positive correlation was indicated.
It is necessary to mention the limitations of the current analysis. First, the sample size of included studies in the current study was relatively small, especially for subgroup analyses. Second, some covariates such as age, tumor stage and grade, and smoking exposure should be added into stratification analysis. However, raw data of the included studies were not available to further evaluate the association between the XPC rs2228001 A/C polymorphism and these factors. Finally, other factors including gene‐gene and gene‐environment interactions are warranted to be considered. As shown in Figure 8, XPC may have the connection of twenty other proteins. Furthermore, TCGA samples have shown that more than 25 genes can participate in the connection of XPC. The AKAP10 gene is the most related gene. There is a significant correlation between XPC and AKAP10 in prostate cancer. However, there is a few research on the further mechanism of this gene, which is warranted to be evaluated in the future studies. Said et al found that XPC rs2228001 A/C variant was not correlated with PCa risk individually; however, combined analysis of rs2228001 A/C and XPC‐PAT variants showed that XPC (A/C + PAT D/D) genotypes were associated with susceptibility of PCa. 27 Additionally, some advantages of our analysis should be considered. First, all eligible data according to the inclusion criteria were summarized to investigate the relationship between XPC rs2228001 A/C polymorphism and PCa risk. The statistical power of the current analysis has been strengthened considerably. Second, no evidence of publication bias was identified in both Begg's funnel plot and Egger's test, indicating that conclusions of our study were stable and trustworthy.
In conclusion, our study suggested that the XPC rs2228001 A/C variant might contribute to elevated PCa risk in Asian patients. The expression of XPC was down‐regulated in PCa with different Gleason scores. In future, more large‐scale and well‐designed studies are warranted to confirm our conclusions in more detail.
CONFLICT OF INTEREST
The authors declare no competing interests.
ETHICAL APPROVAL
Not applicable.
ACKNOWLEDGMENT
Not applicable.
Yan Y, Xu J, Xu B, et al. Effects of Xeroderma pigmentosum group C polymorphism on the likelihood of prostate cancer. J Clin Lab Anal. 2020;34:e23403 10.1002/jcla.23403
Yan and Xu are equal contributors.
Contributor Information
Li Zuo, Email: urology2019@sina.com.
Guoqiang Lv, Email: leoguo88@hotmail.com.
Yunfeng Shi, Email: fzy8353@163.com.
DATA AVAILABILITY STATEMENT
All data generated and analyzed during this study are included in this published article. Please contact the author for data requests.
REFERENCES
- 1. Luján M, Páez A, Angulo JC, et al. Prostate cancer incidence and mortality in the Spanish section of the European Randomized Study of Screening for Prostate Cancer (ERSPC). Prostate Cancer Prostatic Dis. 2014;17(2):187‐191. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10‐29. [DOI] [PubMed] [Google Scholar]
- 3. Gomez SL, Noone AM, Lichtensztajn DY, et al. Cancer incidence trends among Asian American populations in the United States, 1990–2008. J Natl Cancer Inst. 2013;105(15):1096‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sartor O. Advanced prostate cancer update 2018. Asia Pac J Clin Oncol. 2018;14(Suppl 5):9‐12. [DOI] [PubMed] [Google Scholar]
- 5. Henegan JC, Sonpavde G. Promising immunotherapy for prostate cancer. Expert Opin Biol Ther. 2018;18(2):109‐120. [DOI] [PubMed] [Google Scholar]
- 6. Boukovala M, Spetsieris N, Efstathiou E. Systemic treatment of prostate cancer in elderly patients: current role and safety considerations of androgen‐targeting strategies. Drugs Aging. 2019;36(8):701‐717. [DOI] [PubMed] [Google Scholar]
- 7. Trzeciak AR, Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines, PC‐3 and DU‐145. Carcinogenesis. 2004;25(8):1359‐1370. [DOI] [PubMed] [Google Scholar]
- 8. Xu Z, Chen ZP, Malapetsa A, et al. DNA repair protein levels vis‐à‐vis anticancer drug resistance in the human tumor cell lines of the National Cancer Institute drug screening program. Anticancer Drugs. 2002;13(5):511‐519. [DOI] [PubMed] [Google Scholar]
- 9. Park CJ, Choi BS. The protein shuffle. Sequential interactions among components of the human nucleotide excision repair pathway. FEBS J. 2006;273(8):1600‐1608. [DOI] [PubMed] [Google Scholar]
- 10. Kusakabe M, Onishi Y, Tada H, et al. Mechanism and regulation of DNA damage recognition in nucleotide excision repair. Genes Environ. 2019;41:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pérez‐Cadahía B, Laffon B, Valdiglesias V, Pásaro E, Méndez J. Cytogenetic effects induced by Prestige oil on human populations: the role of polymorphisms in genes involved in metabolism and DNA repair. Mutat Res. 2008;653(1–2):117‐123. [DOI] [PubMed] [Google Scholar]
- 12. Sankhwar M, Sankhwar SN, Bansal SK, Gupta G, Rajender S. Polymorphisms in the XPC gene affect urinary bladder cancer risk: a case‐control study, meta‐analyses and trial sequential analyses. Sci Rep. 2016;6:27018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun HY, Zuo L, Zou JG, et al. Current evidence on XPC rs2228001 A/C polymorphism and bladder cancer susceptibility. Int J Clin Exp Med. 2016;9(2):2881‐2888. [Google Scholar]
- 14. Chen C, Wang L, Liao Q, et al. Association between six genetic polymorphisms and colorectal cancer: a meta‐analysis. Genet Test Mol Biomarkers. 2014;18(3):187‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lawania S, Singh N, Behera D, Sharma S. XPC Polymorphism and risk for lung cancer in North Indian patients treated with platinum based chemotherapy and its association with clinical outcomes. Pathol Oncol Res. 2018;24(2):353‐366. [DOI] [PubMed] [Google Scholar]
- 16. He BS, Xu T, Pan YQ, et al. Nucleotide excision repair pathway gene polymorphisms are linked to breast cancer risk in a Chinese population. Oncotarget. 2016;7(51):84872‐84882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirata H, Hinoda Y, Tanaka Y, et al. Polymorphisms of DNA repair genes are risk factors for prostate cancer. Eur J Cancer. 2007;43(2):231‐237. [DOI] [PubMed] [Google Scholar]
- 18. Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, Stanford JL. Genetic variation in DNA repair genes and prostate cancer risk: results from a population‐based study. Cancer Causes Control. 2010;21(2):289‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Y, Chen Z, Wei Q, et al. Poly (AT) polymorphism in the XPC gene and smoking enhance the risk of prostate cancer in a low‐risk Chinese population. Cancer Genet. 2012;205(5):205‐211. [DOI] [PubMed] [Google Scholar]
- 20. Mandal RK, Gangwar R, Kapoor R, Mittal RD. Polymorphisms in base‐excision & nucleotide‐excision repair genes & prostate cancer risk in north Indian population. Indian J Med Res. 2012;135:64‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mittal RD, Mandal RK. Genetic variation in nucleotide excision repair pathway genes influence prostate and bladder cancer susceptibility in North Indian population. Indian J Hum Genet. 2012;18(1):47‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sorour AF, Talaat IM, Youssif TMA, Atta MA. Detection of polymorphisms of DNA repair genes (XRCC1 and XPC) in prostate cancer. J Cancer Ther. 2013;4:1499‐1505. [Google Scholar]
- 23. Mirecka A, Paszkowska‐Szczur K, Scott RJ, et al. Common variants of xeroderma pigmentosum genes and prostate cancer risk. Gene. 2014;546(2):156‐161. [DOI] [PubMed] [Google Scholar]
- 24. Zhang XJ, Liu P, Zhu F. Polymorphisms of DNA repair‐related genes with susceptibility and prognosis of prostate cancer. Genet Mol Res. 2014;13(2):4419‐4424. [DOI] [PubMed] [Google Scholar]
- 25. Kahnamouei SA, Narouie B, Sotoudeh M, et al. Association of XPC Gene polymorphisms with prostate cancer risk. Clin Lab. 2016;62(6):1009‐1015. [DOI] [PubMed] [Google Scholar]
- 26. Wang M, Li Q, Gu C, et al. Polymorphisms in nucleotide excision repair genes and risk of primary prostate cancer in Chinese Han populations. Oncotarget. 2017;8(15):24362‐24371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Said R, Bougatef K, Setti Boubaker N, et al. Polymorphisms in XPC gene and risk for prostate cancer. Mol Biol Rep. 2019;46(1):1117‐1125. [DOI] [PubMed] [Google Scholar]
- 28. Perloy A, Schouten LJ, van den Brandt PA, Godschalk R, van Schooten FJ, Hogervorst JGF. The role of genetic variants in the association between dietary acrylamide and advanced prostate cancer in the Netherlands cohort study on diet and cancer. Nutr Cancer. 2018;70(4):620‐631. [DOI] [PubMed] [Google Scholar]
- 29. Xiao M, Xiao S, Straaten TV, et al. Lu X. Genetic polymorphisms in 19q13.3 genes associated with alteration of repair capacity to BPDE‐DNA adducts in primary cultured lymphocytes. Mutat Res. 2016;812:39‐47. [DOI] [PubMed] [Google Scholar]
- 30. Cornetta T, Festa F, Testa A, Cozzi R. DNA damage repair and genetic polymorphisms: assessment of individual sensitivity and repair capacity. Int J Radiat Oncol Biol Phys. 2006;66(2):537‐545. [DOI] [PubMed] [Google Scholar]
- 31. Henríquez‐Hernández LA, Valenciano A, Foro‐Arnalot P, et al. Single nucleotide polymorphisms in DNA repair genes as risk factors associated to prostate cancer progression. BMC Med Genet. 2014;15:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barry KH, Koutros S, Andreotti G, et al. Genetic variation in nucleotide excision repair pathway genes, pesticide exposure and prostate cancer risk. Carcinogenesis. 2012;33(2):331‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He J, Shi TY, Zhu ML, Wang MY, Li QX, Wei QY. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta‐analysis. Int J Cancer. 2013;133(8):1765‐1775. [DOI] [PubMed] [Google Scholar]
- 34. Wu H, Lv Z, Wang X, Zhang L, Mo N. Lack of association between XPC Lys939Gln polymorphism and prostate cancer risk: an updated meta‐analysis based on 3039 cases and 3253 controls. Int J Clin Exp Med. 2015;8(10):17959‐17967. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated and analyzed during this study are included in this published article. Please contact the author for data requests.


