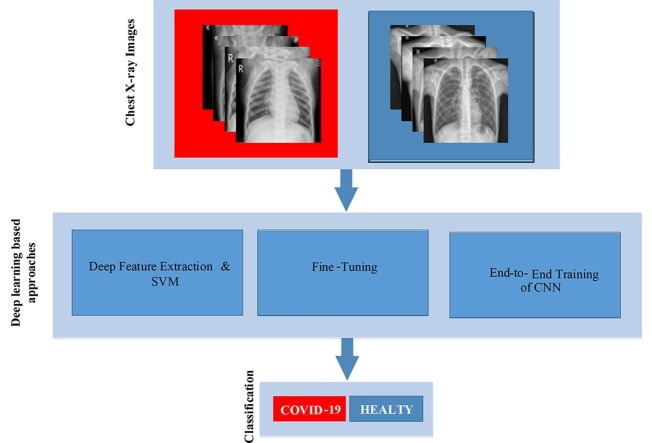
Graphical abstract
Keywords: COVID-19, Chest X-ray images, Deep learning, Convolutional neural networks, Local texture descriptors
Highlights
-
•
A novel application is introduced, X-ray chest image based Covid-19 detection.
-
•
Deep learning approaches are used for Covid-19 detection.
-
•
Local texture descriptors are also used in Covid-19 detection.
Abstract
COVID-19 is a novel virus that causes infection in both the upper respiratory tract and the lungs. The numbers of cases and deaths have increased on a daily basis on the scale of a global pandemic. Chest X-ray images have proven useful for monitoring various lung diseases and have recently been used to monitor the COVID-19 disease. In this paper, deep-learning-based approaches, namely deep feature extraction, fine-tuning of pretrained convolutional neural networks (CNN), and end-to-end training of a developed CNN model, have been used in order to classify COVID-19 and normal (healthy) chest X-ray images. For deep feature extraction, pretrained deep CNN models (ResNet18, ResNet50, ResNet101, VGG16, and VGG19) were used. For classification of the deep features, the Support Vector Machines (SVM) classifier was used with various kernel functions, namely Linear, Quadratic, Cubic, and Gaussian. The aforementioned pretrained deep CNN models were also used for the fine-tuning procedure. A new CNN model is proposed in this study with end-to-end training. A dataset containing 180 COVID-19 and 200 normal (healthy) chest X-ray images was used in the study’s experimentation. Classification accuracy was used as the performance measurement of the study. The experimental works reveal that deep learning shows potential in the detection of COVID-19 based on chest X-ray images. The deep features extracted from the ResNet50 model and SVM classifier with the Linear kernel function produced a 94.7% accuracy score, which was the highest among all the obtained results. The achievement of the fine-tuned ResNet50 model was found to be 92.6%, whilst end-to-end training of the developed CNN model produced a 91.6% result. Various local texture descriptors and SVM classifications were also used for performance comparison with alternative deep approaches; the results of which showed the deep approaches to be quite efficient when compared to the local texture descriptors in the detection of COVID-19 based on chest X-ray images.
1. Introduction
Chest X-ray images are known to have potential in the monitoring and examination of various lung diseases such as tuberculosis, infiltration, atelectasis, pneumonia, and hernia. COVID-19, which manifests as an upper respiratory tract and lung infection, was first investigated in the Wuhan province of China in late 2019, and is mostly seen to affect the airway and consequently the lungs of those infected. The virus has since spread rapidly to become a global pandemic (World Health Organization, 2020), with numbers of cases and associated deaths still increasing on a daily basis (Worldmeter, 2020). Chest X-ray images have been shown to be useful in following-up on the effects that COVID-19 causes to lung tissue (Radiology Assistant, 2020). Consequently, chest X-ray images may also be used in the detection of COVID-19.
In the literature, there have been various deep-learning-based approaches that employ chest X-ray images for disease detection. Kesim, Dokur, and Olmez (2019) proposed a new Convolutional Neural Network (CNN) model for chest X-ray image classification. The authors developed a small-sized CNN architecture due to pretrained CNN models being known to present difficulties in practical applications. A 12-class chest X-ray image dataset was used by the authors, with an 86% accuracy score reported in their testing. Liu et al. (2017) proposed a deep-learning-based approach for the detection of tuberculosis. In their approach, the authors developed a novel CNN model that used chest X-ray images as input, and transfer learning. The authors mentioned that shuffle sampling was used to handle the unbalanced dataset problem, and that an 85.68% accuracy score was obtained using the shuffle sampling method. Dong, Pan, Zhang, and Xu (2017) constructed a vast dataset containing chest X-ray images to which they applied deep CNN models for binary and multilevel classifications. Transfer learning with pretrained AlexNet, ResNet, and VGG16 models were used with the constructed dataset. While an 82.2% accuracy score was reported for the binary classification, over 90% accuracy scores were reported for the other classification tasks. Xu, Wu, and Bie (2018) developed an approach for abnormality detection using chest X-ray images. The authors proposed a hierarchical-CNN model named CXNet-m1 to overcome the over-fitting problem associated with transfer learning. The developed CNN models were shallower than the pretrained CNN models. A novel loss function and optimization of the CNN kernels were also proposed in their work, with a 67.6% accuracy score reported by the authors. Chouhan et al. (2020) detected pneumonia in chest X-ray images using five new deep-transfer-learning-based models applied as an ensemble. The authors reported a 96.4% accuracy score using their developed ensemble deep model. Rajpurkar et al. (2018) developed a 121-layered CNN architecture named CheXNeXt for the classification of 14 different pathologies based on chest X-ray images. The authors trained a developed CNN model using the ChestX-ray8 dataset. Using the area-under-curve (AUC) measure for performance measurements, the authors reported that the proposed model produced AUC values between 0.704 and 0.944. Li et al. (2019) used Multi-Resolution CNN (MR-CNN) for lung nodule detection, with a patch-based MR-CNN employed to extract features, and then various fusion methods were used in the process of classification. The authors used FAUC and R-CPM metrics for performance evaluation, and reported values of 0.982 and 0.987, respectively. Bhandary et al. (2020) modified the AlexNet model for the detection of lung abnormalities based on chest X-ray images. More specifically, the authors used a deep learning approach for the detection of pneumonia. A new “threshold filter” was introduced and a feature ensemble strategy was also defined which produced a 96% classification accuracy rate. Uçar and Uçar (2019) used Laplacian of Gaussian filters to increase the classification performance of a CNN using chest X-ray images, with a reported 82.43% classification accuracy from their new approach.
Woźniak et al. (2018) proposed a new approach, based on using chest X-ray images for the detection of lung carcinomas. In their proposed method, momentum and probabilistic neural networks (PNN) were used. The nodules in the chest X-ray images were initially segmented and the momentum of the contours of segmented nodules then used as features. PNN was used in the classification of the nodules, and a 92% classification accuracy was reported by the authors. Ho and Gwak (2019) opted to use feature concatenation for the efficient classification of 14 thoracic diseases that used deep features and four local texture descriptors (SIFT, GIST, LBP, and HOG). In the classification stage, GDA, k-NN, Naïve Bayes, SVM, Adaboost, Random forests, and ELM were used, and an 84.62% accuracy score was reported. Souza et al. (2019) developed an automatic approach for lung segmentation in chest X-ray images. The main aim of the work was to determine tuberculosis regions in lungs based on chest X-ray images. For segmentation, two deep CNN-based approaches were employed, with a 94% segmentation accuracy reported by the authors.
In the current study, and unlike the methods proposed in the literature, deep learning approaches are proposed for the detection of COVID-19 based on chest X-ray images. Whilst various lung diseases (e.g., tuberculosis, pneumonia, and lung carcinomas) have been detected from chest X-ray images, the current study is limited to the detection of COVID-19 versus normal (healthy) cases using chest X-ray images. In the first deep learning approach, a new CNN model was proposed that was trained end-to-end. In a second approach, pretrained CNN models were used for deep feature extraction using SVM classifiers with various kernel functions (i.e., Linear, Quadratic, Cubic, and Gaussian) used for the purposes of COVID-19 classification. In a third approach, different pretrained CNN models were further trained (or “fine-tuned”) using chest X-ray images for the detection of COVID-19. A chest X-ray image dataset composed of 180 COVID-19 samples and 200 healthy (normal) samples was first constructed (GitHub, 2020, Kaggle, 2020a, Kaggle, 2020b). The ethical issues pertaining to the collection and usage of the images were reported to have been addressed according to the websites from where the images were retrieved.
Classification accuracy was used for the performance evaluation of the proposed methods. The pretrained deep CNN models used in the current study were ResNet18, ResNet50, ResNet101, VGG16, and VGG19. From the study’s experimentation, the deep features model (ResNet50) and SVM with Linear kernel function produced a 94.7% accuracy score, which was the highest among all the results obtained. Test achievements for the fine-tuning of the ResNet50 model and end-to-end training of the developed CNN model were found to be 92.6% and 91.6%, respectively. For comparative purposes, various local texture descriptors were considered; namely, Local Binary Patterns (LBP) (Ahonen, Hadid, & Pietikainen, 2006), Frequency Decoded LBP (FDLBP) (Dubey, 2019), Quaternionic Local Ranking Binary Pattern (QLRBP) (Lan, Zhou, & Tang, 2015), Binary Gabor Pattern (BGP) (Zhang, Zhou, & Li, 2012), Local Phase Quantization (LPQ) (Ojansivu & Heikkilä, 2008), Binarized Statistical Image Features (BSIF) (Kannala & Rahtu, 2012), CENsus TRansform hISTogram (CENTRIST) (Wu & Rehg, 2010), and Pyramid Histogram of Oriented Gradients (PHOG) (Bosch, Zisserman, & Munoz, 2007). From the local texture descriptors, the BSIF with SVM classifier produced a 90.5% accuracy score.
The contribution of this paper is therefore as follows:
-
•
A novel application of a deep learning model is used for the detection of COVID-19 based on chest X-ray images.
The remainder of this paper is arranged as follows. Section 2 details the materials and methods used in the study, including CNN, transfer learning, end-to-end training and SVM theories, as well as a brief introduction to the methodology used. Section 3 describes the experimental works and their results, and Section 4 provides the conclusions of the study.
2. Materials and methods
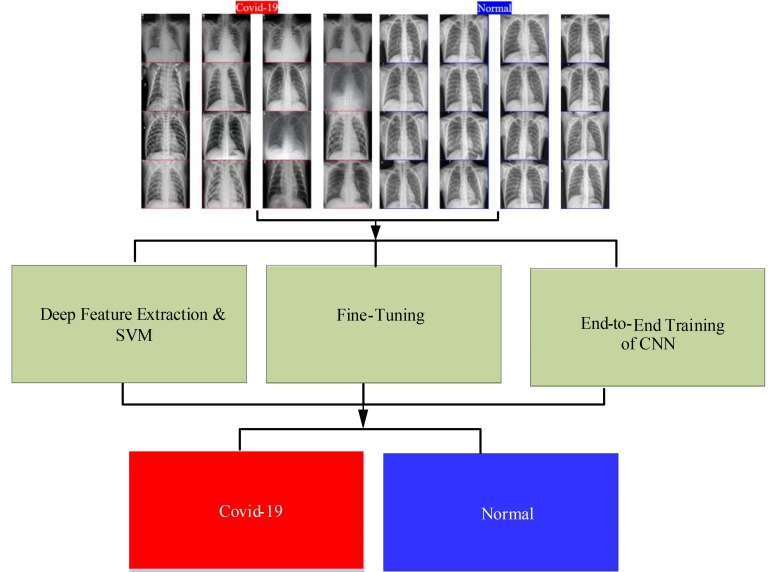
The proposed method is illustrated as shown in Fig. 1 , in which chest X-ray images are used as input to the proposed COVID-19 detection method. The input chest X-ray images are initially resized to 224 × 224 pixels for compatibility with the CNN models. As previously mentioned, three deep learning approaches were considered, namely deep feature extraction from pretrained deep networks, fine-tuning of a pretrained CNN model, and end-to-end training of a CNN model.
Fig. 1.
Illustration of Proposed Methodology for COVID-19 Detection.
For both deep feature extraction and fine-tuning procedures, the ResNet18, ResNet50, ResNet101, VGG16, and VGG19 models were used. For the training of the deep features, SVM classifier was used with various kernel functions, namely Linear, Quadratic, Cubic, and Gaussian. A 21-layered new CNN model was also proposed and trained end-to-end. The proposed network model starts with an input layer, then there are five convolutions layers, five ReLU layers, and five batch normalization layers, respectively. Two pooling layers are used after the first and second ReLU layers, respectively. A fully-connected layer, softmax layer, and classification layer are also used at the end of the model.
2.1. Deep transfer learning
The procedure of deep feature extraction and fine-tuning of pretrained CNN models is defined as deep transfer learning (DTL). With a limited number of training images, DTL helps in the deep learning process for the image classification task of this proposed method (Pan & Yang, 2009). The DTL concept transfers knowledge from a source domain where there are many training samples, to a target domain where there are comparatively much fewer samples. Thus, efficient image classification can be achieved with the support of the larger dataset from the source domain. In terms of the deep learning perspective, especially in the case of a CNN, DTL is defined as transferring certain layers of a pretrained CNN model previously trained with millions of images. More specifically, the task-dependent layers of the CNN model such as the fully-connected layers and the classification output layer are removed from the network architecture and the remaining layers saved for application to the new classification task (Deniz et al., 2018).
2.2. Convolution neural networks (CNNs)
A sequence of convolution, normalization, and pooling layers are used to construct the main building blocks of a CNN architecture (Omar, Sengur, & Al-Ali, 2020). While the convolution layers are responsible for the extraction of the local features, the normalization and pooling layers are responsible for the normalization of the local features and for the down-sampling of the local features, respectively.
The output feature map is obtained in Eq. (1), where shows the local features obtained from the previous layers, and and denote the adjustable kernels and training bias, respectively. Bias is used to prevent overfitting during the training of the CNN (Başaran, Cömert, & Çelik, 2020);
| (1) |
where, and denotes the input map selection and activation function, respectively. As previously mentioned, the pooling layer is employed for the down-sampling of the feature maps. There are various pooling techniques applied, namely average and maximum. Pooling layers are responsible for decreasing the computational nodes and for preventing the issue of overfitting within the CNN architecture (Xu et al., 2019). The pooling process is defined as shown in Equation (2);
| (2) |
where, the down-sampling is shown by the function. It is worth noting here that down-sampling provides an abstract of the local features for the next layer. The fully-connected (FC) layers have full connections to all of the activations in the previous layer. The FC layer provides discriminative features for the classification of the input image into its various classes.
Similar to the traditional machine learning techniques, the training of the CNN was carried out using an optimization process. Stochastic Gradient Descent with Momentum (SGDM), and Adaptive Moment Estimation (ADAM) are two well-known training methods utilized for neural networks.
2.3. Support Vector Machines (SVMs)
Support Vector Machines were developed by Vapnik (Widodo & Yang, 2007), and are well-known as classifiers based on the structural risk reduction principle categorized in the supervised statistical learning theorem. The main idea of the SVM classifier is to determine an optimum hyperplane between positive and negative samples (Qi, Tian, & Shi, 2013). The linear separation of the positive and negative samples can be handled using Equation (3) (Adaminejad & Farjah, 2013);
| (3) |
where indicates the weight vector and is bias value used to determine the position of the hyperplane. A kernel trick is employed to transfer the input data to another hyperplane where the data is more convenient for linear separation. The best hyperplane can be determined by using Eq. (4) (Cheng & Bao, 2014);
| (4) |
3. Experimental works and results
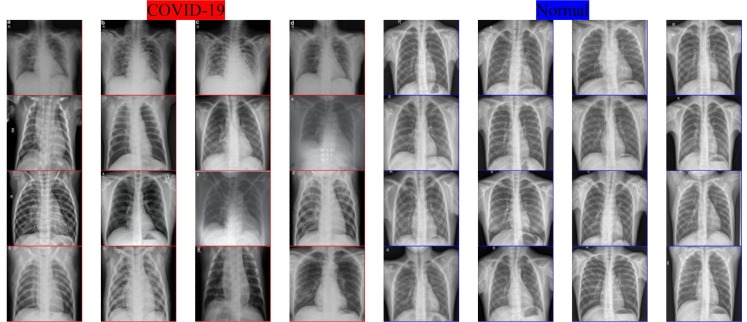
All coding were carried out with MATLAB software on a workstation equipped with the NVIDIA Quadro M4000 GPU with 8 GB RAM. The chest X-ray images were collected from three different sources (GitHub, 2020, Kaggle, 2020b, Radiology Assistant, 2020), with the labelling of the chest X-ray images conducted by specialist doctors. A total of 180 COVID-19 and 200 normal (healthy) chest X-ray images were collected. In the study’s experiments, a random 75% selection of the dataset was used for the purposes of training, and the remaining 25% was used for testing the proposed method. Fig. 2 shows some sample COVID-19 and normal (healthy) chest X-ray images.
Fig. 2.
Various Chest X-ray Images from COVID-19 and Normal Cases.
The chest X-ray images were initially resized to 224 × 224 pixels for compatibility requirements as input to the CNN models. Some of the collected images were in grayscale and these images were converted to color format by copying the grayscale image to all R, G, and B channels. No other preprocessing methods were applied to the chest X-ray images. The hyperparameters of the proposed methods were selected heuristically during the experimentation.
3.1. Deep features and SVM
Five pretrained CNN models, namely VGG16, VGG19, ResNet18, ResNet50, and ResNet101 were used in the study’s experiments. Moreover, SVM method was used for the purposes of classification with four kernel functions, namely Linear, Quadratic, Cubic, and Gaussian. The epsilon value of the SVMs with the Linear and Quadratic kernel functions were set to 0.04 and 0.02, respectively, whilst both the Cubic kernel and Gaussian functions’ epsilon value was set as 0.01. The classification accuracy score was used in the performance evaluation of the study’s testing, and Table1 presents the obtained accuracy scores. While the rows of Table1 show the SVM kernel types, the columns show the pretrained CNN models. The last row and column show the average accuracy scores.
Table 1.
Achievements of pretrained deep CNN models and SVM classifiers on COVID-19 detection.
| Accuracy (%) |
||||||
|---|---|---|---|---|---|---|
| Method: SVM | ResNet18 | ResNet50 | ResNet101 | VGG16 | VGG19 | Average |
| Linear Kernel | 86.3 | 94.7 | 88.4 | 89.5 | 88.4 | 89.5 |
| Quadratic Kernel | 87.4 | 91.6 | 89.5 | 89.5 | 87.4 | 89.1 |
| Cubic Kernel | 89.5 | 90.5 | 91.6 | 90.5 | 89.5 | 90.3 |
| Gaussian Kernel | 86.3 | 93.7 | 88.4 | 89.5 | 87.4 | 89.1 |
| Average | 87.4 | 92.6 | 89.5 | 89.8 | 88.1 | |
From Table1, it can be seen that the ResNet50 model produced the highest average accuracy score, with an average accuracy score of 92.6%, whilst the VGG16 model produced an average accuracy score of 89.8% as the second best score. The ResNet101 model produced an average accuracy score of 89.5%, whilst the VGG19 and ResNet18 models produced average accuracy scores of 88.1% and 87.4%, respectively.
When the results are examined in terms of the kernel functions, it can be seen that the best average accuracy score was 90.3%, produced by the Cubic kernel function. A second-best average accuracy score of 89.5% was produced by the Linear-kernel-based SVM classifier. The SVM classifiers based on the Quadratic kernel and the Gaussian kernel produced identical 89.1% accuracy scores.
From Table1, it can also be observed that the ResNet50 features and Linear kernel SVM classifier produced a 94.7% accuracy score, which was the highest individual accuracy score overall. The second-best individual accuracy score was 93.7%, which was also produced by the ResNet50 model, but with the Gaussian-kernel-based SVM classifier. It is worth mentioning here that the lowest accuracy score was 86.3%, and was produced by the ResNet18 model both with the Linear kernel and Gaussian kernel functions.
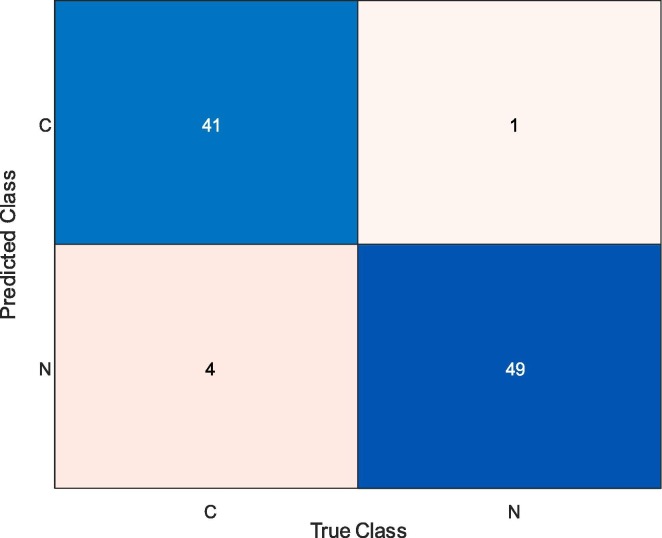
Fig. 3 presents the confusion matrix for the ResNet50 features with the Linear SVM classifier. The labels “C” and “N” represent the COVID-19 and normal (healthy) cases, respectively. From Fig. 3, it can also be observed that while 41 COVID-19 and 49 normal (healthy) samples were correctly classified, four COVID-19 samples and one normal (healthy) sample were misclassified. Therefore, the rate of correct classification of COVID-19 samples was 91.11%, whilst it was 98.0% for the normal (healthy) samples.
Fig. 3.
Confusion Matrix Obtained for ResNet50 and Linear SVM Classifier.
3.2. Fine-tuning
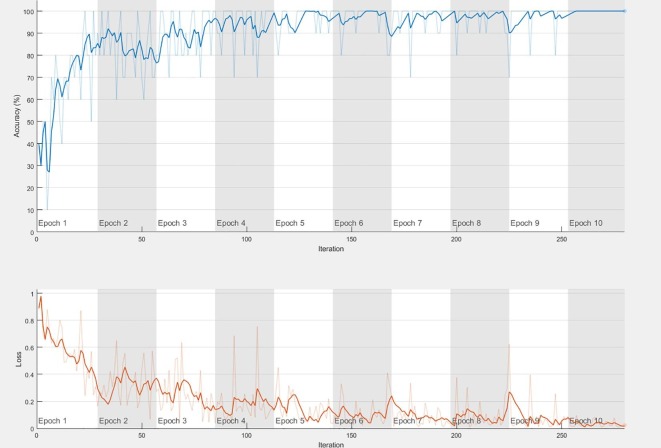
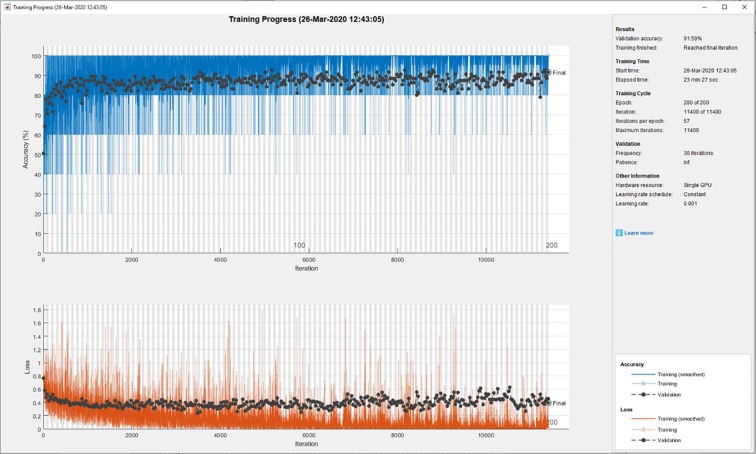
Fig. 4 illustrates the training process applied to fine-tune the ResNet50 model. Data augmentation was conducted both for fine-tuning and for end-to-end training, and was carried out by randomly rotating, shifting, and flipping the training images. The upper graph in Fig. 4 shows the training and average training accuracies (with light-blue = training [smoothed], and blue = testing), whilst the lower graph in Fig. 4 shows the loss value for the training samples (orange = training [smoothed], and light-orange = training). Table2 details the achievements of the fine-tuned pretrained deep CNN models on COVID-19 classification.
Fig. 4.
Fine-tuning of ResNet50 Model for COVID-19 Classification.
Table 2.
Achievements of Fine-Tuning Of Pretrained Deep CNN models on COVID-19 classification.
| Fine-tuning | Accuracy (%) |
|---|---|
| VGG16 | 85.26 |
| ResNet18 | 88.42 |
| ResNet50 | 92.63 |
| ResNet101 | 87.37 |
| VGG19 | 89.47 |
As can be seen in Table2, all of the fine-tuned deep CNN models achieved classification accuracy scores above 85%. The highest accuracy score was 92.63%, produced by the ResNet50 model, whilst the second-best accuracy score of 89.47% was obtained from the VGG19 model. In addition, 88.42%, 87.37%, and 85.26% accuracy scores were obtained from the ResNet18, ResNet101, and VGG16 models, respectively.
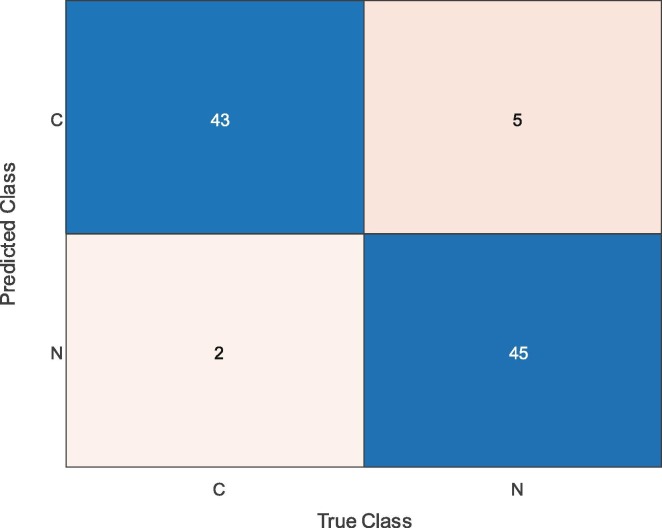
Fig. 5 presents the confusion matrix for the fine-tuned ResNet50 model. As can be seen from Fig. 5, while 43 COVID-19 samples and 45 normal (healthy) samples were classified correctly, two COVID-19 and five normal (healthy) samples were misclassified. The rate of correct classification of COVID-19 samples was therefore 95.56%, whilst it was 90.0% for the normal (healthy) samples.
Fig. 5.
Confusion Matrix Obtained by Fine-tuning ResNet50 Model.
3.3. End-to-end training
In the final experiments of the current study, a novel CNN model was constructed and end-to-end trained for the purposes of COVID-19 classification. The developed CNN model is illustrated as shown in Fig. 6 , and was composed of 21 layers. The network started with an input layer, followed by five convolutions layers, namely conv_1, conv_2, conv_3, conv_4, and conv_5, with batch normalization and ReLU layers following each convolution layer. There were also two pooling layers, pool_1 and pool_2, which followed after the ReLu_1 and ReLu_2 layers, respectively. A fully-connected layer, softmax layer, and classification layer were also used for the purposes of classification. The conv_1, conv_2, and conv_3 layers contained 64, 32, 16, 8, and 4 filters that were size 3 × 3 pixels. The max operator function was used in the pooling layers.
Fig. 6.
Developed CNN Model for COVID-19 Detection.
Details of the end-to-end CNN architecture covering descriptions of the layers, activations, and learnable weights are presented in Table3 . The training of the end-to-end CNN model was conducted using the “SGDM” optimizer, with an initial learning rate of 0.001, and the network was trained over 300 iterations.
Table 3.
Analysis of proposed CNN model for COVID-19 detection.
| 1 | “input” | 224 × 224 × 3 images |
| 2 | “conv_1” | 64 3 × 3 × 3 convolutions with stride [1 1] and padding “same” |
| 3 | “BN_1” | Batch normalization |
| 4 | “relu_1” | ReLU |
| 5 | “pool_1” | 2 × 2 max pooling with stride [2 2] and padding [0 0 0 0] |
| 6 | “conv_2” | 32 3 × 3 × 8 convolutions with stride [1 1] and padding “same” |
| 7 | “BN_2” | Batch normalization |
| 8 | “relu_2” | ReLU |
| 9 | “pool_2” | 2 × 2 max pooling with stride [2 2] and padding [0 0 0 0] |
| 10 | “conv_3” | 16 3 × 3 × 16 convolutions with stride [1 1] and padding “same” |
| 11 | “BN_3” | Batch normalization |
| 12 | “relu_3” | ReLU |
| 13 | “conv_4” | Eight 3 × 3 × 16 convolutions with stride [1 1] and padding “same” |
| 14 | “BN_4” | Batch normalization |
| 15 | “relu_4” | ReLU |
| 16 | “conv_5” | Four 3 × 3 × 16 convolutions with stride [1 1] and padding “same” |
| 17 | “BN_5” | Batch normalization |
| 18 | “relu_5” | ReLU |
| 19 | “fc” | Five fully-connected layers |
| 20 | “softmax” | Softmax |
| 21 | “Classification” | crossentropyex with “0” and nine other classes |
The end-to-end training procedure of the proposed CNN model is shown in Fig. 7 . The upper graph in Fig. 7 shows the training and testing accuracies (blue = training, and black = testing), whilst the lower graph in Fig. 7 shows the loss values for both training and test samples (orange = training, and black = testing). The obtained accuracy score was 91.58%, and the training procedure was completed in 11,400 iterations.
Fig. 7.
End-to-end Training of Developed CNN Model for COVID-19 Classification.
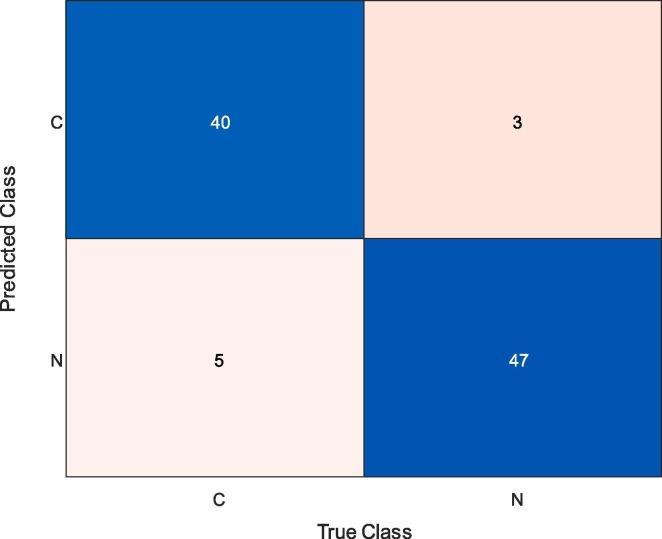
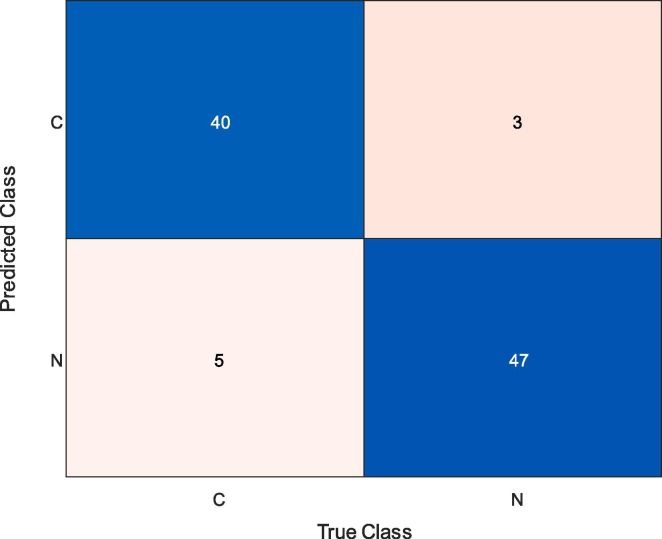
Fig. 8 presents the confusion matrix obtained for the end-to-end trained CNN model. While 40 COVID-19 samples and 47 normal (healthy) samples were classified correctly, five COVID-19 and three normal (healthy) samples were misclassified. Therefore, the rate of correct classification of COVID-19 samples was 88.89%, whilst it was 94.0% for the normal (healthy) cases.
Fig. 8.
Confusion Matrix Obtained by End-to-end Training of Developed CNN Model.
Additional shallow CNN models were then used to investigate their potential in terms of COVID-19 classification accuracy. The shallow networks were also end-to-end trained, and the obtained results are presented in Table4 .
Table 4.
Shallow CNN models and their achievements.
 |
As can be seen in Table 4, the first model contains 21 layers, with one input layer, five convolution layers, five batch normalization layers, five ReLU layers, two pooling layers, one fully-connected layer, one softmax layer, and one classification layer. The second model contains 17 layers, with one input layer, four convolution layers, four batch normalization layers, four ReLU layers, one pooling layer, one fully-connected layer, one softmax layer, and one classification layer. Finally, the third model contains 15 layers, with an input layer, three convolution layers, three batch normalization layers, three ReLU layers, two pooling layers, one fully-connected layer, one softmax layer, and one classification layer. As previously mentioned, the first model produced an accuracy score of 91.58%, which was the best amongst all the shallow CNN model’s achievements. The second and third models produced 88.42% and 86.32% accuracy scores, respectively. From these results, it can be seen that the deep CNN model (model one, with 21 layers) produced the highest accuracy score.
As can be seen from these results, the deep feature extraction and deep transfer learning approach produced better accuracy scores than the end-to-end training of the proposed CNN models. This result was considered reasonable as pretrained deep CNN models were used for both deep feature extraction and deep transfer learning. These models were trained with 25 million images that made the filters of the convolution layers more efficient on new applications. In addition, the depth of these CNN models were quite extensive, which shows that the depth of the CNN model significantly affects the accuracy of the application.
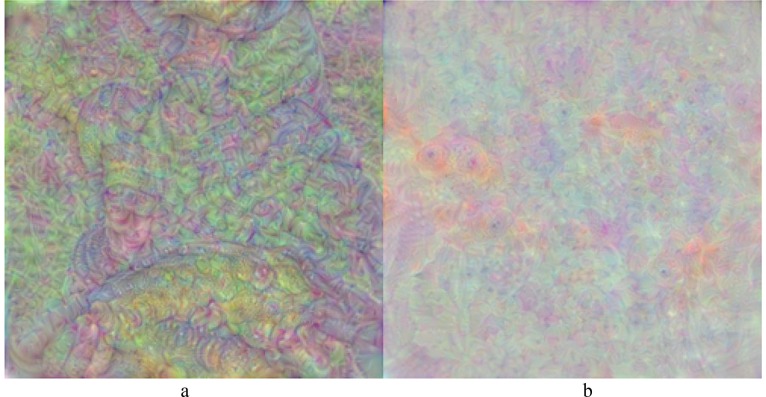
Fig. 9 presents images generated from the fully-connected layer of the further trained ResNet50 model. The first image (Fig. 9a) corresponds to the COVID-19 class, whilst the second image (Fig. 9b) corresponds to the normal (healthy) class.
Fig. 9.
Output Images of Fully-connected Layer of Further Trained ResNet50 Model.
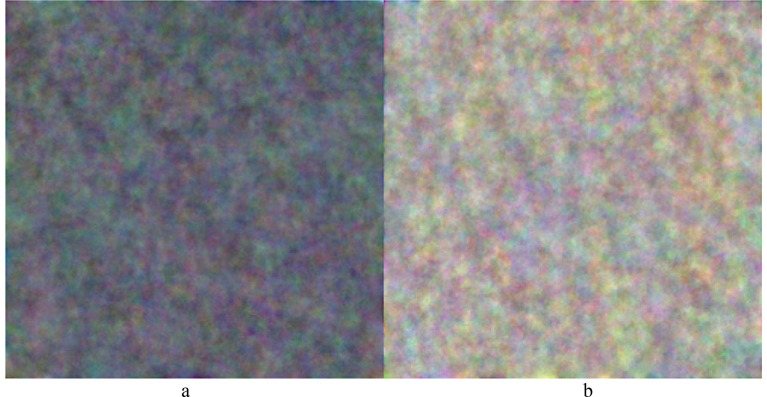
In addition, Fig. 10 presents output images of the fully-connected layer of the proposed CNN model. As a comparison, the images from the fully-connected layer of the further trained ResNet50 model (see Fig. 9) were found to be more regular and meaningful than those from the proposed CNN model (see Fig. 10).
Fig. 10.
Output Images of Fully-connected Layer of Proposed CNN Model.
3.4. Local texture descriptors
For comparative purposes, various local descriptors were also used to extract features from the chest X-ray images for the purposes of COVID-19 detection. In total, eight well-known local texture descriptors were considered, namely LBP (Ahonen et al., 2006), FDLBP (Dubey, 2019), QLRBP (Lan et al., 2015), BGP (Zhang et al., 2012), LPQ (Ojansivu & Heikkilä, 2008), BSIF (Kannala & Rahtu, 2012), CENTRIST (Wu & Rehg, 2010), and PHOG (Bosch et al., 2007).
LBP summarizes local structures of images efficiently by comparing each pixel with its neighboring pixels. FDLBP improves the LBP by applying the decoder concept of multi-channel decoded local binary pattern over the multi-frequency patterns. QLRBP works on the quaternionic representation (QR) of the color image that encodes a color pixel using a quaternion. BGP was developed in order to amalgamate the advantages of both Gabor filters and LBP. LPQ was designed by quantizing the Fourier transform phase in the local neighborhoods of a given pixel. Thus, a robust structure was obtained to add distinction to blurred and low-resolution images. BSIF was produced by the binarization of the responses to linear filters learned from natural images and independent component analysis. CENTRIST was designed similar to the LBP. After coding the pixel values, histogram was used as CENTRIST features. PHOG was developed to encode an image utilizing its local shape at various scales with the help of distributing the direction of intensity and edges.
In the classification phase, the SVM classifier with Linear, Quadratic, Cubic, and Gaussian kernel functions was applied. The obtained results with the local descriptors and SVM classifier are as presented in Table 5 .
Table 5.
Results of first experiment using local feature descriptors and SVM classifiers.
| Local descriptors | Accuracy scores (%) |
||||
|---|---|---|---|---|---|
| Linear SVM | Quadratic SVM | Cubic SVM | Gaussian SVM | Average Accuracy (%) | |
| LBP | 81.1 | 77.9 | 70.5 | 81.1 | 77.7 |
| BGP | 84.2 | 83.2 | 80.0 | 83.2 | 82.7 |
| BSIF | 90.5 | 90.5 | 90.5 | 88.4 | 90.0 |
| CENTRIST | 84.2 | 84.2 | 81.1 | 84.2 | 83.4 |
| PHOG | 85.3 | 83.2 | 83.2 | 84.2 | 84.0 |
| LPQ | 85.0 | 84.2 | 82.6 | 85.0 | 84.2 |
| FDLBP | 87.4 | 87.4 | 87.4 | 87.4 | 87.4 |
| QLRBP | 82.1 | 82.1 | 82.1 | 82.1 | 82.1 |
| Average Accuracy (%) | 85.0 | 84.1 | 82.2 | 84.5 | |
The rightmost column of Table 5 shows the average accuracy scores for each of the local descriptors, whilst the last row of Table 5 shows the average accuracy scores of each kernel function of the SVM classifier. According to the results shown in Table 5, various observations are notable. For example, the BSIF local feature descriptor obtained the highest average classification accuracy score of 90.0%, showing that the BSIF method outperformed all other applied local feature descriptors. Similarly, the Linear SVM technique outperformed all other kernel functions with an average accuracy score of 85.0%. The LBP approach produced a 77.7% average accuracy score, which was the lowest of the accuracy scores. Both the Linear and Gaussian kernel functions produced an 81.1% accuracy score with LBP features, which was the highest of the accuracy scores obtained using LBP features. The BGP local descriptors produced an 82.7% average accuracy score, whilst the best BGP accuracy score of 84.2% was produced by the Linear kernel function.
The best accuracy score overall was 90.5%, and was produced by BSIF with the Linear, Quadratic, and Cubic kernel functions. The CENTRIST technique produced accuracy scores of 84.2% for Linear, Quadratic, and Gaussian kernel functions, and 81.1% for the Cubic kernel function. PHOG features produced the fourth-best average accuracy score with 84.0%, with the LPQ technique slightly ahead with an 84.2% average accuracy score as the third-best. The FDLBP techniques’ achievement was 87.4%, which was the second-best average accuracy score with identical 87.4% accuracy scores across all kernel functions. The QLRBP method produced an average accuracy score of 82.1% which was similar to the FDLBP technique, and the QLRBP method also produced identical accuracy scores across all kernel functions.
A final performance comparison of the used methods is presented in Table6 , which clearly shows that the deep learning approach outperformed the local descriptors.
Table 6.
Performance comparison of the applied methods.
| Method | Accuracy (%) |
|---|---|
| ResNet50 Features + SVM | 94.7 |
| Fine-tuning of ResNet50 | 92.6 |
| End-to-end training of CNN | 91.6 |
| BSIF + SVM | 90.5 |
Additional experiments were then conducted with newly released COVID-19 chest X-ray images (Kaggle, 2020a). However, it should be noted that only the COVID-19 chest X-ray images were selected from this dataset for use in these additional experiments. The obtained results are presented in Table7 . As can be seen from Table7, the ResNet50 Features + SVM, Fine-tuning ResNet50, and BSIF + SVM methods each yielded small improvements, whereas the end-to-end training of the CNN method produced a slightly lower accuracy score compared to the main results (see Table6). However, an overall general improvement can be seen. In order to realize further improvement, normal (healthy) chest X-ray images are also needed in addition to the COVID-19 samples.
Table 7.
Results using additional COVID-19 chest X-ray images.
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | F1 score (%) | AUC |
|---|---|---|---|---|---|
| ResNet50 Features + SVM | 95.79 | 94.00 | 97.78 | 95.92 | 0.9987 |
| Fine-tuning of ResNet50 | 92.63 | 88.00 | 97.78 | 92.63 | 0.9973 |
| End-to-end training of CNN | 90.53 | 88.00 | 93.33 | 90.72 | 0.9920 |
| BSIF + SVM | 91.58 | 90.00 | 93.33 | 91.84 | 0.9933 |
Table8 presents the results obtained from the randomly selected half of the dataset that was used for training purposes, with the remainder used for testing (50% training and 50% test). When compared to Table7, it can be seen that when the number of samples in the training set became reduced, the results were also seen to reduce.
Table 8.
Results Using Additional Samples in Test Set (50% training/50% test).
| Method | Accuracy (%) | Sensitivity (%) | Specificity (%) | F1 score (%) | AUC |
|---|---|---|---|---|---|
| ResNet50 Features + SVM | 94.74 | 91.00 | 98.89 | 94.79 | 0.9990 |
| Fine-tuning of ResNet50 | 89.47 | 90.00 | 88.89 | 90.00 | 0.9889 |
| End-to-end training of CNN | 86.84 | 87.00 | 86.67 | 87.44 | 0.9827 |
| BSIF + SVM | 85.79 | 87.00 | 84.44 | 86.57 | 0.9798 |
Further comparisons of the proposed method were then made to other studies recently published on COVID-19 detection. Toğacar, Ergen, and Cömert (2020) used deep feature extraction and feature selection for COVID-19 detection based on chest X-ray images, with a reported accuracy level of 99.27%. Ozturk et al. (2020) used darknet and yolo for the detection of COVID-19, and reported classification accuracy of 98.08%. Ucar and Korkmaz (2020) used deep Bayes-SqueezeNet for COVID-19 detection based on chest X-ray images, and reported accuracy as being 76.37% for the multiclass case. Das, Kumar, Kaur, Kumar, and Singh (2020) used a fine-tuned pretrained deep model for the detection of COVID-19, with a reported accuracy score of 97.04%. Hemdan, Shouman, and Karar (2020) developed COVIDX-Net for the automated detection of COVID-19 using chest X-ray images, obtaining a COVID-19 classification accuracy of 91%. Asnaoui, Chawki, and Idri (2020) presented a comparative study of eight transfer learning techniques for the classification of COVID-19 pneumonia, with the MobileNet-V2 and Inception-V3 models providing a 96% successful rate of classification accuracy. As can be seen, the proposed deep methods on chest X-ray-based COVID-19 detection produced accuracy scores ranging from 90% to 100%.
Table 9 shows the computational efforts of each model. As can be seen in Table 9, the deep features and SVM accomplished its run in 48.9 s, which was the shortest runtime of the models tested. The second best runtime was 61.76 s, and was achieved for the local texture descriptors and SVM classifier. The longest runtime was 1407.2 s, which was for the End-to-end training of the CNN model.
Table 9.
Computational efforts of the examined methods.
| Method | Computational Time (s) |
|---|---|
| ResNet50 Features + SVM | 48.9 |
| Fine-tuning of ResNet50 | 126.4 |
| End-to-end training of CNN | 1407.2 |
| BSIF + SVM | 61.76 |
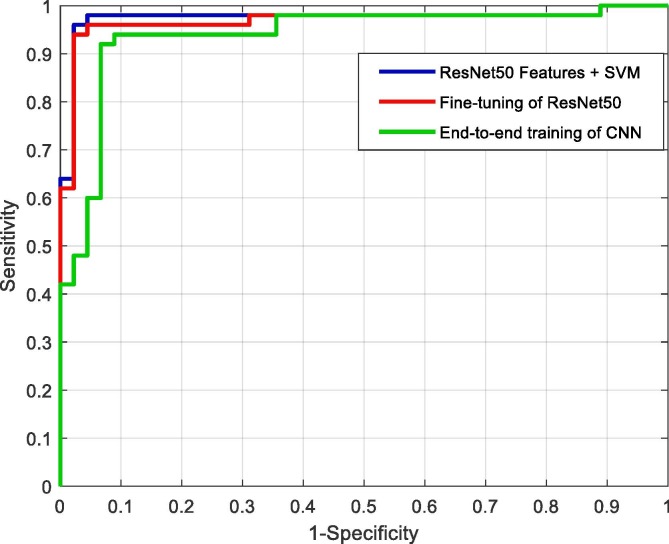
The ROC curves for deep CNN approaches are shown in Fig. 11 . The blue, red, and green colors show the “deep feature extraction and SVM classifier,” “fine-tuning of the pretrained model,” and “end-to-end training of the CNN model,” respectively. From the ROC curves, it can be seen that the “deep feature extraction and SVM classifier” model performed better than the other deep approaches.
Fig. 11.
ROC curves for the deep CNN approaches.
We also compared the achievement of the proposed method with a recently published method proposed by Toğacar et al. (2020). This comparison utilized by the codes shared by the authors. Table 10 presents the comparison according to the accuracy scores, and shows that the proposed method performed almost 2% better.
Table 10.
Performance comparison of the proposed method with Toğacar et al. (2020)’s method.
| Method | Accuracy (%) |
|---|---|
| ResNet50 Features + SVM | 94.7 |
| Toğacar et al. (2020) | 92.6 |
4. Conclusions
The current study applied three deep CNN approaches in the detection of COVID-19 based on chest X-ray images. More specifically, two transfer learning approaches, namely deep feature extraction and fine-tuning, as well as an end-to-end trained new CNN model were experimented. The deep features were classified with SVM classifier, accompanied with different kernel functions. Eight well-known local descriptors are then considered, and the obtained results revealed the following conclusions;
-
1.
The deep learning approaches outperformed the local descriptors. Especially, deep features and SVM classifier performed better than the other approaches.
-
2.
Fine-tuning and end-to-end training requires much more time than deep feature extraction and local feature descriptor extraction.
-
3.
The Cubic kernel function generally outperformed all other kernels in deep feature classification. The ResNet50 model generally produced better results than the other pretrained CNN models.
-
4.
For end-to-end training, deep CNN models produced better results than shallow networks.
In future works, additional COVID-19 chest X-ray images will be collected and deeper CNN models investigated for the detection of COVID-19. Additionally, other lung diseases will also be included in the researchers’ future studies. With the COVID-19 disease presenting different stages of evolution and with different imagistic patterns, this feature will aim to be addressed in future studies, as well as the development of a GUI to help radiologists detect COVID-19.
CRediT authorship contribution statement
Aras M. Ismael: Data curation. Abdulkadir Şengür: Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Adaminejad H., Farjah E. An algorithm for power quality events core vector machine based classification. The Modares Journal of Electrical Engineering. 2013;12(4):50–59. [Google Scholar]
- Ahonen T., Hadid A., Pietikainen M. Face description with local binary patterns: application to face recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2006;28(12):2037–2041. doi: 10.1109/TPAMI.2006.244. [DOI] [PubMed] [Google Scholar]
- Asnaoui, K. E., Chawki, Y., & Idri, A. (2020). Automated methods for detection and classification pneumonia based on x-ray images using deep learning. arXiv preprint arXiv:2003.14363. Retrieved from https://arxiv.org/abs/2003.14363.
- Başaran E., Cömert Z., Çelik Y. Convolutional neural network approach for automatic tympanic membrane detection and classification. Biomedical Signal Processing and Control. 2020;56:101734. doi: 10.1016/j.bspc.2019.101734. [DOI] [Google Scholar]
- Bhandary A., Prabhu G.A., Rajinikanth V., Thanaraj K.P., Satapathy S.C., Robbins D.E., Shasky C., Zhang Y.-D., Tavares J.M.R.S., Raja N.S.M. Deep-learning framework to detect lung abnormality – A study with chest X-Ray and lung CT scan images. Pattern Recognition Letters. 2020;129:271–278. [Google Scholar]
- Bosch A., Zisserman A., Munoz X. Representing shape with a spatial pyramid kernel. ACM; New York, NY: 2007. pp. 401–408. [Google Scholar]
- Cheng L., Bao W. Remote sensing image classification based on optimized support vector machine. TELKOMNIKA Indonesian Journal of Electrical Engineering. 2014;12(2):1037–1045. [Google Scholar]
- Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P., Moreira C.…de Albuquerque V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Applied Sciences. 2020;10(2) Article 559. [Google Scholar]
- Deniz E., Şengür A., Kadiroğlu Z., Guo Y., Bajaj V., Budak Ü. Transfer learning based histopathologic image classification for breast cancer detection. Health Information Science and Systems. 2018;6(1) doi: 10.1007/s13755-018-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Pan Y., Zhang J., Xu W. Proceedings, 2017 IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies. IEEE; 2017. Learning to read chest X-ray images from 16000+ examples using CNN; pp. 51–57. [Google Scholar]
- Dubey S.R. Face retrieval using frequency decoded local descriptor. Multimedia Tools and Applications. 2019;78(12):16411–16431. [Google Scholar]
- GitHub. (2020). COVID-19 [dataset]. Retrieved March 10, 2020 from https://github.com/ieee8023/covid-chestxray-dataset/tree/master/images.
- Hemdan, E. E. D., Shouman, M. A., & Karar, M. E. (2020). Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055. Retrieved from https://arxiv.org/abs/2003.11055.
- Ho T.K.K., Gwak J. Multiple feature integration for classification of thoracic disease in chest radiography. Applied Sciences. 2019;9(19) Article 4130. [Google Scholar]
- Kaggle. (2020a). Covid-19 X-ray chest and CT [dataset]. Retrieved April 20, 2020 from https://www.kaggle.com/bachrr/covid-chest-xray.
- Kaggle. (2020b). X-ray chest [dataset]. Retrieved March 10, 2020 from https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- Kannala J., Rahtu E. Proceedings of the 21st international conference on pattern recognition (ICPR2012) IEEE; 2012. Binarized statistical image features; pp. 1363–1366. [Google Scholar]
- Kesim E., Dokur Z., Olmez T. 2019 scientific meeting on electrical-electronics & biomedical engineering and computer science (EBBT) IEEE; 2019. X-ray chest image classification by a small-sized convolutional neural network. https://doi.org/10.1109/EBBT.2019.8742050. [Google Scholar]
- Lan R., Zhou Y., Tang Y.Y. Quaternionic local ranking binary pattern: a local descriptor of color images. IEEE Transactions on Image Processing. 2015;25(2):566–579. doi: 10.1109/TIP.2015.2507404. [DOI] [PubMed] [Google Scholar]
- Li X., Shen L., Xie X., Huang S., Xie Z., Hong X., Yu J. Multi-resolution convolutional networks for chest X-ray radiograph based lung nodule detection. Artificial Intelligence in Medicine. 2019;103 doi: 10.1016/j.artmed.2019.101744. [DOI] [PubMed] [Google Scholar]
- Liu C., Cao Y., Alcantara M., Liu B., Brunette M., Peinado J., Curioso W. 2017 IEEE international conference on image processing (ICIP) IEEE; 2017. TX-CNN: Detecting tuberculosis in chest X-ray images using convolutional neural network; pp. 2314–2318. [Google Scholar]
- Ojansivu V., Heikkilä J. Blur insensitive texture classification using local phase quantization. In: Abderrahim E., Lezoray O., Nouboud F., Mammass D., editors. International conference on image and signal processing. Springer; Berlin, Germany: 2008. pp. 236–243. [Google Scholar]
- Omar N., Sengur A., Al-Ali S.G.S. Cascaded deep learning-based efficient approach for license plate detection and recognition. Expert Systems with Applications. 2020;149:113280. doi: 10.1016/j.eswa.2020.113280. [DOI] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S.J., Yang Q. A Survey on Transfer Learning. IEEE Transactions on Knowledge and Data Engineering. 2009;22(10):1345–1359. [Google Scholar]
- Qi Z., Tian Y., Shi Y. Robust twin support vector machine for pattern classification. Pattern Recognition. 2013;46(1):305–316. [Google Scholar]
- Radiology Assistant. (2020). X-ray Chest images [dataset]. Retrieved March 23, 2020 from https://radiologyassistant.nl/chest/lk-jg-1.
- Rajpurkar P., Irvin J., Ball R.L., Zhu K., Yang B., Mehta H.…Patel B.N. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Medicine. 2018;15(11) doi: 10.1371/journal.pmed.1002686. Article e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza J.C., Bandeira Diniz João.Otávio., Ferreira J.L., França da Silva G.L., Corrêa Silva A., de Paiva A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Computer Methods and Programs in Biomedicine. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Computers in Biology and Medicine. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucar F., Korkmaz D. COVIDiagnosis-Net: deep bayes-squeezenet based diagnostic of the coronavirus disease 2019 (COVID-19) from X-Ray images. Medical Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uçar M., Uçar E. Computer-aided detection of lung nodules in chest X-rays using deep convolutional neural networks. Sakarya University Journal of Computer and Information Sciences. 2019;2(1):41–52. [Google Scholar]
- Widodo A., Yang B.-S. Support vector machine in machine condition monitoring and fault diagnosis. Mechanical Systems and Signal Processing. 2007;21(6):2560–2574. [Google Scholar]
- World Health Organization. (2020). Coronavirus disease (COVID-19) Pandemic. Retrieved from https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Worldmeter. (2020). COVID-19 Coronavirus Pandemic. Retrieved March 23, 2020 from https://www.worldometers.info/coronavirus.
- Woźniak M., Połap D., Capizzi G., Sciuto G.L., Kośmider L., Frankiewicz K. Small lung nodules detection based on local variance analysis and probabilistic neural network. Computer Methods and Programs in Biomedicine. 2018;161:173–180. doi: 10.1016/j.cmpb.2018.04.025. [DOI] [PubMed] [Google Scholar]
- Wu J., Rehg J.M. Centrist: A visual descriptor for scene categorization. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2010;33(8):1489–1501. doi: 10.1109/TPAMI.2010.224. [DOI] [PubMed] [Google Scholar]
- Xu C., Yang J., Lai H., Gao J., Shen L., Yan S. UP-CNN: Un-pooling augmented convolutional neural network. Pattern Recognition Letters. 2019;119:34–40. [Google Scholar]
- Xu S., Wu H., Bie R. CXNet-m1: Anomaly detection on chest X-rays with image-based deep learning. IEEE Access. 2018;7:4466–4477. [Google Scholar]
- Zhang L., Zhou Z., Li H. 2012 19th IEEE International Conference on Image Processing. IEEE; 2012. Binary gabor pattern: An efficient and robust descriptor for texture classification; pp. 81–84. [Google Scholar]