Abstract
The emergence of the global pandemic caused by the novel SARS-CoV-2 virus has motivated scientists to find a definitive treatment or a vaccine against it in the shortest possible time. Current efforts towards this goal remain fruitless without a full understanding of the behavior of the virus and its adaptor proteins. This review provides an overview of the biological properties, functional mechanisms, and molecular components of SARS-CoV-2, along with investigational therapeutic and preventive approaches for this virus. Since the proteolytic cleavage of the S protein is critical for virus penetration into cells, a set of drugs, such as chloroquine, hydroxychloroquine, camostat mesylate have been tested in clinical trials to suppress this event. In addition to angiotensin-converting enzyme 2, the role of CD147 in the viral entrance has also been proposed. Mepolizumab has shown to be effective in blocking the virus's cellular entrance. Antiviral drugs, such as remdesivir, ritonavir, oseltamivir, darunavir, lopinavir, zanamivir, peramivir, and oseltamivir, have also been tested as treatments for COVID-19. Regarding preventive vaccines, the whole virus, vectors, nucleic acids, and structural subunits have been suggested for vaccine development. Mesenchymal stem cells and natural killer cells could also be used against SARS-CoV-2. All the above-mentioned strategies, as well as the role of nanomedicine for the diagnosis and treatment of SARS-CoV-2 infection, have been discussed in this review.
Keywords: SARS-CoV-2, COVID-19, Global pandemic, Virus mechanism, Vaccine development, Investigational drugs
1. Introduction
The harm wreaked by infectious agents, particularly viruses, among the world's population has a very long history and has periodically challenged human life every few years. Viral infections (especially respiratory viruses) have accounted for a large proportion of epidemics and pandemics to date. Discovering an effective vaccine and implementing a correct vaccination program has led many of these viruses to be eradicated or at least severely restricted. Following the outbreak of any widespread viral disease, studies from around the world should be initiated to undertake the design of an effective vaccine. One of the most important viral families, which have always been a major concern for researchers in vaccine design, is the Coronaviridae family. Viruses of this family can periodically infect humans, causing mainly severe respiratory syndromes, ranging from Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV), to the novel pandemic coronavirus SARS-CoV-2, the causative agent of Coronavirus Disease 2019 (COVID-19).
Coronaviridae family contains RNA viruses that usually infect the respiratory tracts of mammals and birds, and can cause several illnesses, which so far have rarely resulted in death. Although coronaviruses have infected different animal species for a long time, the first coronavirus with the ability to infect humans was only identified in the 1960s [1]. Further studies revealed that two human coronaviruses, HCoV-229E, and HCoV-OC43 were able to cause a common cold syndrome with symptoms akin to colds caused by rhinoviruses [2]. The severe morbidity of coronaviruses in humans became more evident after the identification of SARS-CoV in 2003, HCoV-NL63 in 2004, HKU1 in 2005 [3], MERS-CoV in 2012 [4], and finally SARS-CoV-2 in 2019 [5], which all cause severe respiratory tract infections with the danger of wide-scale mortality.
The COVID-19 and its causative infectious agent, SARS-CoV-2, became a global problem in early 2020, and this horrible disease has threatened the survival of millions of people around the world with a mortality rate of approximately 5–10% [6]. So, there is an urgent need to find ways either to confine the spread of the virus or effectively treat its complications. Among our options, vaccination seems to the best route to our salvation. Due to the genomic and proteomic features of the virus such as the “template switching” (i.e. viral RNA mutations even in the amino acid stage) and despite numerous international efforts to design a vaccine to prevent the infection by this novel human virus, we still face many challenges to supply an effective vaccine, especially on a global scale [7]. The ongoing research into COVID-19 vaccines could light the road ahead for further studies aimed at finding an effective and affordable vaccine for preventing this novel dreadful disease. The present review focuses on these efforts and provides several insights into the accomplishments, failures, and risks of developing SARS-CoV-2 vaccines.
2. Origin and evolution of highly pathogenic coronaviruses
Coronaviruses are zoonotic viruses that naturally infect animals, but can be transmitted from animals to humans and have a powerful ability to infect human cells. Taxonomically, the highly pathogenic coronaviruses (SARS-CoV, MERS-CoV, and SARS-CoV-2) are classified into the Nidovirales order, Coronaviridae family, Coronavirinae sub-family, and also into the Betacoronavirus genus (International Committee on Taxonomy of Viruses).
Human angiotensin-converting enzyme II (ACE2) is the main receptor for SARS-CoV, by which the SARS-CoV S protein enters the host cells. The viral attachment protein or Spike protein binds to ACE2 via the receptor-binding domain (RBD) located on the surface of S protein [8]. Interestingly, structural analysis of the RBDs from S proteins derived from different strains of SARS-CoV has shown that the RBDs have a different affinity for the ACE2 receptor in several animal models. For example, the strain hTor02 of SARS-CoV (the epidemic strain) contains RBDs which have a high affinity for human ACE2, and enable the virus to infect human cells easily [9].
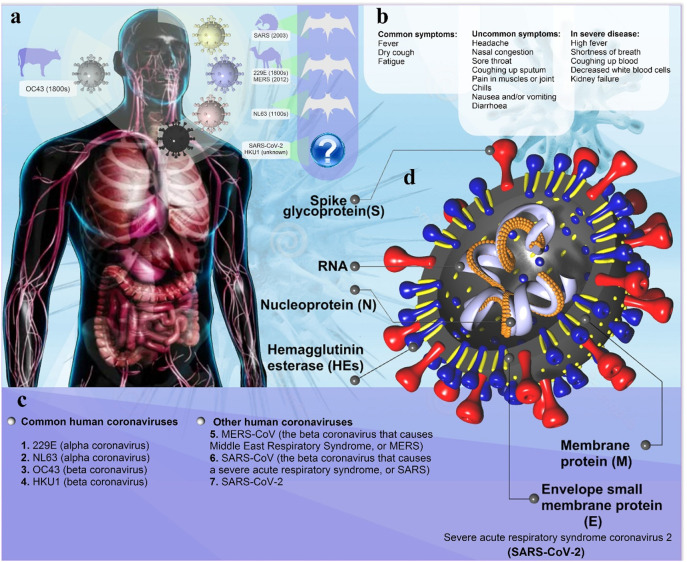
The structural and genomic analysis of SARS-CoV-2 with the viruses isolated from other different species showed that another probable host of the SARS virus might be the pangolin. However, it is hard to be sure whether bats are the primary host or pangolins (Fig. 1a) [10].
Fig. 1.
Coronavirus pathophysiology. (a) Animal (natural and intermediate hosts) origin of human coronaviruses; Pangolins may be intermediate hosts for transmission of the new SARS-CoV-2 from bats to humans. Although cats can be infected with the SARS-CoV-2, and can spread it to each other, dogs have only a low susceptibility to this virus. However, the existence of intermediate animal host(s) of SARS-CoV-2 is still likely. (b) Clinical presentation of patients with SARS-CoV-2, including common, uncommon, and severe symptoms of SARS-CoV-2. (c) Human Coronavirus Types: common human coronaviruses; 229E (alpha coronavirus), NL63 (alpha coronavirus), OC43 (beta coronavirus), HKU1 (beta coronavirus) and other human coronaviruses; MERS-CoV (the beta coronavirus that causes Middle East Respiratory Syndrome, or MERS), SARS-CoV (the beta coronavirus that causes the severe acute respiratory syndrome, or SARS), SARS-CoV-2 (the novel coronavirus that causes coronavirus disease 2019, or COVID-19); d) Diagram of coronavirus virion structure showing genome and structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N).
3. Structural and immunological characterization
The complete structure of the coronavirus virion, as the largest known RNA virus, contains a positive-sense, non-segmented, single-stranded RNA combined with the nucleocapsid (N) proteins assembled into a helical shape. A phospholipid bilayer structure similar to a mammalian cell membrane covers the RNA, and a high number of M proteins and S proteins are located in this layer. The membrane (M) and envelop (E) proteins can be found among these S proteins (Fig. 1d) [11]. When this virus infects a human cell, the immune system is triggered into action to eliminate the virions, and destroy infected cells. As the first line of defense against viral infections, the innate immune response starts its fight against the virus by producing inflammatory cytokines and chemokines. The most important innate immune response mediators involved in the initial defense against coronavirus include RIG-I-like receptors (RLRs), C-type lectin-like receptors (CLRs), toll-like receptors (TLRs), NOD-like receptors (NLRs), and also cytoplasmic receptors such as cGAS, IFI16, STING, and DAI [11]. However, although the activation of the innate immune response is designed to clear infected tissue from the virus, it can also be dangerous and harmful for healthy tissues [12]. Natural killer (NK) cells are the crucial immune cells of the innate immune response, with the ability to deal with the viral infections and kill infected cells by producing perforin or inducing IFN-γ. It has been reported that NK cells are decreased in the serum level of patients with SARS-CoV infection [13]. To prove this concept, a mouse model of SARS was used to show that NK cells were not necessary for the clearance of SARS-CoV [14]. On the other hand, plasma analysis of SARS patients showed that mannose-binding lectins (MBLs) and serum amyloid A (both acute-phase proteins) were elevated in a calcium-dependent manner and MBLs could bind to the S proteins of SARS-CoV to exert their protective effects [15]. The production of IFNs is an essential antiviral mechanism functioning as a chemo-attractant mechanism to attract immune cells to eliminate infected cells, as well as to protect non-infected-cells. SARS-CoV inhibits the function of IFN via hindering its related pathways; for example, this virus increases the nuclear transport of IFN regulatory factor 3 (IRF3) to repress the IFN response [16]. Regarding the inflammatory functions of macrophages and dendritic cells (DCs), SARS-CoV non-specifically infects these immune cells, as well as peripheral blood mononuclear cells (PBMCs) giving rise to the production of several chemokines, including IFN-inducible protein 10 (IP-10), RANTES (CCL5), macrophage inflammatory protein 1 α (MIP-1α), and monocyte chemoattractant protein 1 (MCP-1), all of which subsequently increase the level of inflammation in SARS-CoV-infections [17,18].
During a viral infection, and especially a coronavirus infection, not only is there an innate immune response, but the adaptive immune response is also activated in the host. Cytotoxic T lymphocytes mostly function during cellular immunity to eliminate the virally infected cells. The S proteins of SARS-CoV have two HLA-A2-restricted T cell epitopes that can activate T cells responses in SARS-positive patients [19]. Surprisingly, lymphopenia has been observed during SARS-CoV infection, and this reduction was more pronounced in CD4+ T cells compared to CD8+ T cells [20]. It has been demonstrated that an IgG against the N protein of SARS-CoV is the first antibody produced after primary infection [21]. However, antibodies against the S protein have been reported to have neutralization effects on SARS-CoV virions [22]. These antibodies could also trigger the phagocytosis of infected-cells by MΦs, leading to an elevated level of proinflammatory cytokines and chemokines, and subsequent tissue injury due to excessive inflammation [23].
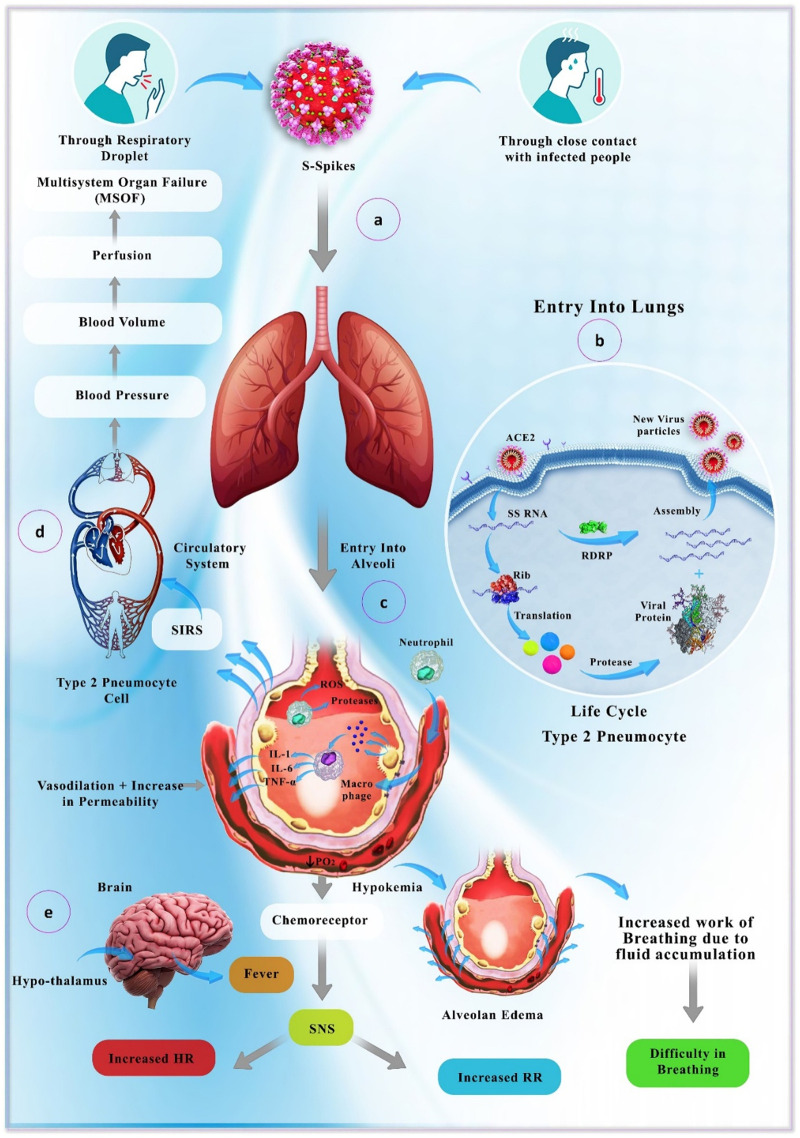
The exact immunopathology mechanisms of SARS-CoV-2 and its related disease, COVID-19, are still under investigation. Indeed, the cytokine storm and incidence of inflammation in lungs have been found to be the leading causes of acute respiratory distress syndrome (ARDS) in COVID-19 patients just like it occurs with SARS patients (Fig. 2 ) [24].
Fig. 2.
Transmission and pathology of the SARS-Cov-2 virus. After transmission via droplets from an infected person (a), the virus particles infect and replicate in type 2 pneumocytes (b), which finally results in inflammation of alveoli (recruitment of inflammatory cells and secretion of inflammatory mediators) (c) and disruption of respiratory and blood circulation systems. Finally, multi-organ dysfunction occurs due to the severe hypoxia and lack of perfusion (d). Reduction in PO2 and fluid accumulation in alveoli further aggravates the clinical condition and leads to pulmonary, as well as cerebral manifestations (e). SIRS: systemic inflammatory response syndrome; RDRP: RNA-dependent RNA polymerase.
4. Cell entry mechanism and therapeutic implications
An understanding of SARS-CoV-2 cell entry mechanisms will facilitate the design of effective therapeutics that could target this critical step in the viral life cycle. The host cell membrane is essential to prevent infection, acting as a barrier between the viral particle and the intracellular site of viral replication [25]. Although not a guarantee of successful infection, the binding and passage of the virus through the cell membrane barrier is a critical step in the life cycle of a virus [26], especially for coronaviruses. Coronavirus entry into a host cell is a dynamic, multi-step cascade process. These viruses access target cells by binding to cell surface receptors, followed by membrane fusion mediated by a multifunctional fusion protein [27,28]. Although there is evidence implicating cellular endocytic pathways for entry of viruses into host cells, the exact mechanisms of entry for many viruses, including coronaviruses, have yet to be fully characterized [29]. Identification of the host cell receptors, the structural binding mechanism, and the virus trafficking pathway will support the development of therapeutic agents against SARS-CoV-2.
4.1. Host membrane proteins render cells susceptible to SARS-CoV-2
In the classical pathway, viruses enter host cells via endocytosis, following binding to cell surface receptors. Viruses can physically penetrate cells by endocytic cellular uptake in a process usually referred to as receptor-mediated endocytosis [30]. Angiotensin-converting enzyme 2 (ACE2) is a critical type 1 integral membrane protein, which is expressed in most human and some animal tissues. ACE2 is highly expressed in the endothelium, the lungs, and the heart [31]. When this cell surface protein was discovered three decades ago, neither of the research groups involved could have appreciated the large number of distinct functions this receptor plays in biology, from viral infection to cardiovascular regulation [32]. ACE2 is the first known host receptor for SARS-CoV-2 [33], and it was found that SARS-CoV-2 does not use other host cell membrane proteins, such as dipeptidyl peptidase 4 (DP IV, CD26) or aminopeptidase N (APN, CD13) [34]. Cao et al. systematically searched for variants of ACE2, which could affect the pathogenesis of SARS-CoV-2 among different populations. Their results showed that was little evidence of genetic variations supporting the existence of susceptibility or resistance in diverse populations. East Asian populations had much higher frequencies in the eQTL allele variants, which may govern different responses to SARS-CoV-2 in different populations [35]. In addition to ACE2, Wang et al. reported that SARS-CoV-2 could enter target cells through a novel interaction of the viral proteins with CD147 [36]. CD147, also known as basigin or extracellular matrix metalloproteinase inducer (EMMPRIN), is expressed in a variety of human cells. CD147 regulates extracellular matrix remodeling during many critical biological processes, including cancer, inflammatory disease, and wound healing [37]. It could be the case that some SARS-CoV-2 receptor variants and expression levels in different patients may be associated with more severe forms of the infection.
Increased viremia (level of viruses in the bloodstream and other bodily fluids) leads to higher severity of infection [38]. During viremia, the human circulatory system facilitates the transport of viruses throughout the entire body. Coronavirus viremia mainly appears one week after the onset of symptoms. Viremia then decreases gradually over a week, becoming undetectable in the bodily fluid samples of convalescent patients [39]. ACE2 is widely expressed in other tissues and cell types, such as cardiomyocytes, cardiofibroblasts, and coronary endothelial cells [40]. CD147, in a similar manner to ACE2, is expressed in many different epithelial, neuronal, lymphoid, and myeloid cell types [41]. Over-expression of these receptors in different tissues and cell types could explain subsequent syndromes such as myocarditis or encephalopathy. Therefore, ACE2/CD147-based therapeutics could inhibit the binding of SARS-CoV-2 to its receptors and prevent the coronavirus from invading its target cells, possibly providing a strategy for the development of anti-SARS-CoV-2 drugs.
4.2. The spike protein of SARS-CoV-2 promotes cell entry
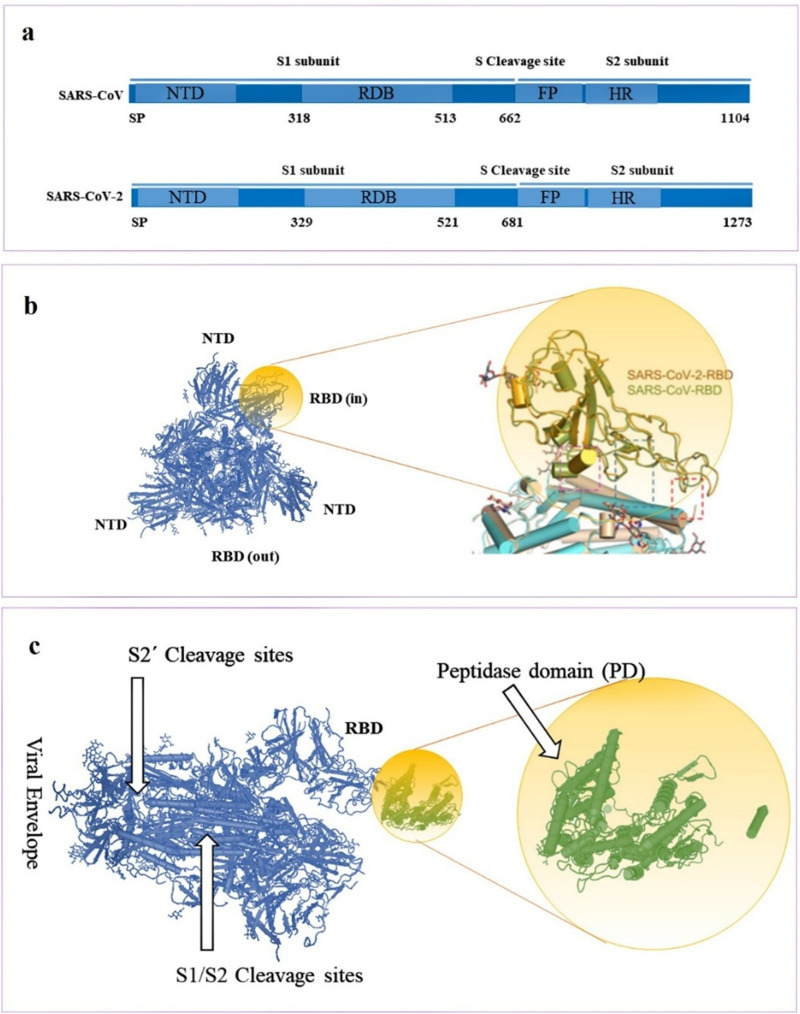
Coronavirus cell entry relies on an interaction between the surface receptor of target cells and the spike (S) proteins of coronaviruses, which mediates viral entry [42]. The Coronavirus S protein is a trimeric type I transmembrane protein with 1160 to 1400 amino acid residues. SARS-CoV and S SARS-CoV-2 proteins are highly glycosylated at 21 to 35 sites, which all have 76.5% identity in amino acid sequences and a high degree of homology (Fig. 3a) [43,44]. These glycoproteins assemble on the coronavirus surface, forming a crown-like array that gives this virus its name (crown = corona). The crystallization of the S protein of SARS-CoV-2 and examination by cryo-electron microscopy showed the role of these sites in the interplay between SARS-CoV-2 S and its target cell receptors [45]. Interestingly, these coronaviruses contain a critical loop with flexible residues. Replacing this loop with other amino acid residues, such as those from SARS-CoV using molecular modeling, showed that the receptor-binding domain has a higher affinity for host cell receptors compared with other coronavirus S proteins (Fig. 3b right) [46].
Fig. 3.
Molecular detail of coronavirus S proteins and host cell ACE2 protein. a) Phylogenetic analysis of SARS-CoV and SARS-CoV-2 S proteins. b) Structural alignment and structure of RBD for the SARS-CoV and SARS-CoV-2 [33]. c) SARS-CoV-2 S protein cleavage sites and its interaction with the PD of ACE2 (Protein Data Bank ID: 6VYB and 1R42).
Recent publications have reported that coronavirus entry is a multi-step process requiring several domains in the S protein [21]. An interplay between a single region of the SARS-CoV-2 S protein called the receptor-binding domain (RBD), and the protease domain (PD) of ACE2 [47] prompts endocytosis of the virus. This interaction then mediates the fusion between the viral particle and the target cell membrane, allowing endocytosis into the cytosol [26] (Fig. 2c). Structures of PD (alone and in complex with the RBD) have revealed the molecular details of the interaction between the RBD and PD [48,49]. Yan et al. demonstrated the three-dimensional structure of ACE2 in a dimeric assembly. Molecular docking studies suggested the simultaneous binding of the ACE2 dimer to two coronavirus S protein trimers [33]. The S protein RGD binds to the RBD at the border of the subdomain (amino acids 437 to 508) [44,50,51]. Residue 479 in SARS-CoV RBD corresponds to residue 394 in the SARS-CoV-2 RBD, and is recognized by the critical residue 31 in the ACE2 enzyme [52,53]. This interaction is now known to trigger a conformational change within the viral S protein, which then mediates fusion of the host cell membrane and the SARS-CoV-2 viral membrane allowing the genetic material to be introduced into the target cell.
4.3. SARS-CoV-2 uses multiple pathways for S protein activation
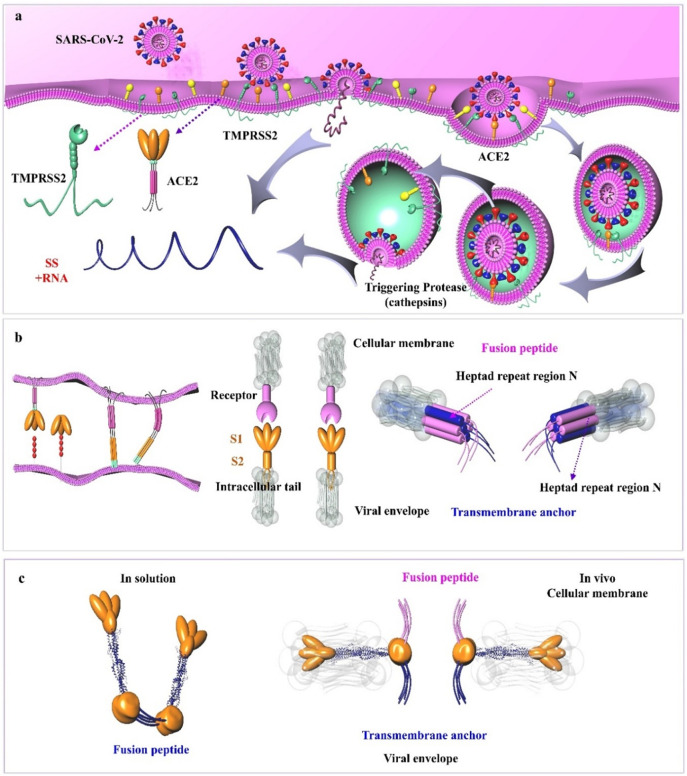
Viruses deliver their genetic material into target cells using a variety of strategies and molecules [54]. The viral S glycoprotein contains multiple cleavage sites. S glycoproteins contain two domains: a C-terminal S2 domain and an N-terminal domain named S1 (Fig. 2a and c). When the S1 subunit binds to ACE2, the S2 cleavage site is then cleaved by host proteases [33]. The coronavirus fusion peptide is located downstream from the S2 N terminus. This critical peptide forms a loop and a short helix, and contains nearly all the hydrophobic residues buried inside the prefusion structure [42,55]. Following host cell binding, the coronavirus S proteins undergo conformational changes exposing hydrophobic domains and the fusion peptide, which becomes embedded into the host cell cytoplasmic membrane. The pre-fusion to post-fusion transition in the S protein is irreversible and is regulated during the cell entry [42,56]. In the next stage, S protein subdomains become refolded into a heptad repeat 1 (HR1), which initiates the endocytosis of coronaviruses (Fig. 4 ) [57].
Fig. 4.
SARS-CoV-2 cell entry mechanisms and subsequent intracellular trafficking. a) Role of host cell proteases in the cellular entry of SARS-CoV-2. Host cell entry of SARS-CoV can proceed via two distinct routes; in the absence of SARS-S-activating protease, the virus is internalized via the binding of SARS-S to ACE2 on the surface of host cells. Within the endosomes, the SARS-S is then cleaved and activated by cathepsin L, a pH-dependent cysteine protease. The SARS-S may also be activated by TMPRSS2 on the membrane surface of host cells when this protease is expressed along with ACE2 allowing the fusion of two membranes (i.e., host and the virus) and viral entrance. b) The role of class I transmembrane proteins expressed on the surface of SARS-CoV-2 in promoting membrane fusion. Conformational changes of these proteins before and after fusion have been shown. c) The conformation of the viral S2 protein has also been indicated in vitro (left) and in vivo (right). Abbreviations: FP, fusion peptide; HR-N, heptad repeat region N; HR-C, heptad repeat region C; IC, intracellular tail; SARS-CoV-2, severe acute respiratory syndrome coronavirus; TM, transmembrane anchor.
Different co-receptors have been identified to be involved in virus entry into the host cells and control the efficiency of cell entry [58]. When S proteins bind to host cell receptors, they encounter cellular co-receptors and activators. These co-receptors and activators may be membrane receptors, transmembrane receptors, proteases, or cations, which facilitate viral fusion protein refolding into an active form that catalyzes host cell membrane coalescence [59]. Many of these molecules are cellular proteases that cleave and activate the S proteins in ways that expose the essential domain for virus fusion [60]. These host cell proteases include trypsin, cathepsins, elastase, thermolysin, furin, the proprotein convertase family, and transmembrane protease/serine (TMPRSS) [61]. TMPRSS11d and TMPRSS2 can both induce coronavirus fusion. When host cells express TMPRSS2, infection of pulmonary cells with coronavirus S-pseudotyped particles was less sensitive to inhibitors of cathepsins B and L. In pulmonary cells, coronavirus S protein employs TMPRSS2 for S protein priming, and the endosomal cathepsins B and L are not essential for viral entry [[62], [63], [64]]. Therefore, further work is needed to assess which co-receptors and activators can enhance the entry of SARS-CoV-2 at the level of S protein.
4.4. SARS-CoV-2 cell entry inhibitors
SARS-CoV-2 entry into cells is a critical step of its life cycle that can be used as a target for treatment. Antiviral molecules that inhibit the host cell entry of coronaviruses have been reported. For example, Adedeji et al. identified compounds that could inhibit coronavirus cell entry through different mechanisms. The first identified inhibitor of SARS-CoV-2 cellular entry was SSAA09E2 (N-[[4-(4-methylpiperazin-1-yl)phenyl]methyl]-1,2-oxazole-5-carboxamide) that acted through prevention of the ACE2–RBD interaction. SSAA09E1 [(Z)-1-thiophen-2-ylethylideneamino]thiourea, was the second identified compound, which inhibited cathepsin L, and SSAA09E3, [N-(9,10-dioxo-9,10-dihydroanthracen-2-yl)benzamide], suppressed the fusion of the viral particles with the target cells [65]. For human coronaviruses, some other peptides have been reported to inhibit host cell entry through different mechanisms. For instance, Struck et al. demonstrated that a hexapeptide that bound to the ACE2 receptor, could block viral infection of host cells [66].
As mentioned, SARS-CoV-2 uses specific receptors, ACE2, and CD147, which are expressed on human airway epithelial cells and lung parenchyma. Compounds that act as angiotensin receptor blockers have been in clinical use since 1995, and are known to be effective antihypertensive agents with excellent tolerability profiles [67]. Many anti-ACE agents that can inhibit the renin-angiotensin system, such as losartan, rifampin, fluconazole, candesartan cilexetil, eprosartan, irbesartan, telmisartan, valsartan, azilsartan medoxomil, and olmesartan medoxomil, have been tested as treatments for hypertension. Other agents that may block the progression of the SARS-CoV-2 infection are angiotensin receptor 1 blockers, such as losartan [68]. Furthermore, anti-CD147 antibodies, such as mepolizumab, could effectively prevent the coronaviruses from invading target cells by blocking the CD147 receptor. These strategies may be reliable and safe without being affected by virus variation and mutation [36]. Interestingly, Hoffmann et al. reported that target cell entry of SARS-CoV-2 could be blocked by camostat mesylate, an inhibitor of TMPRSS2, which is employed for S protein priming [62]. A summary of the clinical trials against SARS-CoV-2, using hydroxychloroquine, chloroquine, losartan, mepolizumab, camostat mesylate, and other compounds is provided in Table 1 .
Table 1.
Selected therapeutic agents as inhibitors of SARS-CoV-2 cell entry currently in clinical trials.
| Phase | Responsible party | Interventions | Recruitment status | Population (enrollment and age) | NCT number |
|---|---|---|---|---|---|
| 2 | GlaxoSmithKline | ● GSK2586881 | Completed | 44 18–80 |
NCT01597635 |
| 2 | University of Minnesota | ● Losartan | Recruiting | 516 ≤18 |
NCT04311177 |
| 4 | Ruijin Hospital | ● Arbidol ● Basic treatment |
Not yet recruiting | 380 18–75 |
NCT04260594 |
| 4 | Beijing YouAn Hospital | ● Carrimycin ● Lopinavir with ritonavir tablets or arbidol or chloroquine phosphate ● Basic treatment |
Not yet recruiting | 520 18–75 |
NCT04286503 |
| 2, 3 | Bassett Healthcare | ● Lopinavir with ritonavir ● Hydroxychloroquine sulfate ● Losartan |
Recruiting | 4000 ≤18 |
NCT04328012 |
| 4 | Instituto de Investigación Marqués de Valdecilla | ● Hydroxychloroquine ● Control group |
Not yet recruiting | 800 18–75 |
NCT04330495 |
| 4 | Wroclaw Medical University | ● Chloroquine phosphate ● Telemedicine |
Not yet recruiting | 400 ≤18 |
NCT04331600 |
| 3 | Massachusetts General Hospital | ● Hydroxychloroquine | Recruiting | 510 ≤18 |
NCT04332991 |
| 1 | University of Washington | ● Hydroxychloroquine sulfate ● Vitamin C |
Not yet recruiting | 2000 18–80 |
NCT04328961 |
| 2 | Asan Medical Center | ● Lopinavir with ritonavir ● Hydroxychloroquine sulfate |
Recruiting | 100 18–99 |
NCT04307693 |
| 2, 3 | Oslo University Hospital | ● Hydroxychloroquine ● Remdesivir ● Standard of care |
Recruiting | 700 ≤18 |
NCT04321616 |
| 3 | Rajavithi Hospital | ● Protease inhibitors, oseltamivir, favipiravir, and chloroquine | Not yet recruiting | 80 18–100 |
NCT04303299 |
| 3 | Shanghai Public Health Clinical Center | ● Hydroxychloroquine | Completed | 30 ≤18 |
NCT04261517 |
| 3 | Population Health Research Institute | ● Azithromycin ● Chloroquine |
Not yet recruiting | 1500 18 |
NCT04324463 |
| 3 | Hospital do Coracao | ● Hydroxychloroquine oral product ● Hydroxychloroquine with azithromycin |
Recruiting | 630 18 |
NCT04322123 |
| 2 | Fundació Institut de Recerca de l'Hospital de la Santa Creu i Sant Pau | ● Tocilizumab ● Hydroxychloroquine ● Azithromycin |
Recruiting | 276 ≤18 |
NCT04332094 |
| 3 | University Hospital, Angers | ● Hydroxychloroquine | Recruiting | 1300 ≤18 |
NCT04325893 |
| 3 | University of Minnesota | ● Hydroxychloroquine | Recruiting | 3000 ≤18 |
NCT04308668 |
| Not applicable | University of Oxford | ● Chloroquine | Not yet recruiting | 10,000 ≤16 |
NCT04303507 |
| 1 | Sanofi | ● Hydroxychloroquine SAR321068 | Recruiting | 210 ≤18 |
NCT04333654 |
| 2 | Fundação de Medicina Tropical Dr. Heitor Vieira Dourado | ● Chloroquine diphosphate | Recruiting | 440 18–100 |
NCT04323527 |
| 3 | Institut National de la Santé Et de la Recherche Médicale, France | ● Remdesivir ● Lopinavir with ritonavir ● Interferon beta-1A ● Hydroxychloroquine ● Standard of care |
Recruiting | 3100 ≤18 |
NCT04315948 |
| 3 | Hospital Israelita Albert Einstein | ● Hydroxychloroquine with azithromycin ● Hydroxychloroquine |
Recruiting | 440 ≤18 |
NCT04321278 |
| 4 | Chronic Obstructive Pulmonary Disease Trial Network, Denmark | ● Azithromycin ● Hydroxychloroquine |
Recruiting | 226 All |
NCT04322396 |
| 2, 3 | Columbia University | ● Hydroxychloroquine | Not yet recruiting | 1600 ≤18 |
NCT04318444 |
| 3 | National Institute of Respiratory Diseases, Mexico | ● Hydroxychloroquine | Recruiting | 400 18 |
NCT04318015 |
| 2 | University of Pennsylvania | ● Hydroxychloroquine Sulfate | Recruiting | 400 ≤18 |
NCT04329923 |
| Early 1 | Rambam Health Care Campus | ● Hydroxychloroquine | Not yet recruiting | 1116 ≤18 |
NCT04323631 |
| 3 | Barcelona Institute for Global Health | ● Hydroxychloroquine | Recruiting | 440 ≤18 |
NCT04331834 |
| Early phase 1 | Azidus Brasil | ● Hydroxychloroquine sulfate ● Azithromycin tablets |
Not yet recruiting | 400 ≤18 |
NCT04329572 |
| 3 | Gangnam Severance Hospital | ● Hydroxychloroquine as post-exposure prophylaxis | Not yet recruiting | 2486 18–99 |
NCT04330144 |
| 3 | National Institute of Respiratory Diseases, Mexico | ● Hydroxychloroquine | Recruiting | 500 18–80 |
NCT04315896 |
| 3 | Centre Hospitalier Universitaire de Saint Etienne | ● Hydroxychloroquine ● Lopinavir and ritonavir |
Recruiting | 1200 ≤18 |
NCT04328285 |
| 2 | Korea University Guro Hospital | ● Ciclesonide metered dose inhaler [Alvesco] ● Hydroxychloroquine |
Not yet recruiting | 141 18–80 |
NCT04330586 |
| 2 | Intermountain Health Care, Inc. | ● Hydroxychloroquine ● Azithromycin |
Recruiting | 300 ≤18 |
NCT04329832 |
| 2 | Oxford University Clinical Research Unit | ● Chloroquine phosphate | Not yet recruiting | 250 ≤18 |
NCT04328493 |
| 3 | University of Calgary | ● Hydroxychloroquine | Recruiting | 1660 ≤18 |
NCT04329611 |
| 3 | Ayub Medical College, Abbottabad | ● Hydroxychloroquine ● Azithromycin ● Dietary supplement: glucose tablets |
Not yet recruiting | 75 18–50 |
NCT04328272 |
| Not applicable | Renmin Hospital of Wuhan University | ● DAS181 | Recruiting | 4 18–70 |
NCT04324489 |
| Not applicable | Neuromed IRCCS | ● ACE inhibitors | Not yet recruiting | 5000 All |
NCT04318418 |
| Not applicable | Istinye University | ● Hydroxychloroquine | Recruiting | 80 20–90 |
NCT04326725 |
| 2 | Ansun Biopharma, Inc. | ● DAS181 | Not yet recruiting | 280 18 |
NCT04298060 |
| 1, 2 | Tang-Du Hospital | ● Meplazumab (a humanized anti-CD147 antibody) for injection | Recruiting | 20 18–75 |
NCT04275245 |
| 3 | Ansun Biopharma, Inc. | ● DAS181 ● DAS181 SARS-CoV-2 ● DAS181 OL |
Recruiting | 250 All |
NCT03808922 |
| 4 | Tongji Hospital | ● Abidol hydrochloride ● Abidol hydrochloride combined with interferon atomization |
Recruiting | 100 18–80 |
NCT04254874 |
| 4 | NCT04255017 | ● Abidol hydrochloride ● Oseltamivir ● Lopinavir with ritonavir |
Recruiting | 400 ≤18 |
NCT04255017 |
5. Small molecule antiviral agents
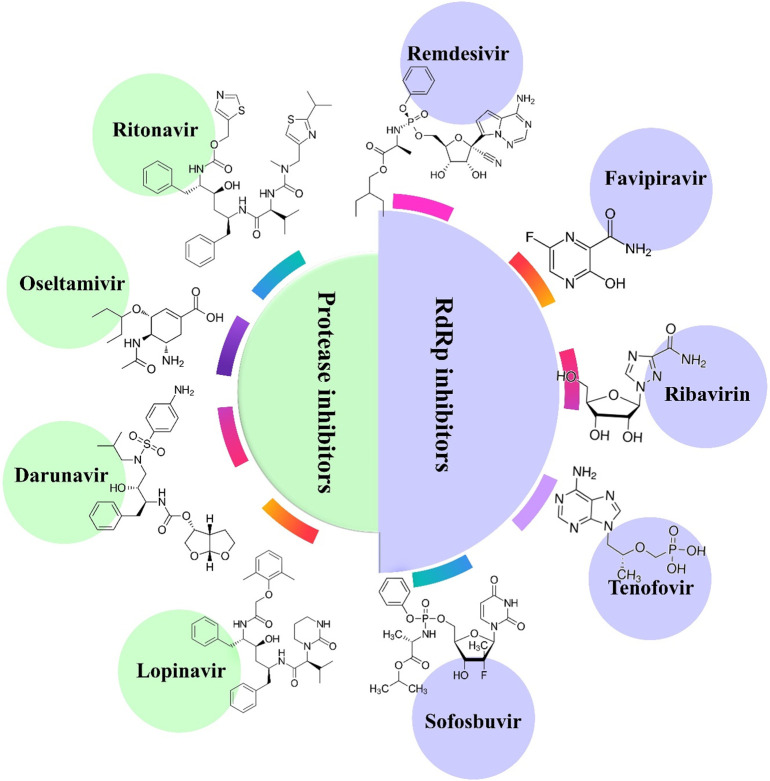
The development of antiviral drugs against SARS-CoV-2 is difficult. Several clinical trials of antiviral agents have been started as of May 2020, now amounting to a total of 306 active trials. Up to now, protease inhibitors, including ritonavir, oseltamivir, darunavir, and lopinavir, have been the most frequently tested class of drugs for the treatment of COVID-19. Lopinavir and ritonavir are HIV protease inhibitors, which have shown some promise in the treatment of SARS-CoV-2 infection [69], and have been tested in trials in COVID-19 patients [70,71]. For example, the third patient diagnosed with SARS-CoV-2 in Korea was treated with lopinavir and ritonavir starting from hospital day 8. After treatment, very low coronavirus titers were observed compared with a control group of untreated patients [72]. Cao et al. conducted a clinical trial using a combination of lopinavir with ritonavir as a potential treatment for hospitalized COVID-19 patients. However, these drugs showed no significant effect beyond standard care [70]. In addition, anti-influenza drugs that are routinely used in clinical practice, including neuraminidase inhibitors (zanamivir, peramivir, oseltamivir, etc.), acyclovir, ribavirin, ganciclovir, and methylprednisolone [73,74], have been studied as anti-SARS-CoV-2 drugs in clinical trials. Darunavir is a protease inhibitor primarily targeting the HIV-1 virus, which is being tested in clinical trials for SARS-CoV-2 treatment. Future clinical trials of protease inhibitors in patients with severe viral respiratory infections may help to exclude or confirm the possibility that they could be beneficial agents.
RNA-dependent RNA polymerase (RdRp) inhibitors are the second most frequently used class of drugs in the treatment of SARS-CoV-2 patients. Compared to the conserved sequence of RdRp in coronaviruses, SARS-CoV-2 and SARS-CoV have similar sequences and structures of RdRp [75]. Nucleoside analogs are generally adenine or guanine derivatives, which block viral RNA synthesis through targeting the RdRp in a broad spectrum of viruses, including human coronaviruses [76,77]. Both approved nucleoside analog drugs in clinical use (sofosbuvir, favipiravir, ribavirin, and tenofovir) and experimental drugs (galidesivir and remdesivir) may have potential therapeutic effects against SARS-CoV-2 RdRp (Fig. 5 ). Remdesivir is an adenosine analog pro-drug with a broad-spectrum antiviral activity that has been shown to inhibit the replication of a wide array of RNA viruses [78,79]. For instance, remdesivir was in clinical trials for the treatment of male Ebola virus disease survivors [80,81]. Remdesivir is presently in clinical trials for the COVID-19 outbreak (Table 2 ), and in one completed clinical trial showed promising antiviral activity against SARS-CoV-2 infection. Although the FDA has approved only a few antiviral combination treatments for a relatively small number of viral diseases, several combinations of antiviral agents with activity against SARS-CoV-2 are currently being assessed (Table 2). Among the clinical trials in progress, some are testing antiviral agents, such as lopinavir plus ritonavir, as the most common drug combination. Overall, among the new antiviral trials that were commenced in 2020, remdesivir has attracted the most attention for the treatment of SARS-CoV-2.
Fig. 5.
Potential protease inhibitors and RdRp inhibitors in clinical trials for SARS-CoV-2.
Table 2.
Selected small molecule therapeutic agents as inhibitors of SARS-CoV-2 in clinical trials.
| Phase | Responsible party | Interventions | Recruitment status | Population (enrollment and age) | NCT number |
|---|---|---|---|---|---|
| 2 | Sunnybrook Health Sciences Centre | ● Lopinavir with ritonavir | Recruiting | 400 ≤6 months |
NCT04330690 |
| – | Gilead Sciences | ● Remdesivir | Available | – ≤18 |
NCT04323761 |
| Not applicable | Peking University First Hospital | ● Favipiravir with tocilizumab ● Favipiravir ● Tocilizumab |
Recruiting | 150 18–65 |
NCT04310228 |
| 3 | St. Michael's Hospital, Toronto | ● Lopinavir with ritonavir | Not yet recruiting | 1220 ≤18 months |
NCT04321174 |
| – | U.S. Army Medical Research and Development Command | ● Remdesivir | Available | – | NCT04302766 |
| 3 | China-Japan Friendship Hospital | ● Remdesivir | Terminated | 453 ≤18 |
NCT04257656 |
| 3 | China-Japan Friendship Hospital | ● Remdesivir | Suspended | 380 ≤18 |
NCT04252664 |
| 3 | Tongji Hospital | ● ASC09F with oseltamivir ● Ritonavir with oseltamivir ● Oseltamivir |
Recruiting | 60 18–55 |
NCT04261270 |
| 3 | Gilead Sciences | ● Remdesivir ● Standard of care |
Recruiting | 600 ≤18 |
NCT04292730 |
| 3 | Shanghai Public Health Clinical Center | ● Darunavir and cobicistat | Recruiting | 30 ≤18 |
NCT04252274 |
| 3 | Gilead Sciences | ● Remdesivir ● Standard of care |
Recruiting | 400 ≤18 |
NCT04292899 |
| 3 | Germans Trias i Pujol Hospital | ● Antiviral treatment and prophylaxis ● Standard public health measures |
Recruiting | 3040 ≤18 |
NCT04304053 |
| 2 | National Institute of Allergy and Infectious Diseases (NIAID) | ● Remdesivir | Recruiting | 440 ≤18 |
NCT04280705 |
| 2 | The University of Hong Kong | ● Lopinavir with ritonavir ● Ribavirin ● Interferon Beta-1B |
Completed | 70 ≤18 |
NCT04276688 |
| 4 | The Ninth Hospital of Nanchang | ● Ganovo with ritonavir with/and interferon nebulization | Completed | 11 18–75 |
NCT04291729 |
| Not applicable | First Affiliated Hospital of Zhejiang University | ● ASC09 with ritonavir group ● Lopinavir with ritonavir group |
Not yet recruiting | 180 18–75 |
NCT04261907 |
| Not applicable | Jiangxi Qingfeng Pharmaceutical Co. Ltd. | ● Lopinavir with ritonavir tablets combined with xiyanping injection ● Lopinavir with ritonavir treatment |
Not yet recruiting | 80 18–100 |
NCT04295551 |
| 1, 2 | University of Aarhus | ● Camostat mesilate | Not yet recruiting | 180 18–110 |
NCT04321096 |
Azithromycin is a 15-membered macrolide antibiotic, that is distinguished from other macrolides by its extensive and rapid penetration into biological compartments, accompanied by an acceptable serum half-life and a prolonged concentration in tissue [82]. Azithromycin has been effective in vitro against Ebola and Zika viruses [[83], [84], [85]], and some other viral infections of the lower and upper respiratory tracts [86]. Gautret et al. evaluated the effect of azithromycin plus hydroxychloroquine on the respiratory SARS-CoV-2 viral load. The results suggested a positive effect of the combination of azithromycin and hydroxychloroquine [87]. Azithromycin is currently under study for treating hospitalized patients with moderate to severe SARS-CoV-2 infection.
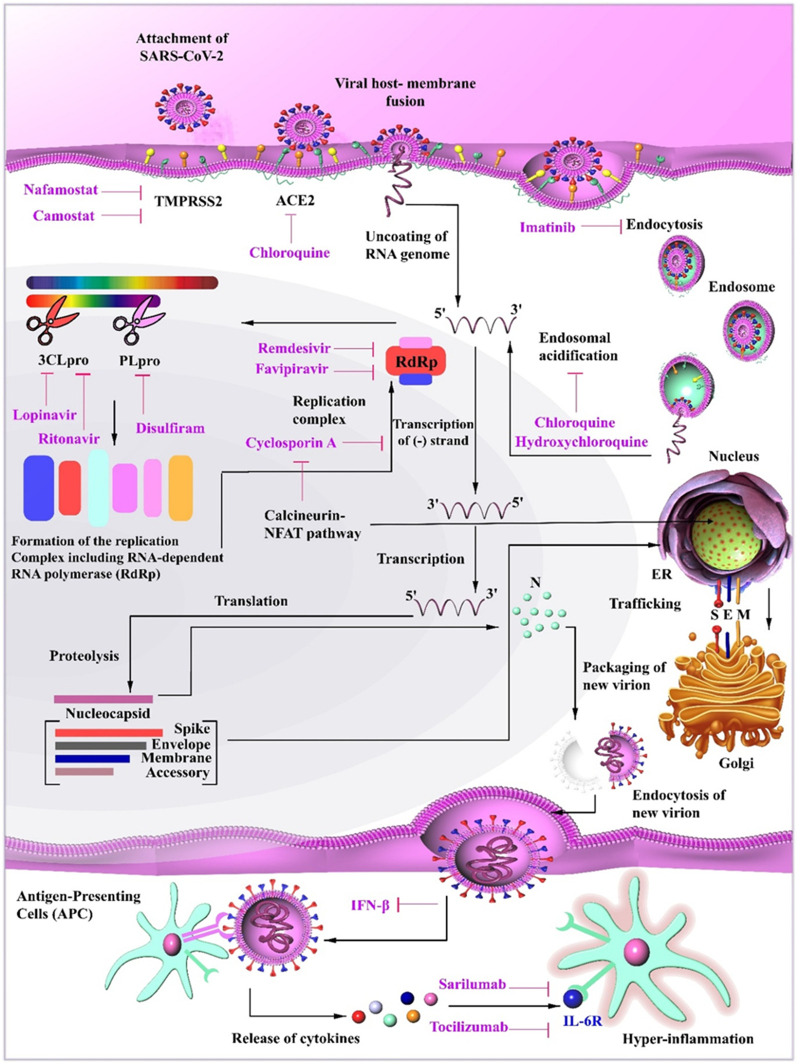
Nevertheless, even after several months from the first appearance of SARS-CoV-2, we still have no definitive drugs to combat the infection. In fact, we are still testing drugs already known to target similar RNA viruses. These drugs have been proposed to interfere with the progression of the SARS-CoV- 2 infections by a multitude of mechanisms. One class of these drugs interferes with the penetration of the virus into cells by inhibiting either membrane attachment (ACE2) or membrane fusion (TMPRSS2). Some other drugs also prevent the formation of endosomes. Nevertheless, after penetration of the viral particles, it is necessary to use agents that inhibit basic biological functions such as protein synthesis (CLpro and PLpro) or DNA replication (RdRp). Furthermore, it may be possible to use modulators of the immune system to increase the antiviral response (e.g., IL-6R). Care should be taken using such drugs as they may worsen clinical symptoms in severely ill SARS-CoV-2 patients, in whom immunosuppressive drugs may actually be more effective (Fig. 6 ). None of these potential drugs (either alone or in combinations) can be considered definitive treatments without passing extensive and well-designed clinical trials, which are fortunately underway.
Fig. 6.
Mechanisms of various drugs proposed to combat SARS-CoV-2 infection. Chloroquine inhibits the attachment of the virus to its receptor ACE2. Nafamostat and Camostat interfere with membrane fusion, which employs TMPRSS2 on the cell surface. Imatinib suppresses endocytosis and hydroxychloroquine induces degeneration of virus-containing endosomes. Remdesivir, Favipiravir, and Cyclosporin A interfere with the replication of the viral genome. Other drugs (Lopinavir, Ritonavir, and Disulfiram) suppress the formation of peptides needed for assembly of virus replicatory machine (RdRp) by deactivating viral proteases (3CLpro and PLpro). Finally, Sarilumab and Tocilizumab mitigate hyper-inflammatory responses by suppressing IL-6 interaction with its receptor and inhibiting signaling pathways.
In recent studies, it has been stated that dexamethasone, a corticosteroid that has been effective in treating autoimmune diseases (e.g. multiple sclerosis, rheumatoid arthritis) as well as inflammatory and hepatic disorders and cancer, may be effective in reducing mortality in patients with COVID-19 infection [88,89].
In fact, this corticosteroid was the first medication that brought hope for saving the lives of severely affected patients with the infection. The efficiency of dexamethasone in improving the clinical condition of COVID-19 patients is currently under investigation along with four other drugs (hydroxychloroquine, azithromycin, lopinavir–ritonavir combination, and tocilizumab) and plasma therapy (the RECOVERY trial) [90]. Giving the ability of nanomaterials to be accumulated in macrophages, Lammers et al. suggested that using nano-forms of dexamethasone may augment its impact on the clinical progression of COVID-19 infection [90,91] a notion which certainly needs more evidence.
6. SARS-CoV-2 vaccine platforms
Several preclinical and clinical trials are now underway testing candidate vaccines against SARS-CoV-2. Vaccination against infectious diseases can be a powerful tool for preventing potential outbreaks of epidemic diseases before they become public health problems [92]. Vaccine strategies are considered as a critical component of SARS-CoV-2 prevention, especially since therapeutic agents are unavailable or ineffective, and that rapid clinical deterioration may limit the effectiveness of any treatment options. The lack of therapeutic vaccines for clinical use against such viruses, makes the coronavirus pandemic a serious global threat [93]. In addition, timely development of SARS-CoV-2 vaccines is needed as soon as possible, not only for controlling the SARS-CoV-2 infection but also for stabilizing the global outlook and bringing the world economy back on track [94]. A number of approaches have been proposed to develop vaccines against coronaviruses [[95], [96], [97], [98]]. SARS-CoV-2 vaccines based on the whole inactivated virus, non-replicating viral vectors, replicating viral vectors, nucleic acids, and subunits, have been tested in preclinical and clinical trials (Table 3, Table 4, Table 5, Table 6 ). The World Health Organization (WHO) has provided an overview of SARS-CoV-2 candidate vaccines in preclinical trials [99]. However, the enthusiasm for a rapid deployment of SARS-CoV-2 vaccines is tempered by the reality of the experience of developing previous coronavirus vaccines. Although vaccine development against SARS-CoV-2 is under development, manufacturing at scale will take a long time, probably at least 12 to 18 months away from scaled-up vaccine production [100]. Hopefully, SARS-CoV-2 vaccine development can be even faster and more efficient compared to previous experience, and possibly using newer technologies. SARS-CoV-2 vaccine development is still in the early stages. To date, very few SARS-CoV-2 vaccine candidates have been tested in clinical trials (Table 3).
Table 3.
Vaccine candidates in clinical trials against SARS-CoV-2.
| Phase | Responsible party | Interventions | Recruitment status | Population (enrollment and age) | NCT number |
|---|---|---|---|---|---|
| 1 | Shenzhen Geno-Immune Medical Institute | ● Pathogen-specific aAPC | Recruiting | 100 6 months to 80 years |
NCT04299724 |
| 3 | Murdoch Childrens Research Institute | ● BCG vaccine | Recruiting | 4170 ≤18 |
NCT04327206 |
| 1, 2 | University of Oxford | ● ChAdOx1 nCoV-19 | Recruiting | 510 18–55 |
NCT04324606 |
| 1 | CanSino Biologics Inc. | ● Recombinant novel coronavirus vaccine (adenovirus type 5 vector) | Active, not recruiting | 108 18–60 |
NCT04313127 |
| 1 | National Institute of Allergy and Infectious Diseases (NIAID) | ● mRNA-1273 | Recruiting | 45 18–55 |
NCT04283461 |
| 1, 2 | Shenzhen Geno-Immune Medical Institute | ● Injection and infusion of LV-SMENP-DC vaccine and antigen-specific CTLs | Recruiting | 100 6 months to 80 years |
NCT04276896 |
| 1, 2 | Shenzhen Geno-Immune Medical Institute | ● Injection and infusion of LV-SMENP-DC vaccine and antigen-specific CTLs | Recruiting | 100 6 months to 80 years |
NCT04276896 |
Table 4.
Recently whole-virus-based vaccine and viral vector-based vaccine candidates against SARS-CoV-2.
| Developer | Platform | Type of candidate vaccine | Current stage |
|---|---|---|---|
| Sinovac | Inactivated | Formaldehyde inactivated with alum | Pre-clinical |
| Codagenix/Serum Institute of India | Live attenuated virus | Deoptimized live attenuated vaccines | Pre-clinical |
| Codagenix/Serum Institute of India | Live attenuated virus | Deoptimized live attenuated vaccines | Pre-clinical |
| GeoVax/BravoVax | Non-replicating viral vector | MVA encoded VLP | Pre-clinical |
| Janssen Pharmaceutical Companies | Non-replicating viral vector | Ad26 (alone or with MVA boost) | Pre-clinical |
| University of Oxford | Non-replicating viral vector | ChAdOx1 | Pre-clinical |
| Altimmune | Non-replicating viral vector | Adenovirus-based NasoVAX | Pre-clinical |
| Greffex | Non-replicating viral vector | Ad5 S (GREVAX™ platform) | Pre-clinical |
| Vaxart | Non-replicating viral vector | Oral vaccine platform | Pre-clinical |
| CanSino Biologics | Non-replicating viral vector | Viral-vectored based | Pre-clinical |
| Zydus Cadila | Replicating viral vector | Measles vector | Pre-clinical |
| Institute Pasteur | Replicating viral vector | Measles vector | Pre-clinical |
| Tonix Pharma/Southern Research | Replicating viral vector | Horse-pox vector | Pre-clinical |
Table 5.
Recently nucleic acid vaccine candidates against SARS-CoV-2.
| Developer | Platform | Type of candidate vaccine | Current stage |
|---|---|---|---|
| Inovio Pharmaceuticals | DNA | DNA plasmid vaccine electroporation device | Pre-clinical |
| Takis/Applied DNA Sciences/Evvivax | DNA | DNA | Pre-clinical |
| Zydus Cadila | DNA | DNA plasmid vaccine | Pre-clinical |
| Fudan University/Shanghai JiaoTong University/RNACure Biopharma | RNA | LNP-encapsulated mRNA cocktail encoding VLP | Pre-clinical |
| Fudan University/Shanghai JiaoTong University/RNACure Biopharma | RNA | LNP-encapsulated mRNA encoding RBD | Pre-clinical |
| China CDC/Tongji University/Stermina | RNA | mRNA | Pre-clinical |
| Moderna/NIAID | RNA | LNP-encapsulated mRNA | Phase 1 |
| Arcturus/Duke-NUS | RNA | mRNA | Pre-clinical |
| Imperial College London | RNA | saRNA | Pre-clinical |
| Curevac | RNA | mRNA | Pre-clinical |
Table 6.
Recent subunit-based vaccine candidates against SARS-CoV-2.
| Developer | Platform | Type of candidate vaccine | Current stage |
|---|---|---|---|
| ExpreS2ion | Protein subunit | Drosophila S2 insect cell expression system VLPs | Pre-clinical |
| WRAIR/USAMRIID | Protein subunit | S protein | Pre-clinical |
| Clover Biopharmaceuticals Inc./GSK | Protein subunit | S-Trimer | Pre-clinical |
| Vaxil Bio | Protein subunit | Peptide | Pre-clinical |
| AJ Vaccines | Protein subunit | S protein | Pre-clinical |
| Generex/EpiVax | Protein subunit | Ii-Key peptide | Pre-clinical |
| EpiVax/Univ. of Georgia | Protein subunit | S protein | Pre-clinical |
| Sanofi Pasteur | Protein subunit | S protein (baculovirus production) | Pre-clinical |
| Novavax | Protein subunit | Full length S trimers/nanoparticle with Matrix M | Pre-clinical |
| Heat Biologics/Univ. Of Miami | Protein subunit | gp-96 backbone | Pre-clinical |
| University of Queensland/GSK | Protein subunit | S protein clamp | Pre-clinical |
| Baylor College of Medicine | Protein subunit | S1 or RBD protein | Pre-clinical |
| iBio/CC-Pharming | Protein subunit | Subunit protein, plant produced | Pre-clinical |
A variety of technological platforms have been exploited in different studies; some of them are briefly described here [101,102] (Fig. 7 ):
Live attenuated vaccines: Modifying the SARS-CoV-2 virus in a way that reduces its pathogenicity and virulence can assist us in producing a live but weakened virus. Codon deoptimization or introducing a mutated E protein is among methods for making incapable viruses [103]. Although this method can draw a fast and potent immune response, it may not be applicable in immunosuppressed individuals.
Viral-vector based vaccines: Other viruses (e.g., adenovirus) can be used as vectors to carry SARS-CoV-2 genes into cells. This method delivers good immunogenicity even in the absence of an adjuvant. A robust cytotoxic T cell (CTL) response is ensured using such vaccines to remove virus-infected cells.
Recombinant protein-based vaccines: In this approach, a recombinant protein is constructed by adjoining SARS-CoV-2 proteins (such as S protein) with adjuvants. Incorporating adjuvants promote the immune response against the viral antigen.
DNA vaccines: Potentially, we can use plasmid DNA to incorporate target viral genes, which are then expressed to SARS-CoV-2 proteins. By using this method, antigens can be efficiently delivered to host cells. Nevertheless, no approved DNA vaccines are currently available to be used in humans.
mRNA vaccines: Transcripts of SARS-CoV-2 genes (i.e., mRNAs) enclosed in structures such as liposomes can carry viral antigens into host cells. However, no approved mRNA vaccines are yet available.
Fig. 7.
Attempts for developing efficient vaccines to cope with the infection of SARS-CoV-2.
An mRNA vaccine by Moderna (NCT04283461) and a vector-based vaccine (using adenovirus type 5) by CanSino Biologicals (NCT04341389) have been developed, which are passing phase I and II clinical trials to confirm their safety and efficiency. Furthermore, recombinant S-protein based vaccines conjugated with conventional adjuvants (AS03 and MF59) have advantages such as enhanced immunogenicity, requiring lower doses, and being applicable in large populations. An efficient vaccine should be able to induce adequate specific antibodies to neutralize the SARS-CoV-2 viruses. As we have learned from studies on the SARS and MERS, vaccines may be potentially associated with unwanted immune enhancement effects. Therefore, enough care should be taken before releasing any COVID-19 vaccine.
We describe below the different platforms of SARS-CoV-2 vaccines based on the WHO landscape and clinical trials.
Interestingly, children appear to suffer from a much less severe form of the SARS-CoV-2 infection. This may be related to differences in innate immunity evident at a young age, as applies to the use of vaccines such as Bacille Calmette-Guerin (BCG) [104,105].
Various strategies have been tried by researches for this purpose. Using alive attenuated virus is one of the options. Alongside this, there are ongoing efforts to develop viral-vector and recombinant protein-based vaccines to deliver viral antigens such as spike (S) protein to antigen-presenting cells. Nucleic acid-based vaccines (viral DNA and mRNA) have also been tried. Because the viral S protein is critical for the entrance of the virus into target cells, this protein has been under attention as an optimal candidate for developing vaccines. To be efficient, a vaccine must be able to trigger the production of adequate anti-virus antibodies. Simultaneously, it should possess a low risk of complications, such as unwanted immune reactions. One potentially threatening phenomenon to be avoided is known as antibody-dependent enhancement (ADE), which can result in exaggerated uptake of viral particles. Furthermore, unprotective Th2 responses, which lead to allergic inflammatory reactions, should be kept minimal following vaccination.
6.1. Whole-virus vaccines
A whole-virus vaccine is based on a physically or chemically inactivated virion, which is the entity that causes the entire disease. The inactivated whole-virus approach offers several advantages, including a good safety profile, cost-effective production, high productivity, and no need for genetic modification [95,106]. An inactivated SARS-CoV vaccine is probably the easiest and most practical for developing a coronavirus vaccine by analogy with available vaccines, including rabies and polio vaccines [107]. Whole vaccines may be more reactogenic to confer protective immunity against coronaviruses [108]. One investigation used an inactivated coronavirus (performed with formaldehyde after preparation in Vero cells) that was intramuscularly injected into rhesus monkeys to promote protective immunity. After three weeks, this vaccine preferentially induced Th1-type inflammatory responses, in addition to other beneficial cellular immune responses [107]. Moreover, a live-attenuated virus vaccine is generated by a variety of techniques to significantly reduce the virulence of a virus while retaining its immunogenicity. Compared with inactivated whole-virus vaccines, live-attenuated virus vaccines can stimulate an adaptive long-term immune response. However, higher immunogenicity is usually associated with a lower safety profile [109,110]. So far, one inactivated virus and five live attenuated whole-virus vaccines, prepared by different developers, have progressed into human pre-clinical trials. Potential whole-virus vaccine candidates against SAR-CoV-2 are summarized in Table 4.
6.2. Viral vector-based vaccines
Multiple vector-based vaccines are in clinical and preclinical trials for developing potential immunity against SARS-CoV-2, and these vaccines could likely be an important tool to control the new coronavirus. These vectors are regarded as powerful tools for vaccination and for gene therapy. In general, their advantages include highly specific delivery of genes to target cells, high-efficiency gene transduction, and induction of robust cellular and humoral responses [111]. Replicating and non-replicating forms of viral vectors that are available include adenoviruses and poxviruses [112]. Zhao et al. found that immunization with a nucleocapsid (N) protein-based vaccine protected mice from this coronavirus through activation of CD4+ T IFN-γ- and cell-dependent immunity [113]. Furthermore, the modified viral vector Ankara was modified to encode the MERS-CoV S protein, and induced CD8+ T cell responses and neutralizing antibodies in pre-clinical studies [114]. The third type of viral vector-based vaccines is adenoviruses, and immunization of mice with a vector expressing S/N proteins led to the production of antibodies [115]. In addition, both Ad5- and Ad41-MERS-CoV S vaccines were shown to induce immune responses in mice [116]. By profiting from lessons learned in previous coronavirus vaccines, vaccine scientists have been working on developing SAR-CoV-2 vaccines within the shortest time frame possible [117]. A number of viral vector-based vaccines have progressed into human pre-clinical trials. The viral vector-based platforms used in SARS-CoV-2 vaccine studies are summarized in Table 4.
6.3. Nucleic acid vaccines
Several nucleic acid-based vaccines for coronavirus have been reported to date. Nucleic acid-based vaccines combine the positive attributes of both subunit vaccines and live-attenuated vaccines, and there has been substantial research into this type of vaccine for diverse diseases, over the last three decades [118]. These vaccines involve direct immunization through the delivery of DNA or RNA sequences encoding the antigen, and have as their main advantages, their purity and the simplicity by which this type of vaccine can be produced [119,120]. In addition, nucleic acid-based vaccines can be manufactured rapidly on a large scale and are relatively low-cost [95,121]. Furthermore, the use of these vaccines that combine the benefits of subunit and inactivated vaccines has been a critical advance [122]. The enhanced humoral and cellular immune response against SARS-CoV were elicited by a DNA-based vaccine encoding S protein, or the S1 fragment. This vaccine induced T-cell responses, as well as neutralizing antibodies [115]. Similarly, a nucleic acid-based vaccine encoding the S protein or the shorter S1 fragment, was developed for MERS-CoV. pVax1™ is a nucleic acid-based vaccine against MERS-CoV, that encodes the S protein plus an IgE leader sequence and a codon to promote expression and mRNA export [123]. Another nucleic acid-based vaccine encoding a full-length S protein against MERS-CoV strain England1, used intramuscular administration and induced neutralizing antibodies in rhesus monkeys [124]. Currently, the safety and immunogenicity of coronavirus nucleic acid-based vaccines are being evaluated in clinical trials. There are a few nucleic acid-based vaccines in the pipeline against SARS-CoV-2 in pre-clinical and clinical trials. For example, an mRNA-based vaccine against SARS-CoV-2 (INO-4800-DNA) was prepared by the US National Institute of Allergy and Infectious Diseases (NIAID) and is currently in phase 1 clinical trials. This vaccine will soon be ready for human testing in additional clinical trials. In addition, it was reported that Stermirna Therapeutics is working to develop an mRNA-based vaccine for human studies [125]. Some potential nucleic acid vaccine candidates against SAR-CoV-2 are summarized in Table 5.
6.4. Subunit vaccines
These vaccines are produced using recombinant or synthetic virus subunits. The viral nucleocapsid (N), spike (S) or envelope (E) subunits are obtained through proteolysis or chemical hydrolysis to prepare the subunit vaccines. By using one viral protein subunit, this type of vaccine activates an immune response without inducing the production of antibodies against unrelated antigens [110]. Although these vaccine platforms have the highest safety profile among all other platforms, they have been considered to be weakly immunogenic [126].
Subunit vaccines are of great interest in the treatment and prevention of coronavirus diseases. Several subunit vaccines have been introduced against coronavirus targeting the S glycoprotein. Of note, the full-length S protein or its fragments, including RBD, NTD, S1 subunit, and S2 subunit, can be used as immunogens for the development of these vaccines against coronaviruses [127]. For example, a polypeptide of the SARS-CoV S glycoprotein has been successfully expressed in baculovirus vectors [128]. The recombinant protein was purified and infused into mice using Ribi or saponin as an adjuvant, and induced higher antibody titers and better protection against SARS-CoV [129]. Modified Ankara virus vaccines were developed to express the full length S protein [130]. RBD in the S1 subunit comprises the critical neutralizing fragment of MERS-CoV S protein without the non-neutralizing immunodominant domain. This type of subunit vaccine is limited to producing RBD-dependent immune responses, and these vaccines are unable to induce harmful nonspecific antibodies [95,124,131]. A sequence engineered RBD-based vaccine allowed the production of three-fold greater neutralizing antibody titers [132,133]. The N protein may provide an ideal target for the development of vaccines against coronavirus. Of note, the N protein cannot elicit antibodies to block the interaction of the virus with host cells and subsequently neutralize viral infection. Nevertheless, it may still induce cellular immune responses and specific antibodies [134,135]. M protein is a major structural protein, which could serve as a potential target for the development of subunit vaccines. In fact, SARS-CoV M subunits have high immunogenicity and can trigger high-titer antibody responses [136]. Several subunit vaccines against SAR-CoV-2 have progressed into human pre-clinical trials. Potential subunit vaccine candidates against SAR-CoV-2 are summarized in Table 6.
7. Passive immunotherapy for SARS-CoV-2
Our current knowledge of specific immune reactions against the novel SARS-CoV-2 is mainly based on previous findings with similar viruses like MERS-CoV [101]. In this regard, it is assumed that pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) will play a central role in sensing viral RNA or its replication intermediates. Within the alveolar epithelium, endosomal single-stranded (ss)RNA, TLR7/8, and the cytosolic double-stranded (ds) RNA sensor; RIG-I/MDA-5, seem to be first PRRs that detect the virus particles. After recognition of the virus by these sensors, MyD88 and MAVS adaptor proteins are activated, which subsequently induce IRF3/7 and NF-κB transcription factors. As a result, the expression of type I interferons (IFN-α and IFN-β) and proinflammatory cytokines (e.g., IL-6 and TNF-α) is increased [137]. On the other hand, the secretion of the inflammatory mediator IL-1β, and the induction of pyroptosis (an inflammatory form of cell death) mediated by the NLRP3 inflammasome, aggravate the inflammatory process. Indeed, the E and 3a proteins derived from the SARS-CoV-2 are involved in the induction of the NLRP3 inflammasome [138]. Our understanding of the recognition mechanisms of the SARS-CoV-2 is still incomplete (Fig. 8 ).
Fig. 8.
Possible immune reactions induced by the SARS-CoV-2. The predictions are based on studies of SARS-CoV and MERS-CoV viruses. Non-specific recognition by innate immune receptors (e.g., RNA sensors, TLR7/8, RIG-I/MDA-5, and NLRP3 inflammasome) seems to be the first effect of the virus within alveolar epithelial cells. The main transcription factors involved in the induction of inflammatory mediators (e.g., IL-1β, IL-6, and type I IFNs) are NF-κB and IRF3/7. The antiviral activity of type I IFNs is augmented by many ISGs such as RNAse L. Cell-based immunity is based on macrophages, B cells, and T cells, which directly eliminate viral particles. However, hyper-inflammation resulting from an unbalanced action of the immune system could exacerbate COVID-19 outcomes.
Immunotherapy potentially overcomes one problem of SARS-CoV-2 treatment. Various host factors in the human immune system are responsible for SARS-CoV-2 progression or regression. Immunotherapy is defined as a therapeutic intervention that targets or manipulates these immune system factors [139]. Numerous investigations have shown that increased amounts of inflammatory factors are associated with pulmonary inflammation and subsequent lung damage, first in SARS-CoV patients [140], next in MERS-CoV infections, and most recently in SARS-CoV-2. These factors include MIP-1A, G-CSF TNFα, MCP-1, IL-7, IL-10, IL-2, and IP-10 [141]. This so-called “cytokine storm” can initiate inflammation-induced lung injury and cause viral sepsis, which leads to acute respiratory distress syndrome (ARDS), respiratory failure, pneumonitis, organ failure, and potentially death [117]. Furthermore, severe cases of SARS-CoV-2 tend to have lower lymphocyte counts, higher leukocyte counts, and an altered neutrophil-lymphocyte-ratio, as well as smaller percentages of eosinophils, basophils, and monocytes. In contrast, the number of both helper T cells and suppressor T cells is significantly decreased in severe cases. However, the percentage of memory helper T cells is reduced, and that of naive helper T cells is increased in severe patients. These patients also have lower levels of regulatory T cells and more noticeable lung damage in acute cases [142]. These immune responses could be modified by drugs, cytokines, monoclonal antibodies, antisera, vitamins and minerals, transplantation, and immunization.
It may be possible to treat SARS-CoV-2 patients using convalescent plasma obtained from recovered patients, and this approach is being considered for several emerging virus outbreaks. A meta-analysis of studies using convalescent plasma for managing severe acute respiratory infections suggests that the appropriate use of these products results in reduced mortality risk [143]. Convalescent plasma was used for treating SARS-CoV patients with potentially promising results. However, in the absence of suitable clinical trials, the results remain controversial [144]. In addition, Zhao et al. published results showed the therapeutic and prophylactic efficacy of camel serum-containing MERS-CoV neutralizing antibodies in reducing weight loss, viral load, and improving pulmonary function in MERS patients [145]. Recently, in a preliminary non-controlled case series of 5 severe patients, the administration of convalescent plasma collected from patients who had recovered from SARS-CoV-2 containing antibodies was followed by an improved clinical outcome [146].
Humoral immune responses to infection, especially the rapid production of neutralizing antibodies, have a protective effect against infection and prevent reinfection. Epitopes of T and B cells were extensively mapped for the main SARS-CoV proteins, N, E, S, and M protein [147]. Furthermore, previous infection with non-SARS-CoV viruses may have caused many people (including children) to already have some levels of protective antibodies against the novel virus [148,149]. For example, Shanmugaraj et al. summarized the potential neutralizing antibody-based therapeutic strategies for SARS-CoV-2 including the neutralizing antibodies against SARS-CoV (80R, CR3014, CR3022, F26G18, F26G19, m396, 1A9, 201, 68 and S230) and MERS-CoV (MERS-4, MERS-27, 4C2, m336, G4, D12, JC57-14, MERS-GD27, MERS-GD33, LCA60, MCA1, CDC2-C2, 7D10, and G2) [150]. A list of possible therapies for SARS-CoV-2 based on neutralizing antibodies, convalescent plasma, and other immunotherapies that have been tested in ongoing and completed human clinical trials, is provided in Table 7 .
Table 7.
Passive immunotherapy for SARS-CoV-2 in clinical trials.
| Phase | Responsible party | Interventions | Recruitment status | Population (enrollment and age) | NCT number |
|---|---|---|---|---|---|
| Early 1 | Tongji Hospital | ● Recombinant human interferon α1β | Not yet recruiting | 328 ≤18 |
NCT04293887 |
| 2 | First Affiliated Hospital of Wenzhou Medical University | ● Thalidomide | Not yet recruiting | 100 ≤18 |
NCT04273529 |
| Not applicable | Tongji Hospital | ● Tocilizumab ● Standard of care Procedure: continuous renal replacement therapy |
Recruiting | 120 18–80 |
NCT04306705 |
| 2 | First Affiliated Hospital of Fujian Medical University | ● Fingolimod | Recruiting | 30 ≤18 |
NCT04280588 |
| 2, 3 | Fasa University of Medical Sciences | ● Levamisole pill with budesonide with formoterol inhaler ● Lopinavir with ritonavir with hydroxychloroquine |
Not yet recruiting | 30 18–100 |
NCT04331470 |
| 2 | First Affiliated Hospital of Wenzhou Medical University | ● Thalidomide | Not yet recruiting | 40 ≤18 |
NCT04273581 |
| 2, 3 | Qilu Hospital of Shandong University | ● Bevacizumab injection | Recruiting | 20 ≤18 |
NCT04275414 |
| 2, 3 | Peking Union Medical College Hospital | ● Methylprednisolone therapy ● Standard care |
Recruiting | 80 ≤18 |
NCT04244591 |
| 3 | Tongji Hospital | ● Sildenafil citrate tablets | Recruiting | 10 ≤18 |
NCT04304313 |
| 4 | Tongji Hospital | ● Methylprednisolone | Recruiting | 100 ≤18 |
NCT04263402 |
| Not applicable | Beijing Chao Yang Hospital | ● Methylprednisolone | Recruiting | 400 ≤18 |
NCT04273321 |
| – | Hudson Medical | ● Eculizumab | Available | – ≤18 |
NCT04288713 |
| 4 | University Hospital, Ghent | ● Usual care ● Anakinra ● Siltuximab ● Tocilizumab |
Not yet recruiting | 342 18–80 |
NCT04330638 |
| 2 | Southeast University, China | ● PD-1 blocking antibody with standard treatment ● Thymosin with standard treatment ● Standard treatment |
Not yet recruiting | 120 ≤18 |
NCT04268537 |
| Not applicable | University of Palermo | Dietary supplement: vitamin C | Recruiting | 500 All |
NCT04323514 |
| Not applicable | Peking Union Medical College Hospital | ● Intravenous immunoglobulin ● Standard care |
Not yet recruiting | 80 ≤18 |
NCT04261426 |
| 2 | Assistance Publique - Hôpitaux de Paris | ● Tocilizumab | Not yet recruiting | 240 ≤18 |
NCT04331808 |
| Not applicable | Shanghai Public Health Clinical Center | ● Inactivated convalescent plasma | Recruiting | 15 All |
NCT04292340 |
| 2 | Xijing Hospital | ● Nitric oxide gas | Not yet recruiting | 104 ≤18 |
NCT04290871 |
| 2 | Massachusetts General Hospital | ● Nitric oxide | Not yet recruiting | 240 ≤18 |
NCT04305457 |
| Not applicable | Foundation IRCCS San Matteo Hospital | ● Hyperimmune plasma | Active, not recruiting | 49 ≤18 |
NCT04321421 |
| 2 | Southeast University, China | ● PD-1 blocking antibody with standard treatment ● Thymosin with standard treatment ● Standard treatment |
Not yet recruiting | 120 ≤18 |
NCT04268537 |
| 2, 3 | Regeneron Pharmaceuticals | ● Sarilumab | Recruiting | 400 ≤18 |
NCT04315298 |
| 4 | Negrin University Hospital | ● Dexamethasone | Not yet recruiting | 200 18 |
NCT04325061 |
| 2 | Università Politecnica delle Marche | ● Tofacitinib | Not yet recruiting | 50 18–65 |
NCT04332042 |
| 3 | Assistance Publique - Hôpitaux de Paris | ● Discontinuation of RAS blocker therapy ● Continuation of RAS blocker therapy |
Not yet recruiting | 554 ≤18 |
NCT04329195 |
| 3 | OncoImmune, Inc. | ● CD24Fc | Not yet recruiting | 230 ≤18 |
NCT04317040 |
| Not applicable | University Health Network, Toronto | ● Ruxolitinib | Not yet recruiting | 64 ≤12 |
NCT04331665 |
| 2 | National and Kapodistrian University of Athens | ● Colchicine ● Standard treatment |
Not yet recruiting | 180 ≤18 |
NCT04326790 |
| 3 | Assistance Publique - Hôpitaux de Paris | ● Usual practice with SYMBICORT RAPIHALER ● Usual practice |
Not yet recruiting | 436 18–75 |
NCT04331054 |
| Not applicable | Wuhan Union Hospital | ● Immunoglobulin of cured patients ● γ-Globulin |
Not yet recruiting | 10 ≤18 |
NCT04264858 |
| 3 | Shanghai Jiao Tong University School of Medicine | ● Recombinant human interferon alpha-1b ● Thymosin alpha 1 |
Recruiting | 2944 18–65 |
NCT04320238 |
| 3 | Misr University for Science and Technology | ●Dietary supplement: natural honey ● Standard care |
Not yet recruiting | 1000 5–75 |
NCT04323345 |
| 1, 2 | Chinese Academy of Sciences | ● CAStem | Recruiting | 9 18–70 |
NCT04331613 |
| 2 | Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins | ● Anti-SARS-CoV-2 plasma ● SARS-CoV-2 non-immune plasma |
Not yet recruiting | 150 ≤18 |
NCT04323800 |
| 2 | Universidad del Rosario | ● Plasma | Not yet recruiting | 10 18–60 |
NCT04332380 |
| 3 | Assistance Publique - Hôpitaux de Paris | ● Naproxen ● Standard of care |
Not yet recruiting | 584 ≤18 |
NCT04325633 |
| 2, 3 | Universidad del Rosario | ● Plasma ● Hydroxychloroquine ● Azithromycin |
Not yet recruiting | 80 18–60 |
NCT04332835 |
| 3 | Estudios Clínicos Latino América | ● Colchicine ● Local standard of care |
Not yet recruiting | 2500 ≤18 |
NCT04328480 |
| Not applicable | Mazandaran University of Medical Sciences | ● Convalescent plasma | Enrolling by invitation | 30 30–70 |
NCT04327349 |
| 2 | Zhongnan Hospital | ● Vitamin C ● Sterile water for injection |
Recruiting | 140 ≤18 |
NCT04264533 |
| 3 | Hospital Sirio-Libanes | ● Dexamethasone | Not yet recruiting | 290 ≤18 |
NCT04327401 |
| 3 | Hospital of Prato | ● Baricitinib | Recruiting | 60 18–80 |
NCT04320277 |
| Not applicable | Jiangxi Qingfeng Pharmaceutical Co. Ltd. | ● Xiyanping injection ● Lopinavir with ritonavir, alpha-interferon nebulization |
Not yet recruiting | 348 18–70 |
NCT04275388 |
| 2, 3 | University of Trieste | ● Methylprednisolone ● Standard care |
Recruiting | 104 18–80 |
NCT04323592 |
| 2 | Mayo Clinic | ● Convalescent plasma | Not yet recruiting | 20 ≤18 |
NCT04325672 |
| 2 | Upinder Singh, Stanford University | ● Peginterferon lambda-1a ● Standard of care treatment |
Not yet recruiting | 120 18–64 |
NCT04331899 |
| 3 | Montreal Heart Institute | ● Colchicine | Recruiting | 6000 ≤40 |
NCT04322682 |
| 2 | Lucio Manenti, Azienda Ospedaliero-Universitaria di Parma | ● Colchicine | Not yet recruiting | 100 18–85 |
NCT04322565 |
| 2 | National Cancer Institute, Naples | ● Tocilizumab injection | Recruiting | 330 All |
NCT04317092 |
| 3 | Hoffmann-La Roche | ● Tocilizumab (TCZ) | Not yet recruiting | 330 ≤18 |
NCT04320615 |
| 2 | Università Politecnica delle Marche | ● Tocilizumab | Not yet recruiting | 30 18–90 |
NCT04315480 |
| 1 | Hospital San Jose Tec de Monterrey | ● Convalescent plasma | Not yet recruiting | 20 ≤18 |
NCT04333355 |
| 2 | Massachusetts General Hospital | ● Nitric oxide gas | Recruiting | 220 18–99 |
NCT04306393 |
| Not applicable | Beijing 302 Hospital | ● Conventional medicines and traditional Chinese medicines granules ● Conventional medicines and lopinavir with ritonavir |
Not Applicable | 150 14–80 |
NCT04251871 |
| 2 | Frederiksberg University Hospital | ● RoActemra iv ● RoActemra sc ● Kevzara sc ● Standard medical care |
Not yet recruiting | 200 ≤18 |
NCT04322773 |
| 2, 3 | Assistance Publique - Hôpitaux de Paris | ● Sarilumab | Recruiting | 240 ≤18 |
NCT04324073 |
| 2 | University of British Columbia | ● Nitric oxide 0.5% with nitrogen 99.5% gas for inhalation | Active, not recruiting | 20 ≤14 |
NCT03331445 |
| 3 | Université de Sherbrooke | ● Vitamin C ● Control |
Recruiting | 800 ≤18 |
NCT03680274 |
| 2, 3 | Swedish Orphan Biovitrum | ● Emapalumab ● Anakinra |
Not yet recruiting | 54 30–79 |
NCT04324021 |
Exaggerated immune and inflammatory responses are considered to be responsible for the severity of symptoms and a poor clinical outcome of coronavirus infections. Interferons have shown to play a crucial role in the defense against coronavirus diseases. A less efficient interferon-mediated immune response can explain the increased mortality rates in the elderly. Earlier induction of interferons in children and their less developed immune system could be the reasons behind their zero or near to zero fatality rate. Administration of interferon-inducing agents could reduce the mortality of SARS at a very early stage of the disease. Adding interferon-γ to an interferon-I, as a synergistic combination therapy, might maximize the benefits [151]. There are currently several interferons employed in clinical settings that could provide a therapy for SARS-CoV-2. Furthermore, nitric oxide (NO) is a selective pulmonary vasodilator and holds promise as an anti-inflammatory agent [152]. NO is a critical cellular signaling molecule synthesized by nitric oxide synthase (NOS). In the pulmonary airways, NOS is present in a variety of cells, including neurons, macrophages, airway epithelial cells, and vascular endothelial cells. NOS activity is critical to mediate smooth muscle relaxation, neurotransmission, mucin secretion, and is also a well-known mediator in the cellular response to microbial infection [153]. Various inflammatory factors, such as cytokines and LPS, can induce high and sustained NO production. Inducible nitric oxide synthase activity can result in anti-inflammatory or pro-inflammatory responses, cytoprotection, or cytotoxicity, depending on the circumstances [154]. Inhaled NO results in a transient improvement in systemic oxygenation. There are no published data from trials that describe the use of pulmonary vasodilators in COVID-19 patients. However, a previous review showed ARDS treatment by inhaled NO had no significant effect on mortality and increased the likelihood of acute kidney injury [155]. Several clinical trials are underway to determine whether inhaled NO can improve oxygenation in SARS-CoV-2 patients (Table 6). In addition, it was proposed that treatment with statins, a multifunctional class of drugs with several potential applications, could inhibit MyD88 signaling and NF-κB response. This could inhibit inflammatory responses that would lead to ameliorated disease progression in COVID19 patients. There is evidence that down-regulation of NF-κB signaling could increase survival in mouse models of SARS-CoV infection [156,157]. A number of immunotherapies that have been proposed as a treatment for SARS-CoV-2 are currently undergoing clinical trials (Table 6).
Melatonin is a neurohormone produced by the pineal gland. This molecule has many beneficial activities, including immunomodulatory, anti-inflammatory, antioxidant properties within the body [158,159]. Because of these functions, some researchers have proposed this agent could be a therapeutic option for treating viral infections and respiratory diseases, including ARDS and acute lung injury (ALI) [160]. The mechanisms of action of melatonin (which has an excellent safety profile) include, at least in part, reducing anxiety, improving sleep, and modulating vascular permeability, which may be useful in improving prognosis of SARS-CoV-2 patients [161].
8. Cell-based therapies
ARDS is a medical condition characterized by severe uncontrolled inflammation in the lungs, which causes disturbances in the surfactant and the pulmonary capillary endothelial cells, resulting in fluid accumulation in the distal parts of the lung [162]. The beneficial effects of using mesenchymal stem cells (MSCs), including their immunomodulatory, antimicrobial, and antiapoptotic properties, have been reported in previous studies. In particular, the immunomodulatory functions of these cells are among their most relevant properties. It has been shown that although histocompatibility complex (MHC) class I molecules are expressed on human MSCs, they lack class II MHC molecules, rendering these cells hypoimmunogenic or “immune-privileged” properties. Through this property, MSCs can escape immune recognition by T helper (CD4+) lymphocytes and immune destruction by natural killer cells. MSCs can regulate the activity of both the innate and adaptive immune responses through either cell-cell interaction, secretion of trophic factors, or activation of regulatory T cells. In animal models and in vitro studies, MSCs have been shown to promote non-specific immune reactions (i.e., innate immunity) through either directly killing bacteria or engaging multiple antimicrobial mediators such as LL-37, lipocalin-2, and beta-defensin-2 via the toll-like receptor 4 signaling pathway [163]. MSCs are also capable of fighting microbial agents by activating tryptophan metabolism by increasing indoleamine 2,3-dioxygenase activity [164]. They are known to support the survival and function of neutrophils and macrophages. This activity is mediated by transforming the macrophage phenotype to the anti-inflammatory (type 2) from the pro-inflammatory (type 1). MSCs mediate many processes, including secretion of growth factors that target vascular cells, hepatocytes, neurons, and other cells, altering the balance of anti/proapoptotic genes, changing mitochondrial biology, and microvesicle transfer [165]. MSCs can trigger antiapoptotic proteins both in vivo (animal models of renal, cerebral, and cardiac injuries) and also in vitro. On the other hand, MSCs can promote autophagy, another form of programmed cell death, through the phosphoinositide 3-kinase/protein kinase B signaling pathway. This function, along with another phenomenon known as mitophagy (i.e., selective degradation of mitochondria), has been shown to be important for MSCs to carry out their protective role against oxidative damage to the lungs. Xu et al. reported the pathological characteristics of a biopsy sample obtained at autopsy from a SARS-CoV-2 patient with severe ARDS [166]. Unfortunately, age-related loss of the capacity of the lung tissue to self-repair may explain the progressive age-related mortality reported in older SARS patients with ARDS [167,168]. No therapeutic drugs have yet been approved for the treatment of ARDS [169]. Hence, current treatment strategies are mainly based on supportive care, such as prone positioning, conservative fluid replacement, and lung-protective ventilation [170]. Therefore, the utilization of stem cells in experimental protocols may be considered for coronavirus diseases, including SARS, MERS, and SARS-CoV-2 in human medicine.
Cell-based therapies, especially stem cells, could have both curative and preventive potential in COVID19. Cell-based therapies may be an innovative treatment for ARDS through several pathways that may augment recovery from lung injury and reduce the magnitude of lung damage [171]. Several types of stem cells have been considered for clinical use, such as cord blood mesenchymal stem cells (CBMSCs) [172], umbilical cord mesenchymal stem cells (HUCMSCs) [172,173], umbilical cord Wharton's Jelly derived-mesenchymal stem cells (UCWJDMSCs) [174], umbilical cord-derived mesenchymal stem cells (UCMSCs) [175], umbilical cord blood mononuclear cells (UCBMCs) [176], and human menstrual blood-derived stem cells (HMBSCs) [177]. MSC-based treatment could help ARDS through inhibition of collagen accumulation, reducing alveolar cell apoptosis, and preventing the collapse of lung airways. Some of the most attractive properties of MSCs are the ability to modulate immune responses, antimicrobial properties, anti-apoptotic effects, and robust regenerative potential (Fig. 9 ). It remains highly controversial as to which of several MSC mediators are involved in the therapeutic response [178]. MSC secreted factor candidates for ARDS treatment, include angiopoietin-1 [179], hepatocyte growth factor (HGF) [180], keratinocyte growth factor (KGF) [181], insulin growth factor (IGF) [182], interleukin 1 receptor antagonist (IL-1RN) [183], interleukin 10 (IL-10) [184], interleukin 6 (IL-6), tumor necrosis factor-stimulated gene 6 (TSG-6) [185], lipoxin A4 [186], prostaglandin E2 (PGE2) [175], lipocalin-2 [187], β-defensin-2 [188], stanniocalcin-1a [189] and extracellular vesicles [190]. Recent studies have suggested that there is a direct correlation between the regulation of these bioactive factors after engraftment of MSCs in the lungs after ARDS [178]. In summary, a large body of preclinical and clinical evidence is accumulating on the use of MSCs for the treatment of ARDS.
Fig. 9.
The therapeutic effects of MSCs and their secreted factors for SARS-CoV-2-related ARDS. The diverse immunological and biological function of mesenchymal stem cells. MSCs promote anti-apoptotic effects mainly by inducing growth factors and directly or indirectly reducing factors that damage cells (ROS, etc.). A primary function of MSCs is to modulate immune responses by activating effector T cells (either CD4+ or CD8+) and regulating the function and proliferation of regulatory T (Tregs) cells. MSCs also affect cellular adaptors of the innate immune response (mainly neutrophils and macrophages). In particular, in response to induction by MSCs, macrophages are phenotypically transformed from a pro- to anti-inflammatory state. All immunomodulatory functions of MSCs finally result in a potent anti-microbial response resulting in microbial clearance in part by activation of lipocaline-2 and cathelicidin. By inducing the secretion of a variety of growth factors, MSCs can potentiate the regeneration of pulmonary alveoli and reduce lung fibrosis. Molecular adaptors of these effects of MSCs are shown. MSCs: mesenchymal stem cells; M1 and M2: M1 and M2 macrophages; B: bacteria; N: neutrophil; D: dendritic cell; NK: natural killer cell; EP: epithelial cell; EN: endothelial cell; EVs: extracellular vesicles; PGE2: prostaglandin E2; HGF: hepatocyte growth factor; IL-1ra: interleukin 1 receptor antagonist; IL-10: interleukin 10; IL-6: interleukin 6; TSG-6: tumor necrosis factor-inducible gene 6 protein; IGF: insulin growth factor; STC-1: stanniocalcin 1; Ang-1: angiopoietin 1; KGF: keratinocyte growth factor; ROS: reactive oxygen species.
Stem cell therapy, especially MSCs, has been shown to reduce pulmonary inflammation and affect pulmonary tissue regeneration. Therefore, it is investigated in ARDS patients. Wilson et al. found that the administration of allogeneic MSCs to ARDS patients resulted in no pre-specified adverse events, including ventricular tachycardia, hypoxemia, and cardiac arrhythmia [191]. Recently, results from Chen et al. suggested that MSCs could notably improve the survival rate of influenza virus subtype H7N9-induced ARDS, which provides a theoretical basis for the treatment of ARDS patients [192]. Of note, coronavirus-induced ARDS shares similar complications and corresponding organ failure to influenza H7N9, so MSCs therapy could be a possible alternative for treating COVID19 patients. Clinical and pre-clinical trials conducted in China have demonstrated that MSCs are naturally resistant to SARS-CoV-2 infection, and transplantation of these cells could improve the outcome in SARS-CoV-2 patients. For instance, Leng et al. investigated whether MSC transplantation affected the outcome of SARS-CoV-2 patients in Beijing Youan Hospital, China. The results showed that MSC therapy could remarkably improve the outcome of SARS-CoV-2 patients without any adverse events (Fig. 10 ). After MSC therapy, the C-reactive protein (CRP) levels decreased, the peripheral blood lymphocytes were increased, and the over-activated cytokine-secreting immune cells CXCR3+ CD8+ T cells and CXCR3+ NK cells CXCR3+ CD4+ T cells disappeared within one week. Moreover, the level of IL-10 increased, while plasma inflammatory factor TNF-α was significantly decreased compared to the control group. Furthermore, MSCs do not express TMPRSS2 and ACE2, explaining why MSCs are resistant to infection by SARS-CoV-2 [193]. Although various clinical trials have been commenced to test the effects of MSCs in severe SARS-CoV-2 patients, convincing positive results have yet to be reported. The upcoming clinical trials to evaluate the safety and effectiveness of MSCs for the treatment of SARS-CoV-2 patients are summarized in Table 8 .
Fig. 10.
MSCs improve the outcome of SARS-CoV-2 patients with ARDS. a) Chest computed tomography images of the severe SARS-CoV-2 patient. b) The pattern of serum cytokine/chemokine/growth factors. c) The profile of the over-activated NK cells and T cells of SARS-CoV-2 patients.
Reproduced with permission from [193].
Table 8.
Selected cell-based therapies against SARS-CoV-2 in clinical trials.
| Phase | Responsible party | Interventions | Recruitment status | Population (enrollment and age) | NCT number |
|---|---|---|---|---|---|
| 1, 2 | Beijing 302 Hospital | ● MSCs ● Saline containing 1% human serum albumin (solution of MSCs) |
Recruiting | 90 18–75 |
NCT04288102 |
| 1 | Beijing 302 Military Hospital | ● MSCs | Recruiting | 20 18–70 |
NCT04252118 |
| Not applicable | Puren Hospital Affiliated to Wuhan University of Science and Technology | ● UC-MSCs | Recruiting | – 18–75 |
NCT04293692 |
| Early 1 | CAR-T (Shanghai) Biotechnology Co., Ltd. | ● Dental pulp MSCs | Not yet recruiting | 24 18–75 |
NCT04302519 |
| 2 | Zhongnan Hospital | Biological: UC-MSCs | Recruiting | 10 18–75 |
NCT04269525 |
| 2 | Tianhe Stem Cell Biotechnologies Inc. | Combination product: stem cells and mononuclear cells isolated by apheresis | Not yet recruiting | 20 18–60 |
NCT04299152 |
| 1 | Ruijin Hospital | ● MSCs-derived exosomes | Not yet recruiting | 30 18–75 |
NCT04276987 |
| 1 | Azidus Brasil | ● NestCell® | Not yet recruiting | 66 ≤18 |
NCT04315987 |
| Not applicable | Wuhan Union Hospital | ● UC-MSCs | Not yet recruiting | 48 18–60 |
NCT04273646 |
| 1, 2 | Chongqing Public Health Medical Center | ● NK cells, IL15-NK cells, NKG2D CAR-NK cells, ACE2 CAR-NK cells and NKG2D-ACE2 CAR-NK cells | Recruiting | 90 ≤18 |
NCT04324996 |
| 1 | Stem Cells Arabia | ● WJ-MSCs | Recruiting | 5 ≤18 |
NCT04313322 |
| 1 | Xinxiang medical university | ● NK cells | Recruiting | 30 18–65 |
NCT04280224 |
9. The emergence of nanomedicine as a new therapeutic strategy in SARS-CoV-2 treatment
Treatment of pulmonary infectious diseases using nanoparticles has recently attracted significant attention [194,195]. Small molecule drugs have disadvantages such as lack of targeting to lungs, difficulties with stability during storage and administration, and considerable expense [196,197]. Upon reaching the lungs, drugs may be enzymatically degraded by pulmonary enzymes. In addition, the pulmonary airways are a mucus-covered epithelial bed, which can act as a barrier preventing the penetration of drugs into the lungs [198]. However, therapeutic nanoparticles may act as an alternative delivery platform to the lungs, depending on physiological parameters (respiratory rate and lung volume) and the pathophysiological state (disease nature). Considering the particle size, the target tissue, and respiratory rate, various mechanisms can be employed to deliver therapeutic agents into the lungs. In the pulmonary alveoli, the clearance rate of particles is mainly determined by the size [199]. Furthermore, vaccines must be equipped with efficient molecules (i.e., new generation composite vaccines) to potentiate their immunogenic and adjuvant activities [200]. To address some of these issues, nano-delivery platforms could provide a viable option as they are designed to allow protection against biological degradation, better stability, and higher efficiency (synergistic effects) [201,202], to improve the effectiveness of therapeutic agents [203]. Therefore, nanoparticles (NPs) are being investigated as carriers for targeting drugs to treat a variety of pulmonary infectious diseases.
To develop effective vaccines for coronavirus diseases (in particular the novel COVID-19), we can use NPs as targeted carriers. Furthermore, NPs have the potential to be employed to develop therapeutic and diagnostic (i.e., biosensor) systems. Recent global pandemics caused by coronaviruses (SARS-CoV, MERS-CoV, and now COVID-19) have underlined the necessity of rapidly developing effective vaccines. Using a novel procedure involving protein-protein micellar NPs, Coleman et al. produced an adjuvant-conjugated vaccine against SARS-CoV S and MERS-CoV S proteins. These researchers used first cultured Baculovirus insect cells to produce the complete S protein, which was then self-assembled into NPs. For potentiating the production of neutralizing antibodies by immune cells, they used adjuvants (aluminum hydroxide (alum) or Matrix M1) [204]. Anti-MERS-CoV immunoglobulins (recognizing MERS-CoV S protein) were detected in the mice treated with MERS-CoV S NPs. The immune response was augmented by using the Matrix-M1 adjuvant triggering the high production of anti-S neutralizing antibody conferring immune protection against MERS-CoV infection in mice [205]. Jung et al. also produced a NP based vaccine with the MERS-CoV S protein gene (Ad5/MERS) and MERS S protein, using a heterologous prime-boost immunization strategy. They used a recombinant adenovirus serotype 5 to conjugate the antigens to the NPs. The vaccines were proved to be able to activate Th1/Th2 lymphocytes in an appropriate ratio [206]. In another study, Roh et al. used a combination of SARS-CoV N protein inhibitors and NP-based RNA oligonucleotides to develop a vaccine against the virus. By applying RNA oligonucleotide conjugated to QDs on a biochip, they showed that the SARS-CoV N protein was effectively suppressed by (−)-catechin gallate and (−)-gallocatechin gallate through a dose-dependent attenuation of its binding affinity [207].
Biosensors are used to detect and quantify biological responses and are now widely used in medical diagnostic procedures as point-of-care instruments [208,209]. Biosensor-based systems are effective, simple, reliable, and relatively inexpensive platforms that can be used in clinical settings. Using these systems, the sensitivity and reproducibility of clinical analysis can be maximized [210,211]. Nanobiosensors were first applied to detect antibody mimicking proteins (AMPs) by Ishikawa et al. who configured In2O3 nanowire based-biosensors with an antimicrobial peptide (fibronectin, Fn). Using bovine serum albumin (44 μM) as the control, this system was able to detect sub-nanomolar levels of SARS-CoV nucleocapsid (N) protein [212]. In addition, point-of-care fluorescent and colorimetric systems have been designed to detect DNA and RNA molecules, especially in less equipped laboratories. In this regard, MERS-CoV DNA was successfully detected using a colorimetric assay (AgNPs were served as a colorimetric agent). In this assay, the viral DNA was detected based on acpcPNA-induced NP aggregation, which was highly sensitive, and did not bind non-complementary nucleic acids (either single, two, or full-length mismatch) [213]. Moreover, Kim et al. developed a colorimetric-based method which was able to detect MERS-CoV DNA (length of 30 bp) as low as 1 pmol/μL using self-complementary double-stranded DNA (dsDNA) shielded with gold NPs [214]. These medical diagnostic strategies could provide cheap and disposable alternatives to detect and screen for the presence of coronaviruses.
Because NPs-based vaccines can effectively trigger the humoral immune response and act as a versatile antigen-presenting system, they can be effectively used to increase immunity against the viruses that cause acute respiratory syndromes [215]. Vaccines that have been developed using mRNAs benefit from advantages, such as mimicking natural infection and triggering a more potent immune response. In addition, there is a possibility to incorporate multiple mRNAs into a single vaccine. The SARS-CoV-2 S protein (140 kDa) is a peptide with 1273 residues [216], and its respective mRNA has been employed to develop an effective vaccine. In this approach, liquid NPs and chemical modification were used to stabilize an injectable form of the mRNA-based vaccine using soluble nanoparticles. This lipid nanoparticle (LNP)-encapsulated mRNA vaccine is being tested in Phase 1 clinical trial (NCT04283461) sponsored by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) in the US. This novel vaccine was designed by Moderna in collaboration with investigators at the NIAID Vaccine Research Center to identify an antigen linked to the SARS-CoV-2 prefusion-stabilized S protein [217]. In February 2020, the Novavax Company, which had already worked on MERS and SARS, also announced the start of animal studies on potential candidates to produce a SARS-CoV-2 vaccine. They used SARS-CoV-2 derived S protein along with Matrix-M adjuvant in a recombinant nanoparticle technology [217]. A trial for the efficacy of adipose mesenchymal stem cell-derived exosomes (MSCs-Exo) (through inhalation of an aerosol) in the treatment of severely ill patients with SARS-CoV-2 infection is underway (NCT04276987). These exosomes are known to mitigate lung inflammation and reduce injury in various pathological conditions, as well as to increase the phagocytic activity and killing potency of macrophages.
10. Concluding remarks
This review has provided an overview of therapeutic agents designed to target SARS-CoV-2 based on an extensive search of ClinicalTrials.gov and medical databases, with a focus on clinical trials. Considering the rapid progression of research in this field, it will be difficult to remain completely up to date. However, the effectiveness of the most recent clinical trials have been collected and potential research areas have been proposed to extend the range of therapeutic candidates against SARS-CoV-2. Most antiviral strategies against SARS-CoV-2, which have been studied in pre-clinical and clinical trials, are already used medicines against other RNA viruses, such as SARS-CoV, MERS-CoV, influenza, HCV, and Ebola. In addition, this review has also included an overview of SARS-CoV-2 biology and antiviral therapies that have exploited several nanoscale agents were explored. Nanomedicine-based therapies (bioengineered and vectored antibodies, cytokines, and vaccines) have shown some promise for the treatment of SARS-CoV-2 infections. We further discussed the complex cellular interactions responsible for SARS-CoV-2 cell entry and replication, as well as drugs that can target these pathways. Vaccine platforms, passive immunotherapy, and cell-based therapies were also covered.
Currently in use or investigational agents for SARS-CoV-2 target either the virus or host-derived molecules (e.g., interferons, glucocorticoids). The combination of lopinavir and ritonavir might treat the specific virus, and SARS-CoV-2 receptor inhibitors may be helpful in reducing lung cell viral entry and improve lung function in COVID19 patients. Some patients infected with SARS-CoV-2 were significantly improved after treatment with the lopinavir in combination with ritonavir. However, other case reports showed that treatment with lopinavir and ritonavir did not significantly alleviate SARS-CoV-2 related pneumonia [218]. Although there were no reports of acute respiratory failure in these patients, it is questionable whether this was solely related to the antiviral drugs or not [69]. Regardless, lopinavir plus ritonavir is still considered a viable treatment for SARS-CoV-2 infection. Cell entry-based therapeutics, such as chloroquine, hydroxychloroquine, anti-ACE2, and anti-CD147 antibodies could also guide us towards the discovery of new treatments for SARS-CoV-2 infection. These therapeutic modalities could also be useful to reveal the fundamental pathways of SARS-CoV-2 replication.
There is also interest in testing whether immunotherapy or biological therapies, such as convalescent plasma and hyperimmune globulin, containing antibodies isolated from blood donated by people who have recovered from COVID19, could shorten the length or reduce the severity of the illness. Furthermore, by studying the protective function of memory immune cells from recovered patients, it may be possible to develop prophylactic and therapeutic approaches to cope with future coronaviruses outbreaks. Treatment of SARS-CoV-2 infection with antibody-based and vaccine-based therapies requires an understanding of how neutralizing antibodies identify and eliminate the virus. The role of humoral immune responses in preventing SARS-CoV-induced lung damage is controversial. In fact, there have been reports of SARS-related mortality in patients who have developed strong neutralizing antibodies. Because these antibodies have been associated with the secretion of proinflammatory cytokines in the lungs, it has been hypothesized that they may be linked to fatal acute inflammatory lung injury. In addition, SARS-CoV-2 may be effectively treated with the administration of MSCs, which are supposed to possess paracrine and immunomodulatory effects on immune cells. It will be necessary to perform randomized clinical trials to assess the effects of MSCs as a potential treatment for COVID19. The immunomodulatory and anti-inflammatory effects of MSCs could be enhanced by pre-exposure to factors such as IFNγ in the presence or absence of TNF-α or IL-1 (i.e., licensing-approach). This is important because T cells are generally poorly activated in patients with SARS-CoV-2 infection, and therefore stimulating factors, such as interferons, may be low in these patients. The cytokine-licensed MSCs can effectively inhibit hyperactive immune responses and promote tissue repair.
In conclusion, despite many studies in humans, there is not yet an optimal treatment for SARS-CoV-2 infection. Detailed laboratory studies and further clinical trials will be required to establish evidence-based treatment for patients with SARS-CoV-2 as well as ARDS.
CRediT authorship contribution statement
F.O. and A.M. wrote the article. A.H., N.H., B.B., H.B.B., M.A.S, M.R.H., H.A.S. contributed in the correction and editing of the manuscript.
Declaration of competing interest
MRH declares the following potential conflicts of interest. Scientific Advisory Boards: Transdermal Cap Inc., Cleveland, OH; BeWell Global Inc., Wan Chai, Hong Kong; Hologenix Inc. Santa Monica, CA; LumiThera Inc., Poulsbo, WA; Vielight, Toronto, Canada; Bright Photomedicine, Sao Paulo, Brazil; Quantum Dynamics LLC, Cambridge, MA; Global Photon Inc., Bee Cave, TX; Medical Coherence, Boston MA; NeuroThera, Newark DE; JOOVV Inc., Minneapolis-St. Paul MN; AIRx Medical, Pleasanton CA; FIR Industries, Inc. Ramsey, NJ; UVLRx Therapeutics, Oldsmar, FL; Ultralux UV Inc., Lansing MI; Illumiheal & Petthera, Shoreline, WA; MB Lasertherapy, Houston, TX; ARRC LED, San Clemente, CA; Varuna Biomedical Corp. Incline Village, NV; Niraxx Light Therapeutics, Inc., Boston, MA. Consulting; Lexington Int, Boca Raton, FL; USHIO Corp, Japan; Merck KGaA, Darmstadt, Germany; Philips Electronics Nederland B.V. Eindhoven, Netherlands; Johnson & Johnson Inc., Philadelphia, PA; Sanofi-Aventis Deutschland GmbH, Frankfurt am Main, Germany. Stockholdings: Global Photon Inc., Bee Cave, TX; Mitonix, Newark, DE.
Acknowledgements
The authors are grateful for financial supports from the Immunology Research Center, Tabriz University of Medical Sciences. M.-A. Shahbazi acknowledges the financial support from the Academy of Finland (grant no. 317316). M.R. Hamblin was supported by US NIH Grants R01AI050875 and R21AI121700.
References
- 1.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005;24(11):S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 2.Geller C., Varbanov M., Duval R.E. Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses. 2012;4(11):3044–3068. doi: 10.3390/v4113044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Hoek L. Human coronaviruses: what do they cause? Antivir. Ther. 2007;12(4 Pt B):651. [PubMed] [Google Scholar]
- 4.de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A. Commentary: Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C.S.G. of the International The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020:1. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.W.H. Organization . 2020, April 5. Coronavirus Disease 2019 (COVID-19): Situation Report-76. [Google Scholar]
- 7.Sola I., Almazan F., Zuniga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annual review of virology. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013;100(1):246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.C.S.M.E. Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandekar A.A., Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.B.G. National Research Project for SARS The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am. J. Clin. Pathol. 2004;121(4):507–511. doi: 10.1309/WPK7Y2XKNF4CBF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass W.G., Subbarao K., Murphy B., Murphy P.M. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173(6):4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 15.Chen J.-H., Chang Y.-W., Yao C.-W., Chiueh T.-S., Huang S.-C., Chien K.-Y., Chen A., Chang F.-Y., Wong C.-H., Chen Y.-J. Plasma proteome of severe acute respiratory syndrome analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proc. Natl. Acad. Sci. 2004;101(49):17039–17044. doi: 10.1073/pnas.0407992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiegel M., Pichlmair A., Martínez-Sobrido L., Cros J., García-Sastre A., Haller O., Weber F. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005;79(4):2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yilla M., Harcourt B.H., Hickman C.J., McGrew M., Tamin A., Goldsmith C.S., Bellini W.J., Anderson L.J. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107(1):93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., Nicholls J.M., Peiris J.M., Lau Y.L. Chemokine up-regulation in sars-coronavirus–infected, monocyte-derived human dendritic cells. Blood. 2005;106(7):2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y.-D., Sin W.-Y.F., Xu G.-B., Yang H.-H., Wong T.-y., Pang X.-W., He X.-Y., Zhang H.-G., Ng J.N.L., Cheng C.-S.S. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J. Virol. 2004;78(11):5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong R.S., Wu A., To K., Lee N., Lam C.W., Wong C., Chan P.K., Ng M.H., Yu L., Hui D.S. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. Bmj. 2003;326(7403):1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsueh P.-R., Huang L.-M., Chen P.-J., Kao C.-L., Yang P.-C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10(12):1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laureti M., Narayanan D., Rodriguez-Andres J., Fazakerley J.K., Kedzierski L. Flavivirus receptors: diversity, identity, and cell entry. Front. Immunol. 2018;9:2180. doi: 10.3389/fimmu.2018.02180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazzon M., Marsh M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Res. 2019:8. doi: 10.12688/f1000research.19694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorley J.A., McKeating J.A., Rappoport J.Z. Mechanisms of viral entry: sneaking in the front door. Protoplasma. 2010;244(1–4):15–24. doi: 10.1007/s00709-010-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulant S., Stanifer M., Lozach P.Y. Dynamics of virus-receptor interactions in virus binding, signaling, and endocytosis. Viruses. 2015;7(6):2794–2815. doi: 10.3390/v7062747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tikellis C., Bernardi S., Burns W.C. Angiotensin-converting enzyme 2 is a key modulator of the renin-angiotensin system in cardiovascular and renal disease. Curr. Opin. Nephrol. Hypertens. 2011;20(1):62–68. doi: 10.1097/MNH.0b013e328341164a. [DOI] [PubMed] [Google Scholar]
- 32.Clarke N.E., Turner A.J. Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012;2012:307315. doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H., Geng J.-J., Wang B., Sun X.-X., Wang C.-F., Yang X., Lin P., Deng Y.-Q., Wei D., Yang X.-M., Zhu Y.-M. 2020. SARS-CoV-2 Invades Host Cells Via a Novel Route: CD147-spike Protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindolet D., Gabison E. Role of CD147 (EMMPRIN/basigin) in tissue remodeling. Anat. Rec. 2020;367:1444–1448. doi: 10.1002/ar.24089. [DOI] [PubMed] [Google Scholar]
- 38.Dolin H.H., Papadimos T.J., Chen X., Pan Z.K. Characterization of pathogenic sepsis etiologies and patient profiles: a novel approach to triage and treatment. Microbiol Insights. 2019;12 doi: 10.1177/1178636118825081. (1178636118825081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W. Antibody response and viraemia during the course of severe acute respiratory syndrome (SARS)-associated coronavirus infection. J. Med. Microbiol. 2004;53:435–438. doi: 10.1099/jmm.0.45561-0. [DOI] [PubMed] [Google Scholar]
- 40.Patel V.B., Zhong J.C., Grant M.B., Oudit G.Y. Role of the ACE2/angiotensin 1-7 axis of the renin-angiotensin system in heart failure. Circ. Res. 2016;118(8):1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grass G.D., Toole B.P. How, with whom and when: an overview of CD147-mediated regulatory networks influencing matrix metalloproteinase activity. Biosci. Rep. 2015;36(1):e00283. doi: 10.1042/BSR20150256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8(1):15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020;177 doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287(12):8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamauchi Y., Greber U.F. Principles of virus uncoating: cues and the snooker ball. Traffic. 2016;17(6):569–592. doi: 10.1111/tra.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shang J., Wan Y., Liu C., Yount B., Gully K., Jian Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16:e1008392. doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4(4):557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobhy H. A comparative review of viral entry and attachment during large and giant dsDNA virus infections. Arch. Virol. 2017;162(12):3567–3585. doi: 10.1007/s00705-017-3497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hantak M.P., Qing E., Earnest J.T., Gallagher T. Tetraspanins: architects of viral entry and exit platforms. J. Virol. 2019;93(6) doi: 10.1128/JVI.01429-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Millet J., Whittaker G. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2005;102:11876–11881. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102(33):11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adedeji A., Severson W., Jonsson C., Singh K., Weiss S., Sarafianos S. Novel inhibitors of SARS-CoV entry acting by three distinct mechanisms. J. Virol. 2013;87 doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Struck A.-W., Axmann M., Pfefferle S., Drosten C., Meyer B. A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antivir. Res. 2012;94:288–296. doi: 10.1016/j.antiviral.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abraham H.M., White C.M., White W.B. The comparative efficacy and safety of the angiotensin receptor blockers in the management of hypertension and other cardiovascular diseases. Drug Saf. 2015;38(1):33–54. doi: 10.1007/s40264-014-0239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020;81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J.Y. Letter to the editor: case of the index patient who caused tertiary transmission of coronavirus disease 2019 in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35(7):e88. doi: 10.3346/jkms.2020.35.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhatnagar T., Murhekar M.V., Soneja M., Gupta N., Giri S., Wig N., Gangakhedkar R. Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: protocol for restricted public health emergency use. Indian J. Med. Res. 2020;151:184–189. doi: 10.4103/ijmr.IJMR_502_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim J., Jeon S., Shin H.Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.W., Kang Y.M., Lee B., Park S.J. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J. Korean Med. Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hama R., Bennett C.L. The mechanisms of sudden-onset type adverse reactions to oseltamivir. Acta Neurol. Scand. 2017;135(2):148–160. doi: 10.1111/ane.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lung J., Lin Y.-S., Yang Y.-H., Chou Y.-L., Shu L.-H., Cheng Y.-C., Liu H.T., Wu C.-Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92:693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 77.De Clercq E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chem. Asian J. 2019;14(22):3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arabi Y.M., Balkhy H.H., Hayden F.G., Bouchama A., Luke T., Baillie J.K., Al-Omari A., Hajeer A.H., Senga M., Denison M.R., Nguyen-Van-Tam J.S., Shindo N., Bermingham A., Chappell J.D., Van Kerkhove M.D., Fowler R.A. Middle East respiratory syndrome. N. Engl. J. Med. 2017;376(6):584–594. doi: 10.1056/NEJMsr1408795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., Swanstrom J., Sheahan T.P., Pickles R.J., Corti D., Randell S.H., Lanzavecchia A., Marasco W.A., Baric R.S. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U. S. A. 2016;113(11):3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Agostini M., Andres E., Sims A., Graham R., Sheahan T., Lu X., Smith E., Case J., Feng J., Jordan R., Ray A., Cihlar T., Siegel D., Mackman R., Clarke M., Baric R., Denison M. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Menzel M., Akbarshahi H., Bjermer L., Uller L. Azithromycin induces anti-viral effects in cultured bronchial epithelial cells from COPD patients. Sci. Rep. 2016;6 doi: 10.1038/srep28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Retallack H., Di Lullo E., Arias C., Knopp K.A., Laurie M.T., Sandoval-Espinosa C., Mancia Leon W.R., Krencik R., Ullian E.M., Spatazza J., Pollen A.A., Mandel-Brehm C., Nowakowski T.J., Kriegstein A.R., DeRisi J.L. Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. U. S. A. 2016;113(50):14408–14413. doi: 10.1073/pnas.1618029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bosseboeuf E., Aubry M., Nhan T., Pina J., Rolain J., Raoult D., Musso D. Azithromycin inhibits the replication of Zika virus. Journal of Antivirals & Antiretrovirals. 2018;10 [Google Scholar]
- 85.Madrid P.B., Panchal R.G., Warren T.K., Shurtleff A.C., Endsley A.N., Green C.E., Kolokoltsov A., Davey R., Manger I.D., Gilfillan L., Bavari S., Tanga M.J. Evaluation of Ebola virus inhibitors for drug repurposing. ACS Infect Dis. 2015;1(7):317–326. doi: 10.1021/acsinfecdis.5b00030. [DOI] [PubMed] [Google Scholar]
- 86.Bacharier L.B., Guilbert T.W., Mauger D.T., Boehmer S., Beigelman A., Fitzpatrick A.M., Jackson D.J., Baxi S.N., Benson M., Burnham C.D., Cabana M., Castro M., Chmiel J.F., Covar R., Daines M., Gaffin J.M., Gentile D.A., Holguin F., Israel E., Kelly H.W., Lazarus S.C., Lemanske R.F., Jr., Ly N., Meade K., Morgan W., Moy J., Olin T., Peters S.P., Phipatanakul W., Pongracic J.A., Raissy H.H., Ross K., Sheehan W.J., Sorkness C., Szefler S.J., Teague W.G., Thyne S., Martinez F.D. Early administration of azithromycin and prevention of severe lower respiratory tract illnesses in preschool children with a history of such illnesses: a randomized clinical trial. Jama. 2015;314(19):2034–2044. doi: 10.1001/jama.2015.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honore S., Colson P., Chabriere E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Theoharides T., Conti P. Dexamethasone for COVID-19? Not so fast. J. Biol. Regul. Homeost. Agents. 2020;34(3):10.23812. doi: 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- 89.Hassan M.E., Hasan H.M., Sridharan K., Elkday A., ElSeirafi M.M. Dexamethasone in severe COVID-19 infection: a case series. Respiratory Medicine Case Reports. 2020:101205. doi: 10.1016/j.rmcr.2020.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lammers T., Sofias A.M., van der Meel R., Schiffelers R., Storm G., Tacke F., Koschmieder S., Brümmendorf T.H., Kiessling F., Metselaar J.M. Dexamethasone nanomedicines for COVID-19. Nat. Nanotechnol. 2020:1–3. doi: 10.1038/s41565-020-0752-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quan L., Zhang Y., Crielaard B.J., Dusad A., Lele S.M., Rijcken C.J., Metselaar J.M., Kostková H., Etrych T.s., Ulbrich K. Nanomedicines for inflammatory arthritis: head-to-head comparison of glucocorticoid-containing polymers, micelles, and liposomes. ACS Nano. 2014;8(1):458–466. doi: 10.1021/nn4048205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gouglas D., Le T., Henderson K., Kaloudis A., Danielsen T., Hammersland N., Robinson J., Heaton P., Røttingen J.-A. Estimating the cost of vaccine development against epidemic infectious diseases: a cost minimisation study. Lancet Glob. Health. 2018;6 doi: 10.1016/S2214-109X(18)30346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.El Zowalaty M.E., Järhult J.D. From SARS to COVID-19: a previously unknown SARS- related coronavirus (SARS-CoV-2) of pandemic potential infecting humans – call for a one health approach. One Health. 2020;9 doi: 10.1016/j.onehlt.2020.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu S. Timely development of vaccines against SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):542–544. doi: 10.1080/22221751.2020.1737580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shehata M.M., Mostafa A., Teubner L., Mahmoud S.H., Kandeil A., Elshesheny R., Frantz R., La Pietra L., Pleschka S., Osman A., Kayali G., Chakraborty T., Ali M.A., Mraheil M.A. Bacterial outer membrane vesicles (OMVs)-based dual vaccine for influenza A H1N1 virus and MERS-CoV. Vaccines (Basel) 2019;7(2) doi: 10.3390/vaccines7020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schindewolf C., Menachery V.D. Middle East respiratory syndrome vaccine candidates: cautious optimism. Viruses. 2019;11(1) doi: 10.3390/v11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1) doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Du Z., Zhu F., Guo F., Yang B., Wang T. Detection of antibodies against SARS-CoV-2 in patients with COVID-19. J. Med. Virol. 2020;92:1735–1738. doi: 10.1002/jmv.25820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Azkur A.K., et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75(7):1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le T.T., Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 103.DeDiego M.L., Álvarez E., Almazán F., Rejas M.T., Lamirande E., Roberts A., Shieh W.-J., Zaki S.R., Subbarao K., Enjuanes L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007;81(4):1701–1713. doi: 10.1128/JVI.01467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao Q., Chen Y.-C., Chen C.-L., Chiu C.-H. SARS-CoV-2 infection in children: transmission dynamics and clinical characteristics. J. Formos. Med. Assoc. 2020;119 doi: 10.1016/j.jfma.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Netea M.G., Schlitzer A., Placek K., Joosten L.A.B., Schultze J.L. Innate and adaptive immune memory: an evolutionary continuum in the host's response to pathogens. Cell Host Microbe. 2019;25(1):13–26. doi: 10.1016/j.chom.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 106.DeZure A., Berkowitz N., Graham B., Ledgerwood J. Whole-inactivated and virus-like particle vaccine strategies for Chikungunya virus. J. Infect. Dis. 2016;214:S497–S499. doi: 10.1093/infdis/jiw352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou J., Wang W., Zhong Q., Wei H., Yang Z., Yuan X., Zhu R., Tang Z., Wang Y., Xian Q., Tang H., Wen L. Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine. 2005;23:3202–3209. doi: 10.1016/j.vaccine.2004.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.See R., Zakhartchouk A., Petric M., Lawrence D., Mok C., Hogan R., Rowe T., Zitzow L., Karunakaran K., Hitt M., Graham F., Prevec L., Mahony J., Sharon C., Auperin T., Rini J., Tingle A., Scheifele D., Skowronski D., Finlay B. Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus. The Journal of general virology. 2006;87:641–650. doi: 10.1099/vir.0.81579-0. [DOI] [PubMed] [Google Scholar]
- 109.Plante K.S., Rossi S.L., Bergren N.A., Seymour R.L., Weaver S.C. Extended preclinical safety, efficacy and stability testing of a live-attenuated Chikungunya vaccine candidate. PLoS Negl. Trop. Dis. 2015;9(9) doi: 10.1371/journal.pntd.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gao S., Song S., Zhang L. Recent progress in vaccine development against Chikungunya virus. Front. Microbiol. 2019;10:2881. doi: 10.3389/fmicb.2019.02881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ura T., Okuda K., Shimada M. Developments in viral vector-based vaccines. Vaccines (Basel) 2014;2(3):624–641. doi: 10.3390/vaccines2030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Robert-Guroff M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007;18(6):546–556. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44(6):1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song F., Fux R., Provacia L.B., Volz A., Eickmann M., Becker S., Osterhaus A.D., Haagmans B.L., Sutter G. Middle East respiratory syndrome coronavirus spike protein delivered by modified vaccinia virus Ankara efficiently induces virus-neutralizing antibodies. J. Virol. 2013;87(21):11950–11954. doi: 10.1128/JVI.01672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Enjuanes L., Dediego M.L., Alvarez E., Deming D., Sheahan T., Baric R. Vaccines to prevent severe acute respiratory syndrome coronavirus-induced disease. Virus Res. 2008;133(1):45–62. doi: 10.1016/j.virusres.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu R.Y., Wu L.Z., Huang B.J., Huang J.L., Zhang Y.L., Ke M.L., Wang J.M., Tan W.P., Zhang R.H., Chen H.K., Zeng Y.X., Huang W. Adenoviral expression of a truncated S1 subunit of SARS-CoV spike protein results in specific humoral immune responses against SARS-CoV in rats. Virus Res. 2005;112(1–2):24–31. doi: 10.1016/j.virusres.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 118.Brito L.A., Kommareddy S., Maione D., Uematsu Y., Giovani C., Berlanda Scorza F., Otten G.R., Yu D., Mandl C.W., Mason P.W., Dormitzer P.R., Ulmer J.B., Geall A.J. In: Advances in Genetics. Huang L., Liu D., Wagner E., editors. Academic Press; 2015. Chapter seven - self-amplifying mRNA vaccines; pp. 179–233. [DOI] [PubMed] [Google Scholar]
- 119.Rezaei T., Khalili S., Baradaran B., Mosafer J., Rezaei S., Mokhtarzadeh A., de la Guardia M. Recent advances on HIV DNA vaccines development: stepwise improvements to clinical trials. J. Control. Release. 2019;316:116–137. doi: 10.1016/j.jconrel.2019.10.045. [DOI] [PubMed] [Google Scholar]
- 120.Jahanafrooz Z., Baradaran B., Mosafer J., Hashemzaei M., Rezaei T., Mokhtarzadeh A., Hamblin M.R. Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today. 2020;25:552–560. doi: 10.1016/j.drudis.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pardi N., Hogan M., Porter F., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17 doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gascón A., Pozo-Rodríguez A., Solinís M. Development of nucleic acid vaccines: use of self-amplifying RNA in lipid nanoparticles. Int. J. Nanomedicine. 2014;9:1833–1843. doi: 10.2147/IJN.S39810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Muthumani K., Falzarano D., Reuschel E., Kraynyak K., Ugen K., Kim P., Maslow J., Kim J.J., Sardesai N.Y., Feldmann H., Weiner D. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in non-human primates. Int. J. Infect. Dis. 2016;45:23. [Google Scholar]
- 124.Wang L., Shi W., Joyce M.G., Modjarrad K., Zhang Y., Leung K., Lees C., Zhou T., Yassine H., Kanekiyo M., Yang Z.-Y., Chen X., Becker M., Freeman M., Vogel L., Johnson J., Olinger G., Todd J., Bagci U., Graham B. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Adnan Shereen M., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Du L., Tai W., Zhou Y., Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert review of vaccines. 2016;15 doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bisht H., Roberts A., Vogel L., Subbarao K., Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology. 2005;334(2):160–165. doi: 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W.-J., Zaki S., Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bisht H., Roberts A.J., Vogel L., Bukreyev A., Collins P., Murphy B., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Du L., Jiang S. Middle East respiratory syndrome: current status and future prospects for vaccine development. Expert. Opin. Biol. Ther. 2015;15:1–5. doi: 10.1517/14712598.2015.1092518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maslow J.N. Vaccines for emerging infectious diseases: lessons from MERS coronavirus and Zika virus. Hum Vaccin Immunother. 2017;13(12):2918–2930. doi: 10.1080/21645515.2017.1358325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Du L., Tai W., Yang Y., Zhao G., Zhu Q., Sun S., Liu C., Tao X., Tseng C.-T.K., Perlman S., Jiang S., Zhou Y., Li F. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu S.J., Leng C.H., Lien S.P., Chi H.Y., Huang C.Y., Lin C.L., Lian W.C., Chen C.J., Hsieh S.L., Chong P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24(16):3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zheng N., Xia R., Yang C., Yin B., Li Y., Duan C., Liang L., Guo H., Xie Q. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine. 2009;27(36):5001–5007. doi: 10.1016/j.vaccine.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.He Y., Zhou Y., Siddiqui P., Niu J., Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2005;43(8):3718–3726. doi: 10.1128/JCM.43.8.3718-3726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen I.-Y., Moriyama M., Chang M.-F., Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Papaioannou N., Beniata O., Vitsos P., Tsitsilonis O., Samara P. Harnessing the immune system to improve cancer therapy. Annals of Translational Medicine. 2016;4 doi: 10.21037/atm.2016.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wong C., Lam K.C., Wu A., Ip W., Lee N., Chan I., Lit L., Hui D., Chan M., Chung S., Sung J. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.-s. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mair-Jenkins J., Saavedra-Campos M., Baillie K., Cleary P., Khaw F.-M., Lim W.S., Makki S., Rooney K., Nguyen-Van-Tam J., Beck C., Xu X. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2014;211 doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stockman L., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao J., Perera R.A.P.M., Kayali G., Meyerholz D., Perlman S., Peiris J.S. Passive immunotherapy with dromedary immune serum in an experimental animal model for MERS coronavirus infection. J. Virol. 2015;89 doi: 10.1128/JVI.00446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. Jama. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., Gao G.F. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antivir. Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Abaturov A.E., Nikulina A.A., Tokareva N.M. vol. 13. 2018. Effect of Treatment With Inosine Pranobex in Acute Respiratory Viral Infections in Children; pp. 490–494. [Google Scholar]
- 149.Wang S.-F., Chen K.-H., Chen M., Li W.-Y., Chen Y.-J., Tsao C., Yen M.-y., Huang J., Chen Y.-M. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24:421–426. doi: 10.1089/vim.2011.0024. [DOI] [PubMed] [Google Scholar]
- 150.Shanmugaraj B., Siriwattananon K., Wangkanont K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 151.Nezhad F., Mosaddeghi P., Negahdaripour M., Dehghani Z., Farahmandnejad M., Moghadami M., Nezafat N., Masoompour S.M. 2020. Therapeutic Approaches for COVID-19 Based on the Dynamics of Interferon-mediated Immune Responses. [Google Scholar]
- 152.Miyoshi H., Inata Y., Takeuchi M. Distinguishing the effects of inhaled nitric oxide and lung recruitment in pediatric acute respiratory distress syndrome: scope for further improvement. Pediatr. Crit. Care Med. 2018;19:505. doi: 10.1097/PCC.0000000000001489. [DOI] [PubMed] [Google Scholar]
- 153.Fang F. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 154.Darwish I., Miller C., Kain K., Liles W. Inhaled nitric oxide therapy fails to improve outcome in experimental severe influenza. Int. J. Med. Sci. 2012;9:157–162. doi: 10.7150/ijms.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Alhazzani W., Møller M.H., Arabi Y.M., Loeb M., Gong M.N., Fan E., Oczkowski S., Levy M.M., Derde L., Dzierba A., Du B., Aboodi M., Wunsch H., Cecconi M., Koh Y., Chertow D.S., Maitland K., Alshamsi F., Belley-Cote E., Greco M., Laundy M., Morgan J.S., Kesecioglu J., McGeer A., Mermel L., Mammen M.J., Alexander P.E., Arrington A., Centofanti J.E., Citerio G., Baw B., Memish Z.A., Hammond N., Hayden F.G., Evans L., Rhodes A. Surviving sepsis campaign: guidelines on the management of critically ill adults with coronavirus disease 2019 (COVID-19) Intensive Care Med. 2020:1–34. doi: 10.1007/s00134-020-06022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Totura A.L., Baric R.S. Reply to "statins may decrease the fatality rate of MERS infection". mBio. 2015;6(5):e01303–e01315. doi: 10.1128/mBio.01303-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.DeDiego M.L., Nieto-Torres J.L., Regla-Nava J.A., Jimenez-Guardeno J.M., Fernandez-Delgado R., Fett C., Castano-Rodriguez C., Perlman S., Enjuanes L. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J. Virol. 2014;88(2):913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xie S., Fan W., He H., Huang F. Role of melatonin in the regulation of pain. J. Pain Res. 2020;13:331–343. doi: 10.2147/JPR.S228577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tao J., Yang M., Wu H., Ma T., He C., Chai M., Zhang X., Zhang J., Ding F., Wang S., Deng S., Zhu K., Song Y., Ji P., Liu H., Lian Z., Liu G. Effects of AANAT overexpression on the inflammatory responses and autophagy activity in the cellular and transgenic animal levels. Autophagy. 2018;14(11):1850–1869. doi: 10.1080/15548627.2018.1490852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Reiter R., Ma Q., Sharma R. Treatment of ebola and other infectious diseases: melatonin “goes viral”. Melatonin Research. 2020;3:43–57. [Google Scholar]
- 161.Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., Liu C., Reiter R. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020:117583. doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Simonson O., Mougiakakos D., Heldring N., Bassi G., Johansson H., Dalén M., Jitschin R., Rodin S., Corbascio M., Andaloussi S., Wiklander O., Nordin J., Skog J., Romain C., Koestler T., Hellgren-Johansson L., Schiller P., Joachimsson P.-O., Hägglund H., Blanc K. In vivo effects of mesenchymal stromal cells in two patients with severe acute respiratory distress syndrome. Stem Cells Transl. Med. 2015;4 doi: 10.5966/sctm.2015-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sung D.K., Chang Y.S., Sung S.I., Yoo H.S., Ahn S.Y., Park W.S. Antibacterial effect of mesenchymal stem cells against Escherichia coli is mediated by secretion of beta-defensin-2 via toll-like receptor 4 signalling. Cell. Microbiol. 2016;18(3):424–436. doi: 10.1111/cmi.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meisel R., Zibert A., Laryea M., Göbel U., Däubener W., Dilloo D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2, 3-dioxygenase–mediated tryptophan degradation. Blood. 2004;103(12):4619–4621. doi: 10.1182/blood-2003-11-3909. [DOI] [PubMed] [Google Scholar]
- 165.dos Santos C.C., Murthy S., Hu P., Shan Y., Haitsma J.J., Mei S.H., Stewart D.J., Liles W.C. Network analysis of transcriptional responses induced by mesenchymal stem cell treatment of experimental sepsis. Am. J. Pathol. 2012;181(5):1681–1692. doi: 10.1016/j.ajpath.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 166.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J.-W., Xia P., Dong J., Zhao J., Wang F.-S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chan M., Kuok I.T., Leung C., Hui K., Valkenburg S., Lau E., Nicholls J., Fang X., Guan Y., Lee J., Chan R., Webster R., Matthay M., Peiris J.S. Human mesenchymal stromal cells reduce influenza A H5N1-associated acute lung injury in vitro and in vivo. Proc. Natl. Acad. Sci. 2016;113:201601911. doi: 10.1073/pnas.1601911113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Matthay M., Ware L., Zimmerman G. The acute respiratory distress syndrome. J. Clin. Invest. 2012;122:2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Silva P., Pelosi P., Rocco P. Personalized pharmacological therapy for ARDS: a light at the end of the tunnel. Expert Opin. Investig. Drugs. 2019;29 doi: 10.1080/13543784.2020.1699531. [DOI] [PubMed] [Google Scholar]
- 170.Guérin C., Reignier J., Richard J.-C., Beuret P., Gacouin A., Boulain T., Mercier E., Badet M., Mercat A., Baudin O., Clavel M., Chatellier D., Jaber S., Rosselli S., Mancebo J., Sirodot M., Hilbert G., Bengler C., P. Group Prone positioning in severe acute respiratory distress syndrome. N. Engl. J. Med. 2013;368:2159. doi: 10.1056/NEJMoa1214103. [DOI] [PubMed] [Google Scholar]
- 171.Curley G., Scott J., Laffey J. Therapeutic potential and mechanisms of action of mesenchymal stromal cells for acute respiratory distress syndrome. Current stem cell research & therapy. 2014;9 doi: 10.2174/1574888x09666140228144812. [DOI] [PubMed] [Google Scholar]
- 172.Lee F.-Y., Chen K.-H., Wallace C., Sung P.-H., Sheu J.-J., Chung S.-Y., Chen Y.-L., Lu H.-I., Ko S.-F., Sun C.-K., Chiang H.-J., Chang H.-W., Lee M., Yip H.-K. Xenogeneic human umbilical cord-derived mesenchymal stem cells reduce mortality in rats with acute respiratory distress syndrome complicated by sepsis. Oncotarget. 2017:8. doi: 10.18632/oncotarget.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang Y., Hu X. Advance on human umbilical cord mesenchymal stem cells for treatment of ALI in severe burns. Zhonghua wei zhong bing ji jiu yi xue. 2017;29:90–96. doi: 10.3760/cma.j.issn.2095-4352.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 174.Moodley Y., Atienza D., Manuelpillai U., Samuel C.S., Tchongue J., Ilancheran S., Boyd R., Trounson A. Human umbilical cord mesenchymal stem cells reduce fibrosis of bleomycin-induced lung injury. Am. J. Pathol. 2009;175(1):303–313. doi: 10.2353/ajpath.2009.080629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhu H., Xiong Y., Xia Y., Zhang R., Tian D., Wang T., Dai J., Wang L., Yao H., Jiang H., Yang K., Liu E., Shi Y., Fu Z., Gao L., Zou L. Therapeutic effects of human umbilical cord-derived mesenchymal stem cells in acute lung injury mice. Sci. Rep. 2017;7:39889. doi: 10.1038/srep39889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mills D.R., Mao Q., Chu S., Falcon Girard K., Kraus M., Padbury J.F., De Paepe M.E. Effects of human umbilical cord blood mononuclear cells on respiratory system mechanics in a murine model of neonatal lung injury. Exp. Lung Res. 2017;43(2):66–81. doi: 10.1080/01902148.2017.1300713. [DOI] [PubMed] [Google Scholar]
- 177.Xiang B., Chen L., Wang X., Zhao Y., Wang Y., Xiang C. Transplantation of menstrual blood-derived mesenchymal stem cells promotes the repair of LPS-induced acute lung injury. Int. J. Mol. Sci. 2017;18(4) doi: 10.3390/ijms18040689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Antebi B., Mohammadipoor A., Batchinsky A., Cancio L. The promise of mesenchymal stem cell therapy for acute respiratory distress syndrome. J. Trauma Acute Care Surg. 2017;84:1. doi: 10.1097/TA.0000000000001713. [DOI] [PubMed] [Google Scholar]
- 179.Ong T., McClintock D., Kallet R., Ware L., Matthay M., Liu K. Ratio of angiopoietin-2 to angiopoietin-1 as a predictor of mortality in acute lung injury patients. Crit. Care Med. 2010;38:1845–1851. doi: 10.1097/CCM.0b013e3181eaa5bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sage E., Thakrar R., Janes S. Genetically modified mesenchymal stromal cells in cancer therapy. Cytotherapy. 2016;18:1435–1445. doi: 10.1016/j.jcyt.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen J., Li C., Gao X., Li C., Liang Z., Yu L., Li Y., Xiao X., Chen L. Keratinocyte growth factor gene delivery via mesenchymal stem cells protects against lipopolysaccharide-induced acute lung injury in mice. PLoS One. 2013;8:e83303. doi: 10.1371/journal.pone.0083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wyse R., Dunbar G., Rossignol J. Use of genetically modified mesenchymal stem cells to treat neurodegenerative diseases. Int. J. Mol. Sci. 2014;15:1719–1745. doi: 10.3390/ijms15021719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xu A., Rodriguez L., Walker K., Mohammadipoor A., Kamucheka R., Cancio L., Batchinsky A., Antebi B. Mesenchymal stem cells reconditioned in their own serum exhibit augmented therapeutic properties in the setting of acute respiratory distress syndrome. Stem Cells Transl. Med. 2019;8 doi: 10.1002/sctm.18-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jerkic M., Masterson C., Ormesher, Gagnon S., Goyal, Rabani, Otulakowski G., Zhang H., Kavanagh, Laffey J. Overexpression of IL-10 enhances the efficacy of human umbilical-cord-derived mesenchymal stromal cells in E. coli pneumosepsis. J. Clin. Med. 2019;8:847. doi: 10.3390/jcm8060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang G., Cao K., Liu K., Xue Y., Roberts A., Li F., Han Y., Rabson A., Wang Y., Shi Y. Kynurenic acid, an IDO metabolite, controls TSG-6-mediated immunosuppression of human mesenchymal stem cells. Cell Death Differ. 2017;25 doi: 10.1038/s41418-017-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fang X., Abbott J., Cheng L., Colby J., Lee J., Levy B., Matthay M. Human mesenchymal stem (stromal) cells promote the resolution of acute lung injury in part through lipoxin A4. Journal of immunology (Baltimore,Md.: 1950) 2015:195. doi: 10.4049/jimmunol.1500244. [DOI] [PubMed] [Google Scholar]
- 187.Gupta N., Krasnodembskaya A., Kapetanaki M., Mouded M., Tan X., Serikov V., Matthay M. Mesenchymal stem cells enhance survival and bacterial clearance in murine Escherichia coli pneumonia. Thorax. 2012;67:533–539. doi: 10.1136/thoraxjnl-2011-201176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sung D., Chang Y.S., Sung S., Yoo H., Ahn S.Y., Park W.S. Antibacterial effect of mesenchymal stem cells against Escherichia coli is mediated by secretion of beta-defensin-2 via toll like receptor 4 signaling. Cell. Microbiol. 2015:18. doi: 10.1111/cmi.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mohammadipoor A., Antebi B., Batchinsky A., Cancio L. Therapeutic potential of products derived from mesenchymal stem/stromal cells in pulmonary disease. Respir. Res. 2018;19 doi: 10.1186/s12931-018-0921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shah T., Predescu D., Predescu S. Mesenchymal stem cells-derived extracellular vesicles in acute respiratory distress syndrome: a review of current literature and potential future treatment options. Clinical and Translational Medicine. 2019;8:25. doi: 10.1186/s40169-019-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wilson J., Liu K., Zhuo H., Caballero L., McMillan M., Fang X., Cosgrove K., Vojnik R., Calfee C., Lee J.-W., Rogers A., Levitt J., Wiener-Kronish J., Bajwa E., Leavitt A., McKenna D., Taylor B., Matthay M. Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial. Lancet Respir. Med. 2014;3 doi: 10.1016/S2213-2600(14)70291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lu C.C., Chen M.Y., Chang Y.L. Potential therapeutic agents against COVID-19: what we know so far. J Chin Med Assoc. 2020;83:534–536. doi: 10.1097/JCMA.0000000000000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Leng Z., Zhu R., Hou W., Feng Y., Yang Y., Han Q., Shan G., Meng F., Du D., Wang S., Fan J., Wang W., Deng L., Shi H., Li H., Hu Z., Zhang F., Gao J., Liu H., Zhao R. Transplantation of ACE2-mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11:216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Oroojalian F., Charbgoo F., Hashemi M., Amani A., Yazdian-Robati R., Mokhtarzadeh A., Ramezani M., Hamblin M.R. Recent advances in nanotechnology-based drug delivery systems for the kidney. J. Control. Release. 2020;321:442–462. doi: 10.1016/j.jconrel.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 195.Oroojalian F., Jahanafrooz Z., Chogan F., Rezayan A.H., Malekzade E., Rezaei S.J.T., Nabid M.R., Sahebkar A. Synthesis and evaluation of injectable thermosensitive penta-block copolymer hydrogel (PNIPAAm-PCL-PEG-PCL-PNIPAAm) and star-shaped poly (CL—CO—LA)-b-PEG for wound healing applications. J. Cell. Biochem. 2019;120(10):17194–17207. doi: 10.1002/jcb.28980. [DOI] [PubMed] [Google Scholar]
- 196.Shubhika K. Nanotechnology and medicine - the upside and the downside. International Journal of Drug Development and Research. 2013;5:1–10. [Google Scholar]
- 197.Rizvi S., Saleh A. Applications of nanoparticle systems in drug delivery technology. Saudi Pharmaceutical Journal. 2018;26:64–70. doi: 10.1016/j.jsps.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lu X., Zhu T., Chen C., Liu Y. Right or left: the role of nanoparticles in pulmonary diseases. Int. J. Mol. Sci. 2014;15:17577–17600. doi: 10.3390/ijms151017577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.van Rijt S., Bein T., Meiners S. Medical nanoparticles for next generation drug delivery to the lungs. Eur. Respir. J. 2014;44 doi: 10.1183/09031936.00212813. [DOI] [PubMed] [Google Scholar]
- 200.Pati R., Shevtsov M., Sonawane A. Nanoparticle vaccines against infectious diseases. Front. Immunol. 2018;9(2224) doi: 10.3389/fimmu.2018.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Soltani F., Parhiz H., Mokhtarzadeh A., Ramezani M. Synthetic and biological vesicular nano-carriers designed for gene delivery. Curr. Pharm. Des. 2015;21(42):6214–6235. doi: 10.2174/1381612821666151027153410. [DOI] [PubMed] [Google Scholar]
- 202.Afsharzadeh M., Hashemi M., Mokhtarzadeh A., Abnous K., Ramezani M. Recent advances in co-delivery systems based on polymeric nanoparticle for cancer treatment. Artificial Cells, Nanomedicine and Biotechnology. 2018;46(6):1095–1110. doi: 10.1080/21691401.2017.1376675. [DOI] [PubMed] [Google Scholar]
- 203.Jackman J., Lee J., Cho N. Nanomedicine for infectious disease applications: innovation towards broad-spectrum treatment of viral infections. Small (Weinheim an der Bergstrasse, Germany) 2015:12. doi: 10.1002/smll.201500854. [DOI] [PubMed] [Google Scholar]
- 204.Coleman C., Liu Y., Mu H., Taylor J., Massare M., Flyer D., Smith G., Frieman M. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32 doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Coleman C., Venkataraman T., Liu Y., Glenn G., Smith G., Flyer D., Frieman M. MERS-CoV spike nanoparticles protect mice from MERS-CoV infection. Vaccine. 2017;35 doi: 10.1016/j.vaccine.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jung S.-Y., Kang K., Lee E.-Y., Seo D.-W., Kim H.-L., Kim H., Kwon T., Park H.-L., Kim H., Lee S.-M., Nam J.-H. Heterologous prime–boost vaccination with adenoviral vector and protein nanoparticles induces both Th1 and Th2 responses against Middle East respiratory syndrome coronavirus. Vaccine. 2018;36 doi: 10.1016/j.vaccine.2018.04.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Roh C. A facile inhibitor screening of SARS coronavirus N protein using nanoparticle-based RNA oligonucleotide. Int. J. Nanomedicine. 2012;7:2173–2179. doi: 10.2147/IJN.S31379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mokhtarzadeh A., Eivazzadeh-Keihan R., Pashazadeh P., Hejazi M., Gharaatifar N., Hasanzadeh M., Baradaran B., de la Guardia M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017;97:445–457. doi: 10.1016/j.trac.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hasanzadeh M., Tagi S., Solhi E., Mokhtarzadeh A., Shadjou N., Eftekhari A., Mahboob S. An innovative immunosensor for ultrasensitive detection of breast cancer specific carbohydrate (CA 15-3) in unprocessed human plasma and MCF-7 breast cancer cell lysates using gold nanospear electrochemically assembled onto thiolated graphene quantum dots. Int. J. Biol. Macromol. 2018;114:1008–1017. doi: 10.1016/j.ijbiomac.2018.03.183. [DOI] [PubMed] [Google Scholar]
- 210.Hassanpour S., Baradaran B., Hejazi M., Hasanzadeh M., Mokhtarzadeh A., de la Guardia M. Recent trends in rapid detection of influenza infections by bio and nanobiosensor. TrAC Trends Anal. Chem. 2018;98:201–215. [Google Scholar]
- 211.Hasanzadeh M., Karimzadeh A., Sadeghi S., Mokhtarzadeh A., Shadjou N., Jouyban A. Graphene quantum dot as an electrically conductive material toward low potential detection: a new platform for interface science. J. Mater. Sci. Mater. Electron. 2016;27(6):6488–6495. [Google Scholar]
- 212.Ishikawa F., Chang H.-K., Curreli M., Liao H.-I., Olson C., Chen P.-C., Zhang R., Roberts R., Sun R., Cote R., Thompson M., Zhou C. Label-free, electrical detection of the SARS virus N-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano. 2009;3:1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Teengam P., Tuantranont A., Vilaivan T., Chailapakul O., Henry C. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89 doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kim H., Park M., Hwang J., Kim J., Chung D.-R., Lee K.-S., Kang M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sensors. 2019;4 doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- 215.Pimentel T.A., Yan Z., Jeffers S.A., Holmes K.V., Hodges R.S., Burkhard P. Peptide nanoparticles as novel immunogens: design and analysis of a prototypic severe acute respiratory syndrome vaccine. Chem. Biol. Drug Des. 2009;73(1):53–61. doi: 10.1111/j.1747-0285.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhang J., Zeng H., Gu J., Li H., Zheng L., Zou Q. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines (Basel) 2020;8(2) doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim J.Y., Choe P.G., Oh Y., Oh K.J., Kim J., Park S.J., Park J.H., Na H.K., Oh M.D. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J. Korean Med. Sci. 2020;35(5):e61. doi: 10.3346/jkms.2020.35.e61. [DOI] [PMC free article] [PubMed] [Google Scholar]