Abstract
It has been reported that microRNA (miRNA/miR)-181b plays an important role in regulating cellular proliferation, invasion and apoptosis in various tumors. However, the role of miR-181b and its molecular mechanisms in colon cancer cells have not yet been elucidated. The present study thus aimed to investigate the mechanisms of miR-181b targeting cylindromatosis (CYLD) to regulate the nuclear factor-κB (NF-κB) signaling pathway, and to determine its role in colon cancer cell proliferation and apoptosis. For this purpose, miR-181b was overexpressed and silenced in the SW480 cell line. The cell proliferation and apoptotic rates were determined using a Cell Counting kit and colony formation assays, and Annexin V-FITC staining, respectively. The expression levels of proteins associated with the NF-κB signaling pathway and apoptosis were detected by western blot analysis. Furthermore, a dual luciferase assay was applied to confirm the interaction between miR-181b and CYLD. CYLD was also overexpressed and silenced in the SW480 cell line using a CYLD overexpression plasmid and siRNA technology, respectively. Transfected cells were used for subsequent experiments. In addition, a nude mouse model was established to measure tumor volume and weight. Immunohistochemistry and a TUNEL assay were performed to detect the Ki67 levels and the cell apoptotic rate, respectively. Compared with the control group, miR-181 silencing or CYLD overexpression significantly attenuated cell proliferation, invasion and migration, and notably increased the proportion of apoptotic cells. Furthermore, the expression levels of Bax and cleaved caspase-3 were markedly increased, whereas those of Bcl-2 were significantly decresaed (P<0.05). In addition, the protein expression levels of p-p65/p65 and p-IκBα/IκBα were significantly down-regulated and upregulated, respectively (P<0.05). Consistent with the results obtained in vitro, in vivo experiments using a nude mouse model yielded similar findings. The aforementioned results indicated that miR-181b down-regulation inhibited human colon cancer cell proliferation by targeting CYLD to attenuate the activity of the NF-κB signaling pathway.
Keywords: microRNA-181b, CYLD, colon cancer, NF-κB signaling pathway
Introduction
Colon cancer is a life-threatening and lethal disease with the highest incidence among solid tumors globally, which is also associated with cancer-related financial stress (1,2). Similar to other malignancies, the development and progression of colon cancer may be caused by mutations in certain key genes (3,4). Therefore, investigating the underlying molecular mechanisms and identifying new targets for the diagnosis and treatment of colon cancer are of utmost importance. These findings will provide novel therapeutic approaches which are beneficial to the patients, and may reduce the harmful effects of the disease.
At present, microRNAs (miRNAs or miRs), types of small non-coding RNAs of 20–24 nucleotides in length, have gained increasing attention due to their regulatory effects on the transcription of their target genes (5). A large number of studies have suggested that miRNAs regulate fundamental biological processes, while their dysfunction may affect the balance of the human body, which in turn induces tissue/organ injuries (5,6). Among the miRNA families, the present study aimed to investigate the role of miR-181b in colon cancer. Several studies have demonstrated the effects of miR-181b on the regulation of cell proliferation, invasion and apoptosis in different types of cancer, such as colorectal (7), ovarian (8) and cervical cancer (9). Furthermore, miR-181 has been shown to be involved in the regulation of tumor growth and the immune system by engaging factors involved in signaling pathways, such as mitogen-activated protein kinase 1 (MAPK1) (10) and nuclear factor-κB (NF-κB) (11). However, the biological functions of miR-181b in regulating the NF-κB signaling pathway in colon cancer remain largely unclear.
The NF-κB pathway regulates the inflammatory responses associated with tumorigenesis (12). Furthermore, the NF-κB signaling pathway is constitutively activated in a number of types of cancer and performs a variety of pro-tumorigenic functions. In thyroid cancer, NF-κB has been shown to regulate the proliferative and anti-apoptotic process (13). The termination of the transcriptional activity of NF-κB is mainly achieved by the upregulation of the expression of inhibitors of the IκB family, particularly IκBα (14). In addition, the negative regulatory factors of the NF-κB pathway are also upregulated, including cylindromatosis (CYLD) (15). CYLD, a K63-specific deubiquitinase, has been shown to inhibit the NF-κB signaling pathway particularly by deconjugating the K63-linked polyubiquitin chains (16). However, the function of miR-181b in regulating CYLD and NF-κB in colon cancer has not yet been fully elucidated; therefore, further studies on this matter are required.
In the present study, in order to gain insight into the role of miR-181b in regulating the NF-κB signaling pathway in colon cancer, its expression levels and functions were determined in colon cancer cells. Furthermore, the association between miR-181b and CYLD was verified.
Materials and methods
Cell culture and treatment
The human colonic cancer cell lines, SW480, HCT116 and LoVo, and the human colonic epithelial HCoEpiC cell line were obtained from the Shanghai Institute of Cell Research. Cells were incubated in a 5% CO2 incubator (Thermo Fisher Scientific, Inc.) at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution (Gibco; Thermo Fisher Scientific, Inc.). Cells were passaged every 2–3 days, and when they reached the log phase, they were used for further experiments.
The expression of miR-181b was analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The levels of miR-181b in the human colon cancer cell lines were significantly higher compared with those in the HCoEpiC cells. In particular, the expression of miR-181b was the highest in the SW480 cells among all the colon cancer cells. The SW480 cells were selected for use in subsequent experiments. The human colonic SW480 cancer cells were passaged and cultured in 6-well plates 24 h prior transfection. Transfection was performed using lentiviral particles according to the manufacturer's instructions. The cells were then divided into the following groups: The blank control group (BC), where cells were not treated; the miR-181b overexpression negative control group (NC1), where cells were transfected with miR-181b scramble; the miR-181b overexpression group (miR-181b), where cells were transfected with miR-181b mimics; the miR-181b silencing negative control group (NC2), where cells were transfected with miR-181b inhibitor negative control; and the miR-181b silencing group (si-miR), where cells were transfected with miR-181b inhibitor. Subsequently, RT-qPCR analysis was performed to detect the miR-181b expression levels in transfected cells.
For further experiments, the SW480 cells were divided into the following groups: The blank control (BC) group; miR-181b silencing (si-miR) group; CYLD overexpression negative control (NC3) group, in which cells were treated with control adenovirus vector containing non-targeting (AdNT, HBAD-GFP, Hanbio Biotechnology Co., Ltd.); CYLD overexpression (CYLD) group, in which cells were treated with adenovirus vector for CYLD (AdCYLD, HBAD-h-CYLD-3*flag-GFP, Hanbio Biotechnology Co., Ltd.); miR-181b silencing + CYLD silencing negative control (NC4) (si-miR + NC4) group, in which cells were transfected with miR-181b inhibitor and scramble control siRNA duplexes using Lipofecamine 2000 transfection reagent; and miR-181b silencing + CYLD silencing (si-C) siRNA (si-miR + si-C) group, in which cells were transfected with miR-181b inhibitor and CYLD specific small-interfering RNA (siRNA) duplexes. RT-qPCR was used to detect the expression of CYLD mRNA in the transfected cells.
Mimic control (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-181b mimic (5′-AACAUUCAUUGCUGUCGGUGGGU-3′), inhibitor control, miR-181b inhibitor (5′-ACCCACCGACAGCAAUGAAUGUU-3′), scramble control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′), siCYLD (5′-GAUUCUGCCUGGCUCUUCUUUGACA-3′) (GenePharma) were transfected into the SW480 cells using Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. All concentrations used were 50 nM. After 24 h, the transfection mixture was removed and fresh complete medium was added. The HBAD-GFP and HBAD-h-CYLD-3*flag-GFP were trans-fected into 293-T cells. The supernatant titer was determined to be 1x108 TU/ml. Subsequently, the lentivirus was used to infect SW480 cells.
RT-qPCR.To determine the mRNA expression levels of CYLD and miR-181b, RT-qPCR was performed. Briefly, total RNA was extracted using TRIzol reagent (Takara Bio, Inc.) and was reverse transcribed into cDNA at 42°C for 50 min using the appropriate kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR reactions were carried out in the Mastercycler® nexus X2 (Eppendorf) under the following conditions: 95°C for 15 sec, 60°C for 1 min and 35 cycles at 72°C for 40 sec. Data were analyzed using the 2−ΔΔCq method (17), and the mRNA expression of U6 and β-actin served as internal control for CYLD and miR-181b expression, respectively. The primer sequences (Shanghai Biotech Engineering Services) used were the following: miR-181b forward, 5′-GCGGATCATTCATTGCTGTCG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-GACCTCTATGCCAACACAGT-3′ and reverse, 5′-AGTACTTGCGCTCAGGAGGA-3′; CYLD forward, 5′-TCTATGGGGTAATCCGTTGG-3′ and reverse, 5′-CAGCCTGCACACTCATCTTC-3′; and β-actin forward, 5′-CAGAGCCTCGCCTTTGCC-3′ and reverse, 5′-GTCGCCCACATAGGAATC-3′.
Cell Counting kit-8 (CCK-8) assay
Cell proliferation was assessed using a CCK-8 assay (Tongren Institute of Chemistry, Japan). Briefly, the cells were plated into 96-well plates, cultured for 24, 48, 72 and 96 h, and the CCK-8 assay was then performed. Subsequently, the cells were further cultured at 37°C for an additional 4 h and the absorbance (OD) at 450 nm was measured in each well using a microplate reader (ELI0A, BioBase).
Cell clone formation assay
Briefly, the cells were digested, plated into 6-well plates at a density of 500 cells/well and cultured with DMEM supplemented with 10% FBS and 1% penicillin-streptomycin solution at 37°C for 2–3 weeks. The culture medium was replaced every 2 days. The cells were then immobilized with 4% paraformaldehyde for 15 min, and soaked in Giemsa stain (G4640, Beijing Solarbio Science & Technology Co., Ltd.) for 30 min at room temperature and the cells were then washed twice with ultra-pure water. Images were acquired using a camera [DSC-HX90, SONY (China), Co. Ltd.].
Transwell migration and invasion assays
The cell invasion ability was assessed using a Transwell assay. Briefly, Matrigel (Beijing Solebao) was diluted 1:1 with pre-chilled DMEM and the solution was evenly spread on the bottom of the Transwell chamber (Corning, Inc.). The cell suspension (density, 5x104 cells; volume, 50 μl) from each group was incubated at 37°C for 4 h. The substratum was supplemented with 600 μl of DMEM for 72 h. The membrane was then removed from the chamber and washed twice with PBS. The transmembrane cells were fixed with 5% glutaraldehyde at 4°C and stained with 0.1% crystal violet (Beijing Solarbio Science & Technology Co., Ltd.) for 30 min at room temperature. Images were acquired under an inverted microscope (Olympus Corporation) and 5 fields (magnification, ×400) were randomly selected to calculate the average number of invaded cells. The migration assay was performed as described above without the Matrigel.
Annexin V-FITC staining assay
An Annexin V-FITC staining assay was performed to determine the cell apoptotic rate. Briefly, following treatment for 24 h, cells were resuspended, 5 μl of Annexin V-FITC and 5 μl of propidium iodide (PI) was added to the cell suspension and the cells were incubated for 15 and 5 min at 37°C, respectively. The cell apoptotic rate was assessed using a flow cytometer (Beckman Coulter, Inc.) and the results were analyzed using CellQuest software (Version 5.1; BD Biosciences).
Western blot analysis for the detection of NF-κB signaling- and apoptosis-related proteins
Cells were lysed and the supernatant was isolated following centrifugation at 850 × g for 20 min at 4°C. The protein concentration was measured with a bicinchoninic acid kit (BSA; Beijing Solarbio Science & Technology Co., Ltd.). Total protein extracts (concentration, 40 μg) were subjected to 8% SDS-PAGE electrophoretic separation and the resolved proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane. Following blocking with 3% bovine serum albumin (BSA) in tris-buffered saline containing Tween-20 (TBST) solution, the membrane was incubated with primary antibodies against Bax (1:500, orb378567), Bcl-2 (1:500, orb10173), cleaved caspase-3 (1:500, orb106556), p65 (1:500, orb344389), p-p65 (1:500, orb14753), IκBα (1:500, orb14528), p-IκBα (1:500, orb14868), CYLD (1:500, orb19453) and β-actin (1:2,000, orb378579; all from Biorbyt Ltd.) overnight at 4°C. Subsequently, the membrane was incubated with a goat anti-rabbit IgG (1:1,000; ab6721; Abcam) secondary antibody for 50 min at 37°C. The protein bands were visualized using an ECL chemiluminescence kit and β-actin was selected as a control to calculate the relative expression of the target proteins. Protein expression levels were semi-quantified using ImageJ software (National Institutes of Health, version 6.0).
Dual luciferase reporter assay
The target gene prediction between miR-181b and CYLD was performed using TargetScan software (www.targetscan.org). The wild-type (wt) and mutant (mut) 3'untranslated region (3'UTR) of CYLD were amplified and subcloned into the pGL3/luciferase vector (Promega Corporation) downstream the luciferase gene. The constructed luciferase reporter plasmid (wt-CYLD or mut-CYLD) was separately co-transfected with miR-181b or NC into SW480 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h. Following co-transfection, cells were cultured for 48 h with DMEM containing with 10% FBS and 1% penicillin-streptomycin solution. The luciferase activity was detected using a dual luciferase reporting system (Promega Corporation) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected cell sample.
Establishment of the subcutaneous tumor transplantation model and animal grouping
A total of 24 male specific pathogen-free (SPF) BALB/c nude mice (age, 4 weeks, weight, 18–20 g) were obtained from Beijing Weitong Lihua Experimental Animal Technology Co., Ltd. [SCXK(Jing)20190006]. Mice were bred in a sterile isolation cage with independent air supply in a room with air laminar flow apparatus, constant temperature (26±1°C) and constant humidity. Mice were free to drink autoclaved water and sterilized food. All animal experiments were conducted in accordance with the National Institute of Health (NIH) guidelines and were approved by the Animal Ethics Committee of Yantaishan Hospital (approval no. 20190456-013). Furthermore, cells were digested with 0.25% trypsin and counted using countess™ cell counting chamber slides (C10228. Thermo Fisher Scientific, Inc.). SW480 cells were transfected with miR-181b inhibitor, CYLD siRNA and AdCYLD, respectively. A total of 4 groups were included, blank control (BC), miR-181b silencing (si-miR), CYLD overexpression (CYLD) and miR-181b silencing + CYLD silencing (si-miR + si-C). The cell concentration was adjusted to 2.5x106 cells/ml with sterile normal saline. Finally, 0.2 ml of the cell suspension were subcutaneously inoculated into the back of the right forelimbs of the nude mice. A total of 24 nude mice were randomly divided into the following groups: The blank control (BC), miR-181b silencing (si-miR), CYLD overexpression (CYLD) and miR-181b silencing + CYLD silencing (si-miR + si-C) groups (n=6 per group). A Vernier caliper was used to measure the long and short diameter of the tumor once a week and the tumor volume was calculated using the following formula: Tumor volume (V)=long diameter × short diameter2/2. The animal health and behaviors were monitored each day, including diet, weight, mental states and none of the mice died before the end of the experiment. There was no death before the end of the experiment. After 28 days, the nude mice were anesthetized with 0.6% sodium pentobarbital (40 mg/kg) by intraperitoneal injection, and sacrificed by decapitation. Cardiac arrest for 5 min confirmed death. The tumor was obtained and weighed. In addition, the tumor tissues were immobilized in 4% paraformaldehyde and embedded in paraffin. A tumor tissue sample was placed in a cryopreservation tube and stored in a refrigerator at −80°C.
Detection of Ki67 using immunohistochemistry
Tissue slices were deparaffinized in xylene, rehydrated in gradient ethanol solution and treated with 3% H2O2. To retrieve the antigen, slices were heated at high temperature in a citrate buffer. Slices were then incubated with a rabbit anti-human Ki67 antibody (1:200, orb389335 Biorbyt) and a secondary goat anti-rabbit IgG (1:1,000, ab6721; Abcam) labeled with horseradish peroxidase. The sections were then dehydrated in gradient ethanol, transparently treated with xylene and sealed with neutral gum. The sections were then observed under a light microscope (CX22, Olympus Corporation) and the Aperio ImageScope software (version 11.1; Leica Microsystems GmbH) was used to count cells.
TUNEL assay
The tumor tissue slices were treated with xylene and gradient ethanol, and cell apoptosis was determined using a TUNEL assay kit (Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.). Apoptotic cells were counted in 5 randomly selected fields under a light microscope (CX22; Olympus Corporation). The apoptotic cells were detected by the presence of tan or brown particles in the cytoplasm and the morphological characteristics of apoptotic cells. The apoptotic rate was calculated as follows: Apoptotic index=(number of positive cells/total number of cells) x100%.
Statistical analysis
SPSS 19.0 software was used to perform the statistical analyses, and data are expressed as the means ± standard deviation (SD). One-way analysis of variance (ANOVA) with a Tukey's post-hoc test was used to analyze the statistical differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Effect of miR-181b on human colon cancer cell proliferation
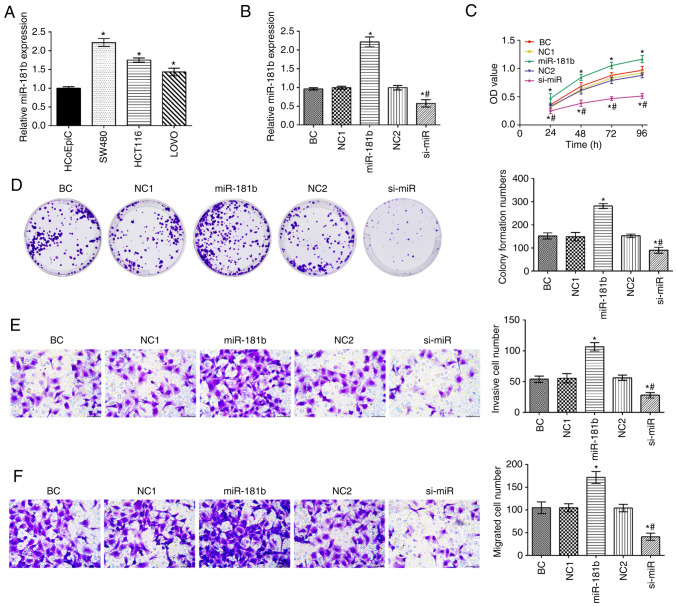
The expression of miR-181b in human colon cancer cell lines was significantly higher compared with that in HCoEpiC cells (P<0.05; Fig. 1A). The expression of miR-181b was the highest in the SW480 cell among colon cancer cells. The SW480 cells were thus selected to assess the expression of miR-181 in the different groups (Fig. 1B). Compared with the BC group, the expression of miR-181b, and the cell proliferative, invasive and migratory capabilities were increased in the miR-181b group. By contrast, miR-181b was downregulated in the si-miR group and this was followed by a decreased cell proliferative, invasive and migratory ability (P<0.05; Fig. 1C-F). Additionally, in contrast to the miR-181b group, the expression of miR-181b and the cell proliferation, invasion and migration rates were significantly decreased in the si-miR group.
Figure 1.
Effect of miR-181b on human colon cancer cell proliferation. (A) Results of RT-qPCR showing miR-181b expression in cells. (B) Results of RT-qPCR showing miR-181b expression in cells following transfection. (C) Results of CCK-8 assay showing the cell proliferative ability. (D) Colony formation assay. (E) Transwell assay results showing cell invasion (magnification, ×400). (F) Transwell assay results showing cell migration (magnification, ×400). *P<0.05, compared with HCoEpiC cells or *P<0.05, compared with the BC group; #P<0.05, compared with the miR-181b group. BC, blank control; NC1, miR-181b overexpression negative control group; NC2, miR-181b silencing negative control group.
Effects of miR-181b on human colon cancer cell apoptosis and the NF-κB signaling pathway
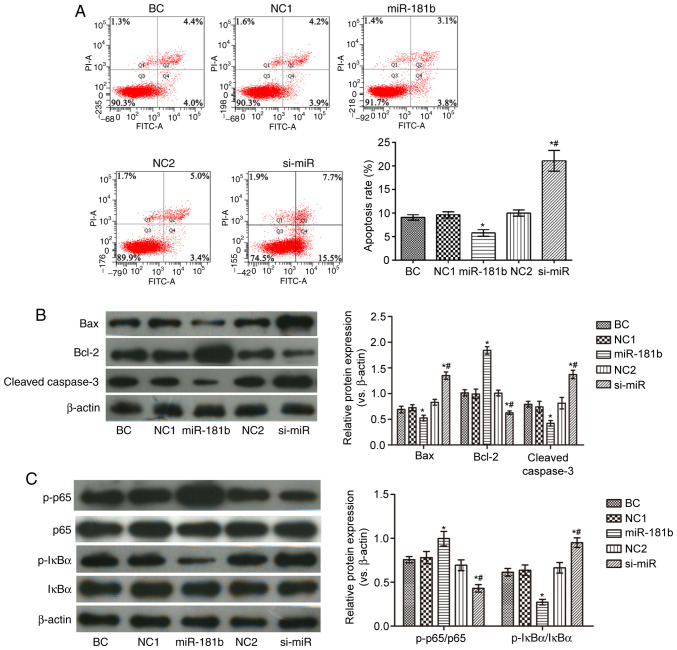
As shown in Fig. 2, compared with the BC group, the proportion of apoptotic cells in the miR-181b group was observably reduced. In addition, the expression levels of Bax, cleaved caspase-3 and p-IκBα/IκBα were decreased, while those of Bcl-2 and p-p65/p65 were markedly increased in the miR-181b group. In the si-miR group, the apoptotic rate and the expression levels of Bax, cleaved caspase-3 and p-IκBα/IκBα were notably increased, while those of Bcl-2 and p-p65/p65 were markedly decresaed, compared with the BC group (P<0.05). Furthermore, compared with the miR-181b group, the proportion of apoptotic cells and the expression of Bax, cleaved caspase-3 and p-IκBα/IκBα were increased, and the expression of Bcl-2 and p-p65/p65 was notably decreased in the si-miR group (P<0.05).
Figure 2.
Effects of miR-181b on cell apoptosis and the NF-κB signaling pathway in SW480 cells. (A) Apoptosis was detected by flow cytometry. (B) The expression levels of the apoptosis-related proteins were detected by western blot analysis. (C) The expression levels of the NF-κB signaling pathway-related proteins were detected by western blot analysis. *P<0.05, compared with the BC group; #P<0.05, compared with the miR-181b group. BC, blank control; NC1, miR-181b overexpression negative control group; NC2, miR-181b silencing negative control group.
miR-181b directly targets CYLD
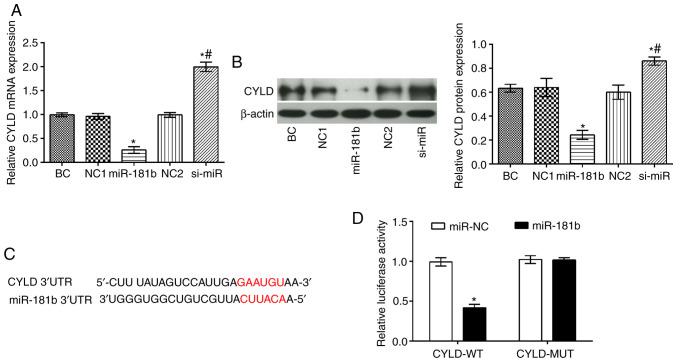
The CYLD mRNA and protein levels were significantly decreased in the miR-181b group compared with the BC group (P<0.05), while the opposite findings were obtained in the si-miR group (P<0.05; Fig. 3). In addition, compared with the miR-181b group, it was apparent that the mRNA and protein expression levels of CYLD were markedly increased in the si-miR group (P<0.05) (Fig. 3A and B). Furthermore, TargetScan (http://www.targetscan.org) software predicted that CYLD was a direct target of miR-181b (Fig. 3C). To further confirm this interaction a dual luciferase assay was performed (Fig. 3C). Therefore, in cells co-transfected with miR-181b mimic and the wt-CYLD 3'UTR, the luciferase activity was markedly attenuated. However, the luciferase activity was unaltered in the cells co-transfected with miR-181b mimic and the mut-CYLD 3'UTR.
Figure 3.
Association between CYLD and miR-181b in SW480 cells. (A) Results of RT-qPCR showing CYLD mRNA expression in SW480 cells. (B) Results of western blot analysis showing the protein expression levels of CYLD in SW480 cells. (C) MiR-181b directly targets CYLD 3'UTR. (D) Results of dual luciferase assay in cells co-transfected with a miR-181b recombinant vector and the CYLD target gene. *P<0.05, compared with the BC group; #P<0.05, compared with the miR-181b group; *P<0.05, compared with the miR-NC group. BC, blank control; NC1, miR-181b overexpression negative control group; NC2, miR-181b silencing negative control group.
Effect of CYLD on human colon cancer cell proliferation
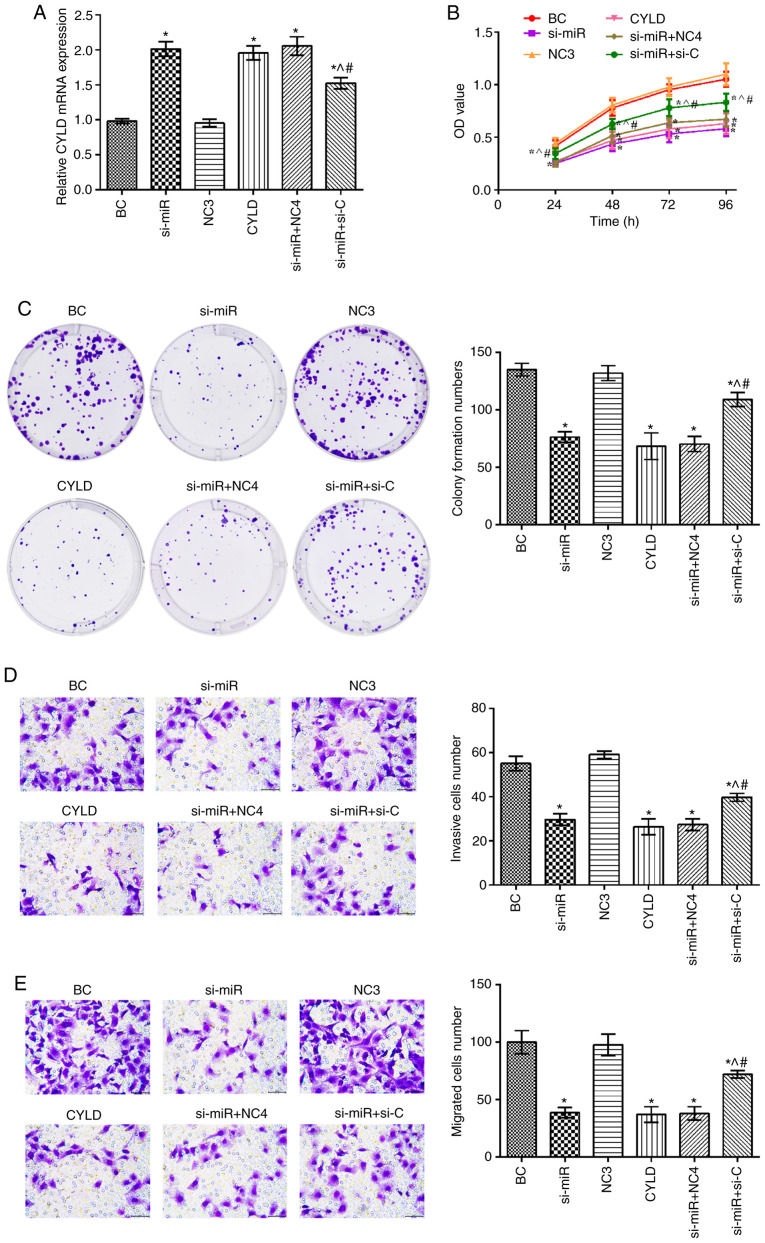
CYLD expression was upregulated in the si-miR, CYLD, si-miR + NC4 and si-miR + si-C groups compared with the BC group (P<0.05; Fig. 4). In addition, the mRNA expression levels of CYLD in the si-miR + si-C group were significantly decreased compared with the si-miR and CYLD groups. Furthermore, compared with the BC group, the cell proliferation, invasion and migration rates were notably suppressed in the si-miR, CYLD, si-miR + NC4 and si-miR + si-C groups (P<0.05). Notably, these rates were elevated in the si-miR + si-C group compared with the si-miR and CYLD groups (P<0.05).
Figure 4.
Effect of miR-181b targeting CYLD on SW480 cell proliferation. (A) Results of RT-qPCR showing CYLD mRNA expression in SW480 cells. (B) Results of CCK-8 assay showing SW480 cell proliferative ability. (C) Results of colony formation assay showing SW480 cell proliferative ability. (D) Results of Transwell assay showing cell invasion (magnification, ×400). (E) Results of Transwell assay showing cell migration (magnification, ×400). *P<0.05, compared with the BC group; ^P<0.05, compared with the si-miR group; #P<0.05, compared with the CYLD group. BC, blank control; NC3, CYLD overexpression negative control; NC4, CYLD silencing negative control.
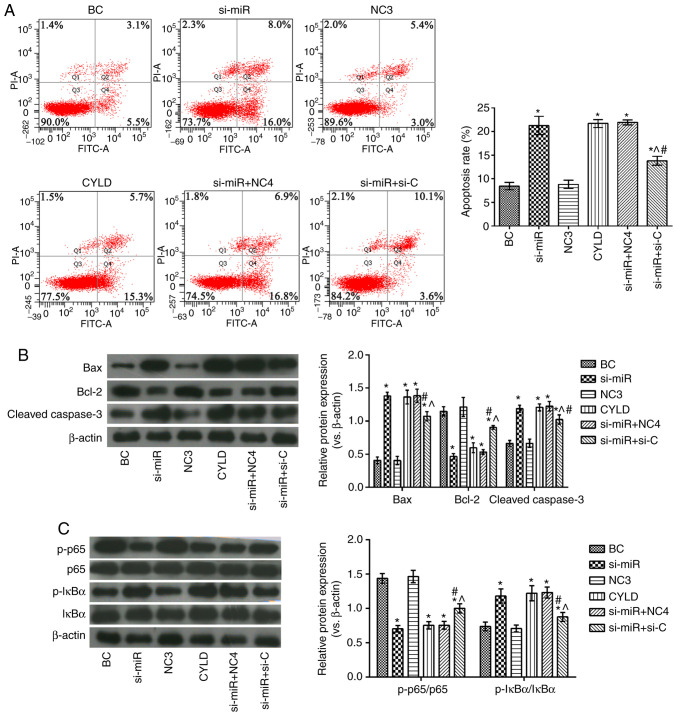
Effects of miR-181b targeting CYLD on human colon cancer cell apoptosis and the NF-κB signaling pathway
As shown in Fig. 5, the si-miR, CYLD, si-miR + NC4 and si-miR + si-C groups exhibited significantly higher apoptotic rates, as well as increased Bax, cleaved caspase-3 and p-IκBα/IκBα expression levels, and decreased Bcl-2 and p-p65/p65 levels compared with the BC group. However, compared with the si-miR and CYLD groups, the proportion of apoptotic cells was evidently decreased in the si-miR + si-C group. In addition, the expression levels of Bax, cleaved caspase-3 and p-IκBα/IκBα were evidently decreased and those of Bcl-2 and p-p65/p65 were notably elevated in the same groups.
Figure 5.
Effects of miR-181b targeting CYLD on SW480 cell apoptosis and NF-κB signaling pathway. (A) Apoptosis was detected by flow cytometry. (B) The expression levels of the apoptosis-related proteins were detected by western blot analysis. (C) The expression levels of the NF-κB signaling pathway-related proteins were detected by western blot analysis. *P<0.05, compared with the BC group; ^P<0.05, compared with the si-miR group; #P<0.05, compared with the CYLD group. BC, blank control; NC3, CYLD overexpression negative control; NC4, CYLD silencing negative control.
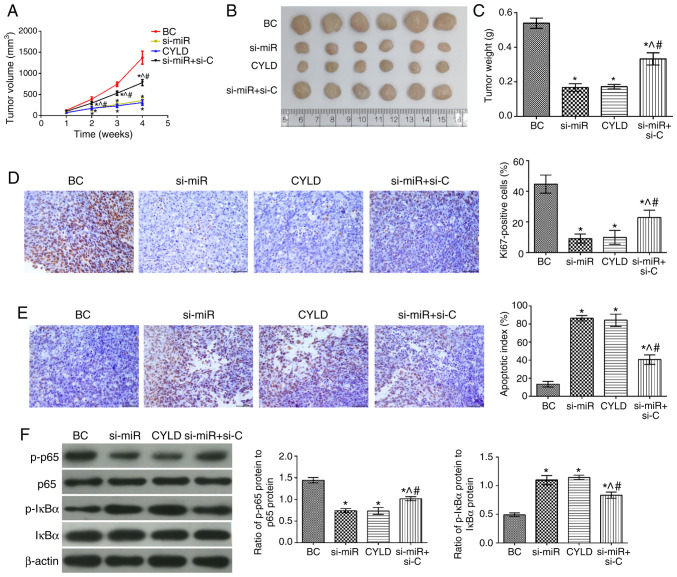
Effects of miR-181b on human colon cancer xenografts
To determine the effects of miR-181b in vivo, colon cancer xeno-grafts were established and evaluated. The tumor growth rate, tumor volume and mass in all groups were significantly attenuated compared with the BC group (Fig. 6). In addition, the number of Ki67-positive cells was also decreased in the tumor tissues, while the apoptotic index was significantly increased. Furthermore, the levels of p-p65/p65 and p-IκBα/IκBα were significantly downregulated and upregulated, respectively (P<0.05). The tumor growth rate, volume and mass were notably higher in the si-miR + si-C group compared with the si-miR and CYLD groups. Moreover, the number of Ki67-positive cells and the expression levels of p-p65/p65 were also increased. Notably, the cell apoptotic rate and the expression levels of p-IκBα/IκBα were markedly attenuated in the tumor tissues (P<0.05).
Figure 6.
Effect of miR-181b targeting CYLD on SW480 cell xenografts. (A) Tumor volume. (B) Tumor image. (C) Tumor weight. (D) Immunohistochemistry results showing Ki67 expression in the transplanted tumors (magnification, ×400). (E) TUNEL assay results showing tumor cell apoptosis (magnification, ×400). (F) Results of western blot analysis showing the expression levels of the NF-κB signaling pathway-related proteins in tumor tissues. *P<0.05, compared with the BC group; ^P<0.05, compared with the si-miR group; #P<0.05, compared with the CYLD group. BC, blank control; NC3, CYLD overexpression negative control; NC4, CYLD silencing negative control.
Discussion
The present study provided a mechanistic insight underlying the miR-181b-mediated development of colon cancer. Firstly, miR-181b was overexpressed and downregulated in the SW480 cells and the results confirmed that miR-181b regulated the cell proliferative, invasive and migratory abilities. Furthermore, the present study also demonstrated that miR-181b downregulation promoted CYLD upregulation and attenuated the activity of the NF-κB signaling pathway, thereby inhibiting cell growth and promoting cell apoptosis both in vitro and in vivo.
The roles of miR-181b in different types of cancer have been widely explored in several studies. It has been reported that the expression levels of miR-181b are associated with the response to chemorapy in non-small cell lung cancer (NSCLC), while miR-181b downregulation is significantly associated with the overall survival of patients with NSCLC (18). Xu et al demonstrated that miR-181b regulated the proliferation of esophageal cancer stem-like cells via the CYLD pathway and that CYLD was directly targeted by miR-181b (19). In colon cancer, a positive feedback loop between miR-181b and STAT3 was identified, which regulated the Warburg effect and xenograft tumor growth (20). It has been well documented that the expression of miR-181b in glioblastoma, where it acts as an onco-suppressor gene, is downregulated via the NF-κB pathway (5). The NF-κB signaling pathway plays a key role in colon cancer cell proliferation and apoptosis (14). The present study revealed that miR-181b was overexpressed in colon cancer cells, while its silencing attenuated colon cancer cell proliferation, invasion and migration abilities. Furthermore, the expression levels of Bax, cleaved caspase-3 and p-IκBα were increased following miR-181b downregulation. These findings suggested that miR-181b played a negative role in the progression of colon cancer and activated the NF-κB signaling pathway.
In unstimulated cells, NF-κB is conjugated to IκBα, which keeps the first in an inactive state in the cytosol (21). Different physiological or pathological stimuli may activate and promote the phosphorylation of IκBα. CYLD is a negative regulatory factor, which inhibits inflammation via the NF-κB pathway (15). Emerging evidence has indicated that the effect of CYLD on targeting NF-κB factors is dependent on the type of cells and activated receptors (22). Moreover, CYLD loss-of-function is involved in NF-κB-associated inflammation, which in turn promotes the development of different types of cancer (23,24). However, the association between CYLD and NF-κB signaling in colon cancer remains unclear. In the present study, CYLD overexpression inhibited cell growth and promoted cell apoptosis in colon cancer cells, while the expression levels of p-p65/p65 were decreased and those of p-IκBα/IκBα were increased. These results indicated that CYLD may be involved in the regulation of the NF-κB signaling pathway in colon cancer.
To verify whether CYLD was a target of miR181b in colon cancer cells, a luciferase reporter assay was performed. Therefore, SW480 cells were co-transfected with plasmids containing wt-CYLD or mut-CYLD 3'UTR and miR-181b or NC mimic. As was expected, miR-181b repressed CYLD expression by directly targeting its 3'UTR. Taken together, the present study confirmed that miR-181b downregulation not only inhibited SW480 cell proliferation, but also promoted apoptosis, partly by targeting CYLD.
In conclusion, the present study suggests that colon cancer may be inhibited through miR-181b downregulation to promote CYLD expression, which in turn attenuates NF-κB activity. At the same time, the present study provides a research basis for the role of miR-181b in colon cancer, indicating that miR-181b may be a novel therapeutic target for patients with colon cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
XY and YS carried out the experimental work and the data collection and interpretation. XY and SH participated in the design and coordination of the experimental work, and acquisition of the data. YS and YZ participated in the study design, data collection, analysis of the data and preparation of the manuscript. XY and YS carried out the study design, the analysis and interpretation of data and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal experiments were conducted in accordance with the National Institute of Health (NIH) guidelines and were approved by the Animal Ethics Committee of Yantaishan Hospital (approval no. 20190456-013).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Orangio GR. The economics of colon cancer. Surg Oncol Clin N Am. 2018;27:327–347. doi: 10.1016/j.soc.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Labianca R, Beretta GD, Kildani B, Milesi L, Merlin F, Mosconi S, Pessi MA, Prochilo T, Quadri A, Gatta G, et al. Colon cancer. Crit Rev Oncol Hematol. 2010;74:106–133. doi: 10.1016/j.critrevonc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, Wu J, Zhang J, Yang Z, Jin W, Li Y, Jin L, Yin L, Liu H, Wang Z. Integrated bioinformatics analysis of key genes involved in progress of colon cancer. Mol Genet Genomic Med. 2019;7:e00588. doi: 10.1002/mgg3.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadun RE, Sachsman SM, Chen X, Christenson KW, Morris WZ, Hu P, Epstein AL. Immune signatures of murine and human cancers reveal unique mechanisms of tumor escape and new targets for cancer immunotherapy. Clin Cancer Res. 2007;13:4016–4025. doi: 10.1158/1078-0432.CCR-07-0016. [DOI] [PubMed] [Google Scholar]
- 5.Indrieri A, Carrella S, Carotenuto P, Banfi S, Franco B. The pervasive role of the miR-181 family in development, neurode-generation, and cancer. Int J Mol Sci. 2020;21:2092. doi: 10.3390/ijms21062092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao LD, Zheng WW, Wang GX, Kang XC, Qin L, Ji JJ, Hao S. Epigenetic silencing of miR-181b contributes to tumorigenicity in colorectal cancer by targeting RASSF1A. Int J Oncol. 2016;48:1977–1984. doi: 10.3892/ijo.2016.3414. [DOI] [PubMed] [Google Scholar]
- 8.Xia Y, Gao Y. MicroRNA-181b promotes ovarian cancer cell growth and invasion by targeting LATS2. Biochem Biophys Res Commun. 2014;447:446–451. doi: 10.1016/j.bbrc.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Wang YL, Liu S, Zhang PP, Chen Z, Liu M, Tang H. miR-181b promotes cell proliferation and reduces apoptosis by repressing the expression of adenylyl cyclase 9 (AC9) in cervical cancer cells. FEBS Lett. 2014;588:124–130. doi: 10.1016/j.febslet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Rezaei T, Amini M, Hashemi ZS, Mansoori B, Rezaei S, Karami H, Mosafer J, Mokhtarzadeh A, Baradaran B. microRNA-181 serves as a dual-role regulator in the development of human cancers. Free Radic Biol Med. 2020;152:432–454. doi: 10.1016/j.freeradbiomed.2019.12.043. [DOI] [PubMed] [Google Scholar]
- 11.Lin Z, Li D, Cheng W, Wu J, Wang K, Hu Y. MicroRNA-181 functions as an antioncogene and mediates NF-κB pathway by targeting RTKN2 in ovarian cancers. Reprod Sci. 2019;26:1071–1081. doi: 10.1177/1933719118805865. [DOI] [PubMed] [Google Scholar]
- 12.Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer. 2013;13:184–199. doi: 10.1038/nrc3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanches JGP, Xu Y, Yabasin IB, Li M, Lu Y, Xiu X, Wang L, Mao L, Shen J, Wang B, et al. miR-501 is upregulated in cervical cancer and promotes cell proliferation, migration and invasion by targeting CYLD. Chem Biol Interact. 2018;285:85–95. doi: 10.1016/j.cbi.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Abdel-Mageed AB, Mondal D, Kandil E. The nuclear factor kappa-B signaling pathway as a therapeutic target against thyroid cancers. Thyroid. 2013;23:209–218. doi: 10.1089/thy.2012.0237. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Meng Q, Qiao W, Ma R, Ju W, Hu J, Lu H, Cui J, Jin Z, Zhao Y, Wang Y. miR-181b/Notch2 overcome chemoresis-tance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res Ther. 2018;9:327. doi: 10.1186/s13287-018-1072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu DD, Zhou PJ, Wang Y, Zhang L, Fu WY, Ruan BB, Xu HP, Hu CZ, Tian L, Qin JH, et al. Reciprocal activation between STAT3 and miR-181b regulates the proliferation of esophageal cancer stem-like cells via the CYLD pathway. Mol Cancer. 2016;15:40. doi: 10.1186/s12943-016-0521-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X, Feng J, Zhu Z, Yao L, Ma S, Hao B, Zhang G. A positive feedback loop between miR-181b and STAT3 that affects Warburg effect in colon cancer via regulating PIAS3 expression. J Cell Mol Med. 2018;22:5040–5049. doi: 10.1111/jcmm.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Huang W, Xu Y, Zhou L, Liang Y, Gao C, Long Y, Xu Y. CYLD deubiquitinase negatively regulates high glucose-induced NF-κB inflammatory signaling in mesangial cells. Biomed Res Int. 2017;2017:3982906. doi: 10.1155/2017/3982906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu RX, Liu RY, Wu CM, Zhao YS, Li Y, Yao YQ, Xu YH. DNA damage-induced NF-κB activation in human glioblastoma cells promotes miR-181b expression and cell proliferation. Cell Physiol Biochem. 2015;35:913–925. doi: 10.1159/000369748. [DOI] [PubMed] [Google Scholar]
- 23.Zehavi L, Schayek H, Jacob-Hirsch J, Sidi Y, Leibowitz-Amit R, Avni D. MiR-377 targets E2F3 and alters the NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol Cancer. 2015;14:68. doi: 10.1186/s12943-015-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.