Abstract
N6-methyladenosine (m6A) is the most common internal modification in eukaryotic mRNA. However, little is known about its role in non-small cell lung cancer (NSCLC). In this study, a total of 1017 NSCLC patients from the cancer genome atlas (TCGA) database with copy number variation (CNV) data were included. Log-rank tests and Cox regression model were used for survival analysis. The relationship between m6A regulators and clinicopathological features was evaluated using the chi-square test. The alteration of m6A regulators were related to T stage. Patients with any CNVs of regulators genes had worse overall survival (OS) than those with diploid genes. The deletion of m6A writer genes was an independent risk factor for poor OS, and the effect synergized with that of copy number gain of eraser genes. High expression of Fat mass-and obesity-associated gene (FTO) was associated with KRAS signaling up. Knockdown of FTO increased m6A content and inhibit proliferation of A549 lung cancer cell. Thus, we identified the genetic changes of m6A regulatory factors in NSCLC for the first time and found a significant relationship between these changes and poor clinical characteristics. FTO might play an important role in promoting NSCLC by decreasing m6A level and activating KRAS signaling.
Keywords: FTO, non-small cell lung cancer (NSCLC), prognosis, m6A, N6-methyladenosine
INTRODUCTION
Lung cancer is one of the most lethal tumors, with a mean survival rate of 18.4% at one year [1], and results in more than 1.3 million deaths per year [2]. NSCLC is the most common pathological type of lung cancer, accounting for about 85% of lung cancer [3]. In recent years, surgery, radiotherapy, chemotherapy, and targeted therapy have been shown to prolong the survival of patients with NSCLC [4]. However, the prognosis of NSCLC patients remains poor, with a 5-year survival rate of only 18% [5]. Therefore, there is an urgent need to find a new and meaningful biomarker or modification in NSCLC cells.
Apart from the genetic elements and proteins that play a vital effect in the occurrence and development of cancer, RNA modifications also play a crucial role in tumorigenesis. Among the RNA modifications, m6A is the most common, participating in multiple biological processes [6–8], such as cell death [9], cancer stem cell formation [10], tumorigenesis [11, 12], as well as contributing to the development of pathological conditions such as obesity and type 2 diabetes [13]. m6A occupies about 0.1–0.4% of the total adenosine residues in cellular RNA [6–8]. The process of m6A modification is mediated by enzymes that function as writers (methylases), erasers (demethylases), and readers [14]. Writers include METTL14, METTL3 and WTAP, and their complex promotes m6A modification in RNA. By contrast, erasers reverse the effect of writers in m6A-modified mRNA. FTO, the first recognized demethylase, has been proven to have cancer-promoting activity in gastric cancer, breast cancer, acute myeloid leukemia (AML), and cervical squamous cell carcinoma [15–19]. The readers enable the instructions of m6A modification to be converted into functional signals, including YTH domain family proteins [20].
In recent years, m6A regulators have been reported to enhance the development of diverse carcinomas, including liver cancer [12], acute myeloid leukemia [21], and glioblastoma [22]. However, m6A regulators also act as tumor suppressors in renal cell carcinoma [21]. Although many studies explain the role of m6A regulators in cancers, little is known about the function of m6A regulators in NSCLC. The present study aimed to understand the functions of m6A regulators in NSCLC by sequencing and analysis of CNV data from TCGA database.
RESULTS
Mutations and CNVs of m6A regulatory genes in NSCLC patients
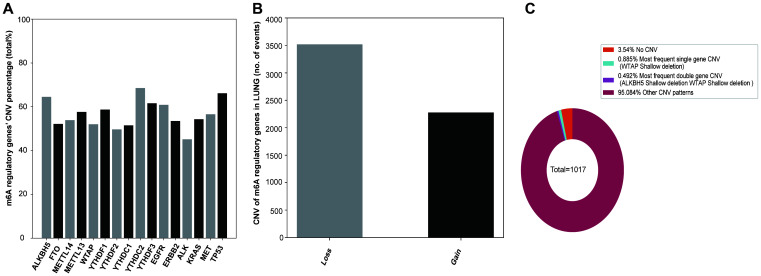
In the sequencing analysis, only 45 out of 408 samples showed mutations in the m6A regulatory genes (Supplementary Table 1). However, among the 1017 NSCLC samples with CNV data, CNVs were often observed in 10 m6A regulatory genes (Figure 1A). From these 10, the frequency of CNVs of YTHDC2 was the highest (68.53% 697/1017), that of YTHDF2 was the lowest (49.66% 505/1017), and that of the other 8 m6A regulatory factor genes was more than 50%. In addition, CNVs of TP53 and EGFR genes were higher in NSCLC patients, about 66.18% and 60.89%, respectively than in controls.
Figure 1.
CNVs of m6A regulatory genes in NSCLC. (A) Percentage of lung cancer samples with CNVs in m6A regulatory factors in TCGA data. (B) Loss and gain of copy number of m6A regulatory factors in patients with NSCLC. (C) The most common CNV mutation pattern of m6A regulators in patients with NSCLC.
Next, we explored the CNV mutation pattern of m6A regulatory factors in NSCLC patients and found that the most frequent CNV type was loss of copy number, and the frequency was similar to that in ccRCC [23] and AML [24] (Figure 1B, Table 1). Because of the high frequency of CNVs of m6A regulatory factors in NSCLC patients, the frequency of CNV of only one regulatory factor or that of two genes at the same time is relatively small. The results showed that the shallow deletion of WTAP is the most frequent CNV of m6A regulatory genes (0.885%) and shallow deletion of WTAP and copy number gain of YTHDF3 were the most frequent double-gene CNV (0.492%) (Figure 1C).
Table 1. Different CNV patterns found in lung cancer samples (n = 1017).
| gene | Diploid | Deep deletion | Shallow deletion | Copy number gain | Amplification | CNV sum | Percentage | |
| Eraser | ALKBH5 | 361 | 9 | 510 | 131 | 6 | 656 | 64.50% |
| FTO | 487 | 7 | 324 | 193 | 6 | 530 | 52.11% | |
| Writer | METTL14 | 469 | 0 | 473 | 75 | 0 | 548 | 53.88% |
| METTL3 | 431 | 4 | 270 | 296 | 16 | 586 | 57.62% | |
| WTAP | 488 | 8 | 382 | 138 | 1 | 529 | 52.02% | |
| Reader | YTHDF1 | 420 | 1 | 106 | 458 | 32 | 597 | 58.70% |
| YTHDF2 | 512 | 4 | 332 | 166 | 3 | 505 | 49.66% | |
| YTHDC1 | 493 | 2 | 374 | 124 | 24 | 524 | 51.52% | |
| YTHDC2 | 320 | 8 | 585 | 102 | 2 | 697 | 68.53% | |
| YTHDF3 | 391 | 3 | 117 | 474 | 32 | 626 | 61.55% | |
| Hot gene | EGFR | 398 | 6 | 85 | 465 | 63 | 619 | 60.87% |
| ERBB2 | 473 | 1 | 148 | 370 | 25 | 544 | 53.49% | |
| ALK | 558 | 5 | 46 | 397 | 11 | 459 | 45.13% | |
| KRAS | 465 | 2 | 128 | 366 | 56 | 552 | 54.28% | |
| MET | 442 | 5 | 130 | 416 | 24 | 575 | 56.54% | |
| TP53 | 344 | 10 | 577 | 85 | 1 | 673 | 66.18% |
Relationship between alterations in m6A regulatory factors and clinicopathological and molecular characteristics
We evaluated the relationship between alterations in m6A regulatory genes (CNV and mutation) and clinicopathological parameters of patients. The results showed that there was a significant correlation between the change in m6A regulatory factors and T stage (p= 0.02) (Table 2). Next, we evaluated the relationship between m6A regulatory gene alterations and the hot genes (EGFR, ERBB2, ALK, MET, TP53 and KRAS) in NSCLC. As expected, there was a significant relationship between the alterations in m6A regulatory factors and alterations in these six genes. The result indicated the patients with mutation and CNV had more the alterations of hot genes than the patients without mutation or CNV (p<0.0001) (Table 3).
Table 2. Clinicopathological parameters of NSCLC patients with or without mutation/CNV of m6A regulatory genes.
| With mutation and/or CNV* | Without mutation and CNV* | P-value | ||
| Age | <=60 | 690 | 7 | 0.417 |
| >60 | 255 | 29 | ||
| gender | Female | 372 | 18 | 0.269 |
| Male | 573 | 18 | ||
| Pathological Stage | I | 479 | 22 | 0.363 |
| II | 267 | 10 | ||
| III | 157 | 3 | ||
| IV | 32 | 0 | ||
| Discrepancy | 10 | 1 | ||
| T stage | T1 | 259 | 18 | 0.02 |
| T2 | 536 | 11 | ||
| T3 | 107 | 6 | ||
| T4 | 41 | 1 | ||
| TX | 2 | 0 | ||
| N stage | N0 | 600 | 30 | 0.162 |
| N1 | 218 | 3 | ||
| N2 | 105 | 3 | ||
| N3 | 7 | 0 | ||
| Nx | 15 | 0 | ||
| M stage | M0 | 709 | 23 | 0.078 |
| M1 | 204 | 0 | ||
| MX | 32 | 13 |
*With mutation or CNV: Cases have mutant or CNV or mutant+CNV, confirmed through TCGA database. Without mutant and CNV: Cases with neither mutant nor CNV, confirmed through TCGA database. Ambiguous variables (Nx, Mx, N/A, discrepancy and Gx) were excluded from chi-square test or non-parametric test.
Table 3. Relationship between molecular characteristics and alteration of m6A regulatory genes in patients with NSCLC.
| Without mutation or CNV* | With mutation and CNV* | X2 | P | |||
| EGFR | N=1017 | wt | 4 | 615 | 36.65 | <0.0001 |
| alteration | 32 | 366 | ||||
| ERBB2 | N=1017 | wt | 0 | 544 | 40.72 | <0.0001 |
| alteration | 36 | 437 | ||||
| ALK | N=1017 | wt | 0 | 459 | 28.84 | <0.0001 |
| alteration | 36 | 522 | ||||
| KRAS | N=1017 | wt | 5 | 547 | 22.87 | <0.0001 |
| alteration | 31 | 434 | ||||
| MET | N=1017 | wt | 2 | 573 | 37.36 | <0.0001 |
| alteration | 34 | 408 | ||||
| TP53 | N=1017 | wt | 1 | 672 | 64.11 | <0.0001 |
| alteration | 35 | 309 |
*With mutation or CNV: Cases have mutant or CNV or mutant+CNV of m6A regulators, confirmed through TCGA database. Without mutant and CNV: Cases with neither mutant nor CNV of m6A regulators, confirmed through TCGA database.
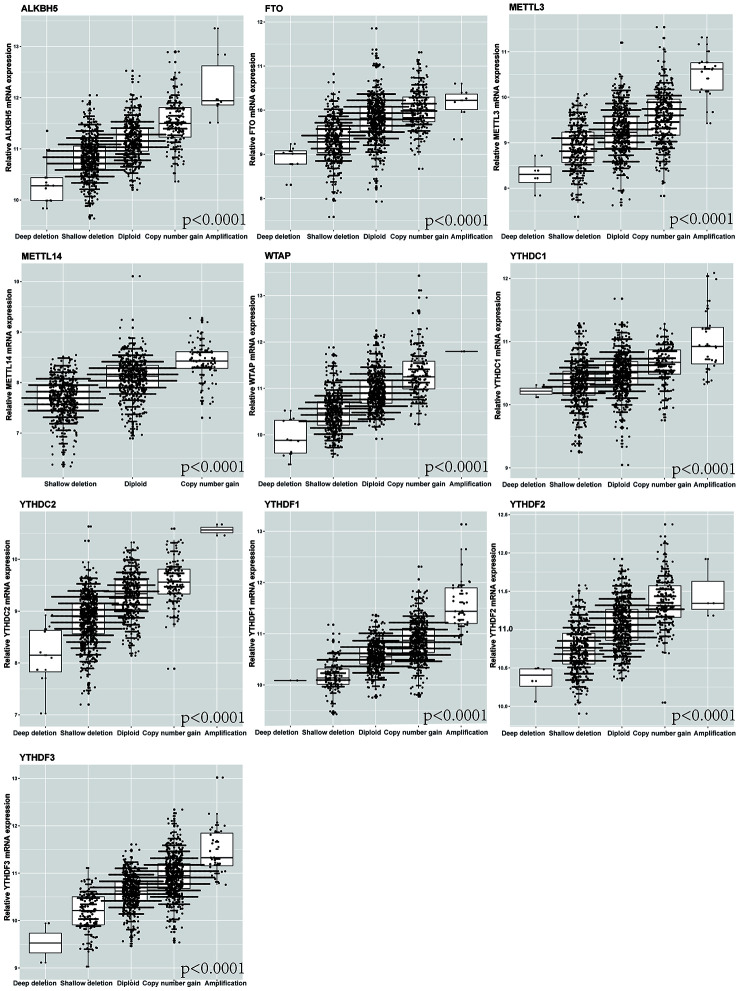
Furthermore, we detected the effect of the alterations of m6A regulatory factors on mRNA expression in NSCLC patients. The results revealed that mRNA expression was significantly correlated with various CNV mutation patterns (p < 0.0001). For all the top 10 genes with CNV, copy number gain was associated with increased mRNA expression, while copy number loss was associated with decreased mRNA expression (Figure 2).
Figure 2.
Correlation between different CNV patterns of 10 m6A regulatory genes and mRNA expression level.
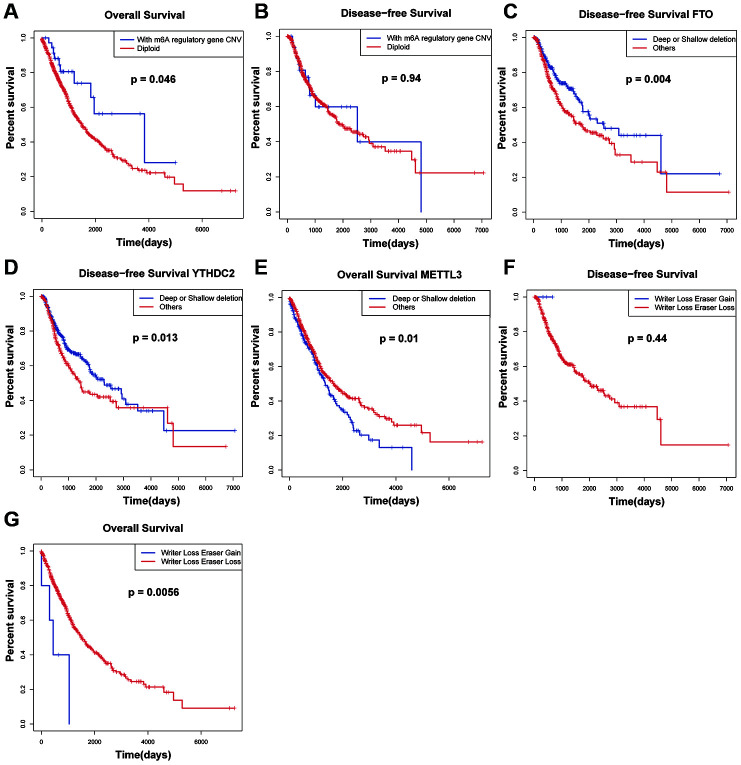
Relationship between CNVs of m6A regulatory genes and survival of NSCLC patients
We also analyzed the relationship between CNVs of m6A regulatory genes and OS and DFS of patients. The results showed that the patients with m6A regulatory gene CNV had better OS than the patients with diploid. (Figure 3A and Figure 3B). Then, the OS and DFS analysis with respect to 10 m6A regulatory genes in NSCLC patients were carried out. The results showed that the patients with FTO and YTHDC2 deletion CNVs had better DFS than those with diploid and copy number gain (Figure 3C and Figure 3D) and patients with METTL3 deletion CNVs had worse OS than those with diploid and copy number gain (Figure 3E). The other genes showed no significant effects. Multivariate Cox regression analysis showed that the alteration of m6A regulatory genes was an independent risk factor for poor OS (Table 4). In addition, the "writers" are a group of methyltransferase genes, and are the most important part of the regulation process of m6A. "Erasers" are a group of demethylase methyltransferase genes. The results show that the "writer" gene down-regulation and "eraser" gene up-regulation may lead to the decline of survival rate of patients.
Figure 3.
Survival rate of patients with CNVs of m6A regulatory factors. (A, B) Relationship between OS, RFS and m6A regulator carrying CNV or diploid in NSCLC patients. (C, D) DFS for NSCLC patients with different CNV patterns of FTO and YTHDC2. (E) OS for NSCLC patients with different CNV patterns of FTO and YTHDC2. (F, G) Relationship between simultaneous changes in m6A regulatory factors: writer genes and eraser genes, and OS, and DFS in NSCLC patients.
Table 4. Clinical Information and risk model univariate/multivariate COX analysis of m6A regulatory genes for overall survival and disease-free survival of patients with NSCLC.
| Variables | OS | DFS | ||||||
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR(95%CI) | p.Value | HR(95%CI) | p.Value | HR(95%CI) | p.Value | HR(95%CI) | p.Value | |
| Age(≥60 vs.<60) | 1.21(0.92-1.57) | 0.168 | 1.04(0.75-1.45) | 0.798 | ||||
| Gender (male vs female) | 1.23(0.97-1.55) | 0.089 | 1.03(0.77-1.38) | 0.835 | ||||
| Pathologic stage (I-II vs III + IV) | 1.86(1.46-2.38) | <0.0001* | 1.19(0.83-1.7) | 0.348 | 1.89(1.35-2.63) | <0.0001* | 1.28(0.82-1.99) | 0.269 |
| M (M1 vs M0) | 1.98(1.21-3.24) | 0.006* | 1.64(0.95-2.83) | 0.078 | 1.37(0.61-3.1) | 0.449 | ||
| N (N1, N2, N3 vs N0) | 1.53(1.23-1.91) | <0.0001* | 1.38(1.07-1.79) | 0.014* | 1.65(1.24-2.2) | 0.001* | 1.51(1.08-2.11) | 0.015* |
| T (T3-T4 vs T1-T2) | 1.74(1.33-2.29) | <0.0001* | 1.55(1.11-2.15) | 0.01* | 1.89(1.32-2.73) | 0.001* | 1.64(1.06-2.53) | 0.026* |
| EGFR (altered vs diploid) | 1.18(0.94-1.48) | 0.148 | 1.3(0.98-1.74) | 0.071 | ||||
| ERBB2 (altered vs diploid) | 0.94(0.76-1.18) | 0.614 | 1.09(0.82-1.45) | 0.549 | ||||
| ALK (altered vs diploid) | 1.27(1.02-1.59) | 0.035* | 1.35(1.07-1.69) | 0.01* | 1(0.75-1.33) | 0.984 | ||
| KRAS (altered vs diploid) | 0.91(0.73-1.13) | 0.391 | 0.9(0.67-1.19) | 0.451 | ||||
| MET (altered vs diploid) | 1.17(0.94-1.47) | 0.156 | 1.34(1.01-1.78) | 0.045* | 1.31(0.97-1.75) | 0.075 | ||
| TP53 (altered vs diploid) | 0.96(0.76-1.22) | 0.74 | 1.06(0.78-1.43) | 0.719 | ||||
| m6A regulator alteration (Writer Loss + Eraser Gain vs others) | 0.27(0.1-0.73) | 0.01* | 0.31(0.11-0.85) | 0.022* | 1212451(0-Inf) | 0.992 | ||
Ambiguous variables (Nx, Mx, N/A, discrepancy and Gx) were excluded
In order to verify our results, we tested the association between CNVs combinations with different patterns of m6A carried by patients and OS and DFS. We found that compared with patients with only writer gene deletion, when the down-regulation of "writer" genes and the up-regulation of "erasers" genes occur simultaneously, the prognosis of patients was worse (Figure 3F and Figure 3G).
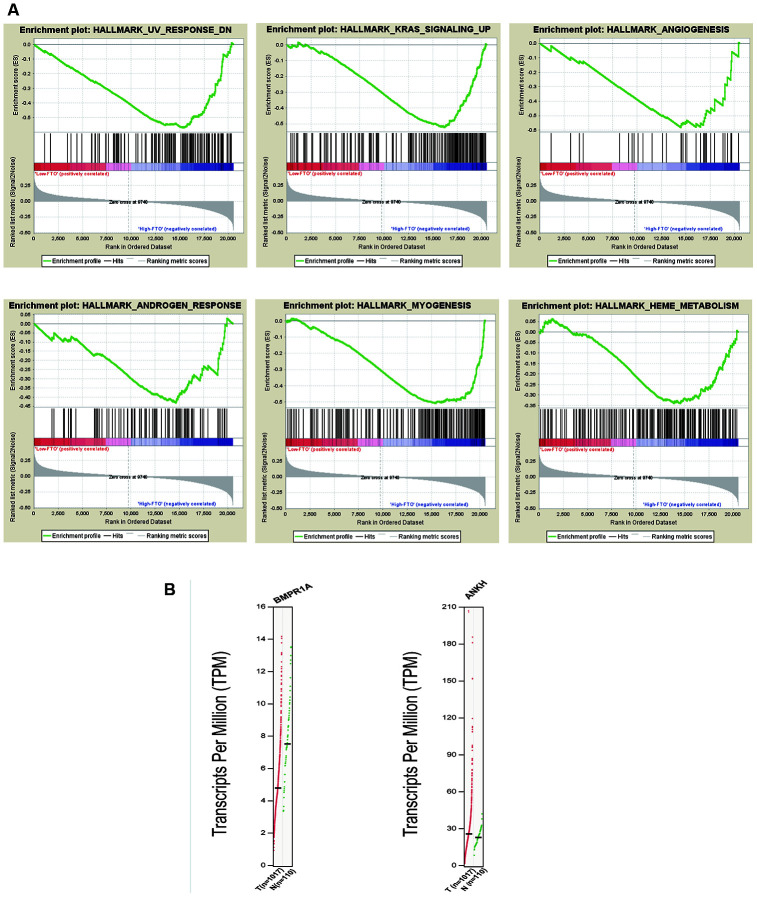
Enrichment analysis of “eraser” genes: FTO
It has been shown that FTO is related to the occurrence and development of NSCLC. Thus, we divided all samples into high and low FTO mRNA expression levels according to the median FTO mRNA expression levels and performed the GSEA to conduct enrichment analysis. The high expression of FTO was significantly enriched in nine related pathways such as UV radiation response, myogenesis, KRAS signaling pathway and TGF-beta signaling (Figure 4A and Supplementary Table 2). Therefore, the genetic alterations of m6A regulatory factors in NSCLC were related to poor survival. To test our findings, we examined gene expression associated with these pathways. The result demonstrated that BMPR1A associated with TGF-beta signal and UV radiation pathway and was downregulated in NSCLC tissues. ANKH associated with androgen response pathway was upregulated in NSCLC tissues (Figure 4B). Besides, several studies have found that FTO could participate in UV radiation response, myogenesis and androgen response, which are consistent with our results [25–27].
Figure 4.
Enrichment analysis of “eraser” genes: FTO. (A) GSEA results of FTO with different expression levels. (B) Expression of genes related to enrichment pathway.
FTO inhibition suppressed the proliferation of NSCLC cells and Increase the level of mRNA m6A in A549
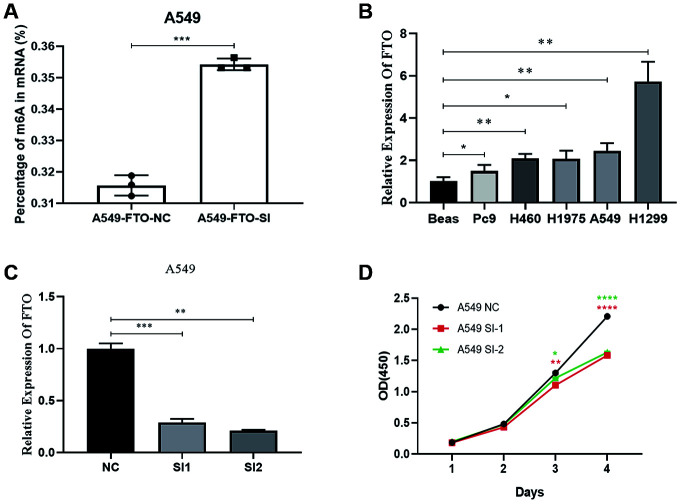
By the EpiQuik m6A RNA Methylation Quantification Kit, we found the m6A content in A549 knocked down the FTO increased (Figure 5A). Furthermore, using qRT-PCR, we found that FTO was highly expressed in A549. Then we used silencer-FTO to knock down the FTO (Figure 5B and Figure 5C). Next, by the CCK-8 assay, we showed knockdown of FTO inhibited the proliferation capacity of A549 lung cancer cell (Figure 5D). This is consistent with our results.
Figure 5.
Effects of silencing FTO on proliferation and mRNA m6A level of lung cancer cells. (A) The mRNA m6A level in human lung cancer cell. (B) The expression of FTO in lung cancer cells. (C) Verification of knockout efficiency. (D) Knockdown of proliferation capacity of FTO inhibited lung cancer cells. (*, p < 0.05; **, p < 0.01; ***, ****, p < 0.001).
DISCUSSION
N6-methyladenosine (m6A) is the most common internal modification in eukaryotic mRNA, the abundance of which varies from 0.1% to 0.4% of total adenosine residues [6–9]. Actually, previous study proved m6A existed in mRNA of more than 7600 genes and over 300 non-coding RNA [28]. It has been reported that m6A is involved in a variety of cellular processes such as cell proliferation, self-renewal, development and cell death [9]. Considering the importance of m6A in transcriptome, some studies have attempted to reveal the role of m6A in cancer and the results shown the regulators of m6A exert enormous functions in cancer development, such as proliferation, migration and invasion [29, 30]. In recent years, m6A regulators have been reported to enhance the development of diverse carcinomas, including liver cancer (11), acute myeloid leukemia [21], and glioblastoma [22]. However, m6A regulators also acted as tumor suppressors in renal cell carcinoma [31]. In our study, the frequency of alterations of the ten m6A related genes was much higher than that shown in AML and ccRCC, suggesting that dysregulation of m6A may play a significant role in the occurrence and development of NSCLC than AML and ccRCC [23, 24]. Thus, having a better understanding and further exploring the underlying mechanism for the m6A regulators in NSCLC is necessary.
Based on the statistics of 408 NSCLC patients, we found the mutations related to m6A regulatory factors in 45 samples. From the 10 m6A regulatory genes, shallow deletion of m6A “writer” gene WTAP and the copy number gain of m6A “reader” gene YTHDF3 were the most frequent double gene alterations, and shallow deletion of m6A “writer” gene WTAP was the most frequent single alteration, implying the importance of m6A writer genes in the process of RNA m6A modification. Due to the fact that EGFR, ERBB2, ALK, MET, TP53 and KRAS played important roles in the pathogenesis of NSCLC, we further evaluated the correlation between m6A regulatory gene variation and the alterations of six hot genes in NSCLC. The result indicated the patients with mutation and CNV of m6A regulators had more the alterations of hot genes than the patients without mutation or CNV (p<0.0001). Previous studies shown that EGFR mutation was related to the level of mRNA methylation in pancreatic cancer [32], and TP53 mutation was related to the level of mRNA methylation in gastric cancer [33].
Upon analyzing the relationship between CNV/mutation in m6A regulators and clinicopathological parameters of patients by chi-squared test, we found that there was a significant correlation between the change of m6A regulatory factors and T stage (p=0.02). Multivariate Cox regression analysis showed that the alteration of m6A regulatory genes was an independent risk factor for poor OS. For all the top 10 genes with CNV, copy number gain was associated with increased mRNA expression, while copy number loss was associated with decreased mRNA expression, revealing that mRNA expression was significantly correlated with various CNV mutation patterns. Besides, the results showed that the patients with m6A regulatory gene CNV had better OS than the patients with diploid. Sun et al. found that NSCLC patients with high expression of METTL3 was associated with better OS [34]. However, Gregory et al. indicated the upregulation of METTL3 could promote the growth, survival, and invasion of human lung cancer cells [35]. In the present study, patients with METTL3 (writer gene) deletion CNVs had worse OS than those with diploid and copy number gain. Moreover, the patients with FTO (eraser gene) and YTHDC2 (reader gene) deletion CNVs had better DFS than those with diploid and copy number gain. Previous study suggested lung cancer patients with depletion of FTO had better prognosis [36]. These results suggested that the down-regulation of m6A level might be associated with poor patient survival. Next, to validate our result, the associations among CNVs combinations with different patterns of m6A carried by patients and OS and DFS were tested, and we found that the "writer" gene down-regulation and "eraser" gene up-regulation may lead to the decline of survival rate of patients. This was consistent with our results.
Furthermore, we found that some studies have discussed the possible mechanisms of action of METTL3 in NSCLC. For example, METTL3 contributes to transforming growth factor-beta-induced epithelial-mesenchymal transition of NSCLC cells through the regulation of JUNB [37]. METTL3 has been found to promote protein translation of oncogenes in lung cancer cells through methyltransferase-independent activity [38]. However, little was known about the mechanisms of action of FTO. Thus, we further explored the correlated pathways with FTO. FTO was originally identified as a fat mass and obesity-associated protein and has been regarded as the first RNA demethylase in recent study [15, 39]. The GSEA analysis suggested that the high FTO expression was significantly enriched in UV radiation response, angiogenesis and KRAS signaling. Moreover, we found that BMPR1A related to TGF-β signaling and UV radiation response was down-regulated in NSCLC tissues and ANKH related to androgen response was up-regulated, which partially validate the GSEA results. OI. Kit et al. analyzed the copy number variation of some gene loci in lung tumor cells extracted by laser capture microdissection and in cell-free DNA in the plasma of patients with lung adenocarcinoma and detected the copy number variation of KRAS and FTO at the same time [40]. GSEA in high expression of FTO showed enrichment of genes up-regulated in KRAS signaling, indicating the activation of KRAS signaling by the upregulation of FTO. In addition, the results have indicated the patients with mutation and CNV had more KRAS alteration than the patients without mutation or CNV (p<0.0001), suggesting there was a significant correlation between KRAS alteration and m6A regulatory gene alterations. KRAS was the most frequently mutated oncogene in NSCLC and KRAS mutations were often associated with low overall survival and resistance to treatment [41, 42]. Previous studies have shown that MEK-ERK depletion reduced the expression of EZH2 in cells with KRAS (G12C) mutation, thereby reducing the level of histone methylation and inhibiting the development of cancer [43]. Therefore, we thought that the eraser gene FTO might down-regulate the m6A level of key molecules in KRAS signaling and further activate KRAS signaling to promote tumor.
Additionally, we found FTO was highly expressed in NSCLC cell A549 and knocking down FTO could inhibit the proliferation of A549, and mRNA m6A content analysis showed that the m6A content in A549 that knocked down the FTO increased. All this partly proved our hypothesis: the down-regulation of m6A level might be associated with poor survival in NSCLC patients. FTO has been reported to participate in the development of NSCLC. Depletion of FTO inhibited the proliferation, invasion, emigration of lung cancer cells [36]. And research has suggested that FTO facilitates lung adenocarcinoma cell progression by activating cell migration through m6A demethylation [44].
In summary, we determined, for the first time, the genetic alterations in m6A regulatory genes in NSCLC and identified a significant relationship between the alterations resulting in decreased m6A level and worse clinical characteristics including survival. Moreover, we found eraser gene FTO might play an important role in promoting NSCLC by decreasing m6A level and activating KRAS signaling. Future studies uncovering the oncogenesis mechanisms of FTO will be required to confirm our findings.
MATERIALS AND METHODS
NSCLC TCGA data
NSCLC-related data, including somatic non-silent mutation (gene-level), phenotype, gene expression, RNA-seq, and copy number (gene-level) data, were downloaded from TCGA (https://cancergenome.nih.gov/).
Mutation of m6A regulatory genes and characterization of CNV in NSCLC patients
The somatic mutation data were used to calculate the somatic mutations of m6A regulatory factors and the copy number (gene level) data were used to calculate the CNV mutation pattern distribution of m6A-related regulatory factors in NSCLC patients. We collected the mutation information of the patients and selected for information of m6A gene. Simultaneously, statistical analyses and visualization were performed to assess the percentage of NSCLC samples with CNV in all patients, the number of samples with amplification and deletion, and the frequency of one or two regulatory factors with CNV.
Analysis of the relationship between the changes in m6A regulatory factors and clinicopathological and molecular characteristics of tumor patients
COX regression analysis was used to explore the correlation between different CNV patterns and the levels of m6A regulatory gene mRNA. The relationship between CNV of m6A-related regulatory factors and clinicopathology was evaluated. Chi-square test was used for analyzing statistical significance.
Association between CNV of m6A regulatory genes and survival of cancer patients
The effects of CNVs on overall survival (OS) and disease-free survival (DFS) of cancer patients and the risk factors were assessed by survival analysis and a univariate/multivariate Cox regression model.
Enrichment analysis of m6A regulatory genes
GSEA was performed to enrich and analyze m6A regulatory genes and the genes that affected cellular pathways were selected.
Quantitative reverse transcription PCR (qRT-PCR) and mRNA m6A level in human NSCLC cells
Total RNA was isolated from cells with RNA Extraction Kit (Aidlab Biotechnologies Co, Ltd, China). The RNA concentrations were detected using a Nanodrop spectrophotometer (Thermo Scientific Ltd., USA). cDNA synthesis was completed using a reverse transcription kit according to the manufacturer’s instructions. qRT-PCR was performed on the CFX ConnectTM Real-Time PCR Detection System (Bio-Rad, USA) using the ChamQTM SYBR® qPCR Master Mix. The expression of GAPDH was used as the reference for normalization. The 2-ΔΔCt method was used to calculate the relative fold change in mRNA expression. The EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (P-9005, Epigentek, USA) was used to measure the m6A content in total RNAs.
Cell proliferation assay
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8; Beyotime, Hangzhou, People’s Republic of China) according to the manufacturer’s instructions. Transfected A549 cells (3000 cells/well) were seeded in 96-well plates with three replicate wells per group. The optical density (OD) was measured at 450 nm every 24 hours using a multimodal plate reader (PE Enspire, USA).
Statistical analysis
The R software was used to analyze all statistics and graphs. Chi-square test was used to analyze the association between m6A regulatory gene CNVs and clinicopathological features. Kaplan-Meier curve analysis and the logarithmic rank test were used to evaluate the predictive value of changes in m6A regulatory genes. Survminer and survival packages in R language were employed for Cox proportional hazard regression model analysis. All statistical results with p <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank everyone who take part in this study. This study was supported by the Independent scientific research project of Wuhan University. (1606-413000202).
Footnotes
AUTHOR CONTRIBUTIONS: Zhe Dong and Jinping Zhao designed the idea; Hongjie Shi, Jinping Zhao, and Linzhi Han collected and analyzed the data, and drafted the paper; Kaijie Wang and Jiajun Shi analyzed the data; Jinping Zhao and Ming Xu revised the final paper. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST: The authors declare no conflicts of interest.
REFERENCES
- 1.Thiru P, Kern DM, McKinley KL, Monda JK, Rago F, Su KC, Tsinman T, Yarar D, Bell GW, Cheeseman IM. Kinetochore genes are coordinately up-regulated in human tumors as part of a FoxM1-related cell division program. Mol Biol Cell. 2014; 25:1983–94. 10.1091/mbc.E14-03-0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127:2893–917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 3.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, D’Amico TA, Demmy TL, Govindan R, Grannis FW Jr, Grant SC, Horn L, Jahan TM, et al. , and National comprehensive cancer network. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw. 2013; 11:645–53. 10.6004/jnccn.2013.0084 [DOI] [PubMed] [Google Scholar]
- 4.Tamura N, Simon JE, Nayak A, Shenoy R, Hiroi N, Boilot V, Funahashi A, Draviam VM. A proteomic study of mitotic phase-specific interactors of EB1 reveals a role for SXIP-mediated protein interactions in anaphase onset. Biol Open. 2015; 4:155–69. 10.1242/bio.201410413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 6.Perry RP, Kelley DE, Friderici K, Rottman F. The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5’ terminus. Cell. 1975; 4:387–94. 10.1016/0092-8674(75)90159-2 [DOI] [PubMed] [Google Scholar]
- 7.Dubin DT, Taylor RH. The methylation state of poly a-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975; 2:1653–68. 10.1093/nar/2.10.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975; 4:379–86. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- 9.Liu N, Pan T. N6-methyladenosine–encoded epitranscriptomics. Nat Struct Mol Biol. 2016; 23:98–102. 10.1038/nsmb.3162 [DOI] [PubMed] [Google Scholar]
- 10.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016; 113:E2047–56. 10.1073/pnas.1602883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, De Braekeleer E, Ponstingl H, Hendrick A, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017; 552:126–131. 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018; 67:2254–2270. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- 13.Klungland A, Dahl JA. Dynamic RNA modifications in disease. Curr Opin Genet Dev. 2014; 26:47–52. 10.1016/j.gde.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 14.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, Bhattarai DP, Zhao YL, Sun BF, Yang YG. Cytoplasmic M6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017; 27:444–47. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011; 7:885–87. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akbari ME, Gholamalizadeh M, Doaei S, Mirsafa F. FTO gene affects obesity and breast cancer through similar mechanisms: a new insight into the molecular therapeutic targets. Nutr Cancer. 2018; 70:30–36. 10.1080/01635581.2018.1397709 [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017; 38:2285–92. 10.3892/or.2017.5904 [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017; 31:127–41. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018; 57:590–97. 10.1002/mc.22782 [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–20. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, Schulman J, Famulare C, Patel M, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017; 23:1369–76. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, He C, Shi Y. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017; 18:2622–34. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, Wang J, Hong B, Ma K, Xie H, Li L, Zhang K, Zhou B, Cai L, Gong K. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging (Albany NY). 2019; 11:1633–47. 10.18632/aging.101856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017; 10:39. 10.1186/s13045-017-0410-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu PH, Zou L, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017; 543:573–76. 10.1038/nature21671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Huang N, Yang M, Wei D, Tai H, Han X, Gong H, Zhou J, Qin J, Wei X, Chen H, Fang T, Xiao H. FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 2017; 8:e2702. 10.1038/cddis.2017.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghafarian-Alipour F, Ziaee S, Ashoori MR, Zakeri MS, Boroumand MA, Aghamohammadzadeh N, Abbasi-Majdi M, Shool F, Asbaghi NS, Mohammadi A, Zarghami N. Association between FTO gene polymorphisms and type 2 diabetes mellitus, serum levels of apelin and androgen hormones among Iranian obese women. Gene. 2018; 641:361–366. 10.1016/j.gene.2017.10.082 [DOI] [PubMed] [Google Scholar]
- 28.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012; 149:1635–46. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol. 2018; 6:89. 10.3389/fbioe.2018.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019; 112:108613. 10.1016/j.biopha.2019.108613 [DOI] [PubMed] [Google Scholar]
- 31.Li X, Tang J, Huang W, Wang F, Li P, Qin C, Qin Z, Zou Q, Wei J, Hua L, Yang H, Wang Z. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget. 2017; 8:96103–16. 10.18632/oncotarget.21726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng Y, Guan R, Hong W, Huang B, Liu P, Guo X, Hu S, Yu M, Hou B. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann Transl Med. 2020; 8:387. 10.21037/atm.2020.03.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, Gong J, Shen L. Reduced m6A modification predicts Malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019; 8:4766–81. 10.1002/cam4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu X, Liu L, Li J, Hu Q, Sun R. Expression and prognostic significance of m6A-related genes in lung adenocarcinoma. Med Sci Monit. 2020; 26:e919644. 10.12659/MSM.919644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016; 62:335–45. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, Zheng H, Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019; 512:479–85. 10.1016/j.bbrc.2019.03.093 [DOI] [PubMed] [Google Scholar]
- 37.Wanna-Udom S, Terashima M, Lyu H, Ishimura A, Takino T, Sakari M, Tsukahara T, Suzuki T. The m6A methyltransferase METTL3 contributes to Transforming Growth Factor-beta-induced epithelial-mesenchymal transition of lung cancer cells through the regulation of JUNB. Biochem Biophys Res Commun. 2020; 524:150–155. 10.1016/j.bbrc.2020.01.042 [DOI] [PubMed] [Google Scholar]
- 38.Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019; 11:1177–87. 10.2147/CMAR.S181058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loos RJ, Yeo GS. The bigger picture of FTO: the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014; 10:51–61. 10.1038/nrendo.2013.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutilin DS, Airapetova TG, Anistratov PA, Pyltsin SP, Leiman IA, Karnaukhov NS, Kit OI. Copy number variation in tumor cells and extracellular DNA in patients with lung adenocarcinoma. Bull Exp Biol Med. 2019; 167:771–78. 10.1007/s10517-019-04620-y [DOI] [PubMed] [Google Scholar]
- 41.Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005; 1756:81–82. 10.1016/j.bbcan.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 42.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003; 3:11–22. 10.1038/nrc969 [DOI] [PubMed] [Google Scholar]
- 43.Riquelme E, Behrens C, Lin HY, Simon G, Papadimitrakopoulou V, Izzo J, Moran C, Kalhor N, Lee JJ, Minna JD, Wistuba II. Modulation of EZH2 expression by MEK-ERK or PI3K-AKT signaling in lung cancer is dictated by different KRAS oncogene mutations. Cancer Res. 2016; 76:675–85. 10.1158/0008-5472.CAN-15-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Y, Qi N, Wang K, Huang Y, Liao J, Wang H, Tan A, Liu L, Zhang Z, Li J, Kong J, Qin S, Jiang Y. FTO facilitates lung adenocarcinoma cell progression by activating cell migration through mRNA demethylation. Onco Targets Ther. 2020; 13:1461–70. 10.2147/OTT.S231914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.