Abstract
Drug resistance and immune escape of tumor cells severely compromise the treatment efficiency of hepatocellular carcinoma (HCC). Long non-coding RNA KCNQ1 overlapping transcript 1 (lncRNA KCNQ1OT1) has been shown to be involved in drug resistance in several cancers. The aim of the present study was to investigate the role of KCNQ1OT1 in sorafenib resistance and immune escape of HCC cells. Reverse transcription-quantitative PCR analysis, western blotting and immunohistochemistry were performed to detect the expression of KCNQ1OT1, miR-506 and programmed death-ligand-1 (PD-L1). Cell Counting Kit-8 assay, flow cytometry and Transwell assays were used to evaluate IC50 value, cell apoptosis and metastasis. ELISA was performed to detect the secretion of cytokines. Dual-luciferase reporter assay was conducted to verify the targeting relationships between miR-506 and KCNQ1OT1 or PD-L1. KCNQ1OT1 and PD-L1 were found to be upregulated and miR-506 was downregulated in sorafenib-resistant HCC tissues and cells. Furthermore, KCNQ1OT1 knockdown reduced the IC50 value of sorafenib, suppressed cell metastasis and promoted apoptosis in sorafenib-resistant HCC cells. Moreover, KCNQ1OT1 knockdown changed the tumor microenvironment and T-cell apoptosis in a sorafenib-resistant HCC/T-cell co-culture model. In addition, it was demonstrated that KCNQ1OT1 functioned as a competing endogenous RNA of miR-506 and increased PD-L1 expression in sorafenib-resistant HCC cells. miR-506 inhibition abolished the effects of KCNQ1OT1 knockdown on sorafenib sensitivity, tumor growth, the tumor microenvironment and T-cell apoptosis. In conclusion, KCNQ1OT1 knockdown inhibited sorafenib resistance and PD-L1-mediated immune escape by sponging miR-506 in sorafenib-resistant HCC cells.
Keywords: hepatocellular carcinoma, sorafenib, KCNQ1 overlapping transcript 1, miR-506, programmed death-ligand-1
Introduction
Hepatocellular carcinoma (HCC) is a type of primary liver cancer characterized by high incidence and mortality rates (1). In recent years, although significant progress has been made in the treatment of HCC, the overall prognosis remains dismal (2). Surgery is the first and most effective method for the treatment of patients with HCC; however, chemotherapy is considered the most appropriate method for some patients with advanced disease (3,4). Sorafenib is a multi-target anti-tumor drug, which is currently the first-line treatment drug for patients with advanced HCC (5). However, due to the development of drug resistance during treatment, sorafenib becomes less effective (6,7). Furthermore, immune escape has been demonstrated to be one of the main obstacles to the treatment of HCC patients (8). Therefore, it is crucial to investigate the molecular mechanisms underlying the development of sorafenib resistance and immune escape in HCC cells.
Long non-coding RNAs (lncRNAs) are a family of transcripts >200 nucleotides in length, which do not have the ability to encode proteins (9). LncRNAs have been verified to be crucial regulators in several biological processes, such as cell metastasis, differentiation, inflammation and immune escape (10–12). Currently, lncRNA KCNQ1 overlapping transcript 1 (KCNQ1OT1) was proven to be associated with drug resistance in diverse types of cancer. For example, Zhang et al reported that KCNQ1OT1 was increased in cisplatin-resistant tongue squamous cell carcinoma (TSCC) and promoted chemoresistance of TSCC cells (13). Ren et al demonstrated that the expression of KCNQ1OT1 was higher in paclitaxel-resistant lung adenocarcinoma (LAD) tissues and cells, and KCNQ1OT1 knockdown enhanced the sensitivity of LAD to paclitaxel (14). However, there are yet no reports on the role and mechanism of action of KCNQ1OT1 in sorafenib resistance and immune escape in HCC cells.
LncRNAs may act as microRNA (miRNA) sponges to regulate the expression and activities of miRNAs (15). As a class of RNA molecules without protein-coding ability, miRNAs contain 18–22 nucleotides and mainly regulate gene expression by recognizing the 3′ untranslated region (UTR) of their target mRNAs (16). In particular, miR-506 has been demonstrated to act as a tumor suppressor in diverse human cancers, such as colorectal (17), cervical (18) and ovarian (19) cancers. Moreover, Zhou et al confirmed that miR-506 was able to reduce oxaliplatin resistance in colorectal cancer cells (20). Wang et al suggested that miR-506 was decreased in the serum of patients with sorafenib-resistant thyroid carcinoma, and that miR-506 overexpression could enhance the sensitivity of thyroid carcinoma cells to sorafenib (21). These findings indicated that miR-506 plays a key role in the development and chemoresistance of several tumors. Moreover, miRNAs can participate in the regulation of a number of biological processes, including immune escape of tumor cells (22). However, the exact regulatory mechanisms of miR-506 in sorafenib resistance and immune escape in HCC remain unclear.
Programmed death-ligand-1 (PD-L1) plays a key role in inhibiting tumor immunity and promoting tumor progression after binding to the receptor programmed death 1 (PD-1), which is expressed on the surface of T lymphocytes (23). To date, a variety of miRNAs have been confirmed to affect the oncogenesis and drug resistance of human cancers by regulating PD-L1 expression. For example, miR-34a reduced chemoresistance of glioma cells by targeting PD-L1 in (24), and the miR-200/PD-L1 axis reduced immunosuppression and metastasis of CD8+ T cells in lung cancer (25). However, whether miR-506 can target PD-L1 in HCC cells remains to be elucidated.
The aim of the present study was to investigate the levels of KCNQ1OT1, miR-506 and PD-L1 in sorafenib-resistant HCC tissues and cells, and to explore the roles of KCNQ1OT1 and miR-506 in sorafenib-resistant HCC cell proliferation, apop-tosis and metastasis. Moreover, the effects of KCNQ1OT1 and miR-506 on the tumor microenvironment and T-cell apoptosis were investigated.
Materials and methods
Tissue collection
Sorafenib-sensitive (n=25) and sorafeni-resistant (n=38) HCC tissue sections were collected from the Sanquan College of Xinxiang Medical University. All the collected samples were immediately placed in liquid nitrogen and then stored in a −80°C refrigerator for RNA extraction. The protocol of the present study was approved by the Ethics Committee of the Sanquan College of Xinxiang Medical University, and all the patients provided written informed consent.
Cell culture
SK-HEP-1 and Huh-7 cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The corresponding sorafenib-resistant HCC cells (SK-HEP-1/sorafenib and Huh-7/sorafenib) were constructed by exposing SK-HEP-1 and Huh-7 cells to gradually increasing concentrations of sorafenib and then continuously culturing in a sorafenib-containing medium for 2 months. The maximum dose of sorafenib was 20 μM (26). All cells were kept in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) in an atmosphere of 5% CO2 and 37°C.
Cell transfection
Short hairpin RNA (shRNA) against KCNQ1OT1 and its negative control (sh-NC), mimics of miR-506 (miR-506) and its matched control (miR-NC), inhibitors of miR-506 (anti-miR-506) and its matched control (anti-NC), KCNQ1OT1 overexpression vector (KCNQ1OT1) and its corresponding control (pcDNA), were synthesized by GenePharma. These oligonucleotides or vectors were transfected into cells using Lipofectamine® 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) based on the manufacturer's instructions.
RNA extraction and quantitative PCR (RT-qPCR) analysis
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from HCC tissues and cells. The concentrations of extracted RNA samples were detected by NanoDrop 2000c (Thermo Fisher Scientific, Inc.). Subsequently, RNAs were reversely transcribed into cDNAs by PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.) or TaqMan Reverse Transcription Reagents (Thermo Fisher Scientific, Inc.). Next, qPCR analysis was conducted using FastStart Universal SYBR Green Master (Roche Diagnostics) on a Real-Time PCR Detection System (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: i) 95°C for 5 min; ii) 40 cycles at 95°C for 30 sec, 60°C for 45 sec and 72°C for 30 sec; iii) dissolving curve at 94°C for 90 sec, 60°C for 180 sec and 94°C for 10 sec. The relative expression of KCNQ1OT1, PD-L1 and miR-506 was examined by the 2−ΔΔCq method (27), with GAPDH or U6 as an internal reference. The sequences of the primers were as follows: KCNQ1OT1 F: 5′-CTTTGCAGCAACCTCCTTGT-3′ and R: 5′-TGGGGTGAGGGATCTGAA-3′; miR-506 F: 5′-ATCCAG TGCGTGTCGTG-3′ and R: 5′-TGCTTAAGGCACCCTTCT-3′; PD-L1 F: 5′-TCCACTCAATGC CTCAAT-3′ and R: 5′-GAAGACCTCACA GAC TCAA-3′; GAPDH F: 5′-CCATGTTCGTCATGGGTGTGAACCA-3′ and R: 5′-GCCAGTAGAGGCAGG GAT GAT GTT C-3′; and U6 F: 5′-AGAGCCTGTGGTGTCCG-3′ and R: 5′-CATCTTCAAAGCACTTCCCT-3′.
Sorafenib resistance assay
Sorafenib resistance was evaluated by the Cell Counting Kit-8 (CCK-8) assay. SK-HEP-1, Huh-7, SK-HEP-1/sorafenib or Huh-7/sorafenib cells were incubated in 96-well plates at a density of 3.0×103cells/well. The cells were then treated with different doses of sorafenib for 48 h. Subsequently, 10 μl CCK-8 solution (5 mg/ml; Beijing Solarbio Science & Technology Co., Ltd.) was added into each well and maintained for 4 h at 37°C. At last, the absorbance at 450 nm was examined using a microplate reader (Bio-Rad Laboratories, Inc.). The value of IC50 was determined by relative survival curve.
Immunohistochemistry (IHC) assay
After being fixed with 4% paraformaldehyde (Beyotime Institute of Biotechnology) for 48 h at room temperature, sorafenib-sensitive or -resistant HCC tissue samples were embedded in paraffin and cut into 4-μm sections. Then, the sections were deparaffinized, hydrated through a graded ethanol series and treated with H2O2 in methanol for 15 min at room temperature. Thereafter, the slides were washed three times (5 min each) using PBS (Beijing Solarbio Science & Technology Co., Ltd.) and interacted with normal goat serum for 20 min followed by incubation with anti-PD-L1 (bs-10159R; 1:2,000; BIOSS) at 4°C overnight and the indicated secondary antibody (bs-0308R; 1:5,000; BIOSS) for 30 min at room temperature. After staining with diaminobenzidine (Beyotime Institute of Biotechnology) for 5 min at room temperature and counterstaining with hematoxylin for 50 sec at room temperature, the slides were photographed using a digital microscope camera (Nikon Corporation) at a magnification of ×100.
Western blot analysis
First, total protein was extracted from SK-HEP-1, Huh-7, SK-HEP-1/sorafenib or Huh-7/sorafenib cells using RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.). Then, extracted protein samples were quantified by BCA Protein Assay kit (Beijing Solarbio Science & Technology Co., Ltd.), separated by 10% SDS-PAGE (Beijing Solarbio Science & Technology Co., Ltd.) and transferred onto PVDF membranes (Pall Life Sciences). The membranes were then blocked with skimmed milk for 1 h at room temperature. Subsequently, the membranes were incubated overnight at 4°C with primary antibody anti-PD-L1 (bs-10159R; 1:2,000; BIOSS) followed by incubation with matched horseradish peroxidase-conjugated secondary antibody (bs-0308R; 1:5,000; BIOSS) for 2 h at room temperature. The protein bands were detected via RapidStep ECL Reagent (EMD Millipore).
Transwell assay
For the detection of cell migration, transfected SK-HEP-1/sorafenib and Huh-7/sorafenib cells (2.0×104 cells) were suspended in serum-free DMEM (Gibco; Thermo Fisher Scientific. Inc.) and seeded into the upper chamber of a Transwell insert (8-μm pore size; Corning Inc.). DMEM (Gibco; Thermo Fisher Scientific. Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific. Inc.) was added into the lower chamber. After incubation for 24 h, cells that did not migrate were removed and cells that migrated were fixed in methanol for 30 min at 37°C and then stained with 0.5% crystal violet solution for 15 min at 37°C (Beijing Solarbio Science & Technology Co., Ltd.). Stained cells were examined under a light microscope (Nikon Corporation) at a magnification of ×100. For the detection of cell invasion, the protocols were the same as those in cell migration assay, except that the upper chamber was pre-coated with Matrigel (Beijing Solarbio Science & Technology Co., Ltd.) for 30 min at 37°C.
Sorafenib-resistant HCC/T-cell co-culture model
First, the separation of peripheral blood mononuclear cells (PBMCs) was conducted by density gradient centrifugation. Separated PBMCs (2.0×106 per well) were seeded into 6-well plates. Subsequently, T cells were activated by incubating PBMCs with SK-HEP-1/sorafenib or Huh-7/sorafenib cell lysates, anti-CD3e (bsm-52386R; 1:2,000; BIOSS) and anti-CD28 (bs-8865R; 1:2,000; BIOSS) for 48 h. Subsequently, activated T cells were co-cultured with SK-HEP-1/sorafenib or Huh-7/sorafenib cells at 37°C for 16 h at a ratio of 10:1. Finally, co-culture media were collected for the following experiments.
ELISA
The levels of tumor necrosis factor (TNF)-α, interferon (IFN)-γ, interleukin (IL)-2, IL-10 and transforming growth factor (TGF)-β secreted by T cells were detected using ELISA Max kits (SEKH-0047; SEKH-0046; SEKH-0018; SEKH-0316; Beijing Solarbio Science & Technology Co., Ltd.) according to the manufacturer's instructions. In brief, cytokine capture antibodies were coated in 96-well plates at 4°C overnight, and the wells were then blocked at room temperature for 1 h. Cell supernatant (100 μl) was added into each well and then incubated for 2 h at 37°C. Next, 100 μl of detection antibody and diluted streptavidin peroxidase were added in succession. Finally, the reaction was stopped by adding 3,3′,5,5′-tetramethylbenzidine substrate (Beijing Solarbio Science & Technology Co., Ltd.) and sulfuric acid (Sigma-Aldrich; Merck KGaA). The optical density value at 450 nm was measured via a microplate reader (Bio-Rad Laboratories, Inc.).
Flow cytometry analysis
At 48 h post-transfection, the apoptosis of SK-HEP-1/sorafenib and Huh-7/sorafenib cells was evaluated by using Annexin V-FITC/PI Apoptosis Detection kit (Vazyme Biotech). In brief, SK-HEP-1/sorafenib and Huh-7/sorafenib cells were harvested and resuspended in binding buffer at a density of 1.0×105 cells/ml. Then, Annexin V-FITC and PI were added to the cell suspension and maintained for 15 min at room temperature in the dark to stain the cells. Finally, flow cytometry (FACSCalibur; Thermo Fisher Scientific, Inc.) was used to detect cell apoptosis. The apoptotic cells were analyzed with software FlowJo 7.6.1 (FlowJo LLC).
To analyze the apoptosis of CD8+T cells, 10 μl/ml anti-PD-L1 (bs-10159R; 1:2,000; BIOSS) was added into the well of sorafenib-resistant HCC/T-cell co-culture media. Then, activated T cells were collected and PE-conjugated, and PD-1, Alexa Fluor 488-conjugated Annexin V and APC-conjugated CD8 antibody were added to stain the PBMCs. The percentage of Annexin V+ cells in the gated PD-1+/CD8+ population was used to evaluate the apoptosis of CD8+ T cells.
Dual-luciferase reporter assay
The sequences of KCNQ1OT1 or 3′UTR of PD-L1 containing the putative complementary sequences of wild-type (WT) or mutant (MUT) miR-506 were amplified and inserted into pcDNA3.1 vector (Guangzhou RiboBio Co., Ltd.) to establish luciferase reporter vectors (WT-KCNQ1OT1, MUT-KCNQ1OT1, PD-L1 3′UTR-WT and PD-L1 3′UTR-MUT, respectively). The indicated luciferase reporter vector and miR-506 or miR-NC were transfected into SK-HEP-1/sorafenib and Huh-7/sorafenib cells using Lipofectamine® 3000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, the luciferase activity was detected by Dual-Luciferase Reporter Assay kit (Vazyme Biotech) and normalized to the Renilla luciferase activity.
Statistical analysis
All analyses were conducted by GraphPad Prism 7 (GraphPad Software, Inc.). The data obtained from three independent experiments are presented as means ± standard deviation. The significance of the differences was analyzed by Student's t-test or one-way ANOVA followed by Tukey's post hoc test. The correlations among KCNQ1OT1, miR-506 and PD-L1 were analyzed by Pearson's correlation analysis. P<0.05 was considered to indicate statistically significant differences.
Results
KCNQ1OT1 and PD-L1 are upregulated and miR-506 is downregulated in sorafenib-resistant HCC tissues
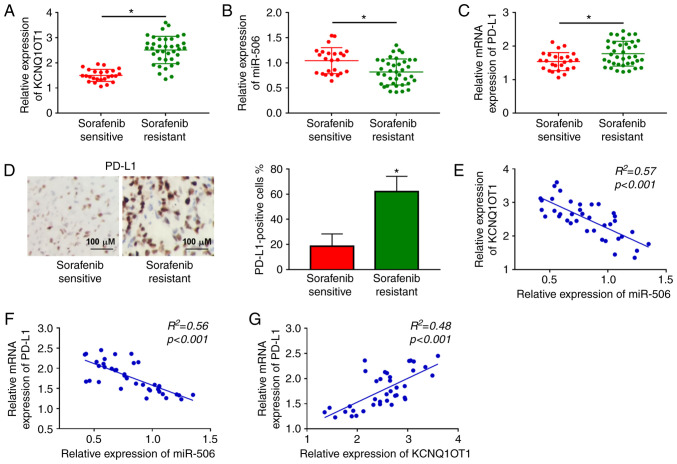
Initially, the relative expression levels of KCNQ1OT1 and miR-506 in 38 sorafenib-resistant HCC tissue samples and 25 sorafenib-sensitive HCC tissue samples were determined by RT-qPCR analysis. The results revealed that the expression level of KCNQ1OT1 was notably increased, and the expression level of miR-506 was markedly decreased in sorafenib-resistant HCC tissues compared to those in sorafenib-sensitive HCC tissues (Fig. 1A and B). Moreover, through RT-qPCR and IHC assays, PD-L1 was shown to be upregulated in sorafenib-resistant HCC tissues in comparison with sorafenib-sensitive HCC tissues (Fig. 1C and D). In addition, the correlations among KCNQ1OT1, miR-506 and PD-L1 were analyzed by Pearson's correlation analysis. The results revealed that miR-506 expression was negatively correlated with KCNQ1OT1 expression and PD-L1 mRNA expression (Fig. 1E and F); moreover, the mRNA expression of PD-L1 was positively correlated with the expression of KCNQ1OT1 in sorafenib-resistant HCC tissues (Fig. 1G). These data suggested that the abnormal expression of KCNQ1OT1, miR-506 and PD-L1 may be associated with sorafenib resistance in HCC.
Figure 1.
The expression levels of KCNQ1OT1 and PD-L1 were increased and the expression level of miR-506 was decreased in sorafenib-resistant HCC tissues. (A and B) The expression levels of KCNQ1OT1 and miR-506 in sorafenib-resistant HCC tissues and sorafenib-sensitive HCC tissues were measured by RT-qPCR. (C and D) RT-qPCR assay and immunohistochemistry were used to evaluate the level of PD-L1. Scale bar, 100 μm. (E-G) The correlations among KCNQ1OT1, miR-506 and PD-L1 were evaluated by Pearson's correlation analysis. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1; PD-L1, programmed death-ligand-1; RT-qPCR, reverse transcription-quantitative PCR analysis.
High expression of KCNQ1OT1 and PD-L1 and low expression of miR-506 is observed in sorafenib-resistant HCC cells
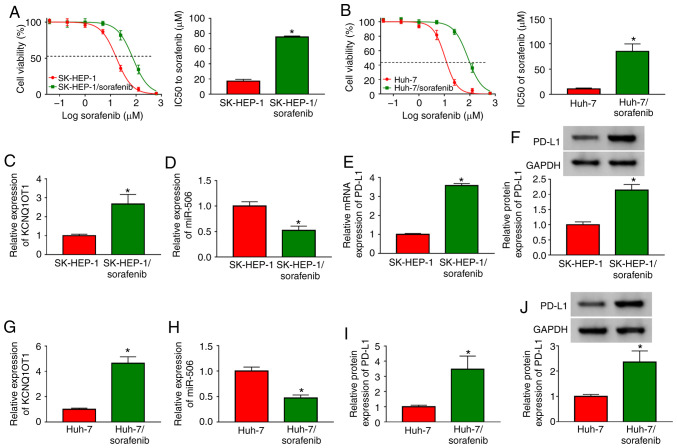
To explore the mechanism of sorafenib resistance in HCC cells, two sorafenib-resistant HCC cell lines (SK-HEP-1/sorafenib and Huh-7/sorafenib) were constructed. Subsequently, the level of sorafenib resistance in SK-HEP-1/sorafenib and Huh-7/sorafenib cells was evaluated by CCK-8 assay after exposing sorafenib-resistant HCC cells to different concentrations of sorafenib for 48 h. The data demonstrated that IC50 values in SK-HEP-1/sorafenib cells and Huh-7/sorafenib cells were notably higher compared with those in SK-HEP-1 and Huh-7 cells (Fig. 2A and B), indicating that sorafenib resistance developed in SK-HEP-1/sorafenib and Huh-7/sorafenib cells. Subsequently, the expression levels of KCNQ1OT1, miR-506 and PD-L1 mRNA in SK-HEP-1/sorafenib cells were assessed by RT-qPCR analysis and the protein expression of PD-L1 was analyzed via western blotting. The results demonstrated that the levels of KCNQ1OT1, and the mRNA and protein expression of PD-L1 were all obviously upregulated, whereas miR-506 was significantly downregulated in SK-HEP-1/sorafenib cells compared with SK-HEP-1 cells (Fig. 2C-F). Similarity, the levels of KCNQ1OT1, and those of PD-L1 mRNA and protein were increased, and the level of miR-506 was decreased in Huh-7/sorafenib cells (Fig. 2G-J). Taken together, the results mentioned above indicate that KCNQ1OT1, miR-506 and PD-L1 may be involved in the sensitivity of HCC cells to sorafenib.
Figure 2.
KCNQ1OT1, miR-506 and PD-L1 were aberrantly expressed in sorafenib-resistant HCC cells. (A and B) SK-HEP-1, SK-HEP-1/sorafenib, Huh-7 and Huh-7/sorafenib cells were treated with different doses of sorafenib for 48 h and cell viability was evaluated by Cell Counting Kit-8 assay. (C-E) The expression levels of KCNQ1OT1, miR-506 and PD-L1 mRNA in SK-HEP-1 and SK-HEP-1/sorafenib cells were determined by RT-qPCR. (F) The protein level of PD-L1 in SK-HEP-1 and SK-HEP-1/sorafenib cells was examined by western blot assay. (G-I) The expression levels of KCNQ1OT1, miR-506 and PD-L1 mRNA in Huh-7 and Huh-7/sorafenib cells were determined by RT-qPCR. (J) The protein level of PD-L1 in Huh-7 and Huh-7/sorafenib cells was examined by western blot assay. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1; PD-L1, programmed death-ligand-1; RT-qPCR, reverse transcription-quantitative PCR analysis.
KCNQ1OT1 knockdown enhances sorafenib sensitivity, promotes apoptosis and inhibits migration and invasion in sorafenib-resistant HCC cells
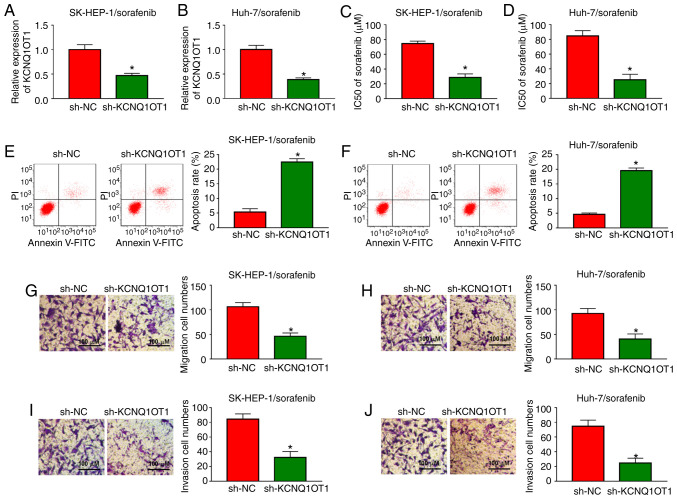
In order to investigate the functional roles of KCNQ1OT1 in sorafenib-resistant HCC cells, a loss-of-function experiment was carried out by transfecting sh-KCNQ1OT1 into SK-HEP-1/sorafenib and Huh-7/sorafenib cells. Knockdown efficiency was evaluated by RT-qPCR and the results demonstrated that sh-KCNQ1OT1 transfection markedly decreased the expression of KCNQ1OT1 in both SK-HEP-1/sorafenib and Huh-7/sorafenib cells (Fig. 3A and B). Then, SK-HEP-1/sorafenib and Huh-7/sorafenib cells were treated with different concentrations of sorafenib and IC50 value was examined by CCK-8 assay. The data demonstrated that the IC50 value of sorafenib was drastically reduced in SK-HEP-1/sorafenib and Huh-7/sorafenib cells transfected with sh-KCNQ1OT1 (Fig. 3C and D), indicating that the sensitivity of SK-HEP-1 and Huh-7 cells to sorafenib was enhanced. Furthermore, cell apoptosis was accelerated by sh-KCNQ1OT1 transfection in SK-HEP-1/sorafenib and Huh-7/sorafenib cells compared with sh-NC transfected cells (Fig. 3E and F). KCNQ1OT1 knockdown distinctly inhibited cell migration and invasion in both SK-HEP-1/sorafenib and Huh-7/sorafenib cells, as illustrated by the Transwell assay (Fig. 3G-J). Taken together, these data demonstrated that knockdown of KCNQ1OT1 sensitized HCC cells to sorafenib, induced cell apoptosis, and suppressed the migration and invasion of sorafenib-resistant HCC cells.
Figure 3.
Sorafenib resistance was reduced, cell apoptosis was increased, and cell migration and invasion were decreased in sh-KCNQ1OT1-treated sorafenib-resistant HCC cells. SK-HEP-1/sorafenib and Huh-7/sorafenib cells were transfected with sh-NC or sh-KCNQ1OT1. (A and B) The expression level of KCNQ1OT1 in SK-HEP-1/sorafenib and Huh-7/sorafenib cells was determined by RT-qPCR. (C and D) SK-HEP-1/sorafenib and Huh-7/sorafenib cells were treated with different concentrations of sorafenib for 48 h and cell viability was evaluated by the Cell Counting Kit-8 assay. (E and F) The apoptosis of SK-HEP-1/sorafenib and Huh-7/sorafenib cells was detected by flow cytometry. (G-J) The migration and invasion of SK-HEP-1/sorafenib and Huh-7/sorafenib cells were evaluated by Transwell assay. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1; RT-qPCR, reverse transcription-quantitative PCR analysis. Scale bar, 100 μm.
KCNQ1OT1 affects cytokine secretion and T-cell apoptosis in a sorafenib-resistant HCC/T-cell co-culture model
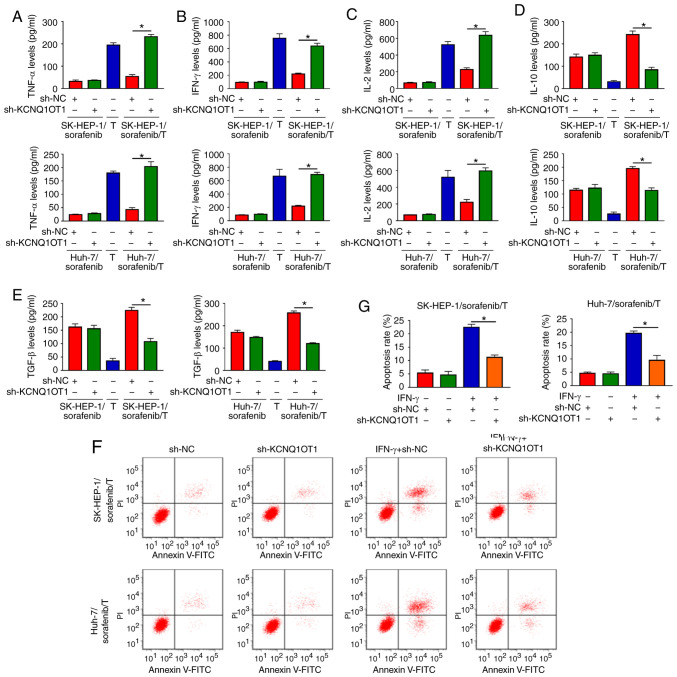
To explore the function of KCNQ1OT1 in the immune surveillance of T cells, two sorafenib-resistant HCC/T-cell co-culture models we established, namely SK-HEP-1/sorafenib/T and Huh-7/sorafenib/T. Subsequently, the levels of cytokines secreted by T cells were detected via ELISA. The results indicated that the levels of TNF-α, IFN-γ and IL-2 were markedly increased, and the levels of IL-10 and TGF-β were markedly decreased in SK-HEP-1/sorafenib cells transfected with sh-KCNQ1OT1 and T-cell co-culture group compared with those in SK-HEP-1/sorafenib cells transfected with sh-NC and T-cell co-culture group (Fig. 4A-E). As predicted, similar results were observed in Huh-7/sorafenib cells transfected with sh-KCNQ1OT1 and co-cultured with T cells (Fig. 4A-E). Furthermore, the apoptosis of CD8+ T cells was evaluated by flow cytometric analysis. We found that co-culture of T cells and SK-HEP-1/sorafenib cells or Huh-7/sorafenib cells treated with IFN-γ and sh-KCNQ1OT1 markedly suppressed the apoptosis of CD8+ T cells when compared to the control group (Fig. 4F and G). These data demonstrated that KCNQ1OT1 knockdown enhanced immune surveillance of T cells in a sorafenib-resistant HCC/T-cell co-culture model.
Figure 4.
KCNQ1OT1 knockdown regulated cytokine secretion and T-cell apoptosis in a sorafenib-resistant HCC/T-cell co-culture model. (A-E) The concentrations of TNF-α, IFN-γ, IL-2, IL-10 and TGF-β in T cells and sh-KCNQ1OT1-transfected SK-HEP-1/sorafenib or Huh-7/sorafenib cells co-cultured with T cells were examined by ELISA. (F and G) The apoptosis of CD8+ T cells and sh-KCNQ1OT1-transfected SK-HEP-1/sorafenib cells or Huh-7/sorafenib cells co-cultured with T cells was detected by flow cytometry. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1; TNF, tumor necrosis factor; IFN, interferon; IL, interleukin; TGF, transforming growth factor.
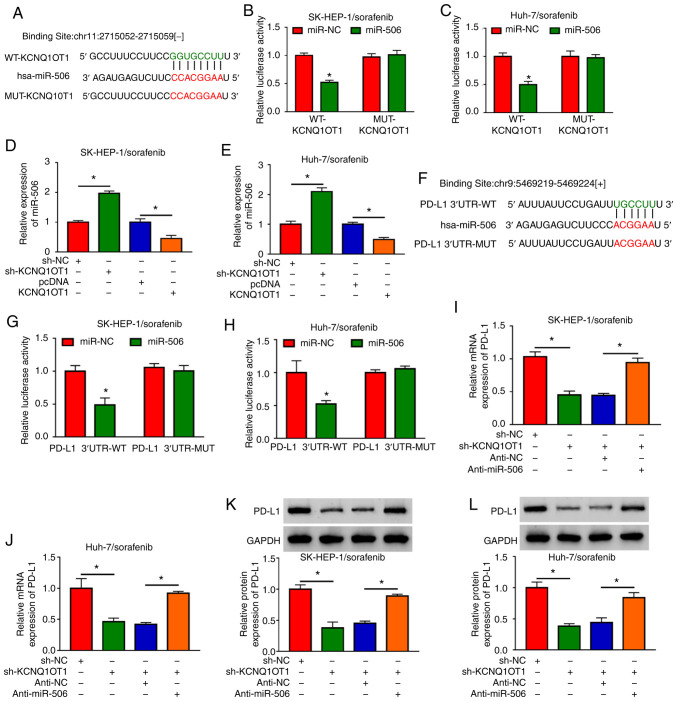
KCNQ1OT1 regulates PD-L1 expression by sponging miR-506
As dysregulated expression of KCNQ1OT1, miR-506 and PD-L1 was observed in sorafenib-resistant HCC tissues and cells, their targeting relationships were predicted using the online software starBase 3.0. As shown in Fig. 5A, miR-506 was predicted to be a target of KCNQ1OT1. miR-506 transfection markedly elevated the expression of miR-506 in SK-HEP-1/sorafenib and Huh-7/sorafenib cells, indicating that miR-506 was successfully transfected (Fig. S1A). Next, dual-luciferase reporter assay was conducted to verify this prediction. The data demonstrated that co-transfection of miR-506 and WT-KCNQ1OT1 significantly suppressed the luciferase activity in both SK-HEP-1/sorafenib and Huh-7/sorafenib cells compared with miR-NC and WT-KCNQ1OT1 co-transfected cells, while co-transfection with miR-506 or miR-NC and MUT-KCNQ1OT1 did not affect the luciferase activity (Fig. 5B and C). After KCNQ1OT1 was successfully transfected into SK-HEP-1/sorafenib and Huh-7/sorafenib cells (Fig. S1B), the impact of KCNQ1OT1 on miR-506 expression was investigated. The results revealed that KCNQ1OT1 knockdown notably increased the expression of miR-506, and KCNQ1OT1 overexpression notably decreased the expression of miR-506 in both SK-HEP-1/sorafenib and Huh-7/sorafenib cells (Fig. 5D and E). However, miR-506 overexpression did not alter the expression of KCNQ1OT1 in SK-HEP-1/sorafenib and Huh-7/sorafenib cells (Fig. S1D). Moreover, PD-L1 was identified as a target of miR-506 using the online software starBase 3.0 and the complementary sequences are shown in Fig. 5F. Dual-luciferase reporter assay revealed that, compared to PD-L1 3′UTR-WT and miR-NC co-transfected groups, the luciferase activity was markedly inhibited in PD-L1 3′UTR-WT and miR-506 co-transfected SK-HEP-1/sorafenib and Huh-7/sorafenib cells, but the luciferase activity was not affected in the PD-L1 3′UTR-MUT groups (Fig. 5G and H). As shown in Fig. S1C, anti-miR-506 transfection markedly decreased the level of miR-506 in SK-HEP-1/sorafenib and Huh-7/sorafenib cells. Subsequently, sh-KCNQ1OT1, sh-NC, sh-KCNQ1OT1+anti-miR-506 or sh-KCNQ1OT1+anti-NC were transfected into SK-HEP-1/sorafenib and Huh-7/sorafenib cells and the expression of PD-L1 was measured. The results of the RT-qPCR and western blot assays indicated that the mRNA and protein expression levels of PD-L1 were notably decreased by sh-KCNQ1OT1, whereas anti-miR-506 reversed this effect in SK-HEP-1/sorafenib cells and Huh-7/sorafenib cells (Fig. 5I-L). All these data demonstrated that KCNQ1OT1 knockdown may inhibit the expression of PD-L1 by targeting miR-506 in sorafenib-resistant HCC cells.
Figure 5.
KCNQ1OT1 regulated PD-L1 expression via sponging miR-506 in sorafenib-resistant HCC cells. (A) The putative binding sites between KCNQ1OT1 and miR-506 were predicted by starBase 3.0. (B and C) The luciferase activity in miR-506 or miR-NC and WT-KCNQ1OT1 or MUT-KCNQ1OT1 co-transfected SK-HEP-1/sorafenib and Huh-7/sorafenib cells was determined by dual-luciferase reporter assay. (D and E) The expression level of miR-506 in sh-KCNQ1OT1, sh-NC, KCNQ1OT1 or pcDNA-transfected SK-HEP-1/sorafenib and Huh-7/sorafenib cells was detected by RT-qPCR. (F) Complementary sequences between miR-506 and PD-Ll. (G and H) The luciferase activity in miR-506 or miR-NC and PD-L1 3′UTR-MUT or PD-L1 3′UTR-WT co-transfected SK-HEP-1/sorafenib and Huh-7/sorafenib cells was determined by dual-luciferase reporter assay. (I-L) The mRNA and protein expression levels of PD-L1 in sh-KCNQ1OT1, sh-NC, sh-KCNQ1OT1+anti-miR-506 or sh-KCNQ1OT1+anti-NC transfected SK-HEP-1/sorafenib and Huh-7/sorafenib cells were detected by RT-qPCR and western blotting, respectively. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1; PD-L1, programmed death-ligand-1; RT-qPCR, reverse transcription-quantitative PCR analysis; WT, wild-type; MUT, mutant; UT, untranslated region.
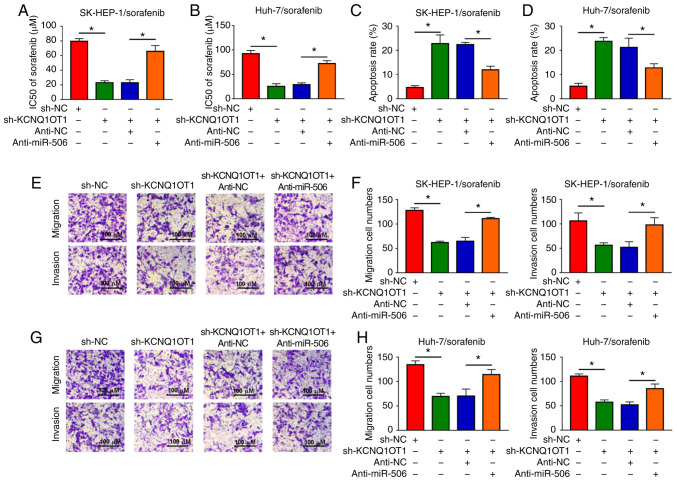
Inhibition of miR-506 partially reverses the effects on sorafenib sensitivity, cell apoptosis, cell migration and cell invasion mediated by KCNQ1OT1 knockdown in sorafenib-resistant HCC cells
Based on the results mentioned above, it was further investigated whether KCNQ1OT1 affects sorafenib sensitivity, cell apoptosis, migration and invasion by targeting miR-506 in sorafenib-resistant HCC cells. It was observed that the reduced IC50 value caused by sh-KCNQ1OT1 was partly abolished following anti-miR-506 transfection in SK-HEP-1/sorafenib and Huh-7/sorafenib cells (Fig. 6A and B). Flow cytometry revealed that downregulation of miR-506 inhibited the apoptosis-promoting effect of KCNQ1OT1 knockdown in both SK-HEP-1/sorafenib and Huh-7/sorafenib cells (Fig. 6C and D). In addition, the migration and invasion of SK-HEP-1/sorafenib and Huh-7/sorafenib cells were suppressed by knocking down the expression of KCNQ1OT1, while these effects were partially reversed by the depletion of miR-506, as shown by the Transwell assay (Fig. 6E-H). These results demonstrated that KCNQ1OT1 affected sorafenib sensitivity, HCC cell apoptosis, migration and invasion via regulating the expression of miR-506.
Figure 6.
KCNQ1OT1 knockdown enhanced sorafenib sensitivity, promoted cell apoptosis, and inhibited cell migration and invasion by targeting miR-506 in sorafenib-resistant HCC cells. sh-KCNQ1OT1, sh-NC, sh-KCNQ1OT1+anti-miR-506 or sh-KCNQ1OT1+anti-NC was transfected into SK-HEP-1/sorafenib and Huh-7/sorafenib cells. (A and B) The sorafenib resistance of SK-HEP-1/sorafenib and Huh-7/sorafenib cells was assessed by Cell Counting Kit-8 assay. (C and D) The apoptosis of SK-HEP-1/sorafenib and Huh-7/sorafenib cells was evaluated by flow cytometry. (E-H) The migration and invasion of SK-HEP-1/sorafenib and Huh-7/sorafenib cells were determined by Transwell assay. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1. Scale bar, 100 μm.
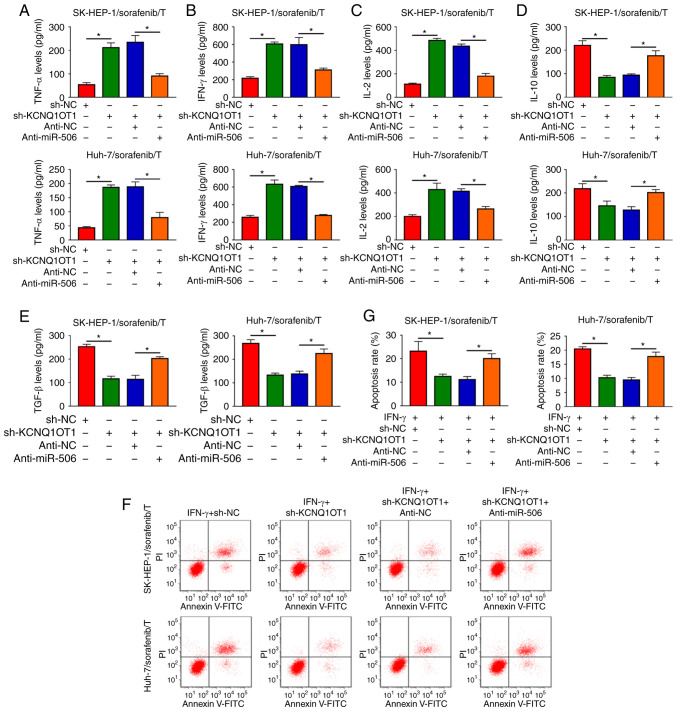
KCNQ1OT1 affects the production of cytokines and the apoptosis of T cells by sponging miR-506 in a sorafenib-resistant HCC/T-cell co-culture model
It was next investigated whether KCNQ1OT1 could regulate the immune escape of sorafenib-resistant HCC cells. The data of ELISA revealed that the inhibitory effects of KCNQ1OT1 knockdown on TNF-α, IFN-γ and IL-2 levels and the promoting effects on IL-10 and TGF-β levels were all partly abolished by miR-506 inhibition in SK-HEP-1/sorafenib/T and Huh-7/sorafenib/T models (Fig. 7A-E). Flow-cytometric analysis revealed that inhibition of miR-506 attenuated the suppression of KCNQ1OT1 knockdown on CD8+ T-cell apoptosis in both SK-HEP-1/sorafenib/T and Huh-7/sorafenib/T models (Fig. 7F and G). Collectively, the data mentioned above indicate that KCNQ1OT1 regulated the production of cytokines and the apoptosis of CD8+ T cells by targeting miR-506 in a sorafenib-resistant HCC/T-cell co-culture model.
Figure 7.
KCNQ1OT1 affected the tumor microenvironment and T-cell apoptosis by sponging miR-506 in a sorafenib-resistant HCC/T-cell co-culture model. SK-HEP-1/sorafenib or Huh-7/sorafenib cells were treated with sh-KCNQ1OT1, sh-NC, sh-KCNQ1OT1+anti-miR-506 or sh-KCNQ1OT1+anti-NC and then co-cultured with activated T cells. (A-E) The levels of TNF-α, IFN-γ, IL-2, IL-10 and TGF-β in SK-HEP-1/sorafenib/T and Huh-7/sorafenib/T models were examined by ELISA. (F and G) The apoptosis of CD8+ T cells in SK-HEP-1/sorafenib/T and Huh-7/sorafenib/T models was detected by flow cytometry. *P<0.05. HCC, hepatocellular carcinoma; KCNQ1OT1, KCNQ1 overlapping transcript 1.
Discussion
Sorafenib is a molecular-targeted drug that is useful for the treatment of patients with HCC. However, patients may develop sorafenib resistance during the treatment process, for which there is currently no effective solution (5). Immune evasion from T-cell surveillance was confirmed to play a key role in tumor development (28). A growing body of literature indicates that lncRNAs are associated with drug resistance and immune escape in human cancers (29). The present study focused on the role and mechanism of action of KCNQ1OT1 in sorafenib resistance and immune escape in HCC. It was observed that the expression of KCNQ1OT1 was markedly increased in sorafenib-resistant HCC tissues and cells. Furthermore, the KCNQ1OT1/miR-506 axis was shown to play a key role in the sensitivity of HCC cells to sorafenib and the apoptosis of T cells mediated by PD-L1.
KCNQ1OT1 has been found to be aberrantly expressed in drug-resistant tumors and to play crucial roles in the regulation of drug resistance. For example, Xian et al reported that KCNQ1OT1 was increased in methotrexate (MTX)-resistant colorectal cancer (CRC) cells, and demonstrated that KCNQ1OT1 knockdown reduced MTX resistance of CRC cells (30). Hu et al suggested that KCNQ1OT1 was highly expressed in oxaliplatin-resistant HCC tissues and cells, and oxaliplatin-resistant HCC cell metastasis was suppressed by knocking down KCNQ1OT1 expression (31). Consistently with these findings, the present study demonstrated that the KCNQ1OT1 level was markedly higher in sorafenib-resistant HCC tissues and cells in comparison with that in sorafenib-sensitive HCC tissues and cells. KCNQ1OT1 knockdown partly reversed sorafenib resistance in sorafenib-resistant HCC cells. Apoptosis is a tightly coordinated cellular process, and dysregulation of the apoptotic pathway alters the course of carcinogenesis; therefore, it appears reasonable for apoptosis to be considered as a strategy of targeted treatment in cancer (32). The cell migration and invasion abilities are essential regulators in tumors and may provide new targets for cancer therapy (33). Thus, the effects of KCNQ1OT1 on HCC cell apoptosis and motility were investigated in the present study. The results demonstrated that KCNQ1OT1 downregulation promoted apoptosis and inhibited the metastasis of sorafenib-resistant HCC cells. These data demonstrated that KCNQ1OT1 knockdown improved sorafenib sensitivity in sorafenib-resistant HCC cells.
LncRNAs were reported to regulate T-cell apoptosis and induce immune escape in tumors (34). It was previously demonstrated that immune escape can accelerate the occurrence and development of HCC, and T-cell apoptosis is an important mechanism through which tumors achieve immune escape (35). CD8+ T-cell exhaustion is commonly observed in the tumor microenvironment (36). Furthermore, the levels of cytokines are associated with T-cell immunity (37,38). Thus, the levels of cytokines produced by T cells and the apoptosis of CD8+ T cells were investigated in a sorafenib-resistant HCC/T-cell co-culture model. The data revealed that TNF-α, IFN-γ and IL-2 were increased, whereas IL-10 and TGF-β were decreased following KCNQ1OT1 knockdown in SK-HEP-1/sorafenib/T and Huh-7/sorafenib/T media. In addition, KCNQ1OT1 knockdown reduced the apoptosis of CD8+T cells in the sorafenib-resistant HCC/T-cell co-culture model. These data indicated that the immune surveillance ability of T cells on sorafenib-resistant HCC cells was enhanced following KCNQ1OT1 knockdown.
miR-506 was identified as a target of KCNQ1OT1, and an inverse correlation was observed between KCNQ1OT1 and miR-506 expression in sorafenib-resistant HCC tissues. Accumulating evidence has confirmed that miR-506 is weakly expressed in drug-resistant tumors, and miR-506 upregulation enhanced the drug sensitivity of tumor cells (20,21). In line with these data, miR-506 was found to be markedly decreased in sorafenib-resistant HCC tissues and cells, whereas miR-506 inhibition abolished the effects of KCNQ1OT1 downregulation on sorafenib resistance, apoptosis and metastasis in sorafenib-resistant HCC cells, demonstrating that miR-506 reduced the resistance of sorafenib-resistant HCC cells to sorafenib. Moreover, the secretion of cytokines and the apoptosis of T cells mediated by KCNQ1OT1 knockdown were reversed by miR-506 inhibition, suggesting that miR-506 inhibition weakens the immune surveillance of T cells on sorafenib-resistant HCC cells.
PD-L1 was verified to be a target gene of miR-506 in sorafenib-resistant HCC cells. Bishop et al found that PD-L1 was upregulated in enzalutamide-resistant prostate cancer cells (39). Liu et al demonstrated that PD-L1 was upregulated in sorafenib-resistant HCC cells (40). In the present study, PD-L1 was notably upregulated in sorafenib-resistant HCC tissues and cells. Moreover, PD-L1 inhibited the activation of T cells, playing a crucial role in the immune escape of tumors (41). However, the exact role of PD-L1 in sorafenib resistance and immune escape of sorafenib-resistant HCC cells requires further investigation.
Based on these data, it was inferred that KCNQ1OT1 knockdown enhances sorafenib sensitivity, induces apoptosis and inhibits the metastasis of sorafenib-resistant HCC cells via targeting miR-506. Moreover, the KCNQ1OT1/miR-506 axis may regulate PD-L1-mediated immune escape of sorafenib-resistant HCC cells. These findings may improve our understanding of the molecular mechanism underlying the role of KCNQ1OT1 in sorafenib resistance in HCC, and may help with designing methods to overcome it. However, in vivo animal experiments must be further conducted to verify the results of the present study.
Supplementary Data
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
Conceptualization and methodology: XZ and XM; formal analysis and data curation: ZY and MH; validation and investigation: JZ and XM; original draft preparation, review and editing: JZ, XZ and XM. All the authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Review Committee of The Sanquan College of Xinxiang Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bruix J, Sherman M. American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cidon EU. Systemic treatment of hepatocellular carcinoma: Past, present and future. World J Hepatol. 2017;9:797–807. doi: 10.4254/wjh.v9.i18.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollebecque A, Malka D, Ferté C, Ducreux M, Boige V. Systemic treatment of advanced hepatocellular carcinoma: From disillusions to new horizons. Eur J Cancer. 2015;51:327–339. doi: 10.1016/j.ejca.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Hsueh KC, Lee TY, Kor CT, Chen TM, Chang TM, Yang SF, Hsieh CB. The role of liver transplantation or resection for patients with early hepatocellular carcinoma. Tumour Biol. 2016;37:4193–4201. doi: 10.1007/s13277-015-4243-z. [DOI] [PubMed] [Google Scholar]
- 5.Colagrande S, Regini F, Taliani GG, Nardi C, Inghilesi AL. Advanced hepatocellular carcinoma and sorafenib: Diagnosis, indications, clinical and radiological follow-up. World J Hepatol. 2015;7:1041–1053. doi: 10.4254/wjh.v7.i8.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow AK, Ng L, Lam CS, Wong SK, Wan TM, Cheng NS, Yau TC, Poon RT, Pang RW. The Enhanced metastatic potential of hepatocellular carcinoma (HCC) cells with sorafenib resistance. PLoS One. 2013;8:e78675. doi: 10.1371/journal.pone.0078675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, Cao Y, Jiang H, Mao A. Autophagy facilitates the sorafenib resistance of hepatocellular carcinoma cells. West Indian Med J. 2013;62:698–700. doi: 10.7727/wimj.2013.040. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, Puigvehi M, Miguela V, Casanova-Acebes M, Dhainaut M, et al. β-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9:1124–1141. doi: 10.1158/2159-8290.CD-19-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 11.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Zhang S, Ma H, Zhang D, Xie S, Wang W, Li Q, Lin Z, Wang Y. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742. doi: 10.1038/s41419-018-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren K, Xu R, Huang J, Zhao J, Shi W. Knockdown of long non-coding RNA KCNQ1OT1 depressed chemoresistance to paclitaxel in lung adenocarcinoma. Cancer Chemother Pharmacol. 2017;80:243–250. doi: 10.1007/s00280-017-3356-z. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao Z, Zhi H, Wang T, Guo Z, Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang M, Xie X, Song X, Gu S, Chang X, Su T, Liang B, Huang D. MiR-506 suppresses colorectal cancer development by inhibiting orphan nuclear receptor NR4A1 expression. J Cancer. 2019;10:3560–3570. doi: 10.7150/jca.28272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R, Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al. miR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2015;34:717–725. doi: 10.1038/onc.2014.9. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Hu L, Zheng H, Bagnoli M, Guo Y, Rupaimoole R, Rodriguez-Aguayo C, Lopez-Berestein G, Ji P, Chen K, et al. MiR-506 inhibits multiple targets in the epithelial-to-mesenchymal transition network and is associated with good prognosis in epithelial ovarian cancer. J Pathol. 2015;235:25–36. doi: 10.1002/path.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Lin C, Zhang Y, Zhang X, Zhang C, Zhang P, Xie X, Ren Z. miR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR 1/P-gp expression. Cell Prolif. 2017;50:e12341. doi: 10.1111/cpr.12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Dai J, Yan J, Zhang Y, Yin Z. Targeting EZH 2 as a novel therapeutic strategy for sorafenib-resistant thyroid carcinoma. J Cell Mol Med. 2019;23:4770–4778. doi: 10.1111/jcmm.14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1 (PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol. 2012;24:207–212. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Wang L. miR-34a attenuates glioma cells progression and chemoresistance via targeting PD-L1. Biotechnol Lett. 2017;39:1485–1492. doi: 10.1007/s10529-017-2397-z. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Igney FH, Krammer PH. Immune escape of tumors: Apoptosis resistance and tumor counterattack. J Leukoc Biol. 2002;71:907–920. [PubMed] [Google Scholar]
- 29.Chen QN, Wei CC, Wang ZX, Sun M. Long non-coding RNAs in anti-cancer drug resistance. Oncotarget. 2017;8:1925–1936. doi: 10.18632/oncotarget.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med. 2019;23:3808–3823. doi: 10.1111/jcmm.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu H, Yang L, Li L, Zeng C. Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem Biophys Res Commun. 2018;503:2400–2406. doi: 10.1016/j.bbrc.2018.06.168. [DOI] [PubMed] [Google Scholar]
- 32.Wong RS. Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eccles SA, Box C, Court W. Cell migration/invasion assays and their application in cancer drug discovery. Biotechnol Annu Rev. 2005;11:391–421. doi: 10.1016/S1387-2656(05)11013-8. [DOI] [PubMed] [Google Scholar]
- 34.Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, Zhao J, Liu J, Lu Y, Xing Y, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19:1112–1125. doi: 10.1038/s41590-018-0207-y. [DOI] [PubMed] [Google Scholar]
- 35.Ghanem I, Riveiro ME, Paradis V, Faivre S, de Parga PM, Raymond E. Insights on the CXCL12-CXCR4 axis in hepatocellular carcinoma carcinogenesis. Am J Transl Res. 2014;6:340–352. [PMC free article] [PubMed] [Google Scholar]
- 36.Li KK, Adams DH. Antitumor CD8+ T cells in hepatocellular carcinoma: Present but exhausted. Hepatology. 2014;59:1232–1234. doi: 10.1002/hep.26779. [DOI] [PubMed] [Google Scholar]
- 37.Barrera L, Montes-Servín E, Barrera A, Ramírez-Tirado LA, Salinas-Parra F, Bañales-Méndez JL, Sandoval-Ríos M, Arrieta Ó. Cytokine profile determined by data-mining analysis set into clusters of non-small-cell lung cancer patients according to prognosis. Ann Oncol. 2015;26:428–435. doi: 10.1093/annonc/mdu549. [DOI] [PubMed] [Google Scholar]
- 38.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, Engblom C, Pfirschke C, Siwicki M, Gungabeesoon J, et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity. 2018;49:1148–1161. doi: 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop JL, Sio A, Angeles A, Roberts ME, Azad AA, Chi KN, Zoubeidi A. PD-L1 is highly expressed in Enzalutamide resistant prostate cancer. Oncotarget. 2015;6:234–242. doi: 10.18632/oncotarget.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Liu Y, Meng L, Liu K, Ji B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol Rep. 2017;38:899–907. doi: 10.3892/or.2017.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. . N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.