Abstract
Colibactin is a genotoxic gut microbiome metabolite long suspected of playing an etiological role in colorectal cancer. Evidence suggests colibactin forms DNA interstrand cross-links (ICLs) in eukaryotic cells and activates ICL repair pathways, leading to the production of ICL-dependent DNA double-strand breaks (DSBs). Here we show that colibactin ICLs can evolve directly to DNA DSBs. Using the topology of supercoiled plasmid DNA as a proxy for alkylation adduct stability, we find that colibactin-derived ICLs are unstable toward depurination and elimination of the 3′ phosphate. This ICL degradation pathway leads progressively to single strand breaks (SSBs) and subsequently DSBs. The spontaneous conversion of ICLs to DSBs is consistent with the finding that non-homologous end joining repair-deficient cells are sensitized to colibactin-producing bacteria. The results herein further our understanding of colibactin-derived DNA damage and underscore the complexities underlying the DSB phenotype.
Graphical Abstract
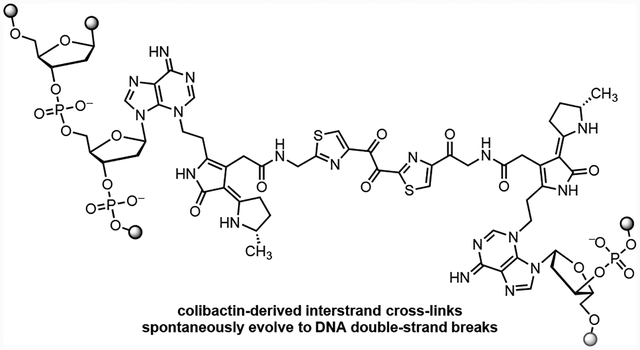
Introduction.
Colibactin is a genotoxic microbiome gut metabolite that has been implicated in the pathogenesis of colorectal cancer.1–5 The biosynthesis of colibactin is encoded in a 54-kb biosynthetic gene cluster (BGC) that is widely distributed among Enterobacteriaceae and is referred to as clb or pks.6 The clb gene cluster has undergone heavy scrutiny by the microbiome community due to its suspected role in colorectal cancer (CRC) progression. Clb+ E. coli are found in 55–67% CRC patients7,8 and accelerate progression from dysplasia to invasive carcinoma in mouse models of intestinal inflammation.7 The structure of colibactin was recently proposed as 1 by isotope labeling and tandem MS.9–12 This proposal was confirmed by chemical synthesis (Fig. 1).9, 10 Colibactin (1) contains two cyclopropane residues, each of which is appended to an α,β-unsaturated imine (blue in 1). The unsaturated imines render the cyclopropane rings electrophilic. Synthetic colibactin (1) forms DNA interstrand cross-links (ICLs) by two-fold ring-opening nucleotide addition to the cyclopropane rings (1→2).13
Fig. 1.
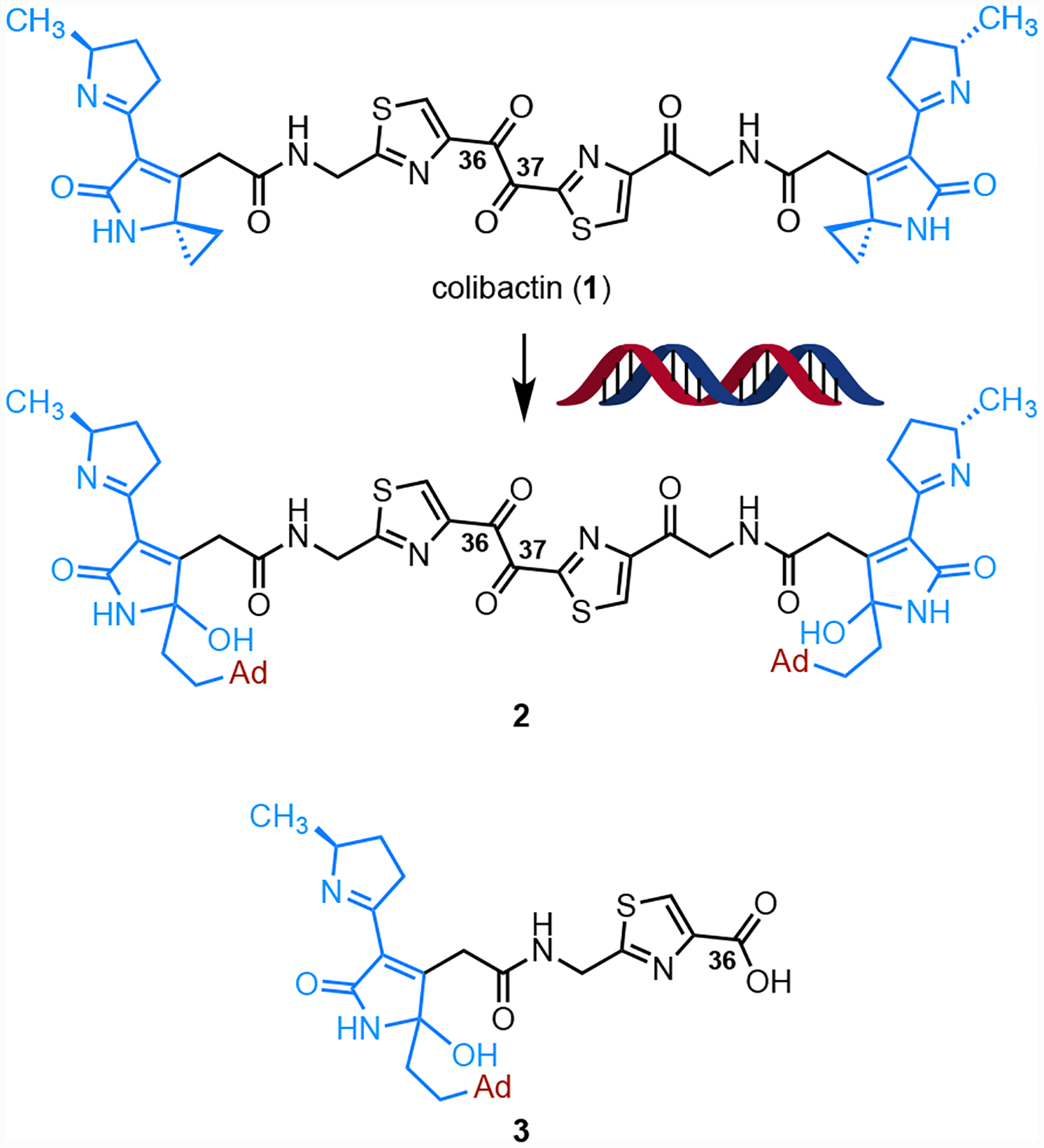
Structure of colibactin (1), the bis(adenine) adduct 2, and the first detected monoadenine adduct 3.
Clb+ E. coli induce ICLs in plasmid and cellular genomic DNA,14 which is consistent with the reactivity subsequently observed with synthetic colibactin (1). It has additionally been reported that clb+ E. coli induce DNA double-strand breaks (DSBs) and activate DNA DSB repair pathways.15, 16 It is likely that the DSB phenotype derives at least in part from ICL repair. ICLs are resolved by the Fanconi Anemia (FA) pathway during S phase17 and repair is initiated by convergence of two replication forks on either side of the ICL. In subsequent steps, the ICL is excised from one parent strand resulting in a DNA DSB. This DSB is resolved by homologous recombination (HR) repair, one of the two primary DSB repair pathways.18 Thus, HR repair markers, such as phospho-Ser139-H2AX (γH2AX), are observable when DSBs are formed during FA repair.19 Foci derived from γH2AX and the FA protein Fanconi anemia protein D2 (FANCD2) formed and colocalized following exposure to clb+ E. coli,14 providing further evidence for FA-dependent DSB formation.
However, cells deficient in non-homologous end joining (NHEJ), the second major DSB repair pathway (which operates throughout the cell cycle), were found to be sensitized to clb+ E. coli.15 Because FA is coupled to HR but not NHEJ, this suggests a mechanism involving the generation of FA-independent DSBs may contribute to the overall cytotoxicity of the bacteria. In our studies of synthetic colibactin mimetics13 and presently colibactin (1) itself, we observed fragmentation of alkylated DNA during denaturing electrophoresis. The mononadenine adduct 3 has been characterized by NMR spectroscopy and was shown as linked through N3 of adenine.20 N3-Adenine alkylation products are susceptible to depurination,21, 22 and the resulting apurinic (AP) sites can progress to SSBs by elimination of the 3′ phosphate.23 Alternatively, AP sites are recognized by apurinic/apyrimidinic endonuclease 1 (APE1), which cleaves the phosphate 5′ to an abasic site, leaving a free 3′ hydroxyl and 5′ deoxyribose phosphate. We hypothesized that 3′ phosphate elimination and/or APE1 activity may contribute to the formation of DSBs outside of S phase. To test this hypothesis, we performed experiments to probe the stability of ICLs generated by clb+ E. coli and synthetic colibactin. Our data suggest DSBs derived from colibactin may arise from a combination of FA repair and depurination of ICLs.
Material and Methods
Bacterial Strains and Growth Conditions.
The E. coli strains used in this study were prepared by the Crawford laboratory at Yale University (New Haven, CT). The BW25113 E. coli. contain either the colibactin gene cluster on pBeloBac11 (clb+), the empty vector alone (clb−), or the vector carrying the clbL point mutant (S179A). The clbL point mutant (S179A) was constructed as previously described.24 Bacteria were grown overnight in LB media containing chloramphenicol (25 μg/mL), and then the grown cultures were diluted into M9-cas-amino acid (M9-CA) medium. To prepare the M9-CA medium, the M9 minimum media (Difco) was supplemented with 0.4% glucose, 2 mM MgSO4, 0.1 mM CaCl2, 12.5 μg/mL chloramphenicol and the following L-amino acid mass composition (5 g/L total): 3.6% Arg, 21.1% Glu, 2.7% His, 5.6% Ile, 8.4% Leu, 7.5% Lys, 4.6% Phe, 9.9% Pro, 4.2% Thr, 1.1% Trp, 6.1% Tyr, 5% Val, 4% Asn, 4% Ala, 4% Met, 4% Gly, 4% Cys, and 4% Ser. Standard growth conditions were 37 °C with 250 rpm agitation.
Synthetic and Commercial Reagents.
Cisplatin was purchased from Biovision® (Milpitas, CA). The synthetic colibactin linear precursor (4) was prepared by a modification of the published procedure.10
In vitro Plasmid Cleavage Assays.
The 2686 bp pUC19 vector (circular) was purchased from New England Biolabs® (Ipswich, MA). For each reaction with E. coli, 1000 ng of circular pUC19 DNA was added to 200 μL (7.7 μM base pairs) of M9-CA medium inoculated with 1.2 × 107 bacteria pre-grown to exponential phase in the M9-CA medium. To prepare the control experiments, 1000 ng of circular pUC19 DNA was added to 200 μL of M9-CA medium, and no bacteria was added. The mixture of DNA and bacteria (or without any bacteria) was incubated at 37 °C for 1 h−9 h, and the bacteria were then pelleted. The DNA was isolated from the supernatant using the polymerase chain reaction (PCR) clean-up kit (New England Biolabs®) and quantified using a nanodrop. The DNA was then stored in −20 °C until electrophoretic analysis or follow-up experiments.
For each reaction with synthetic colibactin linear precursor (4), 200 ng of circular pUC19 DNA (15.4 μM base pairs) was incubated with 4 in concentrations from 100 μM−1 nM, in a total volume of 20 μL. 4 was diluted in DMSO such that each reaction contained 5% DMSO concentration. Reactions were conducted in 10 mM sodium citrate buffer (pH 5.0). Pure DMSO vehicle was used as negative control. A control employing 200 ng of circular pUC19 DNA (15.4 μM base pairs) was treated with pure DMSO vehicle in 10 mM sodium citrate (pH 5) buffer with a final DMSO concentration of 5% was prepared simultaneously. Reactions proceeded for 4 h at 37 °C, and the reacted DNA was stored at −20 °C until electrophoretic analysis or follow-up experiments.
DNA Cross-linking Assays.
Linearized pUC19 DNA was used for all DNA cross-linking assays. To prepare the linearized DNA, the 2686 bp pUC19 vector (New England Biolabs®) was linearized with 20 units/μg EcoRI-HF® (New England Biolabs®) and the linearized DNA was purified using a PCR clean-up kit (New England Biolabs®), eluted into 10 mM Tris (pH 8.0), and quantified using a nanodrop. For each reaction with E.coli, 800 ng of linearized pUC19 DNA was added to 200 μL (6.2 μM base pairs) of M9-CA medium inoculated with 1.2 × 107 bacteria pre-grown to exponential phase in the M9-CA medium. The mixture of DNA and bacteria was incubated at 37 °C for 4 h, and the bacteria were then pelleted. The DNA was isolated from the supernatant using the PCR clean-up kit (New England Biolabs®) and quantified using a nanodrop. The DNA was then stored at −20 °C until electrophoretic analysis or follow-up experiments.
For each reaction with synthetic colibacin linear precursor (4), 200 ng of linearized pUC19 DNA (15.4 μM base pairs) was incubated with 4 in concentrations ranging from 100 μM−1 nM in a total volume of 20 μL. 4 was diluted in DMSO such that each reaction contained a fixed 5% DMSO concentration. Reactions were conducted in 10 mM sodium citrate buffer (pH 5.0). Pure cisplatin (Biovision®) and DMSO vehicle were used as positive and negative controls, respectively. Stock solutions of cisplatin in DMSO were prepared immediately before use. A control containing 200 ng of circular pUC19 DNA (15.4 μM base pairs) was treated with 100 μM cisplatin or DMSO vehicle in 10 mM sodium citrate (pH 5) buffer with a final DMSO concentration of 5% was prepared simultaneously. Reactions were conducted for 4 h at 37 °C, The DNA was stored at −20 °C until electrophoretic analysis or follow-up experiments.
Plasmid Linearization Test
The pUC19 DNA isolated from plasmid cleavage assays with E. coli. was used for the plasmid linearization test. To set up the linearization reactions, 20 units of EcoRI-HF® (New England Biolabs®) was mixed with 500 ng of isolated DNA (40 units/μg DNA) in CutSmart® buffer (New England Biolabs®), pH 7.9, in a total volume of 50 μL for 30 min at 37 °C. The CutSmart® buffer (New England Biolabs®) contains 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, and 100 μg/mL BSA. To set up the negative control reactions, 500 ng of isolated DNA was treated with CutSmart buffer® (New England Biolabs®), pH 7.9, in a total volume of 50 μL for 30 min at 37 °C. The reacted DNA was then purified using PCR clean-up kit (New England Biolabs®) and quantified using the nanodrop. The DNA was then stored at −20 °C before electrophoretic analysis. The clb+ cross-linked linearized pUC19 DNA was used as positive control for analysis.
Endonuclease IV Stability Test
The pUC19 DNA isolated from plasmid cleavage and DNA cross-linking assays with E. coli. was used for the Endonuclease IV (EndoIV) stability tests. The pUC19 DNA exposed to synthetic compounds in plasmid cleavage or DNA cross-linking assays was directly diluted and used for EndoIV stability tests. To set up each reaction, 50 ng of processed DNA was mixed with 20 units of EndoIV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, in a total volume of 20 μL for 16 h−20 h (unless otherwise noted) at 37 °C. The NEBuffer 3.1® (New England Biolabs®) contained 100 mM sodium chloride, 50 mM Tris-HCl, 10 mM magnesium chloride, and 100 μg/ml BSA. To set up each negative control, 50 ng of processed DNA was mixed with NEBuffer 3.1® (New England Biolabs®), pH 7.9, in a total volume of 20 μL for 16 h−20 h (otherwise otherwise noted) at 37 °C. Following completion of the experiment, the DNA was stored at −20 °C before electrophoretic analysis.
Gel Electrophoresis
For each DNA sample (except the samples employed in the EndoIV stability tests), the DNA concentration was pre-adjusted to 10 ng/μL. Five microliters (50 ng) of DNA was removed and mixed with 1.5 μL of 6 × purple gel loading dye; no SDS (New England Biolabs®), and loaded onto 1% agarose Tris Borate EDTA (TBE) gels. For the samples employed in the EndoIV stability tests, the DNA solution (20 μL) was mixed with 4 μL of 6 × purple gel loading dye; no SDS (New England Biolabs®) and directly loaded onto 1% agarose Tris Borate EDTA (TBE) gels. For denaturing gels, 5 μL (50 ng) of DNA was removed each time and separately mixed with 15 μL of 0.2% denaturing buffer (0.27% sodium hydroxide, 10% glycerol, and 0.013% bromophenol blue) or 0.4% denaturing buffer (0.53% sodium hydroxide, 10% glycerol, and 0.013% bromophenol blue) in an ice bath. The mixed DNA samples were denatured at 4 °C for 10 min and then immediately loaded onto 1% agarose Tris Borate EDTA (TBE) gels. All gel electrophoresis was conducted at 90 V for 2 h (unless otherwise noted). The gel was stained with SybrGold (Thermo Fisher) for 2 h.
Quantification of Gel Images
The gel images were quantified using Adobe®Photoshop® CC 2018. The pixel intensity of each gel band was carefully measured, and the average background pixel intensity was deducted from each gel band’s raw measurement. For each individual set of DNA stability experiment, the net pixel intensity values of each target gel band were normalized by the net pixel intensity of the supercoiled pUC19 DNA band at 1 h incubation time point for that DNA experiment condition, to measure the percent DNA pixel remain in that set of experiment. A standard curve of the DNA quantity vs. net pixel intensity is included in the Supporting Information.
Results.
To probe for SSB and DSB formation we began by evaluating the effect of clb+ BW25113 E. coli (clb+) on circular pUC19 plasmid DNA. Nicking (SSB) of the plasmid results in relaxation of the supercoiled structure and lower mobility on an electrophoretic gel. Cleaved (also known as linearized) plasmid DNA, resulting from DSB formation, also has a distinct mobility. Control experiments included plasmid DNA exposed to E. coli lacking the pathway (clb–) or E. coli mutated in the gene clbL (S179A). ClbL is an amidase that has been proposed to mediate a heterodimerization in the final steps of colibactin (1) biosynthesis.10–12 Thus we anticipated that the DNA ICL phenotype would be abrogated in this strain if colibactin (1) were the causative agent. We also carried out a control experiment wherein the plasmid DNA was exposed to M9-cas amino acid media (M9-CA) alone. Following 1–9 h of incubation, the DNA samples were purified and analyzed by native gel electrophoresis (Fig. 2). Trace amounts of DNA damage were observed in the control experiment containing media alone, however this damage did not increase during the course of the experiment (Fig. S1). A background level of nicking and cleavage was observed in the clb− and the clbL mutant (S179A) strains. The amount of DNA damage was markedly increased in the clb+ culture after incubating for 4 h (Lane 11), and all of the circular plasmid had converted to nicked (SSB) or linearized (DSB) DNA after 6 h (Lane 12).
Fig. 2.
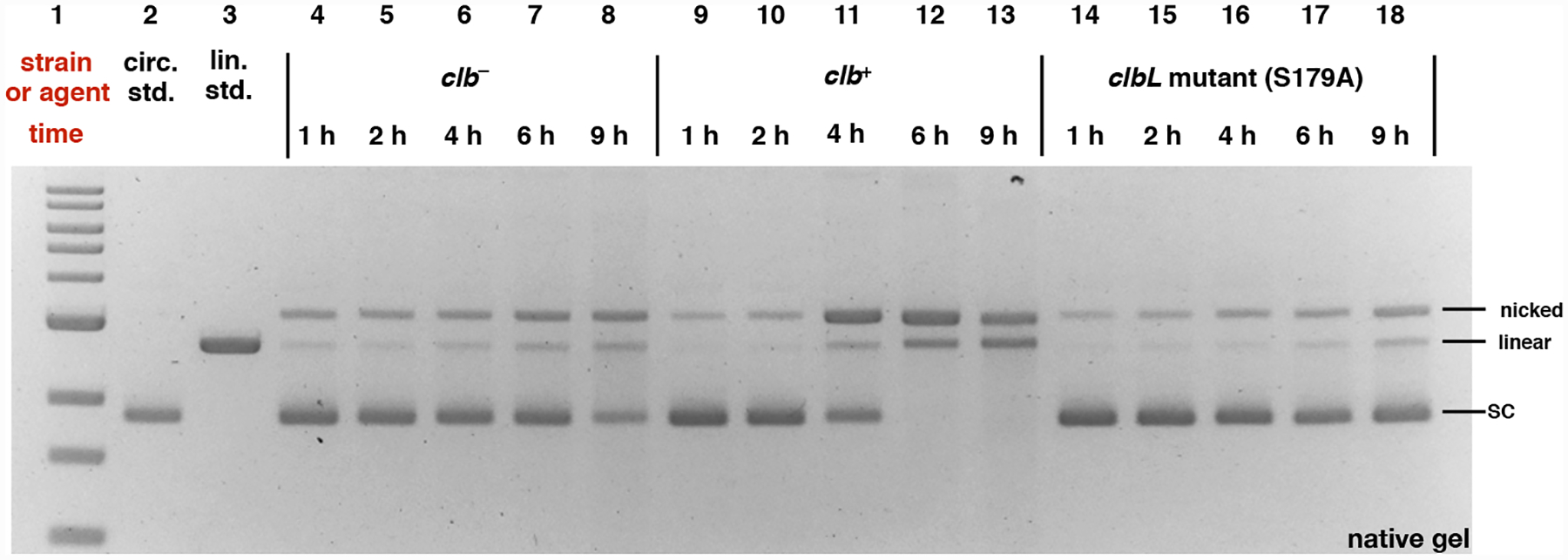
DNA plasmid cleavage assay employing circular pUC19 DNA and E. coli and analysis by neutral gel electrophoresis. DNA ladder (Lane #1); circular pUC19 DNA standard (Lane #2); linearized pUC19 DNA standard (Lane # 3); clb− BW25113 E. coli 1 h (Lane #4), 2 h (Lane #5), 4 h (Lane #6), 6 h (Lane #7), and 9 h (Lane #8); clb+ BW25113 E. coli 1 h (Lane #9), 2 h (Lane #10), 4 h (Lane #11), 6 h (Lane #12), and 9 h (Lane #13); clbL mutant (S179A) BW25113 E. coli 1 h (Lane #14), 2 h (Lane #15), 4 h (Lane #16), 6 h (Lane #17), and 9 h (Lane #18). Conditions (Lane #4–#18): clb−BW25113 E. coli (Lane #4–#8), clb+ BW25113 E. coli (Lane #9–#13): and clbL mutant (S179A) BW25113 E. coli (Lane #14–#18), circular pUC19 DNA (7.7 μM in base pairs), M9-CA media, 37 °C, 1 h, 2 h, 4 h, 6 h, or 9 h. The DNA was isolated, purified, and analyzed by native agarose gel electrophoresis (90 V, 2 h). Order of elution (bottom to top): supercoiled plasmid DNA, linearized DNA, nicked DNA. SC = supercoiled, nicked = SSB, linear = DSB.
Re-analysis of the samples by denaturing electrophoresis revealed a different DNA migration pattern and a significant amount of degradation of the clb+-treated DNA after incubating for 4 h (Fig. 3, Lane 11). Complete degradation of the plasmid was observed after 9 h (Lane 13).
Fig. 3.
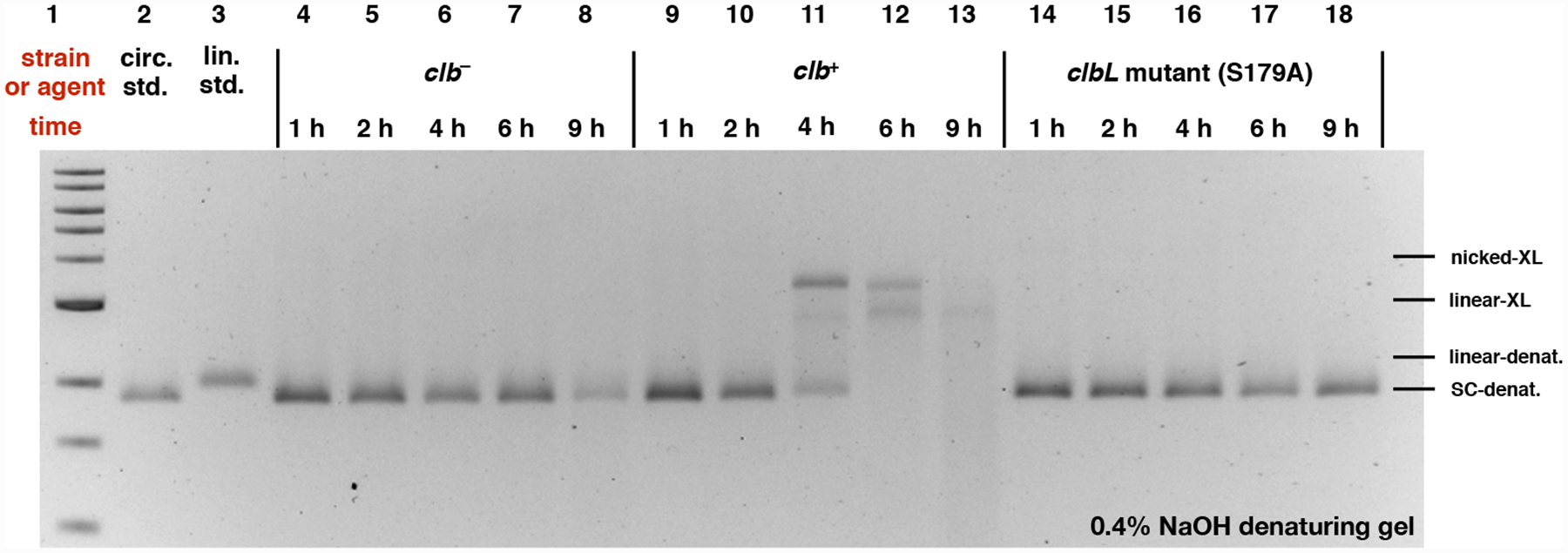
DNA plasmid cleavage assay employing circular pUC19 DNA and E. coli and analysis by denaturing electrophoresis. DNA ladder (Lane #1); circular pUC19 DNA standard (Lane #2); linearized pUC19 DNA standard (Lane # 3); clb− BW25113 E. coli 1 h (Lane #4), 2 h (Lane #5), 4 h (Lane #6), 6 h (Lane #7), and 9 h (Lane #8); clb+ BW25113 E. coli 1 h (Lane #9), 2 h (Lane #10), 4 h (Lane #11), 6 h (Lane #12), and 9 h (Lane #13); clbL mutant (S179A) BW25113 E. coli 1 h (Lane #14), 2 h (Lane #15), 4 h (Lane #16), 6 h (Lane #17), and 9 h (Lane #18). Conditions (Lane #4–#18): clb− BW25113 E. coli (Lane #4–#8), clb+ BW25113 E. coli (Lane #9–#13), and clbL mutant (S179A) BW25113 E. coli (Lane #14–#18), circular pUC19 DNA (7.7 μM in base pairs), M9-CA media, 37 °C, reaction proceed for 1 h, 2 h, 4 h, 6 h, and 9 h. The DNA was isolated, purified, and analyzed by 0.4% NaOH denaturing agarose gel electrophoresis (90 V, 2 h). SC-denat. = supercoiled DNA in denaturing form, linear-denat. = DSB/linearized DNA in denaturing form, nicked-XL = SSB DNA cross-linked by colibactin, linear-XL = DSB DNA cross-linked by colibactin.
We could not accurately quantify the amount of SSBs and DSBs due to degradation of the clb+-treated DNA. As an alternative, we measured the decrease in intensity of the supercoiled DNA band to assess the extent of DNA damage. This approach does not account for the formation of stable alkylation and cross-link products, but does provide an approximate assessment of the relative rates of DNA degradation. The total amount of DNA remaining was expressed as the ratio of the intensity of the supercoiled plasmid band at a given time point to the intensity of this band at 1 h. As shown in Fig. 4, this ratio decreased most dramatically in the clb+ cultures (green series) and to a lesser extent in the clb− or clbL mutant cultures (blue and orange series’, respectively). DNA was essentially unchanged when exposed to media alone (red series).
Fig. 4.
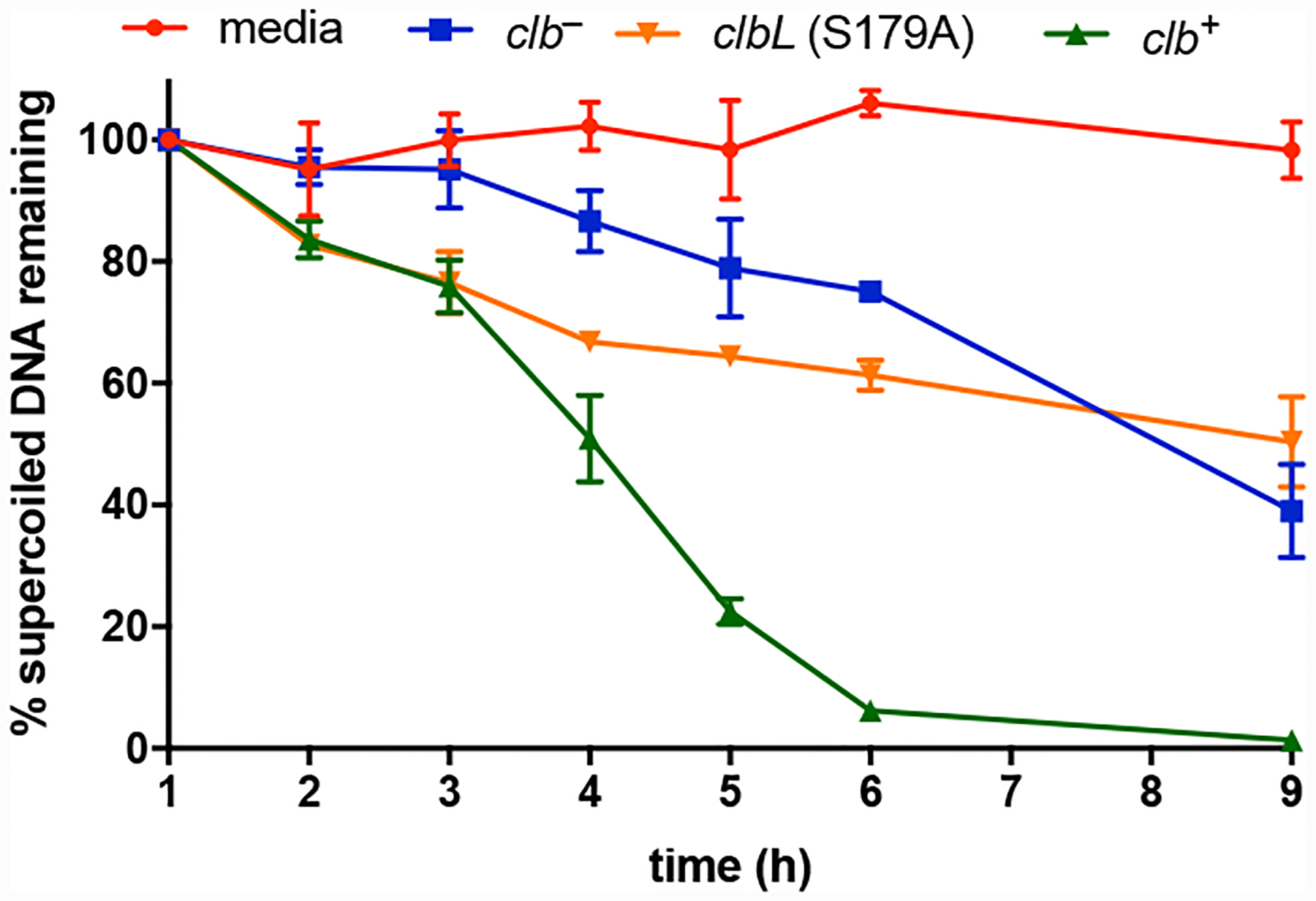
Percent supercoiled DNA remaining after treatment with media, clb− BW25113 E. coli, clbL mutant (S179A) BW25113 E. coli, or clb+ BW25113 E. coli. Percent DNA is defined as (In/I1)*100% where I = the intensity of the supercoiled DNA band at a given time point. Three biological replicates. Error bars represent standard error of the mean.
Based on our previous preparations of mass spectrometry samples, 4 h is the optimum incubation time for preparing cross-linked linearized pUC19 DNA using clb+ E. coli.9, 10, 25 To confirm that these circular pUC19 DNA contained ICLs, we incubated the plasmid with clb+ E. coli for 4 h, and then linearized the DNA using the EcoRI restriction enzyme. The DNA was re-purified and then analyzed using both native and denaturing gel electrophoresis. Fig. 5A shows that linearization of either the clb+ or clb− treated DNA, followed by native gel electrophoresis, produced the expected duplexed DNA (Lanes 7 and 10, compare to Lanes 3 and 4). By comparison, Fig. 5B shows that when the same sample was analyzed by denaturing electrophoresis, only the clb+-treated DNA remained covalently connected, as evidenced by co-migration with pUC19 DNA that was linearized prior to treatment with clb+ E. coli (compare Lanes 10 and 4). ICLs were not detected in the clbL mutant (Fig. S3).
Fig. 5.
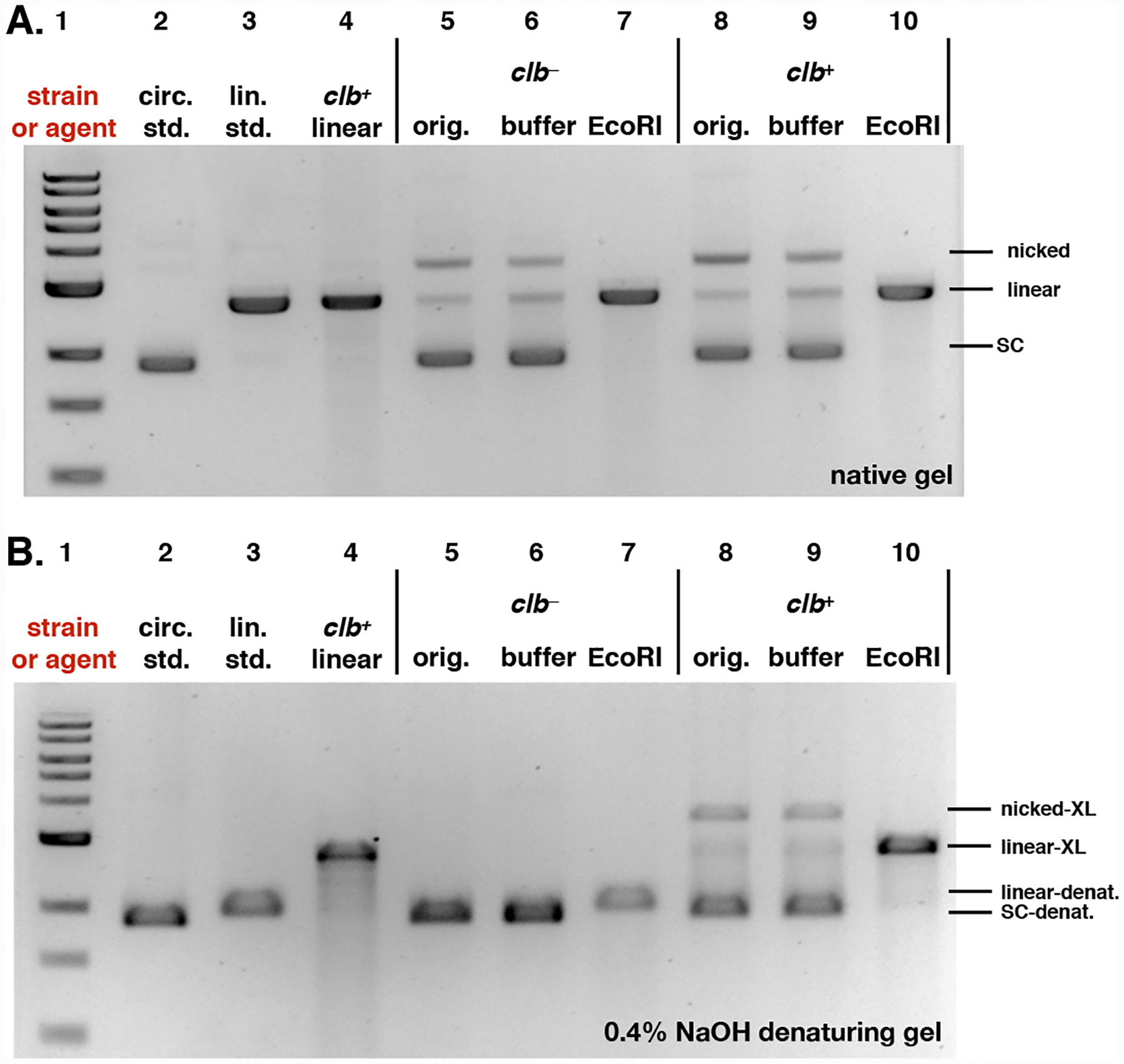
Analysis of pUC19 DNA following treatment with clb− or clb+ E. coli and linearization with the restriction enzyme EcoRI. The cross-linked linearized pUC19 DNA isolated from a co-culture with clb+ BW25113 E. coli was used a positive control. A. Analysis of DNA by native gel electrophoresis. B. Analysis of DNA by denaturing gel electrophoresis. For both A and B: DNA ladder (Lane #1); circular pUC19 DNA standard (Lane #2); linearized pUC19 DNA standard (Lane # 3); linearized pUC19 DNA co-cultured with clb+ BW25113 E. coli (Lane #4); circular pUC19 DNA isolated from co-culture with clb− BW25113 E. coli (Lane #5), reacted with buffer (Lane #6), reacted with EcoRI restriction enzyme (Lane #7); circular pUC19 DNA isolated from co-culture with clb+ BW25113 E. coli (Lane #8), reacted with buffer (Lane #9), reacted with EcoRI restriction enzyme (Lane #10). Conditions (Lane #4): linearized pUC19 DNA, clb+ BW25113 E. coli, M9-CA media, 4 h at 37 °C. Conditions (Lane #5–#7): circular pUC19 DNA isolated from co-culture with clb− BW25113 E. coli in M9-CA media for 4 h at 37 °C (Lane #5); the DNA (15.4 μM base pair) was reacted with CutSmart Buffer® (New England Biolabs®), pH 7.9, at 37 °C for 30 minutes (Lane #6); the DNA (15.4 μM base pair) was reacted with 20 units of EcoRI-HF restriction enzyme in CutSmart Buffer® (New England Biolabs®), pH 7.9, at 37 °C for 30 minutes (Lane #7). Conditions (Lane #8–#10): circular pUC19 DNA isolated from co-culture with BW25113 clb+ E. coli. in in M9-CA media for 4 h at 37 °C (Lane # 8); the DNA (15.4 μM base pair) was reacted with CutSmart Buffer® (New England Biolabs®), pH 7.9, at 37 °C for 30 minutes (Lane #9); the DNA (15.4 μM base pair) was reacted with 20 units of EcoRI-HF restriction enzyme in CutSmart Buffer® (New England Biolabs®), pH 7.9, at 37 °C for 30 minutes (Lane #10). The DNA was isolated and analyzed by native (Fig. 5A) or 0.4% NaOH denaturing (Fig. 5B) agarose gel electrophoresis (90 V, 1.5 h). SC = supercoiled, nicked = SSB, linear = DSB, SC-denat. = supercoiled DNA in denaturing form, linear-denat. = DSB/linearized DNA in denaturing form, nicked-XL = SSB DNA cross-linked by colibactin, linear-XL = DSB/linerized DNA cross-linked by colibactin.
The data shown in Figs. 2–5 can be explained by initial formation of an ICL, depurination, and elimination of the 3′ phosphate to sequentially form SSBs and DSBs (Fig. 6). This model is supported by the rates of depurination and subsequent 3′ phosphate elimination in vitro. The reported half-life for depurination of 3-methyladenosine in rat liver DNA is 24 h (pH 7.0, 37 °C).21, 22 The reported half-life for elimination of the 3′ phosphate from an AP site in circular phage PM2 DNA is 190 h (pH 7.4, 37 °C).23, 26, 27 Additionally it is possible that APE1 introduces additional nicks at AP sites in tissue culture. We note that the site of adenine alkylation on the right hand-side of colibactin has not been rigorously established but alkylation at N3 would be consistent with the structure of 3 and the reactivity observed here. N7A adducts are labile toward depurination (t1/2 = 3 h), while N6A adducts are stable.23
Fig. 6.
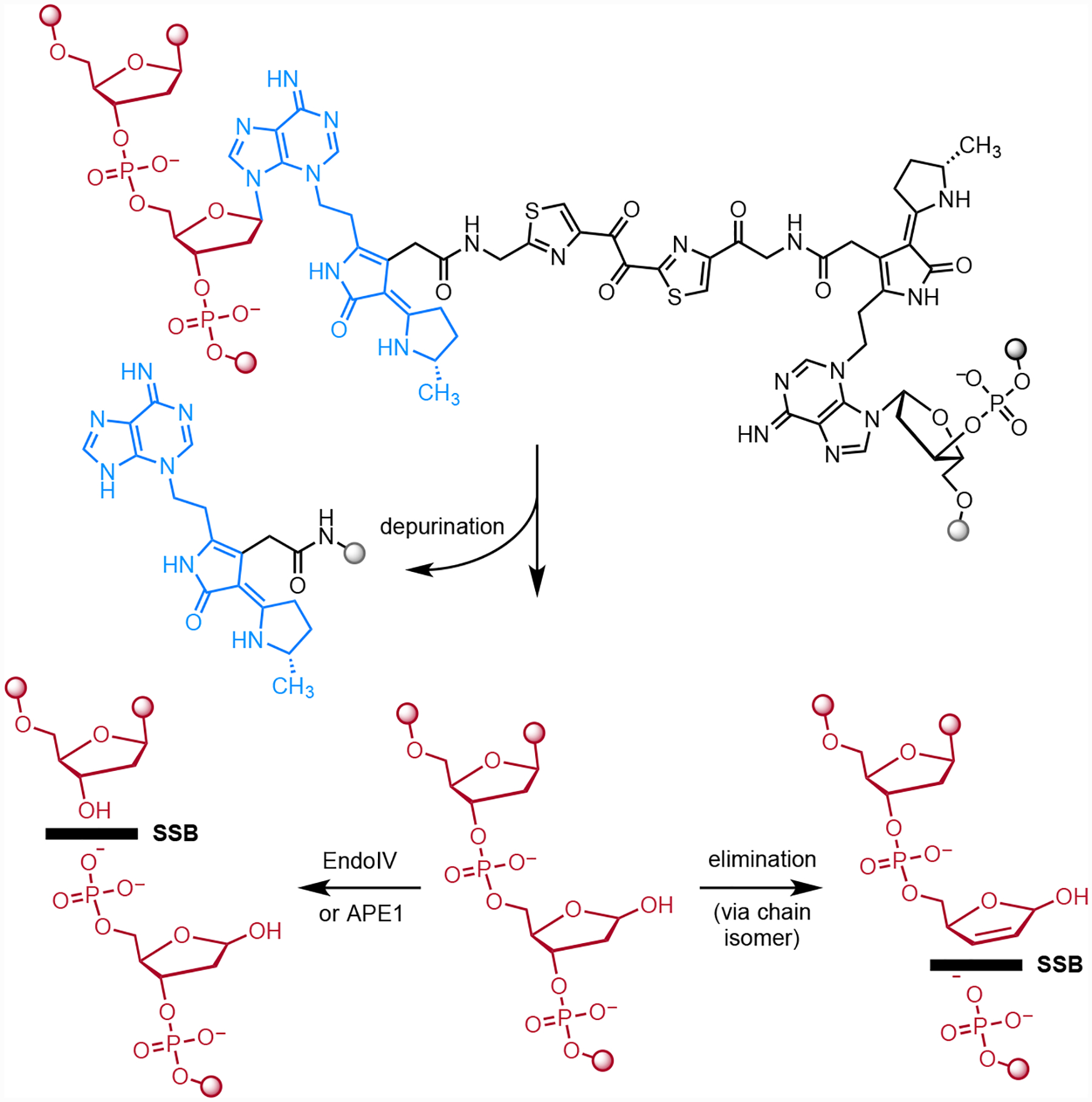
Mechanism of SSB and DSB induction following formation of ICLs. Depurination followed by slower elimination of the 3′ phosphate would introduce a SSB. Alternatively, hydrolysis of the AP site by APE1 in tissue culture would introduce a break in the DNA. Repetition of either pathway at the remaining intact alkylated nucleotide would form a DSB.
According to this model and the relative rates of depurination and 3′ phosphate elimination, the DNA should be accumulating AP sites. To test this, we exposed the clb+-treated plasmid DNA to Endonuclease IV (EndoIV) which recognizes AP sites and cleaves the phosphodiester backbone 5′ to the AP site, resulting in a one nucleotide gap flanked by a 3′ hydroxyl and 5′ deoxyribose phosphate.28, 29 If AP sites are accumulating, one would expect EndoIV treatment to amplify the amount of DNA breaks. As shown in Fig. 7, all of the plasmid DNA was converted to nicked (SSBs) and linearized (DSBs) DNA after treatment with EndoIV, and further degraded DNA fragments were observable (Lane 9). By comparison, DNA that had been exposed to buffer alone partially converted to nicked (SSBs) DNA, and retained a smaller amount of intact plasmid DNA (Lane 8) compared to the untreated sample (Lane 7). This is consistent with the mechanism outlined in Fig. 6 where SSBs and DSBs derive from spontaneous depurination, followed by phosphate elimination. The clb− and clbL mutant (S179A) strains did not display measurable changes in DNA topology on treatment with Endo IV or buffer alone within 20 h of incubation at 37 °C. DNA cleavage increased as the amount of time the clb+-treated circular or linearized pUC19 DNA was exposed to buffer or EndoIV was increased (Fig S4−S5). However, the circular and linearized untreated pUC19 DNA, or the DNA treated with the 100 μM cisplatin (which forms more stable23 N7G ICLs30) did not show any further damage over time in the presence or absence of EndoIV (Fig S6−S9). These results indicate that the colibactin-cross-linked DNA undergoes depurination and then evolves more slowly to strand breaks. These breaks are amplified by exposure to Endo IV.
Fig. 7.
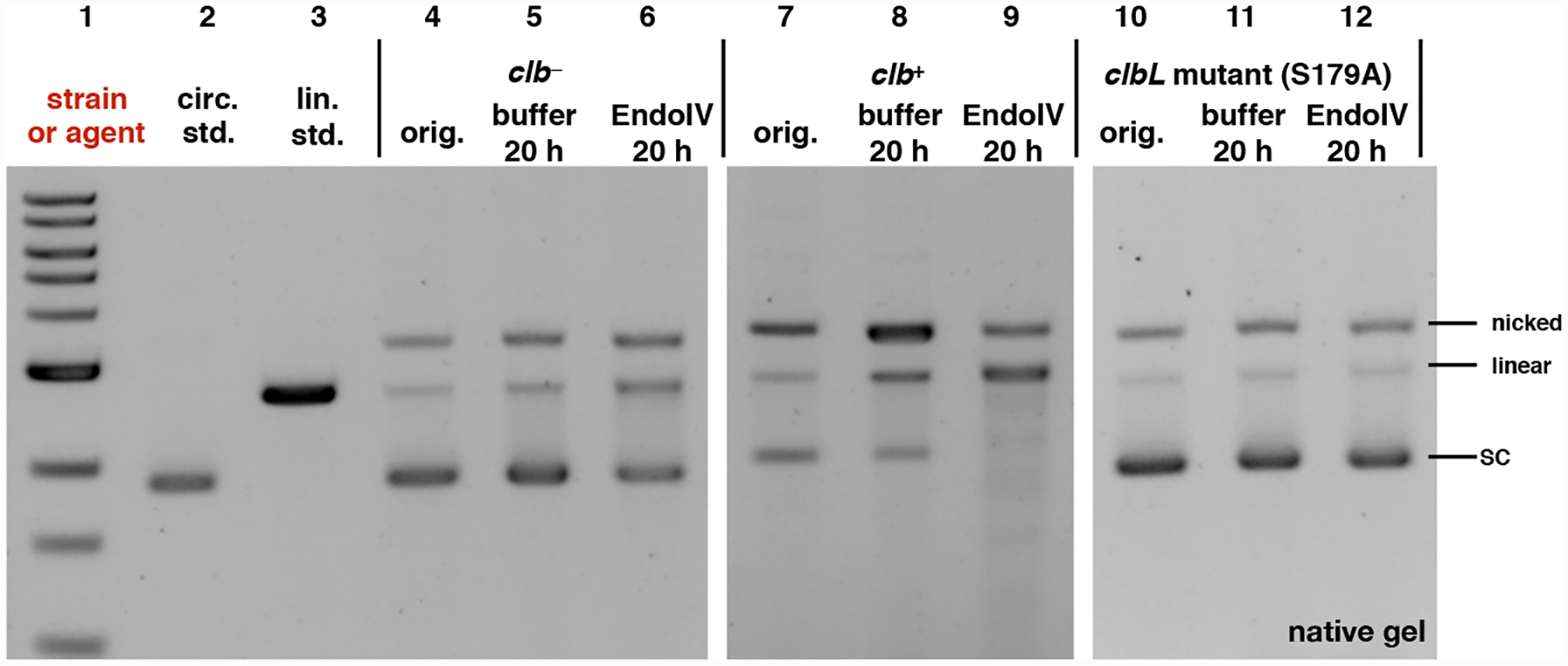
Exposure of pUC19 plasmid DNA to clb+, followed by incubation with Endo IV leads to consumption of undamaged plasmid and formation of nicked and linearized DNA. This is not observed in the clb− or clbL mutant controls. DNA ladder (Lane #1); circular pUC19 DNA standard (Lane #2); linearized pUC19 DNA standard (Lane # 3); circular pUC19 DNA isolated from co-culture with clb− BW25113 E. coli (Lane #4), reacted with buffer (Lane #5), reacted with Endonuclease IV (Lane #6); circular pUC19 DNA isolated from co-culture with clb+ BW25113 E. coli (Lane #7), reacted with buffer (Lane #8), reacted with Endonuclease IV (Lane #9); circular pUC19 DNA isolated from co-culture with clbL mutant (S179A) BW25113 E. coli (Lane #10), reacted with buffer (Lane #11), reacted with Endonuclease IV (Lane #12). Conditions (Lane #4–#6): circular pUC19 DNA from co-culture with clb− BW25113 E. coli in M9-CA media for 4 h at 37 °C (Lane # 4); the DNA (3.9 μM base pair) was reacted with NEBuffer 3.1 (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #5); the DNA (3.9 μM base pair) was further reacted with 20 units of Endonuclease IV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #6). Conditions (Lane #7–#9): circular pUC19 DNA isolated from co-culture with clb+ BW25113 E. coli. in in M9-CA media for 4 h at 37 °C (Lane #7); the DNA (3.9 μM base pair) was reacted with NEBuffer 3.1 (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #8); the DNA (3.9 μM base pair) was reacted with 20 units of Endonuclease IV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #9). Conditions (Lane #10–#12): circular pUC19 DNA isolated from co-culture with clbL mutant (S179A) BW25113 E. coli. in M9-CA media for 4 h at 37 °C (Lane #10); the DNA (3.9 μM base pair) was reacted with NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #11); the DNA (3.9 μM base pair) was reacted with 20 units of Endonuclease IV in NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 20 hours (Lane #12). The DNA was not re-purified and was directly analyzed by native agarose gel electrophoresis (90 V, 1.5 hr).
We then sought to determine if these effects were recapitulated by synthetic material. Because the carbon–carbon bond within the α-diketone function of colibactin (1) is susceptible to nucleophilic cleavage,31 we employed the enediol 4 in our studies (Fig. 8). We have shown that oxidation to the α-dicarbonyl occurs spontaneously under air,31 and that cyclodehydration in aqueous buffer occurs rapidly to form the DNA warheads of 1.13 We evaluated the effects of 4 on DNA. The DNA was completely degraded at 100 μM 4 (Fig. 9A Lane 5; Fig. 9B, Lane 3). Fig. 9A shows that ICLs are formed when linearized pUC19 DNA is exposed to 10 μM–100 nM of 4 for 4 h (compare Lanes 6–8 to Lane 4). Lanes 3–5 of Fig. 9B show that most of the circular pUC19 DNA remains intact following exposure to 10 or 1 μM 4 for 3 h and analysis by native gel electrophoresis. However, on treatment with EndoIV, significant amounts of nicked and linearized DNA are produced (Lanes 8 and 9). The degradation of DNA was qualitatively higher than that obtained when incubating in buffer alone (Lanes 6 and 7). These studies show that synthetic 4 recapitulates the activity of clb+ E. coli in this assay.
Fig. 8.
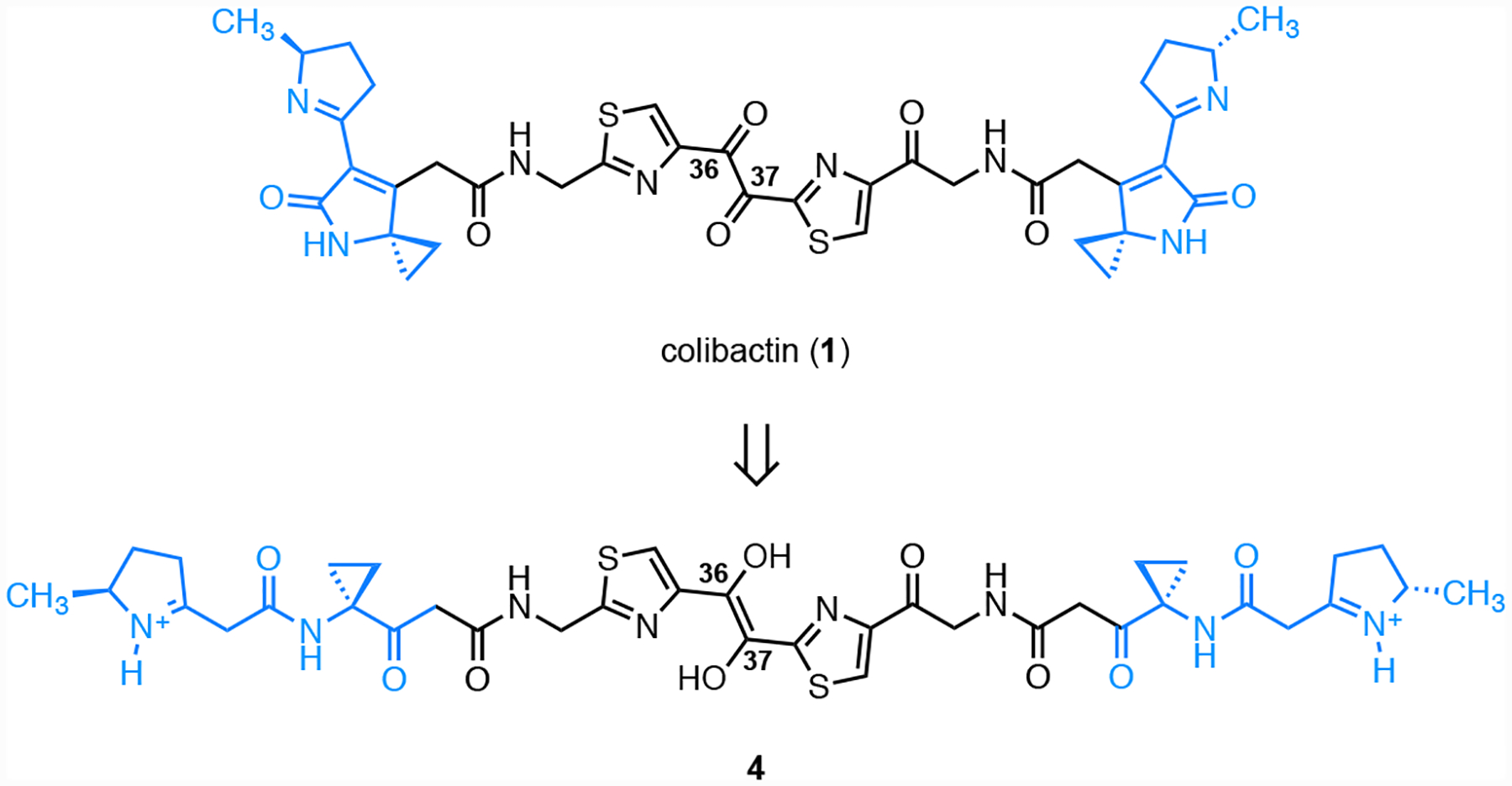
Structure of the colibactin precursor 4.
Fig. 9.
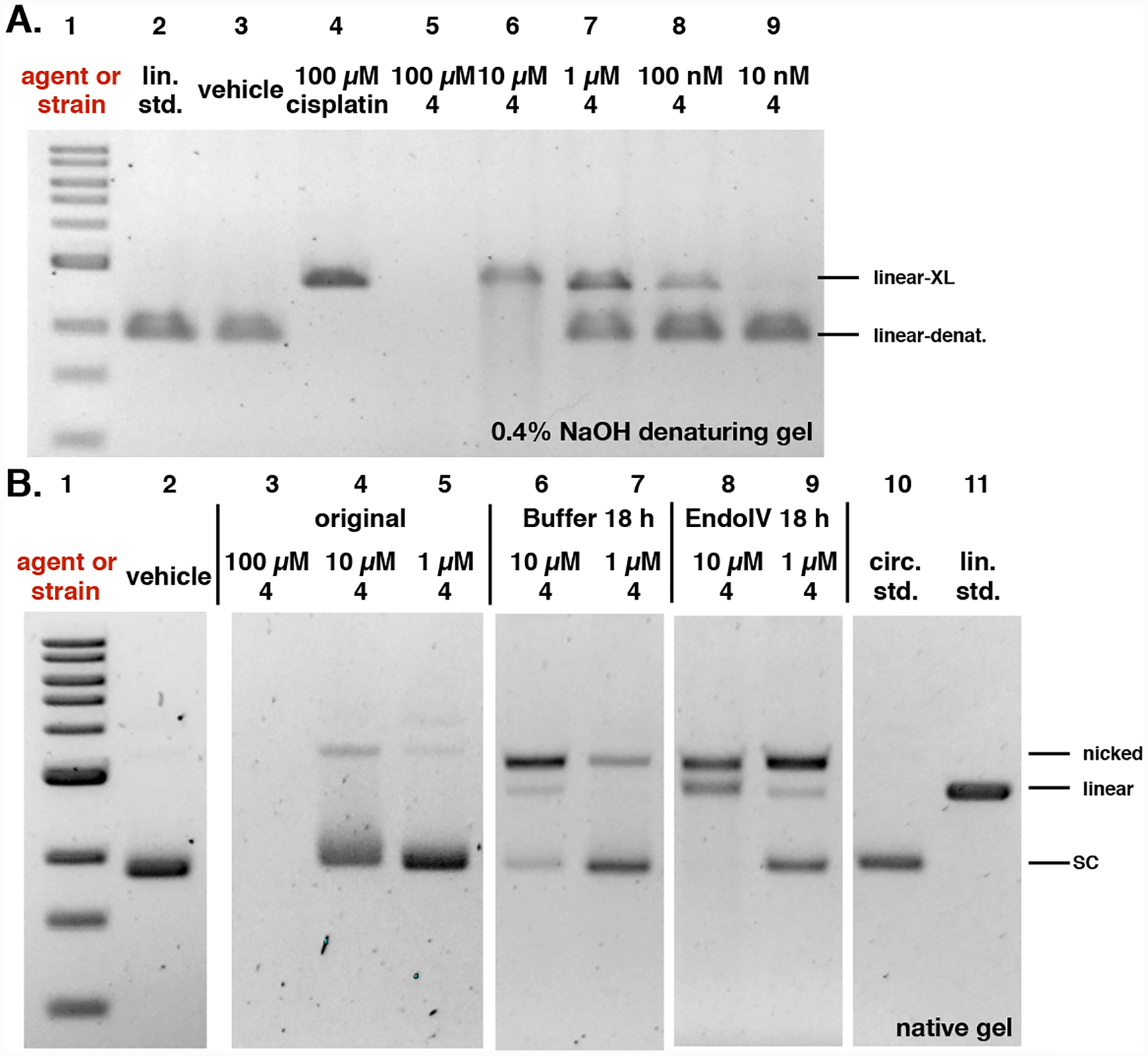
Analysis of induction of AP sites by the colibactin precursor 4. A. Incubation of plasmid pUC19 DNA exposed to 4 in buffer for 18 h results in minor nicking and cleavage. B. Addition of EndoIV increases the amount of nicked and cleaved plasmid. Conditions: A. 5% DMSO was used as vehicle (negative control), and 100 μM cisplatin was used as positive control. DNA ladder (Lane #1); linearized pUC19 DNA standard (Lane #2); 5% DMSO (Lane #3); 100 μM cisplatin (Lane #4); 100 μM 4 (Lane #5); 10 μM 4 (Lane #6); 1 μM 4 (Lane #7); 100 nM 4 (Lane #8); 10 nM 4 (Lane #9). Conditions (Lane #3): linearized pUC19 DNA (15.4 μM in base pairs), 5% DMSO (vehicle), 10 mM citric buffer, pH 5.0, 4 h, 37 °C. Conditions (Lane #4): linearized pUC19 DNA (15.4 μM in base pairs), 5% DMSO (vehicle), 100 μM cisplatin, 10 mM citric buffer, pH 5.0, 4 h, 37 °C. Conditions (Lanes #5–#9): circular pUC19 DNA (15.4 μM in base pairs), 4 (100 μM–10 nM), 5% DMSO, 10 mM citric buffer, pH 5.0, 4 h, 37 °C. The DNA was analyzed by 0.4% NaOH denaturing agarose gel electrophoresis (90 V, 1.5 h). B. 5% DMSO was used as vehicle. DNA ladder (Lane #1); 5% DMSO (Lane #2); 100 μM 4 (Lane #3); 10 μM 4 (Lane #4); 1 μM 4 (Lane #5); post buffer-reacted after 10 μM 4 (Lane #6); post buffer-reacted after 1 μM 4 (Lane #7); post EndoIV-reacted after 10 μM 4 (Lane #8); post EndoIV-reacted after 1 μM 4 (Lane #9); circular pUC19 plasmid standard (Lane #10); linearized pUC19 plasmid standard (Lane #11). Conditions (Lane #2): circular pUC19 DNA (15.4 μM in base pairs), 5% DMSO (vehicle), 10 mM citric buffer, pH 5.0, 4 h, 37 °C. Conditions (Lanes #3–#5): circular pUC19 DNA (15.4 μM in base pairs), 4 (100 μM–1 μM), 5% DMSO, 10 mM citric buffer, pH 5.0, 3 h, 37 °C. Conditions (Lanes #6–#7): 4 (10 μM–1 μM)-treated circular pUC19 DNA (3.9 μM in base pairs), NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 18 h. Conditions (Lanes #8–#9): 4 (10 μM–1 μM)-treated circular pUC19 DNA (3.9 μM in base pairs), 20 units of Endonuclease IV (New England Biolabs®), NEBuffer 3.1® (New England Biolabs®), pH 7.9, at 37 °C for 18 h. The DNA was analyzed by native agarose gel electrophoresis (90 V, 2 h). Linear-denat. = DSB/linearized DNA in denaturing form, linear-XL = DSB/linerized DNA cross-linked by colibactin or cisplatin, SC = supercoiled, nicked = SSB, linear = DSB.
Discussion.
A prior study established that clb+ E. coli form ICLs and activate the FA ICL repair pathway.14 FA repair leads to the generation of DNA DSBs, which in turn are resolved by the HR DNA DSB repair pathway.17 Thus the generation of ICLs is consistent with the earlier determination that clb+ E. coli lead to accumulation of the HR repair factor γH2AX in eukaryotic cells, fragmentation of genomic DNA in a neutral comet assay,15 and co-localization of the FA repair protein Fanconi anemia protein D2 (FANCD2) with γH2AX.14
However cells deficient in Ku80, a factor in the non-homologous end joining (NHEJ) DSB repair pathway,18 are sensitized to clb+ E. coli.16 Because NHEJ is not coupled to FA, this suggests a potential additional mechanism for DSB generation, presumably through DSBs directly induced by colibactin. We had observed that DNA alkylation products derived from synthetic colibactin analogs and colibactin (1) itself were unstable.9, 10, 13 We hypothesized that this inherent ICL instability could explain the sensitization of Ku80 deficient cells to colibactin genotoxicity. To probe this in more detail, we studied the stability of alkylation products derived from circular pUC19 DNA. Here we used the topology of the supercoiled plasmid DNA as a proxy for the stability of colibactin alkylation products. Depurination of a single alkylation product followed by elimination of the 3′ phosphate leads to a SSB (nick) and relaxation of the supercoiled plasmid. Repetition of the same sequence on the complementary strand leads to linearization of the plasmid.
Our data suggests that the colibactin-derived ICLs are unstable and undergo spontaneous depurination and subsequent elimination of the 3′ phosphate. The amount of nicking and cleavage of DNA was increased on treatment with EndoIV, suggesting the accumulation of AP sites, followed by slower strand cleavage. This is consistent with the relative rates of depurination and 3′ phosphate elimination (t1/2 = 24 and 190 h respectively, vide supra). Because the rate of 3′ phosphate elimination is slow, we speculate that the recognition of AP sites by endonucleases, such as APE1, may be contribute to DNA break formation in tissue culture.
This work provides several other insights into colibactin-mediated DNA damage. First, as shown in Fig 7, EndoIV treatment did not increase the number of SSBs and DSBs in DNA exposed to the clbL mutant (S179A). We have previously shown that a clbL mutant (S179A) alkylates but does not cross-link DNA.32 This suggests that the alkylated bases within the colibactin ICL are less stable than those derived from monoalkylation products. This may derive from distortion of the DNA duplex, as observed with other ICL agents such as cisplatin.30 Additionally while the site of DNA alkylation by colibactin (1) has not been rigorously established, the monoalkylation product 3 is linked through N3A.20 Both N3 and N7 adenine adducts are unstable,23 suggesting ICLs are formed by alkylation at N3,N3 or N3,N7.
Conclusion.
In summary, these data provide molecular and mechanistic knowledge of colibactin-induced DNA damage. We have shown that colibactin-derived DNA cross-links are inherently unstable and provided a molecular model that accounts the evolution of colibactin ICLs to DSBs. Our data further our understanding of how clb+ E. coli can induce ICL and DSB phenotypes simultaneously.
Supplementary Material
Acknowledgement.
Financial support from the National Institutes of Health (R01CA215553) is gratefully acknowledged. We thank Emilee Shine (Yale University) for providing the clbL mutant.
Footnotes
Supporting information available: Figs S1–S17
Competing interests. The authors declare no competing interests.
References.
- (1).Balskus EP (2015) Colibactin: Understanding an elusive gut bacterial genotoxin. Nat. Prod. Rep 32, 1534–1540. [DOI] [PubMed] [Google Scholar]
- (2).Trautman EP, and Crawford JM (2015) Linking biosynthetic gene clusters to their metabolites via pathway-targeted molecular networking. Curr. Top. Med. Chem 16, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Taieb F, Petit C, Nougayrede JP, and Oswald E (2016) The enterobacterial genotoxins: Cytolethal distending toxin and colibactin. EcoSal Plus 7, doi: 10.1128/ecosalplus.ESP-0008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Healy AR, and Herzon SB (2017) Molecular basis of gut microbiome-associated colorectal cancer: A synthetic perspective. J. Am. Chem. Soc 139, 14817–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Faïs T, Delmas J, Barnich N, Bonnet R, and Dalmasso G (2018) Colibactin: More than a new bacterial toxin. Toxins 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, Oelschlaeger TA, Oswald E, Forestier C, Hacker J, and Dobrindt U (2009) Genetic structure and distribution of the colibactin genomic island among members of the family enterobacteriaceae. Infect. Immun 77, 4696–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, and Jobin C (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, and Bonnet R (2013) High prevalence of mucosa-associated e. Coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8, e56964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Xue M, Kim CS, Healy AR, Wernke KM, Wang Z, Frischling MC, Shine EE, Wang W, Herzon SB, and Crawford JM (2019) Structure elucidation of colibactin. bioRxiv, 574053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Xue M, Kim CS, Healy AR, Wernke KM, Wang Z, Frischling MC, Shine EE, Wang W, Herzon SB, and Crawford JM (2019) Structure elucidation of colibactin and its DNA cross-links. Science 365, eaax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jiang Y, Stornetta A, Villalta PW, Wilson MR, Boudreau PD, Zha L, Balbo S, and Balskus EP (2019) The reactivity of an unusual amidase may explain colibactin’s DNA cross-linking activity. bioRxiv, 567248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Jiang Y, Stornetta A, Villalta PW, Wilson MR, Boudreau PD, Zha L, Balbo S, and Balskus EP (2019) Reactivity of an unusual amidase may explain colibactin’s DNA cross-linking activity. J. Am. Chem. Soc 141, 11489–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Healy AR, Nikolayevskiy H, Patel JR, Crawford JM, and Herzon SB (2016) A mechanistic model for colibactin-induced genotoxicity. J. Am. Chem. Soc 138, 15563–15570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, and Nougayrede JP (2018) The colibactin genotoxin generates DNA interstrand cross-links in infected cells. MBio 9, e02393–02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, and Oswald E (2006) Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851. [DOI] [PubMed] [Google Scholar]
- (16).Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, and Nougayrède J-P (2010) Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 107, 11537–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Clauson C, Schärer OD, and Niedernhofer L (2013) Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspect. Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Scully R, Panday A, Elango R, and Willis NA (2019) DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol 20, 698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Michl J, Zimmer J, and Tarsounas M (2016) Interplay between fanconi anemia and homologous recombination pathways in genome integrity. The EMBO Journal 35, 909–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carra A, Brennan CA, Chun E, Ngo L, Samson LD, Engelward BP, Garrett WS, Balbo S, and Balskus EP (2019) The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Margison GP, and O’Connor PJ (1973) Biological implications of the instability of the n-glycosidic bond of 3-methyldeoxyadenosine in DNA. Biochim. Biophys. Acta, Nucleic Acids Protein Synth 331, 349–356. [DOI] [PubMed] [Google Scholar]
- (22).Fujii T, Saito T, and Nakasaka T (1989) Purines. Xxxiv. : 3-methyladenosine and 3-methyl-2’- deoxyadenosine : Their synthesis, glycosidic hydrolysis, and ring fission. Chem. Pharm. Bull 37, 2601–2609. [Google Scholar]
- (23).Gates KS (2009) An overview of chemical processes that damage cellular DNA: Spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol 22, 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Trautman EP, Healy AR, Shine EE, Herzon SB, and Crawford JM (2017) Domain-targeted metabolomics delineates the heterocycle assembly steps of colibactin biosynthesis. J. Am. Chem. Soc 139, 4195–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Xue M, Shine E, Wang W, Crawford JM, and Herzon SB (2018) Characterization of natural colibactin-nucleobase adducts by tandem mass spectrometry and isotopic labeling. Support for DNA alkylation by cyclopropane ring opening. Biochemistry 57, 6391–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lindahl T, and Andersson A (1972) Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 11, 3618–3623. [DOI] [PubMed] [Google Scholar]
- (27).Crine P, and Verly WG (1976) A study of DNA spontaneous degradation. Biochim Biophys Acta 442, 50–57. [DOI] [PubMed] [Google Scholar]
- (28).Ljungquist S (1977) A new endonuclease from escherichia coli acting at apurinic sites in DNA. J. Biol. Chem 252, 2808–2814. [PubMed] [Google Scholar]
- (29).Levin JD, Johnson AW, and Demple B (1988) Homogeneous escherichia coli endonuclease iv. Characterization of an enzyme that recognizes oxidative damage in DNA. J. Biol. Chem 263, 8066–8071. [PubMed] [Google Scholar]
- (30).Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, and Zelwer C (1999) Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 a resolution: Hydration at the platinated site. Nucleic Acids Res. 27, 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Healy AR, Wernke KM, Kim CS, Lees NR, Crawford JM, and Herzon SB (2019) Synthesis and reactivity of precolibactin 886. Nat. Chem 11, 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Shine EE, Xue M, Patel JR, Healy AR, Surovtseva YV, Herzon SB, and Crawford JM (2018) Model colibactins exhibit human cell genotoxicity in the absence of host bacteria. ACS Chem. Biol 13, 3286–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


