Abstract
Purpose:
Carboplatin dose is calculated based on kidney function, commonly estimated with imperfect creatinine-based formulae. Iohexol is used to measure glomerular filtration rate (GFR) and allows calculation of a more appropriate carboplatin dose. To address potential concerns that iohexol administered during a course of chemotherapy impacts that therapy, we performed in vitro and in vivo pharmacokinetic drug-drug interaction evaluations of iohexol.
Methods:
Carboplatin was administered IV to female mice at 60 mg/kg with or without iohexol at 300 mg/kg. Plasma ultrafiltrate, kidney and bone marrow platinum was quantitated by atomic absorption spectrophotometry. Paclitaxel microsomal and gemcitabine cytosolic metabolism as well as metabolism of CYP and UGT probes was assessed with and without iohexol at 300 μg/mL by LC-MS/MS.
Results:
In vivo carboplatin exposure was not significantly affected by iohexol co-administration (platinum AUC combination vs alone: plasma ultrafiltrate 1,791 vs 1,920 μg/mL•min; kidney 8,367 vs 9,757 μg/g•min; bone marrow 12.7 vs 12.7 μg/mg-protein•min). Paclitaxel microsomal metabolism was not impacted (combination vs alone: 6-α-OH-paclitaxel 38.3 versus 39.4 ng/mL/60min; 3-p-OH-paclitaxel 26.2 versus 27.7 ng/mL/60min). Gemcitabine human cytosolic elimination was not impacted (AUC combination vs gemcitabine alone: dFdU 24.1 versus 23.7 μg/mL/30min). Iohexol displayed no relevant inhibition of the CYP and UGT enzymes in human liver microsomes.
Conclusions:
Iohexol is unlikely to affect the clinical pharmacokinetics of carboplatin, paclitaxel, gemcitabine, or other agents used in combination with carboplatin treatment. Measuring GFR with iohexol to better dose carboplatin is unlikely to alter the safety or efficacy of chemotherapy through pharmacokinetic drug-drug interactions.
Keywords: iohexol, drug-drug interaction, carboplatin, gemcitabine, paclitaxel, renal function, glomerular filtration rate, pharmacokinetics, mouse, optimal dosing
INTRODUCTION
Carboplatin is a chemotherapeutic agent that is widely used across many disease types for both primary and recurrent disease. The current dosing approach for carboplatin targets an plasma ultrafiltrate area under the concentration time curve (AUC). Carboplatin has a clear exposure- response relationship with increasing AUC resulting in increased antitumor activity, which plateaus, and with further AUC increase merely leading to more toxicity [1,2].
Carboplatin clearance depends largely on kidney function, and because of this, the milligram dose of carboplatin needed to achieve a target carboplatin AUC has historically been calculated using the Calvert equation, which requires imputing a value for glomerular filtration rate (GFR) [3]. The gold standard reference method to obtain measured GFR (mGFR) is to determine constant infusion inulin urinary clearance at steady-state, a prohibitively difficult approach. Alternative mGFR approaches, such as measuring urinary or plasma clearance of exogenous markers, including DTPA, EDTA, iohexol, or iothalamate, have also not been widely adopted in clinical practice. mGFR determination by [51Cr]-EDTA, which requires dosing and handling of radioactive substances, is only used in Europe, and is not practical. Consequently, as a surrogate for GFR, creatinine clearance (CrCl) is estimated with the Cockcroft-Gault (CG) formula, which then is assumed to be equal to GFR and imputed into the Calvert equation. Creatinine clearance is not only important in the calculation of carboplatin dose, but is also used in treatment decisions of many other anticancer drugs [4].
CrCl is often different from GFR, because the serum concentration of endogenous filtration markers is influenced by non-GFR determinants as well as by GFR. For creatinine, these non-GFR determinants include diet, muscle mass, tubular secretion, and non-renal elimination in the gut [5]. The creatinine-based GFR estimation formulas incorporate easily measured covariates (age, sex, race and body size) in an attempt to account for the effect of the non-GFR determinants. The current approach to dosing carboplatin based on CG uses serum creatinine and muscle mass [6] (ignoring body type[7], impact of diet[8], creatinine assay bias[9,10], and assay recalibration in 2010[11]) to estimate creatinine clearance [6]. Furthermore, the CG formula was derived in a biased sample of all white, mostly male patients with an R2 of only 0.69. The value for creatinine clearance is then imputed into the Calvert equation, which is also biased, having been derived in only 31 mostly female patients with an R2 of 0.76 [3]. The calculated dose may be capped by the truncation of GFR at 125 mL/min [12] (incorrectly assumed to be the physiological maximum [13]), or setting a lower limit value for SCr. It is therefore not a surprise that we do not accurately and precisely achieve the target AUC. A recently published study targeting an AUC=6 reported an average observed AUC of 7.9 (SD 2.9, N=45) corresponding to a +31% bias and a 37% coefficient of variation [14] despite all the patient specific calculations.
Recently, the concerns around using historic formulae such as CG to estimate kidney function, and the potential for new approaches have received increasing attention [4,15]. An ongoing clinical trial utilizes iohexol to measure GFR to enable more accurate carboplatin dose calculations (ClinicalTrials.gov Identifier: NCT03997370). Iohexol is a non-ionic contrast medium, mainly used for computed tomography (CT), catheter-based angiography and interventions. Importantly for a GFR marker, iohexol had no effect on the GFR as measured by [51Cr]-EDTA [16-18]. Protein binding was reported to be only 1.5%, allowing this marker to be freely filtrated through the glomerulus. Extra-renal clearance of iohexol is low, based on studies of plasma clearance in anephric patients (2-3 mL/min/1.73m2), and studies of the difference between plasma and urinary clearances in healthy subjects (extra renal clearance of 0-6 mL/min/1.73m2). To address the potential concern that iohexol administered during a course of chemotherapy might impact that therapy by pharmacokinetic drug-drug interactions, we evaluated iohexol as a perpetrator on carboplatin disposition. Because carboplatin clearance is predominantly kidney function dependent, we evaluated this potential interaction in vivo in mice. Carboplatin is used as a single agent as well as in combination regiments, such as with gemcitabine and paclitaxel. We therefore also assessed the ability of iohexol to affect the disposition of these agents. Because the clearance of gemcitabine and paclitaxel is predominantly liver enzyme dependent, we evaluated these interactions in subcellular fractions. For gemcitabine, we investigated the cytosolic metabolism by cytidine deaminase of gemcitabine to dFdU, and for paclitaxel we investigated the microsomal metabolism of paclitaxel by CYP2C8 to 6-alpha-hydroxy paclitaxel (6-α-OH-paclitaxel), and by CYP3A4 to 3-para-hydroxy paclitaxel (3-p-OH-paclitaxel) [19-22]. Lastly, we generalized our assessment by examining the potential of iohexol to affect various microsomal enzymes by using a panel of CYP450 and UGT substrates in microsomal incubations in the presence of iohexol.
EXPERIMENTAL
Chemicals and reagents
Sterile sodium chloride (0.9% NaCl) and phosphate-buffered saline (PBS) were purchased from Baxter Healthcare Corporation (Deerfield, IL). Iohexol powder, paclitaxel, and gemcitabine were purchased from Toronto Research Chemicals Inc (Toronto, ON, Canada). Iohexol dosing solution was manufactured by GE Healthcare, (Shanghai, China) and obtained from the Hillman Cancer Research Pharmacy. All in vitro drug elimination incubations with iohexol were performed at 300 μg/mL which is at the upper range of the reported plasma concentrations [23]. [D5]-iohexol (IS) was purchased from ALSACHIM (Graffenstaden, France). Carboplatin for dosing was manufactured by Ben Venue Labs Inc (Bedford, OH) and obtained from the Hillman Cancer Research Pharmacy. NADPH tetrasodium salt was purchased from Enzo Life Sciences (Farmingdale, NY). Carboplatin powder, MgCl2, alamethicin, UDPGA sodium salt, phenacetin, dextromethorphan and midazolam methanolic solution was obtained from Sigma (St Louis, MO). [D3]-dextrorphan and [D4]-1-hydroxy midazolam were purchased from Sigma (St. Louis MO). 1-hydroxy midazolam, 4-hydroxy diclofenac, 4-hydroxy mephenytoin, dextrorphan tartrate, s-mephenytoin, SN-38, trifluoperazine, mycophenolic acid and naloxone were purchased from Cayman chemicals (Ann Arbor, MI).
In Vitro Stability of Carboplatin and Iohexol
To assure we could co-administer carboplatin and iohexol without these components interacting in the dosing solution, we assessed the stability of carboplatin in the presence of iohexol. Carboplatin (6 mg/mL), iohexol (30 mg/mL) and the combination were prepared in normal saline. Over the course of a 4 h incubation at room temperature (the projected time the dosing solutions would be used in the animal study), samples were taken for the determination of iohexol to assess any loss in iohexol. Because possible reaction products of iohexol and carboplatin would still be small molecular and filter through a 30 kD filter used to obtain ultrafiltrate, we needed to evaluate loss of active carboplatin in another manner. We therefore diluted 30 μL of the 4 h combination saline incubation sample 60-fold in plasma and further incubated at room temperature for 168 h and did the same with a fresh carboplatin saline sample. After this incubation, ultrafiltrate was generated by centrifuging at 12,000 x g for 5 min at 4 °C through an Amicon Ultra 30k centrifugal filter unit (Merk Millipore Ltd. Tullagreen, Carrigtwohill Co. CORK IRL) and analyzed by atomic absorption spectrophotometry. Any loss in active carboplatin through reaction with iohexol would result in a permanently ultrafilterable component and an increase in ultrafilterable fraction after incubation in plasma.
Iohexol and Carboplatin In Vivo DDI Studies
Specific pathogen-free female CFW mice (Swiss-Webster, 9-11 weeks of age) were purchased from Charles River (Wilmington, MA) associated with an approved University of Pittsburgh IACUC protocol (Protocol#: 17111628). Mice were allowed to acclimate to the University of Pittsburgh Animal Facility for at least 1 week before studies were initiated as described previously [24]. Mice were stratified based on body weights into treatment arm and time point groups to eliminate statistical differences in body weight. Mice (N=3/time point) were dosed with 60 mg/kg carboplatin, 300 mg/kg iohexol, or the combination formulated in normal saline, or with vehicle only and euthanized by CO2 inhalation at the following time points after a 30 s IV bolus administration (0.01 mL/g exact body weight): (1) Carboplatin arm: 5, 10, 15, 30 min, 1, 2, 4, and 6 h; (2) Iohexol arm: 2.5, 5, 10, 15, 30 min, 1, and 2 h; and (3) Combination arm: 2.5, 5, 10, 15, 30 min, 1, 2, 4, and 6 h. Each arm included vehicle dosed animals euthanized at 5 min. Blood was collected by cardiac puncture with EDTA flushed syringes and plasma was isolated by centrifugation at 12,000 x g for 4 min. Plasm ultrafiltrate was generated with Amicon Ultra 30k centrifugal filter units (Merk Millipore Ltd. Tullagreen, Carrigtwohill Co. CORK IRL) and centrifuged at 12,000 x g for 10 min at 4 °C. Kidneys were collected, weighed, snap frozen in liquid N2, and stored at −80 °C until analysis. Bone marrow was obtained by flushing both femurs from each mouse with PBS. The bone marrow from each mouse was isolated by centrifugation at 12,000 x g followed by removal of the supernatant except for approximately 50 μL. Prior to analysis of the bone marrow, samples were resuspended, sonicated, and a 20 μl aliquot taken for determination of protein concentration using the Bio-Rad Protein assay using the manufacturer’s instructions, with bovine serum albumin as standards.
In Vitro Metabolism of Paclitaxel and Gemcitabine
Human microsome incubation with paclitaxel:
Microsomal metabolism of paclitaxel was studied as previously reported [25] in the absence and presence of 300 μg/mL iohexol. Paclitaxel stock at 10 mg/ml in methanol was diluted with 0.1 M Na2KPO4 buffer to 20 μM as working concentration. Paclitaxel (10 μM final concentration) was incubated with NADPH (3 mM final concentration) and pooled human liver microsomes (0.1 mg/ml, Cat# M0317 ,Sigma, St. Louis, MO) for periods ranging from 0 to 60 min at 37 ° C, alone or in the presence of iohexol (300 μg/mL final concentration) and 0.1 M Na2KPO4 (pH 7.4) in a total volume of 100 μl. Metabolism of paclitaxel was also evaluated in the presence of 10 μM ketoconazole (CYP3A) or 3 μM Montelukast (CYP2C8). At 0, 5, 15, 30 and 60 min incubation, aliquots of 10 μL were diluted with 90 μl human plasma on ice, and stored at −80 °C until analysis.
Human cytosol incubation with gemcitabine:
Gemcitabine (50 μg/ml final concentration) was incubated with pooled, mixed gender human liver cytosol: (0.1 mg/ml, Xenotech, Lot# 1610027, Kansas City, KS) for periods rangingfrom 0 to 60 min at 37 ° C alone or in the presence of iohexol (300 μg/mL final concentration) and 50 mM tris-acetate (pH 7.4) in a total volume of 100 μl. At 0, 5, 10, 15, 30, 45 and 60 min incubation, aliquots of 10 μL were diluted with 90 μl plasma containing 100 μg/mL THU before analysis for dFdU concentrations.
In Vitro Production of CYP and UGT Specific Metabolic Products of Probe Substrates
CYP cocktail probes and Iohexol interaction:
CYP probe cocktail consisting of phenacetin (5 μM) for CYP1A2, diclofenac (2.5 μM) for CYP2C9, S-mephenytoin (30 μM) for CYP2C19, dextromethorphan (5 μM) for CYP2D6 and midazolam (2.5 μM) for CYP3A4, were incubated with human liver microsomes (pool of 50, mixed gender, Lot # 1610016, Sekisui Xenotech, Kansas City, KS) at a concentration of 0.25 mg/mL, based on earlier published protocols [26]. The concentrations of all the probes used were below their Km values to increase sensitivity of the iohexol inhibition under linear metabolite formation conditions. The incubation mixture also contained 3.3 mM MgCl2 and the reactions were carried out in phosphate buffer (pH 7.4). The reactions were initiated by addition of 1 mM NADPH. The reactions were carried out at 37 °C in open Eppendorf tubes for 15 min. The total reaction volume was 500 μL and the reactions were stopped by addition of 300 μL MeOH containing [D4]-acetaminophen (1 μM), [D4]-4-hydroxy mephenytoin (1 μM), [D3]-dextrorphan ~80 nM) and [D4]-1-hydroxy midazolam (~60 nM). The reactions were carried out with and without iohexol (300 μg/mL). Metabolism of midazolam was also evaluated in the absence and presence of 1 μM of the known CYP3A inhibitor ketoconazole.
UGT cocktail probes and Iohexol interactions:
UGT cocktail probes consisting of SN-38 (0.5 μM) for UGT1A1, trifluperazine (0.5 μM) for UGT1A4, mycophenolic acid (0.2 μM) for UGT1A9 and naloxone for UGT2B7 (1 μM) were incubated with human liver microsomes (pool of 50, mixed gender, Lot # 1610016, Sekisui Xenotech, Kansas City, KS) at a concentration of 0.25 mg/mL, based on earlier published protocols [26]. The concentrations of all the probes used were below their Km values to increase sensitivity of the iohexol inhibition under linear metabolite formation conditions. The incubation mixture also contained 10 mM MgCl2 and the reactions were carried out in Tris-HCl buffer (pH 7.4). The incubation mixture was preactivated for 15 min on ice by alamethicin (25 μg/mL) followed by initiation by addition of 5 mM UDPGA. The reactions were carried out at 37 °C in open Eppendorf tubes for 60 min. The total reaction volume was 500 μL and the reactions were stopped by addition of 250 μL of acetonitrile containing terfenadine (IS, 5 ng/mL). Metabolism of trifluoperazine and naloxone was also evaluated in the absence and presence of 100 μM the known UGT1A4 and UGT2B7 inhibitor diclofenac.
Bioanalysis
Platinum:
Platinum was quantitated by Atomic Absorbance Spectroscopy (AAS) with Zeeman background correction using a Perkin-Elmer AAS 600 atomic absorption spectrophotometry equipped with an AS-800 autosampler (Perkin-Elmer, Perkin Life and Analytical Science, Shelton, CT 06484). The AAs was operated per user manual with the Pt cathode lamp operated at 25 mA with a 0.7 nm slit, and wavelength set at 265.9 nm. Plasma samples (10 μL) were diluted 10-fold with 50/50 plasma/plasma ultrafiltrate (v/v). Kidneys were homogenized in 3 parts PBS (v/g), and 200 μL incubated overnight with 1 part volume of concentrated nitric acid in a glass tub at 60 °C. Digested sample (50 μl) was diluted with 50 μl of 50/50 plasma/ultrafiltrate. Bone marrow was lysed with 100 μl of 0.25% Triton-100, 10 μl of sample was analyzed for protein using a Bio-Rad kit, while 50 μl of sample was diluted with 50 μl of 50/50 plasma/plasma ultrafiltrate. Platinum standard at 50, 100, 200, 500, 1,000, 2,000, 5,000 ng/ml were diluted with 50/50 plasma/ultrafiltrate. 100 μl of diluent buffer (0.5% ammonium dihydrogen phosphate and 0.03% magnesium nitrate 6-hydrate) were added to each sample and standard before AAS. The sample volume was 10 μl. Based on QC samples at 75, 750, and 4000 ng/mL, accuracy (102.5-106.4%) and precision (6.4-11.0%CV) was acceptable.
Iohexol:
Iohexol was quantitated with an assay previously validated in plasma to FDA guidance [27], which was both accurate (101.3–102.1 %) and precise (<3.4 %CV) over a range of 1-500 μg/mL. The assay was cross validated for use with kidney homogenate by quantitating control kidney homogenate spiked at QC levels (accuracy (89.7-109.2%) and precision (0.9-2.1%CV) was acceptable), and thereafter the plasma assay was used to quantitate iohexol in kidney homogenate.
Paclitaxel metabolites:
6-α-OH-paclitaxel, and 3-p-OH-paclitaxel were quantitated with an assay validated in plasma to FDA guidance, as described previously [28], , which was both accurate (94.3–110.4%) and precise (<11.3%CV) from 10–10,000 ng/mL for paclitaxel and 1–1000 ng/mL for both metabolites. Samples were analyzed after dilution with 4 parts control human plasma and analyzed using a calibration curve prepared in a matrix of 20% media and 80% plasma.
Gemcitabine metabolite:
dFdU was analyzed with a previously described assay for plasma samples containing THU to halt ex vivo conversion of gemcitabine to dFdU by cytidine deaminase [29]. Samples were analyzed after dilution with 9 parts control human plasma containing THU. The assay was accurate (103.9-104.8%) and precise (3.0-6.0%CV).
CYP probes:
The bioanalysis of the metabolites of the CYP probes from the human liver microsomal incubations was performed by LC-MS/MS. The analysis was performed using previously validated method with minor modifications [30]. The validated method was developed for hepatocytes and utilized solid phase extraction for sample processing and because the present study involved a cleaner matrix of human liver microsomes, protein precipitation extraction method was employed. Detection and quantification of probes utilized a Xevo TQS Waters triple quadrupole mass spectrometer using mass transitions that produced the best response. In addition, increased sensitivity of the mass spectrometer allowed a wider linear dynamic range of the calibration curves for most analytes in the present study compared to the previously developed assay. The calibration ranges and transitions are detailed as follows: 1-hydroxy midazolam (m/z 341.9>203.0; D4: 345.8>140.0) ranged from 10-2,500 nM, 4-hydroxy diclofenac (m/z 311.9>230.0; using D4-1-hydroxy midazolam as IS) and dextrorphan (m/z 257.9>157.1; D3: 260.9>133.5) from 10-1,000 nM, 4-hydroxy mephenytoin (m/z 234.9>150.3; D3: 238.0>150.1) from 50-5,000 nM, and acetaminophen (m/z 152.1>110.2; D4: 156.3>114.2) from 10-5,000 nM. The LC method consisted of Solvent A (95% water, 5% acetonitrile, 0.1% formic acid, 2 mM ammonium acetate) and Solvent B (5% water, 95% acetonitrile, 0.1% formic acid, 2 mM ammonium acetate), pumped through an Acquity UPLC BEH C18 column (1.7 μm, 2.1 x 150 mm) at a constant flow rate of 0.2 mL/min and a column temperature of 40 °C. The gradient changed solvent B from 5% to 95% over 5 min, followed by a return to initial conditions to allow for re-equilibration for 2 min, with a total run time of 8 min. Calibration curves for 4-hydroxy diclofenac (accuracy 95.6-97.7%, precision <4.2%CV), dextrorphan (accuracy 85.2-105%, precision <1.5%CV), 4-hydroxy mephenytoin (accuracy 83.2-103%, precision <14%CV), and acetaminophen (accuracy 91.6-107%, precision <6.1%CV) were fitted to straight line with a weighting of 1/x2 except 1-hydroxy midazolam (accuracy 93.3-102%, precision <5.7%CV) that was fitted to a second order equation with the same weighting. All the calibration curves displayed r2 values above 0.99.
UGT probes:
Microsomal samples from UGT probe studies were analyzed for metabolites based on the assay described previously [26]. The UGT assay was implemented on an Agilent (Palo Alto, CA, USA) 1200 SL autosampler, with binary pump and thermostatted column compartment and an ABI SCIEX (Concord, ON, Canada) 4000Q hybrid linear ion trap tandem mass spectrometer with electrospray ionization in positive multiple reaction monitoring (MRM) mode. Standard curves were fit by linear regression with weighting by 1/y2, followed by back-calculation of concentrations. The assay proved linear from 1 to 300 ng/mL for SN-38 glucuronide (accuracy 89.2-99.3%, precision <4.5%CV) and naloxone glucuronide (accuracy 90.8-110%, precision <14.4%CV) and was linear from 10 to 3,000 ng/mL for mycophenolic acid glucuronide (accuracy 93.9-120%, precision <17.9%CV) and trifluoperazine glucuronide (accuracy 96.7-106.5%, precision <6.6%CV).
Pharmacokinetics
Non-compartmental pharmacokinetic parameters were derived from the average concentrations per time point (N=3 per time point) by non-compartmental methods using PK Solutions 2.0 (Summit Research Services, Montrose, CO; www.summitPK.com). Data used for carboplatin and iohexol PK parameters were limited to those time-points as described for the respective single agent sample time points (see above) so that each single agent-combination comparison was based on the same time points.
Statistics
CYP and UGT specific probe metabolite production, as well as paclitaxel and gemcitabine metabolite production were compared non-parametrically by a 2-sided Wilcoxon exact signed rank test. Carboplatin and iohexol PK were analyzed statistically using AUC0-t values comparing single agent PK versus the combination. After log-transformation, these ratios were subjected to a two-sided z-test, under the null hypothesis of log (AUC-ratio)=0, as described previously [31,32]. A value of p<0.05 was considered statistically significant.
RESULTS
In Vitro Stability of Carboplatin and Iohexol
As can be seen in Suppl.Figure 1A, there was no change in iohexol concentration with and without carboplatin in the dosing formulation over the time period required to dose mice. Variability in sample preparation was likely responsible for the small and relatively constant difference observed across the entire time course. Overall, there was a mean (SD) of 97.0% (2.5%) remaining for iohexol alone and 92.7% (2.0%) with carboplatin. Conversely, the impact of iohexol on carboplatin stability was also assessed. Carboplatin was able to platinate plasma proteins, and this was undiminished by the presence of iohexol for 4 h, as can be seen in Suppl.Figure 1B.
Pharmacokinetics of Carboplatin in Female Mice
Ultrafilterable plasma, total plasma, kidney, and bone marrow platinum concentration versus time profiles with and without co-administration of iohexol are depicted in Fig. 1A-D and pharmacokinetic parameters are listed in Table 1.
Fig. 1.
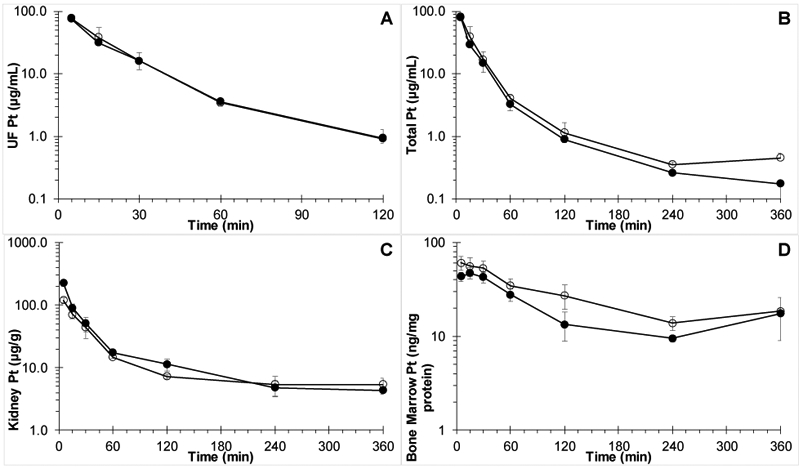
Platinum concentration versus time curves in A) plasma ultrafiltrate (P=0.94); B) plasma; C) kidney; and D) bone marrow after IV dosing of female Swiss Webster mice with 60 mg/kg carboplatin alone (○) or co-administered with (●) 300 mg/kg iohexol.
Table 1.
Platinum non-compartmental plasma and tissue pharmacokinetic parameters after IV administration to female Swiss Webster of 60 mg/kg carboplatin, or the combination with 300 mg/kg iohexol.
| Carboplatin (mg/kg) | 60 | 60 |
| Iohexol (mg/kg) | - | 300 |
| Parameter | ||
| Exact dose (mg/kg) | 59.97 | 56.86 |
| Plasma (ultrafilterable) | ||
| Cmax (μg/mL) | 77.7 (7.5) | 75.5 (5.9) |
| Half-life (min) | 31.3 | 30.9 |
| AUC0-t (μg/mL•min) | 1878 | 1749 |
| AUC0-inf (μg/mL•min) | 1920 | 1791 |
| Vss (mL/kg) | 346 | 368 |
| CL (mL/min/kg)* | 16.4 | 16.7 |
| Plasma (total) | ||
| Cmax (μg/mL) | 81.5 (2.9) | 79.2 (7.4) |
| Half-life (min) | 97.5 | 102 |
| AUC0-t (μg/mL•min) | 2116 | 1794 |
| AUC0-inf (μg/mL•min) | 2179 | 1820 |
| Vss (mL/kg) | 642 | 554 |
| CL (mL/min/kg) | 14.5 | 16.4 |
| Kidney | ||
| Cmax (μg/g) | 120 (12) | 226 (25) |
| Tmax (min) | 5 | 5 |
| Half-life (min) | 558 | 174 |
| AUC0-t (μg/g•min) | 5421 | 7,277 |
| AUC0-inf (μg/g•min) | 9,,757 | 8,367 |
| Bone marrow | ||
| Cmax (ng/mg prot) | 60.8 (10.1) | 43.5 (4.9) |
| Tmax (min) | 5 | 15 |
| Half-life (min) | 134 | 243 |
| AUC0-t (ng/mg•min) | 9,134 | 6,537 |
| AUC0-inf (ng/mg•min) | 12,692 | 12,687 |
AUC0-t, area under the plasma concentration versus time curve till the last observed time point; AUC0-inf, AUC extrapolated to infinity; CL, total body clearance; Cmax, maximum plasma concentration (mean with standard deviation); Tmax, time of Cmax; Vss, volume of distribution at steady-state.
P=0.94
Analysis of dosing solutions showed platinum concentrations of 3.15 and 2.99 μg/mL of platinum for the carboplatin without, and with iohexol, respectively, corresponding to a carboplatin dose of 6.00 and 5.69 mg/kg, respectively. As can be seen in Table 1, platinum PK parameters were similar after carboplatin alone or in combination with iohexol, with ultrafilterable plasma carboplatin clearance, being nearly identical with values of 16.4 and 16.7 mL/min/kg, respectively. Ultrafilterable platinum AUC (corrected for exact dose per quantitative analysis of dosing solutions) was not significantly different as assessed according to Bailer (P=0.94)[31].
Pharmacokinetics of Iohexol in Female Mice
Plasma and kidney iohexol concentration versus time profiles with and without co-administration of carboplatin are depicted in Fig. 2A-B and pharmacokinetic parameters are listed in Table 2. Iohexol PK parameters were similar after iohexol alone or in combination with carboplatin, with iohexol clearance, the most important parameter, being nearly identical with values of 18.7 and 18.6 mL/min/kg, respectively. Plasma AUC (corrected for exact dose per quantitative analysis of dosing solutions) was not significantly different as assessed according to Bailer‘s method of comparing AUC (P=0.67)[31].
Fig. 2.
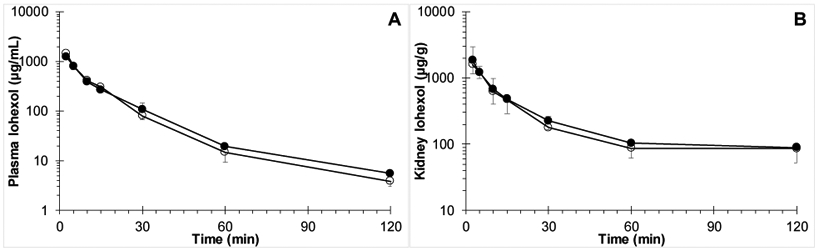
Iohexol concentration versus time curves in A) plasma (P=0.67); and B) kidney after IV dosing of female Swiss Webster mice with 300 mg/kg iohexol alone (○) or co-administered with (●) 60 mg/kg carboplatin.
Table 2.
Iohexol non-compartmental plasma and tissue pharmacokinetic parameters after IV administration to female Swiss Webster of 300 mg/kg iohexol or the combination with 60 mg/kg carboplatin.
| Carboplatin (mg/kg) | - | 60 |
| Iohexol (mg/kg) | 300 | 300 |
| Parameter | ||
| Exact dose (mg/kg) | 304.3 | 295.4 |
| Plasma | ||
| Cmax (mg/mL) | 1.43 (0.19) | 1.23 (0.05) |
| Half-life (min) | 30.6 | 32.9 |
| AUC0-t (mg/mL•min) | 16.1 | 15.7 |
| AUC0-inf (mg/mL•min)* | 16.2 | 15.9 |
| Vss (mL/kg) | 259 | 316 |
| CL (mL/min/kg) | 18.7 | 18.6 |
| Kidney | ||
| Cmax (mg/g) | 1.60 (0.45) | 1.83 (1.14) |
| Tmax (min) | 5 | 5 |
| Half-life (min) | 100 | 76 |
| AUC0-t (mg/mL•min) | 29.4 | 32.5 |
| AUC0-inf (mg/mL•min) | 41.9 | 42.2 |
Some of the iohexol PK parameters after administration of iohexol alone were published previously [27].
AUC0-t, area under the plasma concentration versus time curve till the last observed time point; AUC0-inf, AUC extrapolated to infinity; CL, total body clearance; Cmax, maximum plasma concentration (mean with standard deviation); Tmax, time of Cmax; Vss, volume of distribution at steady-state.
P=0.67
Microsomal Metabolism of Paclitaxel
Paclitaxel metabolism by human liver microsomes in the absence and presence of iohexol is depicted in Fig. 3A. Iohexol did not impact the production of CYP2C8 metabolite 6-α-OH-paclitaxel (39.4 versus 38.3 ng/mL/60min; P=0.69), or CYP3A4 metabolite 3-p-OH-paclitaxel (27.7 versus 26.2 ng/mL/60min; P=0.69). Ketoconazole successfully inhibited 3-p-OH-paclitaxel production to below the limit of quantitation at less than 30% of controls, while montelukast successfully inhibited 6-α-OH-paclitaxel production to less than 26% compared to controls.
Fig. 3.
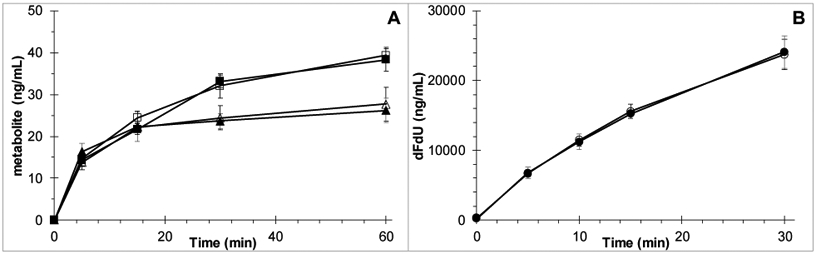
A) Human liver microsomal production of 6-α-OH-paclitaxel in the absence (□) and presence (■) of iohexol (P=0.69), and 3-p-OH-paclitaxel in the absence (△) and presence (▲) of iohexol (P=0.69) (mean of N=4 with SD error bars). B) Human liver cytosolic production of dFdU in the absence (○) and presence (●) of iohexol (P=0.83) (mean of N=3 with SD error bars).
Cytosolic Metabolism of Gemcitabine
Gemcitabine metabolism by human liver cytosolic cytidine deaminase (CDA) in the absence and presence of iohexol is depicted in Fig. 3B. Iohexol did not impact the conversion of gemcitabine by CDA to dFdU (dFdU 23.7 versus 24.1 μg/mL/30min; P=0.83 at 30 min). THU successfully inhibited dFdU production to below the limit of quantitation at less than 1% of controls.
CYP and UGT probe metabolite production
Iohexol did not display significant inhibition of the various selected CYP isoforms in the human liver microsomes. The rates of the metabolite formation for all the CYP probe drugs were comparable in the presence and absence of iohexol (Fig. 4A). Ketoconazole successfully inhibited midazolam metabolism to less than 10% of control. In the case of UGT isoforms, iohexol displayed a small statistically significant inhibition (6.3% decrease in activity compared to control; P=0.029) of UGT1A9, while no effect was detected for the remaining UGT isoforms (Fig. 4B). Diclofenac successfully inhibited trifluoperazine and naloxone glucuronidation to less than 30% compared to controls.
Fig. 4.
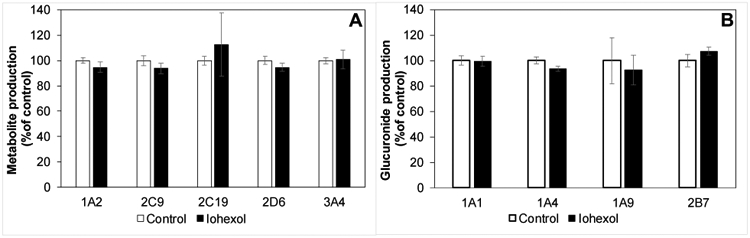
A) Lack of relevant impact of iohexol on CYP1A2, 2C9, 2C19, 2D6, or 3A4 as measured by production of CYP specific metabolite production of corresponding probe substrates phenacetin, diclofenac, mephenytoin, dextromethorphan, and midazolam, respectively in the absence (open bars) or presence of (solid bars) 300 μg/mL iohexol (mean, standard deviation). No significant difference by Wilcoxon exact rank test. B) Lack of relevant impact of iohexol on UGT1A1, 1A4, 1A9, or 2B7 as measured by production of UGT specific glucuronide metabolite production of corresponding probe substrates SN38, trifluoperazine, mycophenolic acid, and naloxone, respectively in the absence (open bars) or presence of (solid bars) 300 μg/mL iohexol (mean, standard deviation). Only UGT1A4 showed a statistically significant difference by Wilcoxon exact rank test (P=0.029; 6.3% decrease in activity, N=4).
DISCUSSION
Iohexol may be used to measure GFR in order to determine a carboplatin dose that is more appropriate than the dose calculated through imperfect creatinine-based formulae. We addressed the potential concern that iohexol is a perpetrator in pharmacokinetic drug-drug interactions with chemotherapeutics. Iohexol has no relevant impact on carboplatin clearance, paclitaxel and gemcitabine metabolism, or common CYP450 and UGT activity.
In humans, iohexol has a reported low volume of distribution (0.16 L/kg) indicating that it is primarily distributed in extracellular water, confirmed by cellular partitioning studies in hepatocytes that suggest it has minimal cellular penetration (data not shown)[33]. To maximize the sensitivity of our systems to detect any potential drug-drug interaction, we therefore chose to carry out our studies in subcellular fractions, as opposed to hepatocytes. Because iohexol does not undergo significant metabolism [33], it is not expected that production of reactive intermediates would be likely to produce time dependent inhibition [34], allowing us to use a simple microsomal system without extensive pre-incubations. The iohexol concentrations used in our studies were close to maximal plasma concentrations observed in clinical studies [23].
Our data suggest that carboplatin and iohexol exposure are unaffected by each other’s presence. Because carboplatin clearance occurs predominantly through renal filtration, this interaction was best studied in vivo. When carboplatin was administered with iohexol, platinum exposure in bone marrow and kidney, target organ of carboplatin toxicity and clearance respectively, was comparable to the exposure after carboplatin alone. Iohexol exposure in kidney, organ of iohexol clearance, appeared unimpacted by carboplatin. Mouse iohexol volume of distribution at 259 mL/kg was very close to the reported mouse extracellular water volume of 232 mL/kg[35]. Mouse iohexol clearance, and therefore the GFR, was 18.7 mL/min/kg or 529 μL/min for our average 28.3 g mouse. This value is close to the 51Cr-EDTA-determined GFR of 388 μL/min previously reported for female Swiss Webster mice [36]. Our data also suggest that in mice, the carboplatin clearance is almost identical to iohexol-measured GFR, while in humans it is higher than GFR as also reflected by the constant in the Calvert equation: carboplatin clearance (mL/min) = GFR+25 [3]. The fast kinetics in mice result in rapid carboplatin clearance by the kidneys, leaving no time for the slow reaction of carboplatin and macromolecules (see Suppl.Figure 1) to meaningfully contribute to carboplatin clearance.
Paclitaxel and gemcitabine are two of the more common partners of carboplatin in combination regimens. Iohexol did not affect paclitaxel metabolism by either CYP3A4 or CYP2C8 and it did not affect gemcitabine metabolism by CDA.
To broaden our evaluation of iohexol as a perpetrator of pharmacokinetic drug-drug interactions beyond specific victim drugs, we investigated the impact of iohexol on the activity of those CYP and UGT isoforms predominantly involved in the metabolism of most commercially available drugs. Iohexol displayed a lack of relevant inhibitory activity for the investigated CYP and the UGT enzymes. Although a small statistically significant inhibition of UGT1A9 was observed in vitro, the poor membrane permeability of iohexol will limit iohexol distribution into hepatocytes, further minimizing the relevance of the observed inhibition.
In summary, iohexol is predicted to have no relevant impact on the clinical pharmacokinetics of carboplatin, paclitaxel, gemcitabine and other chemotherapeutics. Iohexol may be used to accurately measure GFR and more accurately calculate carboplatin dose, without altering the safety or efficacy of carboplatin containing chemotherapy regimens through pharmacokinetic drug-drug interactions.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants UM1 CA186690, U24CA247643, and R50 CA211241. This project used the UPMC Hillman Cancer Pharmacokinetics and Pharmacodynamics Facility (CPPF) and Animal Facility (AF) and was supported in part by award P30 CA47904.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Jodrell DI, Egorin MJ, Canetta RM, Langenberg P, Goldbloom EP, Burroughs JN, Goodlow JL, Tan S, Wiltshaw E (1992) Relationships between carboplatin exposure and tumor response and toxicity in patients with ovarian cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 10 (4):520–528 [DOI] [PubMed] [Google Scholar]
- 2.Company B-MS (2010) Package Insert - PARAPLATIN® (carboplatin) for Injection, USP. [Google Scholar]
- 3.Calvert AH, Newell DR, Gumbrell LA, O'Reilly S, Burnell M, Boxall FE, Siddik ZH, Judson IR, Gore ME, Wiltshaw E (1989) Carboplatin dosage: prospective evaluation of a simple formula based on renal function. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 7 (11):1748–1756. doi: 10.1200/JCO.1989.7.11.1748 [DOI] [PubMed] [Google Scholar]
- 4.Casal MA, Nolin TD, Beumer JH (2019) Estimation of Kidney Function in Oncology. Clinical Journal of the American Society of Nephrology. doi: 10.2215/cjn.11721018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levey AS, Inker LA, Coresh J (2014) GFR estimation: from physiology to public health. American journal of kidney diseases : the official journal of the National Kidney Foundation 63 (5):820–834. doi: 10.1053/j.ajkd.2013.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cockcroft DW, Gault MH (1976) Prediction of creatinine clearance from serum creatinine. Nephron 16 (1):31–41 [DOI] [PubMed] [Google Scholar]
- 7.Winter MA, Guhr KN, Berg GM (2012) Impact of various body weights and serum creatinine concentrations on the bias and accuracy of the Cockcroft-Gault equation. Pharmacotherapy 32 (7):604–612. doi: 10.1002/j.1875-9114.2012.01098.x [DOI] [PubMed] [Google Scholar]
- 8.Preiss DJ, Godber IM, Lamb EJ, Dalton RN, Gunn IR (2007) The influence of a cooked-meat meal on estimated glomerular filtration rate. Annals of clinical biochemistry 44 (Pt 1):35–42. doi: 10.1258/000456307779595995 [DOI] [PubMed] [Google Scholar]
- 9.Andreev E, Koopman M, Arisz L (1999) A rise in plasma creatinine that is not a sign of renal failure: which drugs can be responsible? J Intern Med 246 (3):247–252 [DOI] [PubMed] [Google Scholar]
- 10.Levey AS, Perrone RD, Madias NE (1988) Serum creatinine and renal function. Annu Rev Med 39:465–490. doi: 10.1146/annurev.me.39.020188.002341 [DOI] [PubMed] [Google Scholar]
- 11.Lawson J, Switchenko JM, McKibbin T, Donald Harvey R (2016) Impact of Isotope Dilution Mass Spectrometry (IDMS) Standardization on Carboplatin Dose and Adverse Events. Pharmacotherapy 36 (6):617–622. doi: 10.1002/phar.1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr M, Maranta AF, Reichegger H, Gillessen S, Cathomas R (2018) Carboplatin dose based on actual renal function: no excess of acute haematotoxicity in adjuvant treatment in seminoma stage I. ESMO Open 3 (3):e000320. doi: 10.1136/esmoopen-2018-000320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wesson L (1969) Physiology of the Human Kidney. Grune & Stratton, New York, NY [Google Scholar]
- 14.Appleman LJ, Beumer JH, Jiang Y, Lin Y, Ding F, Puhalla S, Swartz L, Owonikoko TK, Donald Harvey R, Stoller R, Petro DP, Tawbi HA, Argiris A, Strychor S, Pouquet M, Kiesel B, Chen AP, Gandara D, Belani CP, Chu E, Ramalingam SS (2019) Phase 1 study of veliparib (ABT-888), a poly (ADP-ribose) polymerase inhibitor, with carboplatin and paclitaxel in advanced solid malignancies. Cancer chemotherapy and pharmacology. doi: 10.1007/s00280-019-03960-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beumer JH, Inker LA, Levey AS (2018) Improving Carboplatin Dosing Based on Estimated GFR. American journal of kidney diseases : the official journal of the National Kidney Foundation 71 (2):163–165. doi: 10.1053/j.ajkd.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delanaye P, Ebert N, Melsom T, Gaspari F, Mariat C, Cavalier E, Bjork J, Christensson A, Nyman U, Porrini E, Remuzzi G, Ruggenenti P, Schaeffner E, Soveri I, Sterner G, Eriksen BO, Back SE (2016) Iohexol plasma clearance for measuring glomerular filtration rate in clinical practice and research: a review. Part 1: How to measure glomerular filtration rate with iohexol? Clinical kidney journal 9 (5):682–699. doi: 10.1093/ckj/sfw070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsson B, Aulie A, Sveen K, Andrew E (1983) Human pharmacokinetics of iohexol. A new nonionic contrast medium. Investigative radiology 18 (2):177–182 [DOI] [PubMed] [Google Scholar]
- 18.Aakhus T, Sommerfelt SC, Stormorken H, Dahlstrom K (1980) Tolerance and excretion of iohexol after intravenous injection in healthy volunteers. Preliminary report. Acta radiologica Supplementum 362:131–134 [PubMed] [Google Scholar]
- 19.Kearns CM, Gianni L, Egorin MJ (1995) Paclitaxel pharmacokinetics and pharmacodynamics. Seminars in oncology 22 (3 Suppl 6):16–23 [PubMed] [Google Scholar]
- 20.Cresteil T, Monsarrat B, Alvinerie P, Treluyer JM, Vieira I, Wright M (1994) Taxol metabolism by human liver microsomes: identification of cytochrome P450 isozymes involved in its biotransformation. Cancer research 54 (2):386–392 [PubMed] [Google Scholar]
- 21.Rahman A, Korzekwa KR, Grogan J, Gonzalez FJ, Harris JW (1994) Selective biotransformation of taxol to 6 alpha-hydroxytaxol by human cytochrome P450 2C8. Cancer research 54 (21):5543–5546 [PubMed] [Google Scholar]
- 22.Beumer JH, Eiseman JL, Gilbert JA, Holleran JL, Yellow-Duke AE, Clausen DM, D'Argenio DZ, Ames MM, Hershberger PA, Parise RA, Bai L, Covey JM, Egorin MJ (2011) Plasma pharmacokinetics and oral bioavailability of the 3,4,5,6-tetrahydrouridine (THU) prodrug, triacetyl-THU (taTHU), in mice. Cancer chemotherapy and pharmacology 67 (2):421–430. doi: 10.1007/s00280-010-1337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaspari F, Perico N, Ruggenenti P, Mosconi L, Amuchastegui CS, Guerini E, Daina E, Remuzzi G (1995) Plasma clearance of nonradioactive iohexol as a measure of glomerular filtration rate. Journal of the American Society of Nephrology : JASN 6 (2):257–263 [DOI] [PubMed] [Google Scholar]
- 24.Kiesel BF, Parise RA, Guo J, Huryn DM, Johnston PA, Colombo R, Sen M, Grandis JR, Beumer JH, Eiseman JL (2016) Toxicity, pharmacokinetics and metabolism of a novel inhibitor of IL-6-induced STAT3 activation. Cancer chemotherapy and pharmacology 78 (6):1225–1235. doi: 10.1007/s00280-016-3181-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai PB, Duan JZ, Zhu YW, Kouzi S (1998) Human liver microsomal metabolism of paclitaxel and drug interactions. European journal of drug metabolism and pharmacokinetics 23 (3):417–424 [DOI] [PubMed] [Google Scholar]
- 26.Lee B, Ji HK, Lee T, Liu KH (2015) Simultaneous Screening of Activities of Five Cytochrome P450 and Four Uridine 5'-Diphospho-glucuronosyltransferase Enzymes in Human Liver Microsomes Using Cocktail Incubation and Liquid Chromatography-Tandem Mass Spectrometry. Drug metabolism and disposition: the biological fate of chemicals. doi: 10.1124/dmd.114.063016 [DOI] [PubMed] [Google Scholar]
- 27.Holleran JL, Parise RA, Guo J, Kiesel BF, Taylor SE, Ivy SP, Chu E, Beumer JH (2020) Quantitation of iohexol, a glomerular filtration marker, in human plasma by LC-MS/MS. Journal of pharmaceutical and biomedical analysis Accepted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christner SM, Parise RA, Ivy PS, Tawbi H, Chu E, Beumer JH (2019) Quantitation of paclitaxel, and its 6-alpha-OH and 3-para-OH metabolites in human plasma by LC-MS/MS. Journal of pharmaceutical and biomedical analysis 172:26–32. doi: 10.1016/j.jpba.2019.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoller R, Schmitz JC, Ding F, Puhalla S, Belani CP, Appleman L, Lin Y, Jiang Y, Almokadem S, Petro D, Holleran J, Kiesel BF, Ken Czambel R, Carneiro BA, Kontopodis E, Hershberger PA, Rachid M, Chen A, Chu E, Beumer JH (2017) Phase I study of veliparib in combination with gemcitabine. Cancer chemotherapy and pharmacology. doi: 10.1007/s00280-017-3409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillai VC, Strom SC, Caritis SN, Venkataramanan R (2013) A sensitive and specific CYP cocktail assay for the simultaneous assessment of human cytochrome P450 activities in primary cultures of human hepatocytes using LC-MS/MS. Journal of pharmaceutical and biomedical analysis 74:126–132. doi: 10.1016/j.jpba.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailer AJ (1988) Testing for the equality of area under the curves when using destructive measurement techniques. Journal of pharmacokinetics and biopharmaceutics 16 (3):303–309 [DOI] [PubMed] [Google Scholar]
- 32.Beumer JH, Franke NE, Tolboom R, Buckle T, Rosing H, Lopez-Lazaro L, Schellens JH, Beijnen JH, van Tellingen O (2010) Disposition and toxicity of trabectedin (ET-743) in wild-type and mdr1 gene (P-gp) knock-out mice. Investigational new drugs 28 (2): 145–155. doi: 10.1007/s10637-009-9234-8 [DOI] [PubMed] [Google Scholar]
- 33.Company GE (2015) Package Insert - OMNIPAQUE™ (iohexol) Injection.
- 34.Grimm SW, Einolf HJ, Hall SD, He K, Lim HK, Ling KH, Lu C, Nomeir AA, Seibert E, Skordos KW, Tonn GR, Van Horn R, Wang RW, Wong YN, Yang TJ, Obach RS (2009) The conduct of in vitro studies to address time-dependent inhibition of drug-metabolizing enzymes: a perspective of the pharmaceutical research and manufacturers of America. Drug metabolism and disposition: the biological fate of chemicals 37 (7): 1355–1370. doi: 10.1124/dmd.109.026716 [DOI] [PubMed] [Google Scholar]
- 35.Durbin PW, Jeung N, Kullgren B, Clemons GK (1992) Gross composition and plasma and extracellular water volumes of tissues of a reference mouse. Health physics 63 (4):427–442. doi: 10.1097/00004032-199210000-00007 [DOI] [PubMed] [Google Scholar]
- 36.Qi Z, Breyer MD (2009) Measurement of glomerular filtration rate in conscious mice. Methods in molecular biology 466:61–72. doi: 10.1007/978-1-59745-352-3_5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


