Abstract
Ras family proteins play an essential role in several cellular functions, including growth, differentiation, and survival. The mechanism of action of Ras mutants in Costello syndrome and cancers has been identified, but the contribution of Ras mutants to Noonan syndrome, a genetic disorder that prevents normal development in various parts of the body, is unknown. Son of Sevenless (SOS) is a Ras guanine nucleotide exchange factor. In response to Ras-activating cell signaling, SOS autoinhibition is released and is followed by accelerative allosteric feedback autoactivation. Here, using mutagenesis-based kinetic and pulldown analyses, we show that Noonan syndrome Ras mutants I24N, T50I, V152G, and D153V deregulate the autoactivation of SOS to populate their active form. This previously unknown process has been linked so far only to the development of Noonan syndrome. In contrast, other Noonan syndrome Ras mutants—V14I, T58I, and G60E—populate their active form by deregulation of the previously documented Ras GTPase activities. We propose a novel mechanism responsible for the deregulation of SOS autoactivation, where I24N, T50I, V152G, and D153V Ras mutants evade SOS autoinhibition. Consequently, they are capable of forming a complex with the SOS allosteric site, thus aberrantly promoting SOS autoactivation, resulting in the population of active Ras mutants in cells. The results of this study elucidate the molecular mechanism of the Ras mutant–mediated development of Noonan syndrome.
Keywords: Ras, SOS, Noonan syndrome, allostery, autoactivation, guanine nucleotide exchange factor (GEF), Ras protein, allosteric regulation, enzyme kinetics, catalysis, autoinhibition, Son of Sevenless
Ras, a small GTPase, plays key roles in many important cell-signaling events, such as cell proliferation, differentiation, apoptosis, and migration (1, 2). Ras isoforms include Kirsten Ras (KRas), neuroblastoma Ras (NRas), and Harvey Ras (HRas); the expression of each one often differs in organs and cell types (3–5). Ras cycles between the active GTP- and inactive GDP-bound form. Active and inactive Ras functions as an on-off switch of its downstream effector, Raf, which further controls the Ras mitogen-activated protein kinase (MAPK) signaling pathway (1, 2). Ras regulators, guanine nucleotide exchange factors (GEFs), and GTPase-activating proteins (GAPs) control the Ras activation cycle. The slow intrinsic Ras nucleotide exchange is enhanced by GEFs, whereas GAPs stimulate the slow intrinsic Ras GTPase activity that hydrolyzes the bound GTP to GDP and free phosphate (6).
Deregulation of the Ras MAPK signaling pathway causes various diseases, including RASopathy syndromes and cancers (7, 8). Noonan syndrome, like Costello syndrome, belongs to the RASopathy syndromes that directly or indirectly alter this pathway (7, 8). However, unlike Costello syndrome, Noonan syndrome is characterized by congenital heart defects (8, 9). Both syndromes characteristically result in short stature and developmental delays (10). A majority of Noonan syndrome cases have been linked to missense mutations in protein tyrosine phosphatase non-receptor type 11 (PTPN11) and Src homology region 2 domain–containing phosphatase-2 (SHP-2) (8, 11–13). Moreover, certain cases of Noonan syndrome reportedly are caused by mutations in KRas and NRas as well as by Raf and GEFs. All these are directly or indirectly involved in control of the Ras MAPK signaling pathway (13). Costello syndrome has been shown to arise solely from HRas mutations. This is unlike the causes linked to Noonan syndrome (11, 12, 14–18).
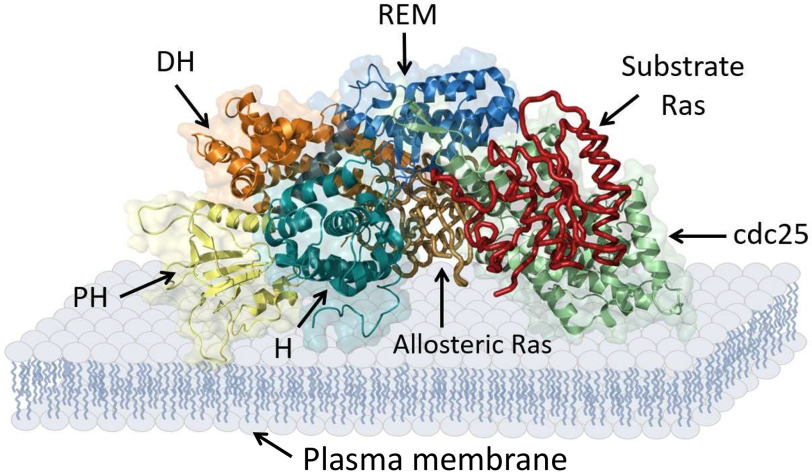
Of the many known Ras GEFs, Son of Sevenless (SOS) is of interest because several of its mutants are implicated in the development of Noonan syndrome (19–22). Although SOS shares the same catalytic domain of the cell division control protein 25 (Cdc25) as other Ras GEFs, some of its regulatory domains differ. Fig. 1 shows that the regulatory domains of SOS encompass the C-terminal proline-rich (PR) domain as well as the Ras-exchanger motif (REM), pleckstrin homology (PH), Dbl-homology (DH), and histone (H) domains (23).
Figure 1.
SOS domains. Multimeric SOS domains with allosteric and substrate Ras-binding sites are shown. The scheme was generated using PyMOL with PDB entry 3KSY.
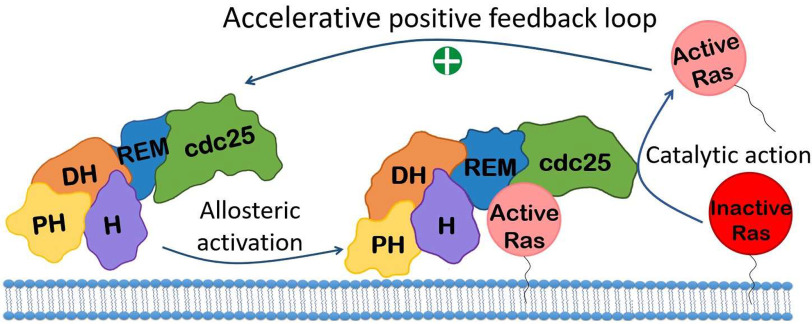
The REM domain serves as a homotropic allosteric site that binds an active Ras (Fig. 1), which in turn activates the SOS catalytic action (23). The SOS allosteric REM domain functions to cooperate with the SOS Cdc25 domain catalytic action, endowing SOS with a positive feedback loop activation (Fig. 2) (24, 25). A recent study further showed that the positive feedback loop activation of SOS is accelerative; hence, it is termed SOS autoactivation (Fig. 2) (26). This allosteric SOS autoactivation is autoinhibited by the action of the DH and PH domains in combination with the H domain of SOS (24–26). Together, these SOS autoinhibition regulatory domains then function to block active Ras from accessing the SOS REM allosteric site (26).
Figure 2.
Model mechanism of SOS autoactivation. The allosteric SOS activation followed by the SOS catalytic action is illustrated. The action of the accelerative positive feedback loop between the SOS allostery and catalysis results in SOS autoactivation.
Although the detailed molecular mechanism remains unknown, we now know that the interaction of SOS with a plasma membrane in the presence of phosphatidylinositol 4,5-bisphosphate (PIP2) results in the release of SOS autoinhibition; this release then allows an active Ras to bind to the SOS allosteric site to autocatalytically activate SOS (26).
Several SOS mutants that deregulate the SOS allosteric function to populate active Ras have been reported (19, 20, 23, 27–29). These mutants include E108K, M269R, R552G, W729L, E846K, and R1131K. All have been shown to relate to the development of Noonan syndrome. Their common mechanistic feature in causing its development is likely to enhance the binding interaction of SOS with the plasma membrane that allosterically activates SOS and thus populates active Ras in cells (19, 20, 23, 27–29).
Ras GAPs include GAP1 and neurofibromatosis type 1 (NF1), among others (2, 6). Deregulation of their catalytic action by Ras mutations attenuates the GTPase activity of Ras, populating active Ras in cells (30). One of the well-characterized Ras activation mechanisms explained by the deregulation of GAP action is that Ras mutations at the site of the p-loop and Switch II residues of Ras—such as Gly-12 and Ala-59—deter the GAP-mediated Ras GTPase action. The perturbation of the GAP action on Ras conserves the active GTP-bound form of Ras and thus populates the active Ras in cells (14). This Ras mutation–driven deregulation of the GAP action applies to nearly all cases of known diseases, such as Costello syndrome and cancers. Intriguingly, contrary to deregulation of the function of Ras GAPs, all the currently known deregulators of the function of SOS attributed to GEFs are because of the mutations in SOS per se, but not those in Ras (see above).
We have established a method that estimates the cellular population of active Ras. Our method is based upon the comprehensive Ras kinetic features that encompass Ras activation and inactivation, with and without the action of Ras regulators GEF and GAP (14, 31). The GEF action defines the core kinetic process of the Ras nucleotide exchange by the catalytic Cdc25 domain, but not the GEF activity regulation. The GAP action also is defined only according to its catalytic function. This approach with the comprehensive Ras kinetic features clarifies that, in addition to the Ras activity regulated by GEF and GAP, the intrinsic Ras activity regulation without GEF and GAP also contributes significantly to establishment of the cellular population of active Ras (14, 31). One use of the comprehensive Ras kinetic features is to determine the kinetic step of the activation or inactivation of Ras mutants with and without GEF and GAP that results in alteration of the overall population of active Ras in cells.
Equipped with knowledge of the comprehensive Ras kinetic features, we have compared and characterized the comprehensive kinetic features of most of the 13 known HRas mutants and of certain oncogenic Ras mutants that include embryonic Ras (ERas) (14, 31). Accordingly, although the degree of their intensity of perturbation differs, the Costello syndrome and oncogenic Ras mutants are all constitutively activated by the perturbation of both the intrinsic and GAP-mediated Ras GTPase activities. Nevertheless, in no single case does a Ras mutant cause deregulation of the core GEF catalytic action on Ras to populate active Ras. Despite this knowledge, unlike with Costello syndrome HRas mutants and ERas and other oncogenic Ras mutants (14, 31), no determination has been made of the role of Ras mutants in the comprehensive kinetic features of the development of Noonan syndrome. These previously uncharacterized Ras mutants include KRas mutants of V14I, T58I, V152G, and D153V Ras as well as NRas mutants of I24N, T50I, and G60E Ras (11, 15–18).
Within the course of this study, we have investigated the comprehensive kinetic features of the previously uncharacterized Noonan syndrome-relevant KRas and NRas mutants (15–18). We have learned that not all these Noonan syndrome Ras mutants alter the comprehensive kinetic features of Ras. The V14I and T58I KRas and G60E NRas are the only ones that do alter the comprehensive Ras kinetic features of the intrinsic and GAP-mediated Ras GTPase activities. Although their mutation formulas are unique, these Ras mutations occur at the vicinal site of the p-loop Gly-12 or the Switch II Ala-59 residue of Ras. The perturbations of the GAP function at the Gly-12 or Ala-59 residue of Ras or vicinal to it are not unusual in serving to alter the comprehensive Ras kinetic features to populate the active Ras (14).
Although other Noonan syndrome Ras mutants, I24N and T50I NRas as well as V152G and D153V KRas, do not alter the comprehensive kinetic features of Ras, these unusual Ras mutants have been found instead to perturb the SOS autoinhibition mechanism; this perturbation causes uncontrolled SOS autoactivation to populate active Ras. This is the first time that the features of their unusual deregulation of SOS activation have been reported. Our structure-based kinetic analysis also proposes molecular mechanisms to explain their role in this deregulation of SOS autoinhibition. Accordingly, our combined findings of the deregulation of the comprehensive Ras kinetic features and of SOS autoactivation by these Ras mutants explain the previously unknown cellular mechanism of this KRas and NRas mutant–mediated development of Noonan syndrome.
Results
The kinetic parameters used to evaluate the comprehensive Ras kinetic features include the intrinsic as well as the core catalytic actions of GEF and GAP on Ras (14, 31). Ras mutants with any alteration of their kinetic parameters associated with the intrinsic and GAP-mediated catalytic functions of Ras were termed “GTPase-affecting Ras mutants.” In contrast, Ras mutants that possess altered kinetic parameters of the GEF-mediated nucleotide exchange of Ras were labeled for convenience as “GEF-affecting Ras mutants.”
To the best of our knowledge, all the previously characterized Ras mutants, such as Costello syndrome and oncogenic Ras, are GTPase-affecting Ras mutants. This is because the constitutive activation mechanisms of these Ras mutants with respect to their comprehensive Ras kinetic features have altered at least one of their GTPase-affecting Ras kinetic parameters (14, 31). To determine whether the previously reported but uncharacterized Noonan syndrome Ras mutants also are the GTPase-affecting Ras mutants, we determined the key GTPase-affecting Ras kinetic parameters of these uncharacterized Ras mutants that elicit Noonan syndrome. These uncharacterized Ras mutants are I24N (18), V14I (11), T50I (17), T58I (11), G60E (17), V152G (15), and D153V (11, 15).
GTPase-affecting kinetic features of Ras mutants associated with Noonan syndrome
Of these Noonan syndrome Ras mutants, V14I, T58I, and G60E Ras possess kinetic features similar, to some extent, to those of the G12V and A59T Ras mutants (see Tables S1 and S2). These results suggest that the constitutive activations of the Noonan syndrome V14I, T58I, and G60E Ras mutants in cells (11, 15, 17, 18) are due to alterations of their GTPase-affecting kinetic parameters that are associated with their intrinsic and GAP-mediated GTPase actions. However, activation of these mutants does not occur because of the perturbation of the core GEF catalytic action on Ras. Thus, these Noonan syndrome Ras mutants, V14I, T58I, and G60E Ras, belong to the GTPase-affecting Ras mutants. In light of this relationship, their constitutive activation mechanism resembles that of the previously studied Costello syndrome and oncogenic Ras mutants.
GEF-affecting kinetic features of Ras mutants associated with Noonan syndrome
None of the GTPase-affecting kinetic features of I24N, T50I, V152G, and D153V mutants deviates significantly from the features of WT Ras (see Tables S1 and S2). Consequently, in contrast to the established GTPase-affecting Noonan syndrome Ras mutants (i.e. V14I, T58I, and G60E Ras; see above), those of the unusual Noonan syndrome—I24N, T50I, V152G, and D153V Ras—are at present termed uncharacteristic Ras mutants. Note that our research results shown below revealed that these uncharacteristic Ras mutants are actually GEF-affecting Ras mutants. This is because they all deregulate the function of SOS, which is one of the GEFs. Nevertheless, it is puzzling how, in light of their differences, these uncharacteristic Ras mutations are linked to the development of Noonan syndrome.
The currently inexplicable kinetic features of these uncharacteristic Ras mutants suggest the existence of a previously unrecognized deregulation mechanism(s) specific to them for Ras activation. One of the unexplored potential kinetic mechanism(s) for this unusual activation is likely the Ras mutant-mediated perturbation of the regulatory function of SOS. This possibility is because Ras itself also functions as a homotropic allosteric regulator of SOS (23). Accordingly, Ras mutants may have the capability to perturb the SOS allosteric function that results in an unusual activation of these mutants in cells. With this possibility in mind, we examined the GEF-affecting kinetic features of SOS autoactivation with the uncharacteristic Ras mutants under various experimental conditions.
Unique kinetic mechanism of SOS activation and its potential relevance to the activation of uncharacteristic Ras mutants
Several kinetic features of the allosteric autoactivation of SOS differ from conventional enzyme catalysis. All the GTPase-affecting kinetic features of Ras belong to the conventional enzyme catalysis. The SOS autoactivation–specific term of the rate and its relevant kinetic parameter (kauto) in SOS autoactivation should be differentiated from the rate and from its relevant rate constant (k) in conventional enzyme catalysis. This is because the rate of SOS autoactivation reflects the speed of SOS activation through the action of the accelerative positive feedback loop between active and inactive SOS (26). In contrast, a reference to a rate in enzyme action describes the speed of enzymatic catalysis (32).
Another key SOS autoactivation-specific kinetic feature is the requirement of a reaction initiator (or trigger) to initiate SOS autoactivation (26). Conventional enzyme catalysis does not require a trigger. The amount of the reaction initiator necessary for SOS autoactivation is defined as the threshold concentration of active Ras (26). This specifies the minimum quantity of active Ras necessary on the SOS allosteric sites to ensure an active population of SOS sufficient to initiate the cycle of SOS autoactivation (26, 33, 34). Besides, the completion of SOS autoactivation results in the production of a steady-state fraction of active SOS. This steady-state active SOS fraction is uniquely upheld yet again by the unique kinetic features of SOS autoactivation. This is because, unlike other conventional kinetic processes, SOS autoactivation continues to operate unless otherwise terminated (26).
Last, autoinhibition of SOS (23) is a factor that affects these three unique SOS autoactivation-specific kinetic features. This is because the autoinhibitory domains of SOS limit active Ras access to the empty SOS allosteric site; this limited access impedes the allosteric autoactivation of SOS (26). Thus, when SOS interacts with the plasma membrane in the presence of PIP2, SOS autoinhibition is released and autoactivation is free to proceed (26).
Deregulation of kinetics of SOS autoactivation with uncharacteristic Ras mutants
Uncharacteristic Ras mutants in cytoplasm and on the plasma membrane with and without PIP2 may play a role in the potential kinetic perturbations or alterations of the combinational action of SOS autoactivation with SOS autoinhibition. However, examination of such a role would have required an SOS construct (SOSmemb-cat) capable of enabling the allosteric SOS autoinhibition in combination with the use of (i) Ras in solution and (ii) Ras tethered to the lipid vesicle and to the PIP2-containing lipid vesicle. Ras in solution mimics the cytosolic Ras. Ras tethered to the lipid vesicle represents Ras on the plasma membrane without PIP2. Ras tethered to the PIP2-containing lipid vesicle mimics Ras on the plasma membrane with PIP2 (see details under “Experimental procedures”).
When necessary, as controls, instead of using the uncharacteristic Ras mutants, we used WT and GTPase-affecting Ras mutants in solution and tethered to the lipid vesicle with and without PIP2. Note that only Ras tethered to the PIP2-containing lipid vesicle generates the SOS autoinhibition–free condition. This contrasts with the capability of Ras in solution and tethered to the lipid vesicle (that lacks PIP2) to create SOS autoinhibition in both conditions.
SOScat that lacks the SOS autoinhibitory function also was used as a control in two instances. The first was to measure the maximal fractional activation of uncharacteristic Ras by SOSmemb-cat. The second was to determine the total binding occupancy of the SOSmemb-cat allosteric site with uncharacteristic Ras mutants under the SOS autoinhibition and autoinhibition-free conditions (see details under “Experimental procedures”).
Unprecedented SOS autoactivation with uncharacteristic Ras mutants that define GEF-affecting Ras mutants
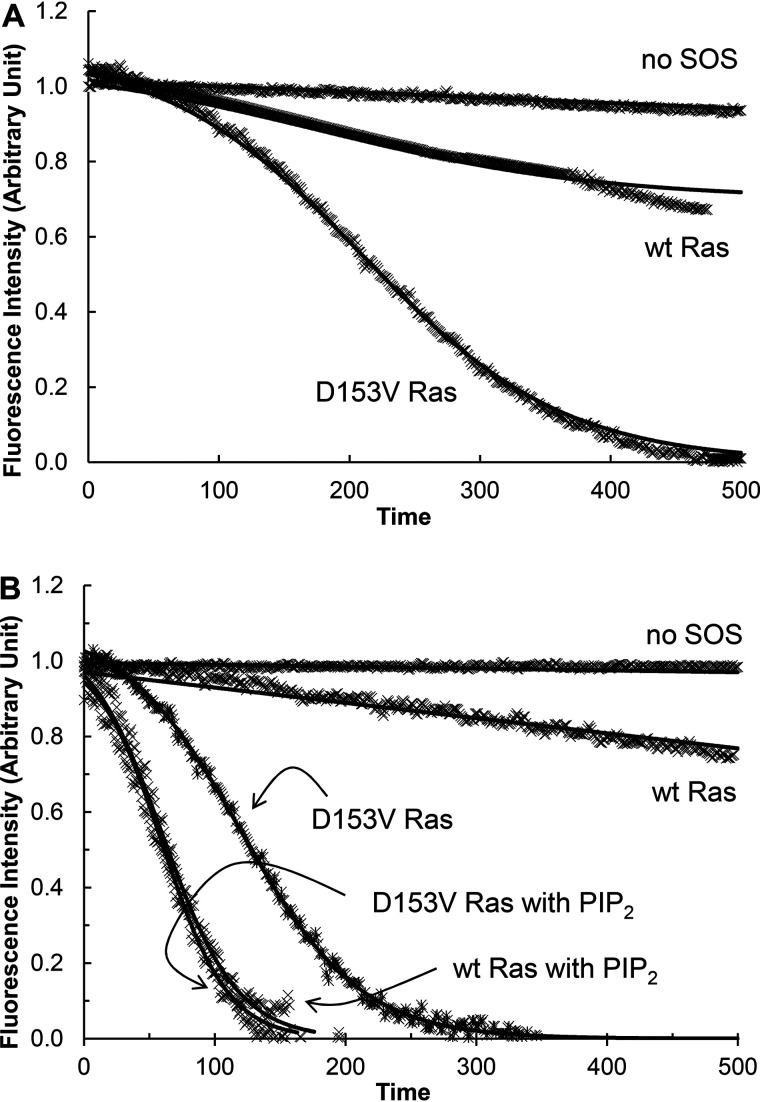
Fig. 3 (A and B) shows that SOSmemb-cat was capable of activating the uncharacteristic D153V Ras mutant under the SOS autoinhibition conditions. To produce the SOS autoinhibition conditions, D153V Ras was in solution and tethered to the lipid vesicle. Other uncharacteristic Ras mutants—V152G, I24N, and T50I Ras—also were activated by SOSmemb-cat under conditions of SOS autoinhibition (not shown). These uncharacteristic Ras-specific kinetic features under the SOS autoinhibition conditions are novel. This is because WT Ras and GTPase-affecting Ras mutants were capable of activating SOSmemb-cat only under the SOS autoinhibition-free conditions (Fig. 3B).
Figure 3.
Autoactivation of SOS with WT and D153V mutant Ras. Fluorescence mant-based SOS autoactivation assays were performed with WT and D153V Ras in solution (A) and tethered to the lipid vesicle (B). Given that a reaction initiator is necessary to start SOS autoactivation, in all assays unless otherwise unnecessary, a GTP analog GppNHp-bound active Ras in solution or tethered to the lipid vesicle (30% of total Ras mole fraction) was added before the assay was initiated. Initiation was by the addition of SOSmemb-cat (1 μm) to an assay cuvette that contained Ras·mdGDP (1 μm) in solution or tethered to the lipid vesicle in the presence of excess GppNHp (1 mm) in an assay buffer. Once the assay was initiated, we monitored the change in the intensity of mant fluorescence over time as described under “Experimental procedures.” When necessary, Ras tethered to the PIP2-containing lipid vesicle, instead of Ras tethered to the lipid vesicle, was also used for the assay (B). The SOS autoactivation data fit was performed as described in the previous study (26) that gave the kauto values of SOSmemb-cat with WT and D153V Ras in solution as >1500 and 219 s, respectively. The kauto values of SOSmemb-cat with WT Ras tethered to the lipid vesicle (PIP2 lacking) and the PIP2-containing lipid vesicle were determined to be >1500 and 129 s, respectively. The kauto values of SOSmemb-cat with D153V Ras tethered to the lipid vesicle (that lacks PIP2) and the PIP2-containing lipid vesicle were determined to be 484 and 133 s, respectively. For all analyses, triplicate experiments were performed, and the graphic figures close to the average values are shown.
Moreover, all these observed activations of SOSmemb-cat by uncharacteristic Ras mutants were hysteretic as a function of time; this indicates these uncharacteristic Ras mutants were capable of initiating autoactivation of SOSmemb-cat over time without regard for its autoinhibition status. In accounting for the notion that the initiation of autoactivation of SOS (26) requires release of SOS autoinhibition, these results suggest that the Ras mutants had some unique capability to evade SOS autoinhibition without action by the membrane with PIP2.
Because these uncharacteristic Ras mutants essentially deregulate the SOS function, they are classified as GEF-affecting Ras mutants. Accordingly, within this article, the uncharacteristic Ras mutants are essentially the same as GEF-affecting Ras mutants.
Unusual kinetic features of SOS autoactivation with GEF-affecting Ras mutants
Table 1 shows that the kauto values of SOSmemb-cat with these GEF-affecting Ras mutants in solution were less than those corresponding values with Ras tethered to the lipid vesicle. These values, in turn, were less than those corresponding values with Ras tethered to the PIP2-containing lipid vesicle. Nevertheless, the kauto values of SOSmemb-cat autoactivation with the GEF-affecting Ras mutants tethered to the PIP2-containing lipid vesicle did not differ significantly from the WT Ras and GTPase-affecting Ras mutants tethered to the PIP2-containing lipid vesicle (Table 1). These results together suggest that nevertheless, under SOS autoinhibition-free conditions, its autoactivation processes were, to some extent, deterred compared with those processes of GEF-affecting Ras mutants as well as those of WT Ras and GTPase-affecting Ras mutants. These results also suggest that, under the SOS autoinhibition-free conditions, no significant differences occur in the process of SOS autoactivation by the GEF-affecting Ras mutants or SOS autoactivation by the WT Ras and GTPase-affecting Ras mutants.
Table 1.
Comparative SOS autoactivation-specific kinetic parameters of WT Ras and various Ras mutants
The results shown were determined as described under “Experimental procedures.” Ras-solution, Ras-lipid vesicle, and Ras-lipid vesicle-PIP2 denote, respectively, Ras in solution, tethered to the lipid vesicle, and tethered to the PIP2-containing lipid vesicle. S.D. values for all the results shown were less than 15% of the values shown.
|
Kauto value (s) |
Threshold concentration (mol % active Ras)a |
Fraction size of active Ras (%)b |
Fraction occupancy of SOS with Ras (%)c |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ras-Solution | Ras-Lipid Vesicle | Ras-Lipid vesicle-PIP2 | Ras-Solution | Ras-Lipid Vesicle | Ras-Lipid vesicle-PIP2 | Ras-Solution | Ras-Lipid Vesicle | Ras-Lipid vesicle-PIP2 | Ras-Solution | Ras-Lipid Vesicle | Ras-Lipid vesicle-PIP2 | |
| HRas | ||||||||||||
| WT | >1500 | >1500 | 129d | 1.5e | 91 | 28f | 22f | 93f | ||||
| G12V | >1500 | >1500 | 140 | 1.4 | 89 | 28 | 19 | 91 | ||||
| A59T | >1500 | >1500 | 127 | 1.7 | 91 | 29 | 18 | 89 | ||||
| KRas | ||||||||||||
| V14I | >1500 | >1500 | 134 | 1.7 | 91 | 29 | 21 | 92 | ||||
| T58I | >1500 | >1500 | 142 | 1.7 | 93 | 25 | 15 | 92 | ||||
| V152G | 508 | 259 | 131 | 26 | 8 | 1.6 | 33 | 21 | 91 | 27 | 19 | 91 |
| D153V | 484d | 219d | 133d | 25 | 7 | 1.5 | 28 | 17 | 91 | 29f | 17f | 88f |
| NRas | ||||||||||||
| I24N | 501 | 238 | 136 | 26 | 9 | 1.5 | 32 | 21 | 90 | 31 | 19 | 91 |
| T50I | 498 | 340 | 133 | 28 | 16 | 1.4 | 35 | 23 | 90 | 29 | 18 | 90 |
| G60E | >1500 | >1500 | 142 | 1.5 | 92 | 26 | 16 | 88 | ||||
aThe SOS autoactivation threshold concentration of Ras depends on the mole percent (mol %) of the active Ras in all Ras in the assay solution (26), so it is expressed as it is.
bAll fraction sizes of active Ras by SOSmemb-cat were normalized against the value of the same active Ras by SOScat that were set at 100%.
cY64A and its derivative Ras mutant forms, such as Y64A/D153V GEF–affecting Ras, were used instead of WT Ras and the original mutants to specifically determine the fraction occupancies of the SOS allosteric site with Ras. The fraction occupancies of SOSmemb-cat with Ras were expressed as normalized values against the values of fraction occupancies of SOScat with WT Ras tethered to the lipid vesicle, which was set at 100%.
dTaken from Fig. 3.
eTaken from a previous study (26).
fTaken from Fig. 4.
It also is noteworthy that the kauto values of SOSmemb-cat with these solution forms of the GEF-affecting Ras mutants in solution are almost indistinguishable from one another (Table 1). However, this similarity was not always conserved when these Ras mutants were tethered to the lipid vesicle. Specifically, the kauto value of SOSmemb-cat with T50I Ras tethered to the lipid vesicle was definitely less than that of all other GEF-affecting Ras mutants tethered to the lipid vesicle. This similarity and difference do not necessarily indicate similarities or differences in their activation mechanisms. Nevertheless, the unique kauto value of SOSmemb-cat with T50I Ras tethered to the lipid vesicle suggests a potential difference in the deregulation mechanism of SOS action by T50I Ras on the plasma membrane compared with that of other GEF-affecting Ras mutants on the plasma membrane.
Distinct threshold concentrations of SOS autoactivation by GEF-affecting Ras mutants
The overall SOSmemb-cat autoactivation initiation threshold concentrations of the GEF-affecting Ras mutants in solution exceeded those of the GEF-affecting Ras mutants tethered to the lipid vesicle (Table 1). In turn, these latter concentrations exceeded those of the GEF-affecting Ras mutants tethered to the PIP2-containing lipid vesicle. However, no significant difference occurred in the SOSmemb-cat autoactivation threshold concentrations of the GEF-affecting Ras mutants tethered to the PIP2-containing lipid vesicle compared with those of the WT Ras and GTPase-affecting Ras mutants tethered to the PIP2-containing lipid vesicle (Table 1). The results suggest that the initiation of SOS autoactivation by these GEF-affecting Ras mutants under autoinhibition conditions is less efficient than initiation of autoactivation by WT Ras and GTPase-affecting Ras mutants under SOS autoinhibition–free conditions. Also, when SOS autoinhibition is released, regardless of the features of Ras mutations, the GEF-affecting and GTPase-affecting Ras mutants as well as WT Ras are all equally efficient in initiation of SOS autoactivation. Note there was no occurrence of SOSmemb-cat autoactivation with WT Ras and GTPase-affecting Ras mutants in solution and tethered to the lipid vesicle. Thus, the SOSmemb-cat autoactivation threshold values of GEF-affecting Ras mutants under SOS autoinhibition conditions were not comparable with those of WT Ras and GTPase-affecting Ras mutants under SOS autoinhibition conditions.
Table 1 shows that the threshold concentrations of the GEF-affecting Ras mutants in solution for the initiation of SOSmemb-cat autoactivation were so similar that they were indistinguishable from one another. However, when GEF-affecting Ras mutants were tethered to the lipid vesicle, the SOSmemb-cat autoactivation threshold concentration of T50I Ras tethered to the lipid vesicle became noticeably higher than that of all other GEF-affecting Ras mutants tethered to the lipid vesicle. This result suggests that the deregulation mechanism of the initiation of the SOS autoactivation by T50I Ras on the plasma membrane in the absence of PIP2 is definitely inefficient compared with that of other GEF-affecting Ras mutants under the same conditions.
Deregulation of steady-state of SOS autoactivation with GEF-affecting Ras mutants
All the above SOS autoactivation analyses are relevant to the time-dependent dynamic feature states of the active GEF-affecting Ras mutants. This is because the process of SOS autoactivation essentially links with the time-dependent production of the active Ras. However, the steady-state features of active Ras are often more relevant to Ras-relevant diseases because they reflect the constitutively activated fraction of Ras in cells. To examine the steady-state content of GEF-affecting Ras mutants by autoactivated SOSmemb-cat, we examined the resultant fractions of active GEF-affecting Ras mutants after the completion of SOSmemb-cat autoactivation with GEF-affecting Ras mutants.
Table 1 shows that the overall fraction sizes of the active GEF-affecting Ras mutants in solution were smaller than those of the active GEF-affecting Ras mutants tethered to the lipid vesicle. In turn, these tethered mutants were also smaller than those of the active GEF-affecting Ras mutants tethered to the PIP2-containing lipid vesicle. However, the fraction sizes of the active GEF-affecting Ras mutants tethered to the PIP2-containing lipid vesicle were similar to the fraction sizes of WT Ras and GTPase-affecting Ras mutants tethered to the PIP2-containing lipid vesicle. When the resultant SOSmemb-cat activity after SOSmemb-cat autoactivation was taken into account, the theoretical fractional values of the active GEF-affecting Ras mutants that were derived by using the previously established formula (14) approximated those of the titration kinetic results (not shown).
These similarities and differences in the fraction sizes of these Ras proteins in solution and tethered to the lipid vesicle were maintained during the experimental time period. This is likely because SOS autoactivation maintains the fraction of the active SOS until termination (26). This maintenance of the active fraction of Ras is the unique feature of SOS autoactivation. The results suggest that, although these GEF-affecting Ras mutants are capable of populating active SOS and thus active Ras under the autoinhibition conditions, their overall steady-state active fractions were less than those occurring under autoinhibition-free conditions.
Table 1 also shows the similarity of the fraction sizes of the active GEF-affecting Ras mutants in solution. However, not all the fraction sizes of the active GEF-affecting Ras mutants tethered to the lipid vesicle were the same. These variations were because the fraction size of the active T50I Ras tethered to the lipid vesicle was the smallest among the nearly uniform fraction sizes of all other active GEF-affecting Ras mutants tethered to the lipid vesicle. This suggests that the overall active fractional population of T50I Ras on a plasma membrane is less than that of other GEF-affecting Ras mutants. The fractional population of active Ras on a plasma membrane is the resultant combinational action of the initiation and SOS autoactivation process. Consequently, this lowest active T50I Ras fraction likely links with the slowest rate of SOS autoactivation by T50I Ras. It also likely links with the highest threshold concentration of T50I Ras for SOS autoactivation when compared with the rates and thresholds of all other GEF-affecting Ras mutants on a plasma membrane.
SOS allostery and its link to unusual autoactivation of SOS with the GEF-affecting Ras mutants
SOS autoactivation is an echo of accelerative feedback homotropic allosteric SOS activation (26). Therefore, the unusual features of SOSmemb-cat autoactivation with these GEF-affecting Ras mutants in solution and tethered to lipid vesicles likely indicate that these GEF-affecting Ras mutants are somehow capable of accessing the SOSmemb-cat allosteric site even under SOS autoinhibition conditions. To examine this possibility, the SOS allosteric site–specific binding features of these GEF-affecting Ras mutants were examined by titrations of SOSmemb-cat with Y64A and Y64A GTPase–affecting Ras mutants (i.e. Y64A/D153V Ras). This was because Y64A and its derivative Ras mutants bind only to the SOS allosteric site (see “Experimental procedures”).
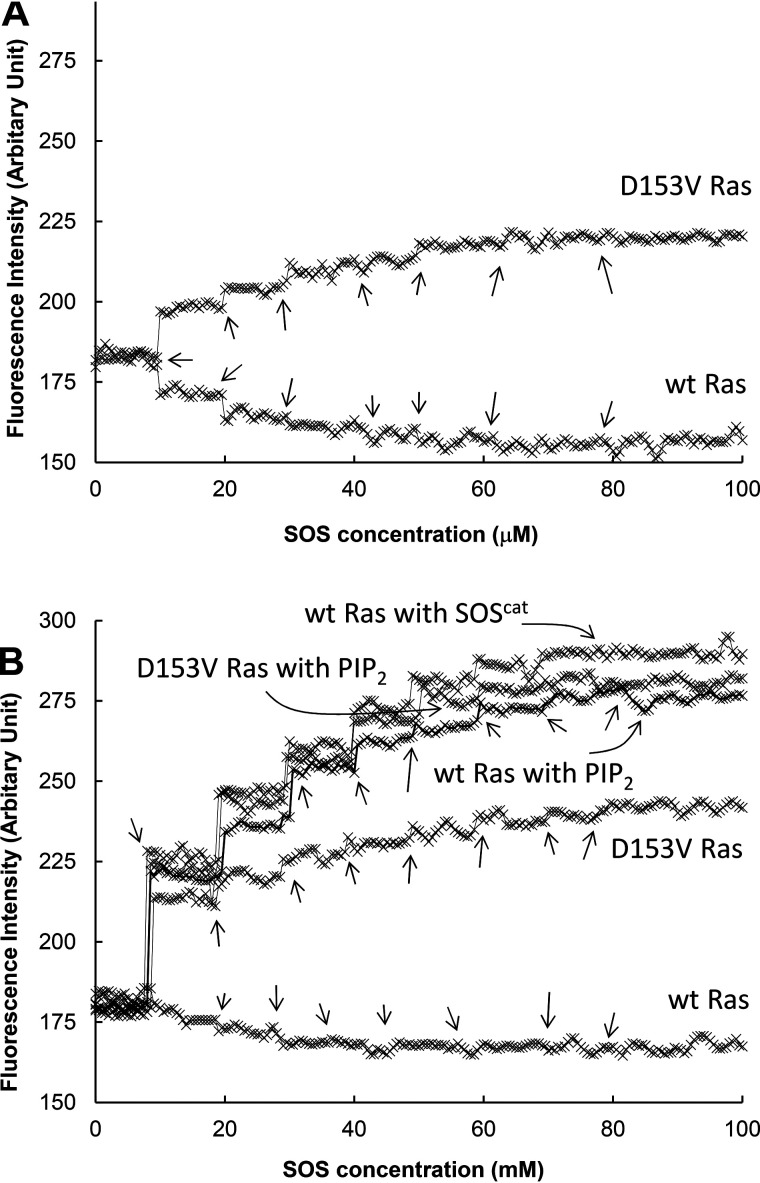
Fig. 4 and Table 1 show that the maximal fractions of the SOSmemb-cat–bound Y64A Ras in solution and when tethered to the lipid vesicle were ∼30 and 20%, respectively. In contrast, the fraction of the SOSmemb-cat–bound Y64A Ras tethered to the PIP2-containing lipid vesicle was ∼90%. Although found in small quantities, the fraction sizes of the SOSmemb-cat–bound Y64A Ras in solution and when tethered to the lipid vesicle were nevertheless unexpected. This is because, under SOS autoinhibition conditions, the SOS allosteric site is known to be inaccessible for the binding of Y64A Ras.
Figure 4.
Fraction occupancies of SOS with WT Ras and various Ras mutants under the SOS autoinhibition conditions. SOSmemb-cat was titrated with Y64A and Y64A Ras (i.e. Y64A/D153V Ras) as described under “Experimental procedures.” For this titration, SOSmemb-cat was the titrant, whereas Ras·mdGDP in solution (A) as well as Ras·mdGDP tethered to the lipid vesicle and tethered to the PIP2-containing lipid vesicle (B) were receptors. The titration was performed by adding the titrant, as indicated with arrows, to the receptor repeatedly until the increase in mant fluorescence ended. As a control, SOScat was also used instead of SOSmemb-cat for the titration with Y64A Ras tethered to the lipid vesicle. The fluorescence-based titration results of these Ras proteins with SOSmemb-cat were normalized against the fluorescence-based titration results of Y64A Ras tethered to the lipid vesicle with SOScat. The normalized fluorescent intensities were plotted against the titrant concentrations of SOS and then fitted to saturation kinetics that gave the maximal fraction occupancy (Bmax) of the SOSmemb-cat allosteric site with Y64A and Y64A/D153V Ras in solution as 28 and 29%, respectively. The Bmax values of the SOSmemb-cat allosteric site with Y64A and Y64A/D153V Ras tethered to the lipid vesicle were determined to be 22 and 17%, respectively. The Bmax values of the SOSmemb-cat allosteric site with Y64A and Y64A/D153V Ras tethered to the lipid vesicle were determined to be 93 and 88%, respectively. All these reported values were averages of the three independent measurements, and the S.D. values are less than 15% of the value reported.
Intriguingly, however, we failed to displace the SOSmemb-cat–bound Y64A Ras in solution or tethered to the lipid vesicle with fresh Y64A Ras in solution. The same was true when it was tethered to the lipid vesicle. The SOSmemb-cat–bound Y64A Ras in solution or when tethered to the lipid vesicle, however, were both successfully displaced with fresh Y64A Ras tethered to the PIP2-containing lipid vesicle. Moreover, we could displace the SOS-bound Y64A Ras tethered to the PIP2-containing lipid vesicle with any form of the fresh Y64A Ras, such as Y64A Ras in solution, whether tethered to the lipid vesicle or tethered to the PIP2-containing lipid vesicle. A common denominator in allowing the displacement of the SOSmemb-cat–bound Y64A Ras with fresh Y64A Ras was the presence of PIP2 in the lipid vesicle, which is known as the key factor that releases SOS autoinhibition.
In taking into account this common denominator as well as Y64A Ras as a version of the SOS allosteric-specific WT Ras, we interpreted these results as follows. (i) The SOS autoinhibition feature does not completely block the access of Y64A Ras to the SOS allosteric site. Although it occurs in a lesser quantity than WT Ras under SOS autoinhibition-free conditions, WT Ras is still capable of binding to the SOS allosteric site under autoinhibition conditions. (ii) The SOS-bound WT Ras under autoinhibition conditions is detained or stuck at the SOS allosteric site unless the SOS autoinhibition is otherwise released by the action of the plasma membrane with PIP2. Discovery of this detained WT Ras SOS autoinhibition does not eliminate the role of the original SOS autoinhibition (that lacks Ras in the SOS allosteric site) in regulation of the SOS activity. This is because a remaining large fraction of WT Ras is unable to bind to the SOS allosteric site because of the feature state of the SOS autoinhibition. As a result, unless SOS autoinhibition is otherwise released, the major fraction of SOS cannot be activated. Note that nearly identical results were observed when GTPase-affecting Y64A Ras mutants were used instead of Y64A Ras (not shown). Thus, the notion of detained WT Ras SOS autoinhibition is applicable to SOS with the GTPase-affecting Ras mutants.
Fig. 4 shows that, as in the case of Y64A Ras, Y64A/D153V Ras—one of the GEF-affecting Y64A Ras mutants—was also capable of binding to the SOSmemb-cat allosteric site when it was either in solution or tethered to the lipid vesicle. The fraction sizes of the SOSmemb-cat–bound Y64A/D153V Ras in solution or tethered to the lipid vesicle resembled those of Y64A Ras in solution or tethered to the lipid vesicle, respectively. However, unlike in the case of the SOSmemb-cat–bound Y64A Ras in solution or tethered to the lipid vesicle, the SOS-bound Y64A/D153V Ras in solution or tethered to the lipid vesicle could be displaced with fresh Y64A/D153V Ras in solution or tethered to the lipid vesicle without the presence of PIP2. The same treatment was applicable with all other GEF-affecting Y64A Ras mutants. The results indicate that, unlike in the case of WT Ras, these GEF-affecting Ras mutants at the SOS allosteric site were not detained but separable under the SOS autoinhibition conditions. This explains the observed SOS autoactivation under SOS autoinhibition conditions in which the dissociation of the SOS-bound GEF-affecting Ras under the autoinhibition conditions allows the binding of a fresh active GEF-affecting Ras to the SOS allosteric site, thus enabling autoactivation of SOS.
Unlocking the detained Ras-mediated SOS autoinhibition by the GEF-affecting Ras mutants
The notion of the occupancy of the SOS allosteric site by the detained WT Ras as well as the GEF-affecting Ras mutants under the SOS autoinhibition conditions is unprecedented. Similarly unprecedented is the notion that the release of the GEF-affecting Ras mutants from the SOS allosteric site also occurs under the SOS autoinhibition conditions. To investigate whether these notions are applicable to the SOS in cells, we examined the feature statuses of the SOS allosteric site occupancy by WT and GEF-affecting Ras in the unstimulated cells that retain the SOS autoinhibition. Notably, any potential unwanted priming of SOS activations in cells (24, 25) should also be restricted within this examination. Accordingly, HEK293T cells were a choice for this examination because they do not express the RasGRPs that produce active WT Ras, which serves as a primer to initiate SOS activation. (24).
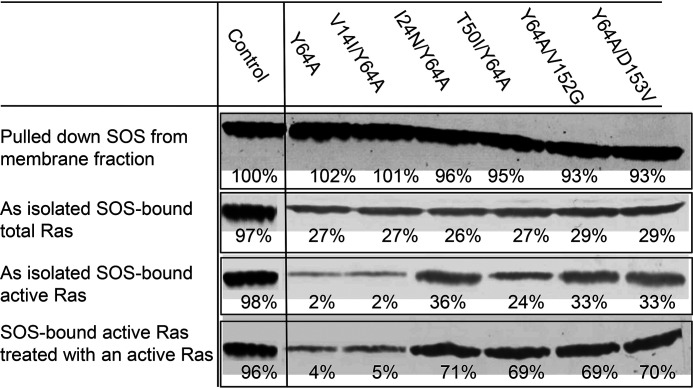
Fig. 5 shows that, under autoinhibition conditions, various forms of Y64A Ras expressed in HEK293T cells were shown to bind to the membrane-bound SOS in HEK293T cells. However, the Y64A Ras fractions bound to SOS were at least less than 30% of the control of the lipid vesicle-tethered active Y64A Ras bound to SOSmemb-cat. These SOS-bound Y64A Ras proteins include WT Ras, the GTPase-affecting V14I mutant, and all the GEF-affecting Ras mutants (Fig. 5). A similar fraction of other Noonan syndrome–relevant GTPase-affecting Y64A Ras proteins (i.e. Y64A/T58I and Y64A/G60E Ras) also was shown to bind to SOS in HEK293T cells under autoinhibition conditions (not shown). These results support the notion that, although fractional, WT, GTPase-affecting, and GEF-affecting Ras proteins listed within this study are, indeed, all capable of binding to the SOS allosteric site in cells under autoinhibition conditions.
Figure 5.
The SOS allosteric site–binding interactions with Y64A Ras mutants under the autoinhibition conditions in cells. Y64A and V14I/Y64A Ras as well as I24N/Y64A, T50I/Y64A, Y64A/V152G, and Y64A/D153V are Ras proteins that are, respectively, versions of SOS allosteric site-specific WT and GTPase-affecting V14I Ras as well as GEF-affecting I24N, T50I, V152G, and D153V Ras proteins. These Ras proteins were expressed in HEK293T cells as described under “Experimental procedures.” The SOS in the membrane fraction was isolated. Then the Ras fractions bound to SOS and their activity were determined as described under “Experimental procedures.” A portion of isolated SOS was pretreated with the lipid vesicle-tethered active Y64A Ras proteins before analysis of the Ras fractions bound to SOS and their activity. For a control, SOSmemb-cat fully loaded with the lipid vesicle–tethered active Y64A Ras was used. All these cell-based analyses were performed three times, and the graphic figures close to the average densitometric analysis values are shown. The figure's embedded triplicate mean values of the results of the densitometric analysis are reported as relative densitometry values compared with the control SOSmemb-cat that was set at 100% as indicated. The S.D. values of these measurements are less than 15% of the values noted.
Moreover, Fig. 5 shows that most of the Y64A Ras bound on SOS in HEK293T cells were inactive, whereas a large fraction of the GEF-affecting Y64A Ras mutants bound on SOS in HEK293T cells were active. Notably, the presence of the fraction of active T50I/Y64A Ras on the membrane-bound SOS was shown to be the least among all the other GEF-affecting Ras mutants present there. This also is consistent with the in vitro kinetic analysis results shown above. Fig. 5 also shows that, although an active Y64A Ras was incapable of displacing the inactive Y64A Ras on SOS, the active GEF-affecting Y64A Ras (e.g. Y64A/D153V Ras) was capable of displacing the GEF-affecting Y64A Ras on SOS.
Overall, the same results were observed with the SOS in cytoplasm, where its Y64A Ras occupancy was less than 30% compared with the control solution Y64A Ras occupancy on SOSmemb-cat. Nevertheless, only the GEF-affecting Y64A Ras proteins—and not Y64A and the GTPase-affecting Y64A Ras proteins—bound to the cytosolic SOS were shown to be mainly active (not shown). The combination of these results supports the notion that, in the case of WT Ras, the bound Ras is detained in its inactive form so that SOS is not activated; thus, cell signaling is under control. However, unlike with WT Ras, the SOS-bound GEF-affecting Noonan syndrome Ras is exchangeable even under autoinhibition conditions, so that SOS is autoactivated and thus cell signaling is deregulated.
Discussion
This study shows that the Ras mutants associated with Noonan syndrome operate through two mechanistic-perturbation modes in deregulating Ras activity. Those that we have grouped as GTPase-affecting Ras mutants produce their constitutively active forms through perturbation of their intrinsic and GAP-mediated Ras GTPase activities. Another group of Noonan syndrome–relevant Ras mutants, termed GEF-affecting Ras mutants, produce their constitutively active forms through perturbation of SOS allostery, resulting in constitutively autoactivated SOS. V14I, T58I, and G60E Ras mutants belong to the GTPase-affecting Noonan syndrome category; their GEF-affecting counterparts include I24N, T50I, V152G, and D153V Ras mutants.
The role of the GTPase-affecting Ras mutants in many diseases such as Costello syndrome and some cancers has been known for a while. However, the role of the GEF-affecting Ras mutants was previously unknown. So far, it is applicable only to the development of Noonan syndrome.
Deregulation mode of the activity of GTPase- and GEF-affecting Noonan syndrome Ras mutants
Many factors could govern the resultant types of diseases, such as KRas, NRas, and HRas specificities and their downstream cascades. However, from a kinetic perspective, the relevant population intensity of the constitutively active form of the mutant Ras in cells could also play a role in several types of diseases. This speculation occurs because the degree of active Ras population directly affects the signaling intensity of the Ras MAPK pathway.
The overall steady-state fraction populations of the active GTPase-affecting Noonan syndrome Ras mutants shown within this study are smaller than those seen in the Costello syndrome and oncogenic Ras mutants (14, 31). Therefore, we speculate that the relatively lesser severity of disease symptoms seen with Noonan syndrome—as compared with those of the Costello syndrome and cancers—is, at least to a limited degree, because of the relatively lesser population intensity of active GTPase-affecting Noonan syndrome Ras mutants.
The overall steady-state fraction of the active Ras populations varies within the GEF-affecting Noonan syndrome Ras mutants. Therefore, the severity of the symptoms of Noonan syndrome may vary according to which of these Ras mutants is involved. Intriguingly, the steady-state cellular populations of active GEF-affecting Noonan syndrome Ras mutants are smaller than those of the Costello syndrome and of the oncogenic and Ras mutants; however, these active Ras steady-state populations are similar to those of the GTPase-affecting Noonan syndrome Ras mutants. Thus, we speculate that the milder symptoms of Noonan syndrome—compared with those of Costello syndrome and cancers—are to some degree because of the lesser concentrations of these disease agents. Nevertheless, further studies are required to link the fraction of the active form of Ras variations within and beyond any potential characteristics of these diseases.
Potential mechanism of unusual SOS autoactivation with the GEF-affecting Ras mutants under the SOS autoinhibition condition
Structure-based analyses of the Ras SOS complex (PDB entry 1NVV) show that GEF-affecting Ras mutants perturb Ras SOS allosteric binding interactions (see supporting information). Accordingly, we have proposed an overall scheme of the features of the Ras SOS-binding interaction as a way to decipher the molecular mechanism of the initiation of the SOS autoactivation by the GEF-affecting Ras mutants under the SOS autoinhibition condition.
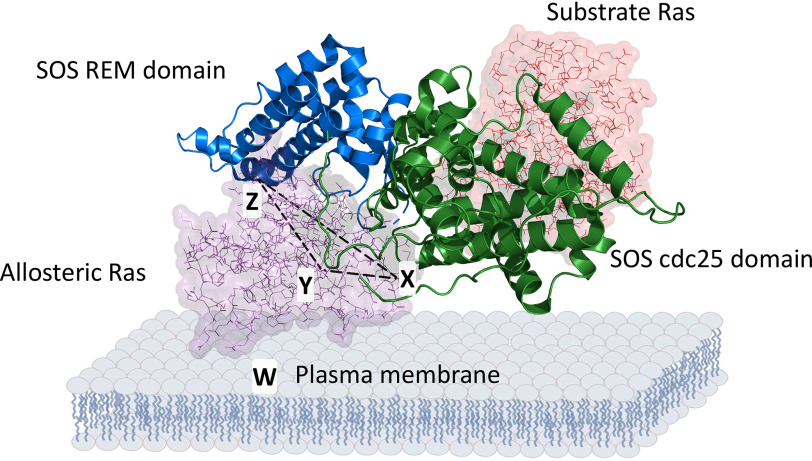
Fig. 6 defines three critical binding interfaces between Ras and SOS: a rigid SOS residue, Ser-921, with Ras Thr-50 (interface X); a flexible SOS gate cluster residue, Gln-755, with Ras Glu-153 (interface Y); and autoinhibitory SOS residues Lys-728 and Lys-724 with Ras Glu-63 (interface Z). When accounting for them as a triangle of X, Y, and Z interfaces, it can be postulated that all three interfaces function to bind Ras securely to the SOS allosteric site. Of these three, interfaces X and Y are identified by the SOS autoactivation kinetic study with GEF-affecting Ras mutants in combination with the structure-based analyses of the SOS-binding interaction with the GEF-affecting Ras mutants (see Figs. S2 and S3). Interface Z was identified by the structural analysis of the binding interfaces between Ras and the SOS allosteric site (see Figs. S1 and S2). An additional fourth binding interface, the Ras SOS complex with the plasma membrane (interface W), should also be considered. This consideration is warranted because it secures the binding of Ras in the SOS allosteric site through the triangular binding interfaces (Fig. 6). In view of this, the cytosolic Ras SOS-binding interactions occur only through the X, Y, and Z interfaces, whereas the membrane-bound Ras SOS-binding interactions happen through the X, Y, Z, and W interfaces.
Figure 6.
Critical Ras SOS allosteric binding interfaces. Ras in the allosteric site and the catalytic site of SOS are shown with space fill structures, whereas SOS is shown with a strand structure. The Ras SOS interfaces with X, Y, and Z residues that also are indicated with arrows. The triangle configuration of the X, Y, and Z interfaces also is shown with a dotted line. An additional W interface is noted below the Ras SOS complex. PyMOL with PDB entry 1NVV was used to produce this scheme.
This study identifies the detained Ras SOS autoinhibition and is an addition to the previously defined conventional SOS autoinhibition that lacks Ras on its allosteric site. We propose that this detention is because none of these critical binding interfaces between WT Ras and the SOS allosteric site is perturbed under the autoinhibition conditions. We also propose, in the case of GEF-affecting Ras mutants instead of WT Ras, that because one of the critical Ras SOS-binding interfaces is perturbed, these mutants are not detained on the SOS allosteric site but rather interchangeable with fresh, active GEF-affecting Ras mutants. For example, when the GEF-affecting D153V Ras mutant binds to the SOS allosteric site, it destabilizes the Y interface of these critical Ras SOS allosteric site–binding interfaces. This destabilization leads to the release of the bound Ras from the SOS allosteric site. The result is that a fresh active GEF-affecting D153V Ras mutant can bind to the SOS allosteric site to initiate SOS autoactivation.
The effect of the presence of the W interface, in addition to that of the presence of the triangle interface of X, Y, and Z, on the SOS autoactivation with the GEF-affecting Noonan syndrome Ras mutants under SOS autoinhibition conditions would be mechanically substantial. This is because the SOS autoactivation-specific kinetic parameters of SOS with GEF-affecting Noonan syndrome Ras mutants tethered to the lipid vesicle (secured with the X, Y, Z, and W interfaces) differ significantly from those of SOS with GEF-affecting Ras mutants in solution (secured only with the X, Y, and Z interfaces). This result also is not unexpected because the presence or absence of the W interface could also mechanically affect the binding interactions of the triangle formed by the X, Y, and Z interfaces. Notwithstanding this structure, because the X interface is rigid whereas the Y interface is flexible (see supporting information), the effect of the presence of the W interface may be asymmetrical on the X and Y interfaces (see below).
The Ras SOS allosteric site interface analyses (see supporting information) also suggest that GEF-affecting I24N and V152G Ras mutants can be grouped with the GEF-affecting D153V Ras that commonly perturbs the Y interface; in contrast, the GEF-affecting T50I Ras mutant has been shown to perturb only the X interface. Intriguingly, the SOS autoactivation kinetic parameters of these GEF-affecting Ras mutants (i.e. I24N, V152G, and D153V) that commonly perturb the Y interface are all similar. However, the common site perturbation does not necessarily produce similar kinetic parameters. Nevertheless, there may be an interface site-specific effect on the perturbation of the Ras SOS allosteric binding interactions.
This speculation is justified because when the GEF-affecting T50I Ras mutant was tethered to the lipid vesicle (the W interface was added to the X, Y, and Z interfaces to produce X, Y, Z, and W interfaces), the SOS autoactivation kinetic parameters of the GEF-affecting T50I Ras mutant certainly differed from those of other GEF-affecting Ras mutants. We speculate that this result is because, without the W interface—and regardless of the configuration of the rigid X and flexible Y interfaces—the perturbation of the X or Y interface releases Ras without any further obstacle. However, when the W interface is present, it upholds the Ras SOS allosteric complex even after the perturbation of the X or Y interface. In this case, the flexibility of interface Y may be a problem because it can be easier to perturb than the rigid X interface. This speculation explains why the autoactivation rate of the SOS autoactivation with the GEF-affecting T50I Ras mutant tethered to the lipid vesicle (that perturbs the rigid X interface) is slower than that of the SOS autoactivation with GEF-affecting D153V, V152G, and I24N Ras mutants tethered to the lipid vesicle (that commonly perturbs the flexible Y interface). Further studies that clarify the mechanisms of these Ras SOS–binding interfaces are necessary to understand the kinetic features of the SOS autoactivation with these GEF-affecting Ras mutants under conditions of SOS autoinhibition.
As we discussed earlier, the fractions of the active GEF-affecting Ras mutants in cells were lesser than those of Costello syndrome and oncogenic Ras mutants. One of the mechanical reasons for such a low fraction of the active GEF-affecting Ras mutants in cells would link with the limited fraction sizes of SOS with the detained WT Ras. Alternatively, it might link with the GEF-affecting Ras mutants in the SOS allosteric site under the SOS autoinhibition conditions. Therefore, we speculate that, at least to some extent, the milder symptoms of Noonan syndrome—compared with the symptoms of Costello syndrome and cancers—associated with these GEF-affecting Ras mutants are rooted in the limited occupancy of the active GEF-affecting Ras mutants in the SOS allosteric site under autoinhibition conditions.
Concluding remarks
The identification of the kinetic modes of the GTPase-affecting Noonan syndrome Ras mutants demonstrates how these V14I, T58I, and G60E Ras mutants constitutively populate their active forms in cells to produce Noonan syndrome. These findings expand on the previously well-studied Ras mutants that cause Costello syndrome and cancers.
The unveiling of the previously unknown kinetic mode of the GEF-affecting Ras mutants provides new insight into the mechanism of the production of constitutively active I24N, T50I, V152G, and D153V Ras mutants in cells and its link to development of diseases such as Noonan syndrome. A structure-based Ras SOS-binding interface analysis explains the unusual SOS activation by these GEF-affecting Ras mutants. This analysis further proposes a novel molecular mechanism for the deregulation of SOS autoinhibition by these GEF-affecting Ras mutants. However, additional mechanistic studies, including computational approaches, are necessary to evaluate and elaborate on the proposed mechanism.
The results of the cell-based pulldown study also support the kinetic mode of the GEF-affecting Noonan syndrome Ras mutants. However, the results require further rigorous verification through more in-depth and definitive cell biological and functional studies. These studies could include localization and functional studies of SOS with the GEF-affecting Noonan syndrome Ras mutants at the subcellular level before and after cell stimulation.
Our knowledge of the development of Noonan syndrome by the GEF-affecting Noonan syndrome Ras mutants still does not explain how these mutants overall alter or deregulate the Ras downstream effectors and thus the Ras-dependent signaling cascades (i.e. MAPK signaling events) in cells. Filling the gaps in our knowledge will require studies of the subcellular localizations, interactions, and functions of the GEF-affecting Ras mutants with Ras downstream effectors. These additional studies will connect the unusual function of the GEF-affecting Ras mutants with the development of Noonan syndrome and thus are essential to advance the development of the treatment of diseases and disorders caused by the GEF-affecting Ras mutants that deregulate SOS autoactivation.
Experimental procedures
Preparation of proteins
All human-origin WT HRas and mutant forms of HRas, KRas, and NRas (residues 1–181) were expressed in, and purified from, Escherichia coli as described in a previous study (14). These bacterially expressed Ras proteins encompass several oncogenic and Noonan syndrome Ras mutants. All were used for the necessary kinetic analyses in solution form and lipid vesicle–tethered form. Although the solution form of Ras (e.g. Noonan syndrome D153V Ras mutant) was used in its original purified form (i.e. Ras in solution), an additional mutation was introduced to the Ras (e.g. Noonan syndrome C118S/D153V Ras mutant) to tether it to the lipid vesicle (i.e. Ras tethered to the lipid vesicle) for the experiments. The lipid vesicle–tethered Ras through the mutated Ras Ser-118 side chain was prepared, with one exception, as described in the published method previously (23, 35). The exception to this method was that we used an Avanti Mini-Extruder with a 0.1 μm membrane to produce lipid vesicles, and we used a Superose 6 column (1.5 × 15 cm; GE Healthcare) instead of a Sepharose CL-4B column to remove unmodified Ras from Ras-tethered lipid vesicles as indicated in our previous study (26). The lipid vesicle Ras surface densities used within this analysis were in the range of ∼3000–3600 molecules/µm2, whereas the lipid vesicle Ras surface densities in the presence of 5% PIP2 were in the range of ∼3200–3500 molecules/µm2. The variation of the lipid vesicle Ras surface densities is slight, so it does not significantly alter the kinetic parameters associated with SOS autoactivation.
Human-origin SOS (SOS1) constructs possessing only the Cdc25 domain (termed SOSCdc25); Cdc25 and REM domains (termed SOScat); and Cdc25, REM, H, DH, and PH domains (termed SOSmemb-cat) were expressed in, and purified from, insect Sf9 cells by using the pIEx vector (Novagen) (26). A full-length p120GAP was also expressed in, and purified from, the same insect Sf9 cells as described in a previous study (14).
Kinetic assays
The assay buffer for all kinetic analyses—including the intrinsic and the SOS and GAP–mediated Ras activation and inactivation assays with various forms of Ras—consisted of 5 mm MgCl, 10 mm MgCl2, and 1 mm EDTA in 10 mm Tris-HCl (pH 7.4).
Unless otherwise noted, all kinetic analyses were performed with Ras using fluorescent 2′/3′-O-(N-methyl-anthraniloyl)–tagged GDP (Ras·mdGDP); Ras·mdGDP can be in solution or tethered to the lipid vesicle. The change in the fluorescence intensity by the intrinsic or SOS-mediated displacement of the Ras·mdGDP with the abundant nonhydrolyzable free GTP analog GppNHp, producing active Ras with GppNHp (Ras·GppNHp), was monitored using an LS 55 fluorescence spectrometer (excitation wavelength of 360 nm and emission wavelength of 440 nm).
Measurement of the autoactivation constant (kauto) of SOSmemb-cat with various forms of Ras was performed essentially as described in the previous study by using Ras·mdGDP (26). Briefly, the assay was initiated by the introduction of SOS to the assay solution that contained an inactive Ras·mdGDP complex in solution, or it was tethered to the lipid vesicle and an excess fresh solution GTP analog GppNHp. SOS facilitates the nucleotide exchange of the inactive Ras-bound mdGDP with a fresh GppNHp over time to produce an active Ras and free mdGDP. The production of the active GppNHp-bound Ras and free mdGDP is inversely coupled with the change in the mant fluorescence intensity over time. This is because the fluorescence tag tied with a heavier molecule (i.e. Ras·mdGDP) gives rise to a higher intensity of mant fluorescence than one tied with a lighter molecule (i.e. the released free mdGDP). Hence, the time-dependent declination of the mant fluorescence intensity that reflects the production of the active GppNHp-bound Ras over time denotes the catalytic action of SOS. The “time-dependent declination of the mant fluorescence intensity” fits into the autoactivation equation (Equation 1) that defines the fraction of SOS active () at a given time (t) with kauto (26).
| (Eq. 1) |
where and in Equation 1 are, respectively, the fraction of inactive and active SOS at time 0, and their ratio () determines the lag time of the process. It also reflects a trigger threshold fraction ratio of the active SOS () over the inactive SOS () for SOS autoactivation (26). Given that active SOS can be produced by the binding of an active Ras to the inactive SOS, either the active SOS (as in the mol % active SOS) or the active Ras (as in the mol % active Ras) can be the equivalent of a trigger of SOS autoactivation.
Determination of the threshold concentrations of various forms of active Ras for the initiation of autoactivation of SOSmemb-cat was also undertaken essentially as described in the previous study that also used Ras·mdGDP in the presence of the GTP analog GppNHp (26). For this analysis, an inactive SOSmemb-cat (0% activated SOSmemb-cat, 1 μm) was added to the assay cuvette that contained either the solution Ras mixture or the lipid vesicle–tethered Ras mixture in the presence of excess GppNHp (1 mm) in an assay buffer. The solution Ras mixture was the solution Ras·mdGDP (1 μm) plus various amounts of the solution Ras·mdGppNHp (the reaction initiator, 0-20 mol % of the total solution Ras). The lipid vesicle–tethered Ras mixture was the lipid vesicle–tethered Ras·mdGDP (∼3.3 × 103 Ras·GDP molecules/µm2) plus the lipid vesicle-tethered Ras·mdGppNHp (the reaction initiator, 0–20 mol % of the total lipid vesicle–tethered Ras). The decline of the intensity of the mant fluorescence upon addition of the inactive SOSmemb-cat to the assay cuvette reflects any SOS activity. This is because only active SOS facilitates exchange of either the solution or of the vesicle-tethered Ras·mdGDP with the abundant free GppNHp to produce either the solution or the vesicle-tethered GppNHp-bound active Ras, respectively, that lowers the mant fluorescence intensity. SOSmemb-cat activities, as in terms of the slope of the decline of fluorescence intensity over time (0–60 s) were monitored and then plotted against the threshold concentrations of Ras·mdGppNHp (the mol % of the reaction initiator Ras·mdGppNHp in the pool of total Ras). Notice that unlike with other titrations, the graphic of the threshold concentration titrations was unusual because SOS was very rapidly activated at a certain concentration of the reaction initiator Ras·mdGppNHp (the threshold of concentration of the reaction initiator Ras·mdGppNHp), which is one of the unique features of the autoactivation of SOS as seen in the previous study (26).
The fraction size of the active Ras produced by SOSmemb-cat after the completion of SOS autoactivation was determined by the comparison of the fraction of the active Ras produced by SOScat after the completion of the SOS autoactivation, which also used Ras·mdGDP in the presence of GppNHp under the same experimental conditions. An identical concentration of SOSmemb-cat and SOScat was used for this comparative analysis. This comparison was performed because SOScat lacks the SOS autoinhibitory domains; consequently, under any experimental conditions, WT Ras produced by SOScat in the assay solution became fully activated after the completion of SOS autoactivation that was set to 100%.
To determine the fractional occupancy of Ras in the SOS allosteric sites under various experimental conditions, such as autoinhibition, Y64A and its derivative Ras mutant forms, such as GEF-affecting Y64A/D153V Ras, were used instead of WT Ras and the original mutants. The Y64A Ras mutant was shown to conserve all WT Ras kinetic features with the exception that it failed to bind to the SOS catalytic site (23). Accordingly, Y64A and its derivative Ras mutants are, respectively, versions of homotropic allosteric WT Ras and Ras mutants that can only bind to the SOSmemb-cat allosteric site. Thus, the use of Y64A Ras mutants restricts specifically to the SOS allosteric site any quantitative measurements of the binding features of WT Ras and GTPase- and GEF-affecting Ras mutants.
Determination of the fractional occupancy of SOS with various forms of these Y64A Ras mutants was performed by fluorescence-based titration of Y64A Ras mutants that were in a form of Ras·mdGDP (receptor; typically 10 μm) with SOSmemb-cat (titrant; 0–100 μm). During the titration, each titration interval exceeded 1 min to allow Y64A Ras mutants in the form of Ras·mdGDP to bind to the SOSmemb-cat allosteric site. For comparison, a control titration with SOScat (instead of Y64A Ras tethered to the lipid vesicle with SOSmemb-cat) was also performed. The result of this control titration was set at 100% and subsequently used to normalize the titration results of various forms of Y64A Ras mutants with SOSmemb-cat for further analysis. This was because, as noted above, SOScat lacks the SOS autoinhibitory domain. As a result, regardless of any experimental conditions, SOScat could be fully occupied with Y64A Ras tethered to the lipid vesicle.
Expressions of Ras mutants in HEK293T cells
The endogenous WT NRas and KRas in HEK293T cells were knocked down by using shRNA plasmids (Santa Cruz Biotechnology, Inc.) according to the vendor's protocol. HEK293T cells were then transfected with Y64A, GTPase-affecting Y64A, or GEF-affecting Y64A Ras mutant by using the retroviral vector pLEGFP-N1 (36) in combination with Lipofectamine 2000 (Invitrogen). Transfected cells were serum-starved for 24 h and then cultured with serum for 1 h. Cells were iced and washed with an extraction buffer that contained 5 mm MgCl2, 1 mm EDTA, 0.5 mm DTT, and 100 mm Tris-HCl (pH 7.4). The cells were then sonicated and centrifuged briefly (500 × g for 10 min at 4 °C) to remove intact cells and cell debris to yield whole-cell lysate. The whole-cell lysate supernatant was further ultracentrifuged (100,000 × g for 1 h at 4 °C) to separate the membrane-bound proteins from the soluble cytosolic proteins. When necessary, before the ultracentrifugation, cell samples were treated with an excess of the lipid vesicle–tethered active Y64A or GAP- or GEF-affecting Y64A Ras mutants (1 mm) for 10 min. The pellet and the supernatant were resuspended in the extraction buffer with 1% Triton X-100. The resuspended pellet and supernatant were then used, respectively, for the pulldown of the cytosolic and membrane-bound SOS1 by using the anti-SOS1 mAb-tagged agarose beads (Thermo Fisher Scientific) as described in the manufacturer's protocol. The SOS1 antibody, pan-Ras antibody, and the Ras-binding domain of Raf-1 were used, respectively, to further analyze the pulled down samples for the quantity of the pulled down SOS, the SOS-bound total Ras, and the SOS-bound active Ras. Except for the step of treating the cell sample treatments with active Y64A or GAP- or GEF-affecting Y64A Ras mutants, all cell sample preparation procedures were performed at 4 °C or in ice.
Data availability
All data are contained within the article and supporting information.
Supplementary Material
This article contains supporting information.
Author contributions—H. G. U. and J. H. conceptualization; H. G. U., K. N., and J. H. formal analysis; H. G. U. and J. H. supervision; H. G. U., H. M. H., H. E. J., and J. H. investigation; H. G. U. and H. M. H. writing-original draft; H. G. U. and J. H. project administration; H. G. U. and J. H. writing-review and editing; H. M. H., H. E. J., and J. H. software.
Funding and additional information—This work was supported by National Institutes of Health Grant 1R15AI096146-01A1 (to J. H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- MAPK
- mitogen-activated protein kinase
- GppNHp
- 5′-guanylyl imidodiphosphate
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- SOS
- Son of Sevenless
- PR
- proline-rich
- REM
- Ras-exchanger motif
- PH
- pleckstrin homology
- DH
- Dbl-homology
- H
- histone
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PDB
- Protein Data Bank.
References
- 1. Bos J. L. (1989) ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 [PubMed] [Google Scholar]
- 2. Geyer M., and Wittinghofer A. (1997) GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr. Opin. Struct. Biol. 7, 786–792 10.1016/S0959-440X(97)80147-9 [DOI] [PubMed] [Google Scholar]
- 3. Barbacid M. (1987) ras genes. Annu. Rev. Biochem. 56, 779–827 10.1146/annurev.bi.56.070187.004023 [DOI] [PubMed] [Google Scholar]
- 4. Kinbara K., Goldfinger L. E., Hansen M., Chou F. L., and Ginsberg M. H. (2003) Ras GTPases: integrins' friends or foes? Nat. Rev. Mol. Cell Biol. 4, 767–776 10.1038/nrm1229 [DOI] [PubMed] [Google Scholar]
- 5. Colicelli J. (2004) Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13 10.1126/stke.2502004re13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 7. Myers A., Bernstein J. A., Brennan M. L., Curry C., Esplin E. D., Fisher J., Homeyer M., Manning M. A., Muller E. A., Niemi A. K., Seaver L. H., Hintz S. R., and Hudgins L. (2014) Perinatal features of the RASopathies: Noonan syndrome, cardiofaciocutaneous syndrome, and Costello syndrome. Am. J. Med. Genet. A 164A, 2814–2821 10.1002/ajmg.a.36737 [DOI] [PubMed] [Google Scholar]
- 8. Gelb B. D., Roberts A. E., and Tartaglia M. (2015) Cardiomyopathies in Noonan syndrome and the other RASopathies. Prog. Pediatr. Cardiol. 39, 13–19 10.1016/j.ppedcard.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prendiville T. W., Gauvreau K., Tworog-Dube E., Patkin L., Kucherlapati R. S., Roberts A. E., and Lacro R. V. (2014) Cardiovascular disease in Noonan syndrome. Arch. Dis. Child. 99, 629–634 10.1136/archdischild-2013-305047 [DOI] [PubMed] [Google Scholar]
- 10. Rauen K. A. (2013) The RASopathies. Annu. Rev. Genomics Hum. Genet. 14, 355–369 10.1146/annurev-genom-091212-153523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schubbert S., Zenker M., Rowe S. L., Boll S., Klein C., Bollag G., van der Burgt I., Musante L., Kalscheuer V., Wehner L. E., Nguyen H., West B., Zhang K. Y., Sistermans E., Rauch A., et al. (2006) Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38, 331–336 10.1038/ng1748 [DOI] [PubMed] [Google Scholar]
- 12. Gelb B. D., and Tartaglia M. (2006) Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum. Mol. Genet. 15 Spec No 2, R220–R226 10.1093/hmg/ddl197 [DOI] [PubMed] [Google Scholar]
- 13. Roberts A. E., Allanson J. E., Tartaglia M., and Gelb B. D. (2013) Noonan syndrome. Lancet 381, 333–342 10.1016/S0140-6736(12)61023-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wey M., Lee J., Jeong S. S., Kim J., and Heo J. (2013) Kinetic mechanisms of mutation-dependent Harvey Ras activation and their relevance for the development of Costello syndrome. Biochemistry 52, 8465–8479 10.1021/bi400679q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carta C., Pantaleoni F., Bocchinfuso G., Stella L., Vasta I., Sarkozy A., Digilio C., Palleschi A., Pizzuti A., Grammatico P., Zampino G., Dallapiccola B., Gelb B. D., and Tartaglia M. (2006) Germline missense mutations affecting KRAS Isoform B are associated with a severe Noonan syndrome phenotype. Am. J. Hum. Genet. 79, 129–135 10.1086/504394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schubbert S., Shannon K., and Bollag G. (2007) Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308 10.1038/nrc2109 [DOI] [PubMed] [Google Scholar]
- 17. Cirstea I. C., Kutsche K., Dvorsky R., Gremer L., Carta C., Horn D., Roberts A. E., Lepri F., Merbitz-Zahradnik T., König R., Kratz C. P., Pantaleoni F., Dentici M. L., Joshi V. A., Kucherlapati R. S., et al. (2010) A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat. Genet. 42, 27–29 10.1038/ng.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Runtuwene V., van Eekelen M., Overvoorde J., Rehmann H., Yntema H. G., Nillesen W. M., van Haeringen A., van der Burgt I., Burgering B., and den Hertog J. (2011) Noonan syndrome gain-of-function mutations in NRAS cause zebrafish gastrulation defects. Dis. Model. Mech. 4, 393–399 10.1242/dmm.007112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roberts A. E., Araki T., Swanson K. D., Montgomery K. T., Schiripo T. A., Joshi V. A., Li L., Yassin Y., Tamburino A. M., Neel B. G., and Kucherlapati R. S. (2007) Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 39, 70–74 10.1038/ng1926 [DOI] [PubMed] [Google Scholar]
- 20. Tartaglia M., Pennacchio L. A., Zhao C., Yadav K. K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W., Ustaszewska A., Martin J., Bristow J., Carta C., Lepri F., et al. (2007) Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 39, 75–79 10.1038/ng1939 [DOI] [PubMed] [Google Scholar]
- 21. Lepri F., De Luca A., Stella L., Rossi C., Baldassarre G., Pantaleoni F., Cordeddu V., Williams B. J., Dentici M. L., Caputo V., Venanzi S., Bonaguro M., Kavamura I., Faienza M. F., Pilotta A., et al. (2011) SOS1 mutations in Noonan syndrome: molecular spectrum, structural insights on pathogenic effects, and genotype-phenotype correlations. Hum. Mut. 32, 760–772 10.1002/humu.21492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cordeddu V., Yin J. C., Gunnarsson C., Virtanen C., Drunat S., Lepri F., De Luca A., Rossi C., Ciolfi A., Pugh T. J., Bruselles A., Priest J. R., Pennacchio L. A., Lu Z., Danesh A., et al. (2015) Activating mutations affecting the Dbl homology domain of SOS2 cause Noonan syndrome. Hum. Mutat. 36, 1080–1087 10.1002/humu.22834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gureasko J., Galush W. J., Boykevisch S., Sondermann H., Bar-Sagi D., Groves J. T., and Kuriyan J. (2008) Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 15, 452–461 10.1038/nsmb.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roose J. P., Mollenauer M., Ho M., Kurosaki T., and Weiss A. (2007) Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol. Cell. Biol. 27, 2732–2745 10.1128/MCB.01882-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jun J. E., Rubio I., and Roose J. P. (2013) Regulation of ras exchange factors and cellular localization of ras activation by lipid messengers in T cells. Front. Immunol. 4, 239 10.3389/fimmu.2013.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoang H. M., Umutesi H. G., and Heo J. (2019) Allosteric autoactivation of SOS and its kinetic mechanism. Small GTPases 1–16 10.1080/21541248.2019.1601954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yadav K. K., and Bar-Sagi D. (2010) Allosteric gating of Son of sevenless activity by the histone domain. Proc. Natl. Acad. Sci. U. S. A. 107, 3436–3440 10.1073/pnas.0914315107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen P. C., Wakimoto H., Conner D., Araki T., Yuan T., Roberts A., Seidman C., Bronson R., Neel B., Seidman J. G., and Kucherlapati R. (2010) Activation of multiple signaling pathways causes developmental defects in mice with a Noonan syndrome-associated Sos1 mutation. J. Clin. Invest. 120, 4353–4365 10.1172/JCI43910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakamura Y., Umeki N., Abe M., and Sako Y. (2017) Mutation-specific mechanisms of hyperactivation of Noonan syndrome SOS molecules detected with single-molecule imaging in living cells. Sci. Rep. 7, 14153 10.1038/s41598-017-14190-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grewal T., Koese M., Tebar F., and Enrich C. (2011) Differential regulation of RasGAPs in cancer. Genes Cancer 2, 288–297 10.1177/1947601911407330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wey M., Lee J., Kim H. S., Jeong S. S., Kim J., and Heo J. (2016) Kinetic mechanism of formation of hyperactive embryonic ras in cells. Biochemistry 55, 543–559 10.1021/acs.biochem.5b00902 [DOI] [PubMed] [Google Scholar]
- 32. Segel I. H. (1993) Enzyme Kinetics, Wiley-Interscience, New York [Google Scholar]
- 33. Frost A. A., and Pearson R. G. (1953) Kinetics and Mechanism, Wiley, New York [Google Scholar]
- 34. Heo J., Halbleib C. M., and Ludden P. W. (2001) Redox-dependent activation of CO dehydrogenase from Rhodospirillum rubrum. Proc. Natl. Acad. Sci. U. S. A. 98, 7690–7693 10.1073/pnas.141230698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gureasko J., Galush W. J., Sondermann H., Groves J. T., and Kuriyan J. (2010) Direct coupling of Ras to preformed maleimide-functionalized lipid membranes. Protocol Exchange 10.1038/nprot.2010.155 10.1038/nprot.2010.155 [DOI] [Google Scholar]
- 36. Shin J. Y., Wey M., Umutesi H. G., Sun X., Simecka J., and Heo J. (2016) Thiopurine prodrugs mediate immunosuppressive effects by interfering with Rac1 protein function. J. Biol. Chem. 291, 13699–13714 10.1074/jbc.M115.694422 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and supporting information.