Abstract
Plants optimize their growth in fluctuating environments using information acquired by different organs. This information is then transmitted through the rest of the plant using both short- and long-distance signals, including hormones and mobile proteins. Although a few of these signals have been characterized, long-distance signaling is not well understood in plants. Recently, the light-regulated transcription factor HY5 was reported to move from the shoot to the root to regulate root growth. We generated a cell-type specifically expressed HY5 fusion protein that could not be detected outside the tissue in which it was targeted. By expressing this DOF-HY5 protein in specific cell types of the hypocotyl, we showed that its local activity was sufficient to regulate hypocotyl growth. We also found that, although DOF-HY5 was expressed specifically in the shoot and not detected in the roots, it could rescue hy5 growth defects in primary roots but not in lateral roots. We therefore conclude that HY5 protein mobility is not required in the hypocotyl or for shoot-to-root communication. Our results indicate that a signal downstream of, or in parallel with, HY5 in the shoot is mobile and links shoot and root growth.
Keywords: HY5, shoot-to-root communication, hypocotyl, root, lateral root, Arabidopsis
Local HY5 activity in specific cell types of the hypocotyl can rescue hy5 primary root growth defects, in the absence of HY5 mobility to the root, and regulate hypocotyl growth. This study suggests that a mobile signal downstream of HY5 can carry out this signaling function during early seedling development.
Introduction
As sessile organisms, plants have developed unique mechanisms that provide them with developmental plasticity to optimize their growth in response to changes in the environment. In many cases, these changes are only sensed by single plant organs; for example, roots sense changes in soil water or nutrient availability, and shoots sense changes in light quantity and quality. Therefore, mechanisms that allow communication between organs and tissues are necessary for plants to develop and grow. In the hypocotyl, mechanical and mobile signals that facilitate communication between tissue layers control growth and gravitropic responses (Kutschera and Niklas, 2007, Savaldi-Goldstein et al., 2007, Kim et al., 2016, Procko et al., 2016). Mechanical signals coordinate processes of the inner stem layer, which tends to grow, with those of the outer layer, which restricts growth (Kutschera and Briggs, 1988, Kutschera and Niklas, 2007). The small-molecule hormone auxin is a mobile signal that participates in many environmental responses. When a neighboring plant is in close proximity, the shaded plant produces auxin in the cotyledons. The newly-produced auxin then travels to the hypocotyl epidermis, where it relaxes the constraints on inner tissue growth, resulting in elongation growth and increased light capture. Blocking the transcriptional response to auxin in the epidermis inhibits elongation growth (Tao et al., 2008, Procko et al., 2014, Procko et al., 2016). Brassinosteroid (BR), another important plant hormone, also regulates hypocotyl growth by acting on the epidermis. Restoring BR perception or biosynthesis, specifically in the epidermis, rescues shoot growth in BR receptor and biosynthetic mutants (Savaldi-Goldstein et al., 2007). In addition, expressing photoreceptor phytochrome B (phyB), specifically in the epidermis, restores the hypocotyl length and gravitropic response of phyB mutants (Endo et al., 2005, Kim et al., 2016). However, the epidermis also signals to the inner cell layers, as the expression of phyB in the epidermis leads to the degradation of PHYTOCHROME INTERACTING FACTOR transcription factors in the vasculature, suggesting that there is a mobile signal downstream of phyB (Kim et al., 2016).
In addition to the communication between shoot cell layers, communication between aerial and underground organs is crucial for coordinating growth and development between these tissues (van Gelderen et al., 2018b). Disrupting this communication by severing aerial and underground portions of a plant leads to decreased growth of both the shoot and roots, and the accumulation of chlorophyll in roots (Kobayashi et al., 2017, Bellstaedt et al., 2019, Gaillochet et al., 2020). However, as mechanical perturbations are quite severe, the combined effects of the wounding response and the loss of all mobile signals make specific effects difficult to discern.
Perhaps the most fundamental aspect of shoot–root long-distance interactions is nutrient transport. The shoot sends photosynthesis-derived energy to roots, and roots transport water and nutrients from the soil to the shoot (Coruzzi and Bush, 2001). In addition, a set of mobile signals, including gibberellin (GA), auxin, abscisic acid, sucrose, and certain proteins, regulate the root architecture (Reed et al., 1998, Bhalerao et al., 2002, Kircher and Schopfer, 2012, Regnault et al., 2015, Chen et al., 2016, van Gelderen et al., 2018b, Ha et al., 2018). Moreover, light can be piped from the shoot through the vascular tissue, thereby activating phyB in roots (Lee et al., 2016), which in turn can also regulate root growth. The lack of nuclear-localized phyB in deeper roots suggests different mechanisms of regulation between shallow and deep root systems (Salisbury et al., 2007). In both cases, however, local phyA and phyB control root growth and lateral root (LR) development by affecting auxin (Salisbury et al., 2007, Lee et al., 2016).
phyB exerts its key regulatory influence by inducing the expression and promoting the stability of the transcription factor ELONGATED HYPOCOTYL 5 (HY5) in the shoot and roots (Lu et al., 2015, Lee et al., 2016). HY5 is a key light-regulated factor that controls hypocotyl elongation, root development, and LR formation (Oyama et al., 1997, Clough and Bent, 1998, Lee et al., 2016, van Gelderen et al., 2018a). Furthermore, HY5 coordinates above-the-ground growth with below-the-ground growth. Recent studies have indicated that HY5 accomplishes this by moving from the shoot to the root. Illumination of the shoot promotes the accumulation of HY5, which then travels to roots where it controls growth and nutrient uptake, and promotes its own local expression (Chen et al., 2016, van Gelderen et al., 2018a).
There is compelling evidence to support the presence of a phytochrome-HY5 signaling axis that controls shoot and root growth as well as LR formation. However, an explanation for the domain-specific function of HY5 in the hypocotyl, as well as the mechanism underlying shoot–root translocation, remain elusive. In this study, we characterize domain-specific hy5 rescue lines and show that HY5 can function in a non-cell-autonomous manner during hypocotyl elongation. We demonstrate that HY5 movement is not required for communication between the shoot and roots, indicating that a mobile signal acts downstream of HY5 during early seedling development in Arabidopsis.
Results and Discussion
HY5 Can Function in a Non-cell-autonomous Manner During Hypocotyl Elongation
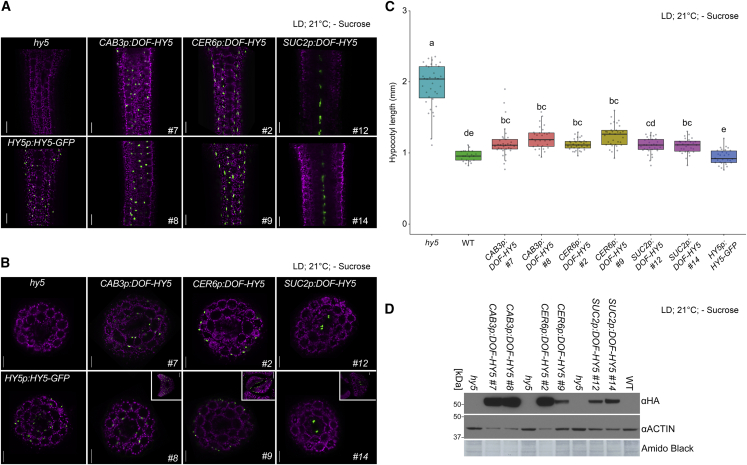
The Arabidopsis hypocotyl is composed of a central cylinder of vascular cells (mainly the xylem and phloem), surrounded by layers of endodermis, cortex, and epidermis. The epidermis has been reported to be the main tissue layer controlling hypocotyl growth. Manipulating epidermal auxin, BR, or light signaling has dramatic effects on hypocotyl growth responses, whereas similar manipulations in other cell layers result in milder, or even opposite, effects on growth (Kutschera and Niklas, 2007, Savaldi-Goldstein et al., 2007, Kim et al., 2016, Procko et al., 2016). As HY5 controls hypocotyl elongation downstream of photoreceptors by regulating auxin and BR levels (Gangappa and Botto, 2016), we aimed to investigate its tissue-specific function in the hypocotyl. We first examined the tissue layers in which HY5 regulates hypocotyl growth. We therefore expressed HY5 fused at its N-terminus to an HA-YFP-HA (DOF) tag in the hy5 mutant background (see Methods) under the control of several tissue-specific promoters. We expressed this fusion protein in the epidermis under the control of the CER6 promoter (CER6p:DOF-HY5), in the green tissue under the control of the CAB3 promoter (CAB3p:DOF-HY5), and in phloem companion cells under the control of the SUC2 promoter (SUC2p:DOF-HY5) (Guyomarc’h et al., 2012, Hooker et al., 2002, Procko et al., 2016, Susek et al., 1993, Truernit and Sauer, 1995). As HY5 was proposed to move between cell types, we examined the mobility of HY5 between cell layers in the hypocotyl. Surprisingly, we were not able to detect DOF-HY5 outside the tissue in which it was targeted (Figure 1A and 1Band Supplemental Figures 1A and 2A). We found that expressing DOF-HY5 in the epidermis, green tissue, or vasculature either partly or fully rescued the long hypocotyl phenotype of hy5 (Figure 1C and Supplemental Figures 1 and 2). Taken collectively, these results showed that local HY5 expression in sub-domains of the hypocotyl is sufficient to regulate growth similar to levels observed in wild-type plants.
Figure 1.
HY5 Movement Is Not Required for Rescue of the hy5 Long Hypocotyl Phenotype.
(A and B) Representative fluorescence images of the hypocotyl (A) and hypocotyl cross-sections (B) of seedlings expressing HA-YFP-HA (DOF)-HY5 (green) under the control of different tissue-specific promoters or HY5p:HY5-GFP seedlings. Inset in (B) shows a cross-section of the leaf primordium. All constructs were expressed in the hy5 mutant background. CAB3p was used to drive DOF-HY5 in the green tissue, CER6p in the epidermis, and SUC2p in phloem companion cells. SUC2p line number 14 displayed occasional weak expression in the epidermis, consistent with previous results (Procko et al., 2016). Magenta, chlorophyll autofluorescence. As these constructs contained different fluorophores that were expressed in different tissues at different levels, image capture settings were different for each image. Scale bars in (A) and (B), 50 μm; inset, 40 μm.
(C) Hypocotyl length of seedlings expressing the indicated constructs in the hy5 mutant background and wild-type seedlings. Boxes indicate the first and third quartiles, whiskers indicate the minimum and maximum values, and black lines within the boxes indicate the median values. Different letters denote statistical differences (p < 0.05) among samples, as assessed by one-way ANOVA and Tukey HSD (n > 22).
(D) Western blots of proteins prepared from shoots isolated from seedlings expressing DOF-HY5 under the control of different tissue-specific promoters in the hy5 mutant background. DOF-HY5 was visualized with an anti-HA antibody, and an anti-ACTIN antibody served as a loading control.
Images and measurements in (A)–(D) were taken of 6-day-old seedlings grown in white light (117 μE) long-day conditions (LD) (16 h light, 8 h dark) at 21°C on media without sucrose.
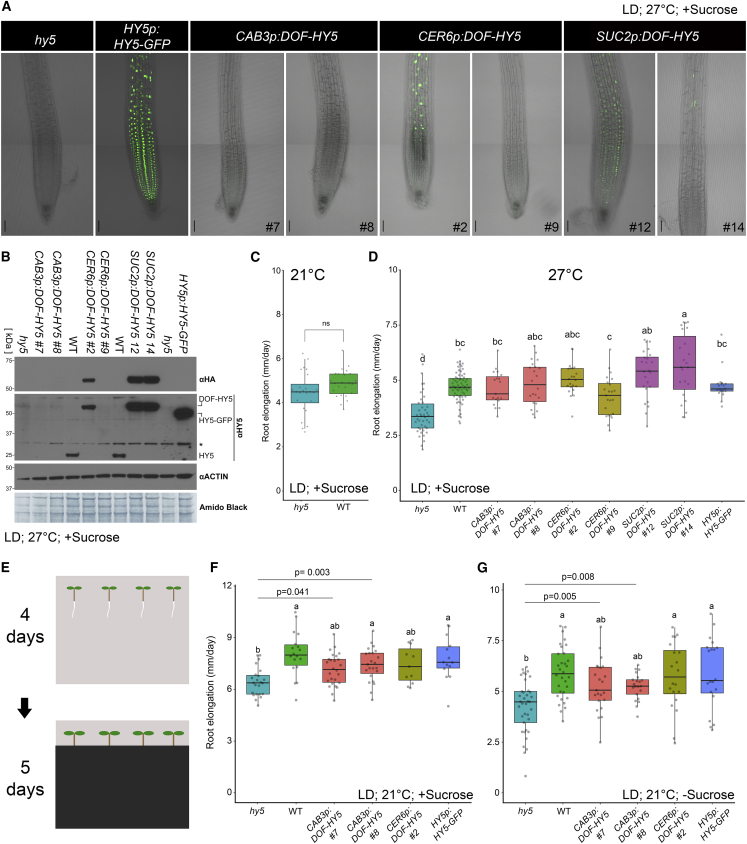
Figure 2.
HY5 Movement from the Shoot to the Root Is Not Required to Rescue the Primary Root Growth of hy5.
(A) Representative fluorescence and bright-field images of primary roots from seedlings expressing DOF-HY5 (green) under the control of different tissue-specific promoters in the hy5 mutant background or HY5p:HY5-GFP hy5. Images were taken of seedlings grown in white light (120 μE) LD at 27°C. Image capture settings were identical for all constructs, except HY5p:HY5-GFP. Scale bars, 50 μm.
(B) Western blots of proteins prepared from roots isolated from seedlings grown in white light LD at 27°C and expressing HY5 under the control of different tissue-specific promoters in the hy5 background. DOF-HY5 was visualized with an anti-HA antibody (upper panel). DOF-HY5, HY5-GFP, and native HY5 were visualized with an anti-HY5 antibody (middle panel), and an anti-ACTIN antibody and amido black staining served as loading controls (lower panels). ∗Cross-reacting band.
(C and D) Root elongation of seedlings grown in white light LD at 21°C (C) or 27°C (D) and expressing the indicated constructs in the hy5 mutant background. Images and final measurements in (A)–(D) were taken of 6-day-old seedlings grown on media with sucrose.
(E–G) Root elongation of seedlings expressing the indicated constructs in the hy5 mutant background grown in white light LD at 21°C on media with sucrose (F) or without sucrose (G) for 4 days and then transferred into dark boxes (shoots in the light, roots in the dark) for another 5 days. (E) Different letters denote statistical differences (p < 0.05) among samples, as assessed by one-way ANOVA and Tukey HSD (n > 19). In (F) and (G), the p-values are from Student's t-test between the indicated genotypes.
We demonstrated that HY5 can coordinate hypocotyl growth from the vasculature, mesophyll, and epidermis with similar efficiency. Interestingly, SUPPRESSOR OF PHYA-105 1 (SPA1), which negatively regulates the HY5 protein level via the COP1/SPA complex, has been reported to function in the phloem where it regulates seedling development, leaf expansion, and flowering time (Ranjan et al., 2011). As both HY5 and SPA1 can regulate seedling development via the phloem, it is tempting to speculate that they can also interact there, which may affect HY5 activity and mobility. These results, however, are surprising, given the necessity and sufficiency of epidermal BR and auxin signaling and the failure of vasculature-expressed phyB to rescue the phenotype of the phyB mutant in regulating hypocotyl elongation (Savaldi-Goldstein et al., 2007, Procko et al., 2016). The epidermis is likely the primary tissue regulating hypocotyl growth; thus, these observations suggest that a signal downstream of HY5 moves to the epidermis where it causes growth arrest (Kutschera and Niklas, 2007, Savaldi-Goldstein et al., 2007, Procko et al., 2016). Given that auxin and BR regulate hypocotyl growth in the epidermis and HY5 regulates both hormones, they likely represent the mobile signals potentiating HY5 activity (Reed et al., 1998, Bhalerao et al., 2002, Sibout et al., 2006, Shi et al., 2011, Li and He, 2016, Gangappa and Kumar, 2017). However, BR is synthesized locally, where it acts on nearby receptors, and blocking the auxin response in inner cells did not inhibit elongation as strongly as in the epidermis, suggesting that they are unlikely to be the HY5-dependent signals (Savaldi-Goldstein et al., 2007, Procko et al., 2016). HY5 also regulates GA and a variety of transcription factors; therefore, we propose that one of these might be the mobile signal that subsequently regulates auxin and BR in the epidermis (Regnault et al., 2015, Gangappa and Kumar, 2017, Burko et al., 2020).
A Mobile Signal Downstream of HY5 in the Shoot Regulates Primary Root Growth
Our HY5 constructs showed no, or undetectable, intercellular movement; therefore, this was a good opportunity to investigate the role of HY5 and its movement in the coordination of shoot–root growth. We examined the ability of DOF-HY5 to move from the shoot to the root; however, we did not detect DOF-HY5 or cleaved HY5 (using an antibody against native HY5) in the roots of plants expressing DOF-HY5 driven by the CAB3 promoter (Figure 2A and 2B and Supplemental Figure 3A and 3B). These results indicate that DOF-HY5 expressed in the shoot could not move to roots, and if it did, the amount was below our limit of detection, which is consistent with the limited intracellular movement of DOF-HY5 in the shoot. When DOF-HY5 was expressed under the control of the CER6 or SUC2 promoter, it could be detected in roots (Figure 2A and 2B and Supplemental Figure 3A and 3B). However, these promoters also promoted expression in roots, as detected by RT–qPCR (Supplemental Figure 3C).
The recent finding that HY5 fused to GFP (HY5-GFP) can transit from the shoot to the root (Chen et al., 2016) has prompted us to suggest several hypotheses for the lack of movement of DOF-HY5 in our experimental settings. (1) The position of the tag (N- versus C-terminus) may affect the fusion protein's ability to move from the shoot to the root, suggesting that HY5 movement is facilitated by interactions with other factors. The different positions of the tags fused to HY5 may prevent or facilitate such interactions. (2) DOF-HY5 (∼51 kDa) is slightly larger than HY5-GFP (∼45 kDa), and as such, the size of the fusion protein may have affected its mobility. It was reported that small, highly expressed proteins have a higher probability of detection in the phloem, with the probability of phloem transport reduced above a molecular mass of approximately 70 kDa (Paultre et al., 2016). However, both HY5 fusion proteins were within the size range of highly abundant proteins in the phloem stream (∼20–70 kDa) (Figure 2B and Supplemental Figure 4B). (3) Lastly, expression levels and expression patterns of the CAB3 promoter fragments chosen for the constructs may have been different. The CAB3 promoter, which was cloned in our lab, lacks 29 bp at the 3′ end (Procko et al., 2016, see Methods) compared with the fragment used by Chen et al. (2016). To test the last point, we compared the HY5 protein level in the shoot and the root of CAB3p:DOF-HY5 (this study) with that of CAB3p:HY5-GFP seedlings (Chen et al., 2016). DOF-HY5 and HY5-GFP were expressed at comparable levels in the shoot (Supplemental Figure 4A and 4B). However, we could not detect any DOF-HY5 in the root, and HY5-GFP was detected in the root, as previously reported (Chen et al., 2016, Supplemental Figure 4A and 4C). These results argue against the hypothesis that the lack of DOF-HY5 mobility in CAB3p:DOF-HY5 plants is a result of the lower expression in the shoot. In summary, the most likely explanation for the different behaviors of these fusion proteins is the N- versus C-terminal orientation of the tags.
These results put forward two major questions. Firstly, is the lack of detection of DOF-HY5 in the root due to the sensitivity limits of the methods we used. Secondly, does endogenous HY5 move from the shoot to the root in wild-type plants. We concluded that the orientation of the tag is the most likely cause of the differential mobility, but we still did not know which orientation recapitulated the characteristics of the native protein. In addition, although HY5 mobility was demonstrated by grafting HY5p:HY5-GFP or CAB3p:HY5-GFP shoots onto the hy5 rootstock (Chen et al., 2016), we consistently found that the level of HY5-GFP in these lines was significantly higher than that of native HY5 (Figure 2B and Supplemental Figures 3B, 4B, and 4C). In terms of grafting, it is possible that the high level of HY5 artificially increased its diffusion to the phloem, enabling its mobility from the shoot to the root. Immunohistochemical staining of native HY5 in the roots of hy5 mutants grafted onto wild-type shoots may directly address these issues.
The HY5 protein in the CAB3p:DOF-HY5 line was only detectable in the shoot, which prompted us to ask whether a signal downstream of HY5, in addition to the HY5 protein itself, could mediate the communication between the shoot and the root. As the effect of hy5 on root growth was weak at 21°C (Figure 2C), we evaluated the rescue of the root growth phenotype at 27°C, a condition in which hy5 roots are shorter than those of wild-type plants (Figure 2D). Importantly, this condition did not affect DOF-HY5 localization in the shoot or the root (compare Figure 1A with Supplemental Figure 1A, and Figure 2B with Supplemental Figure 3B). CER6p:DOF-HY5, SUC2p:DOF-HY5, and HY5p:HY5-GFP, which were all expressed in both the shoot and the root, partly or fully rescued the hy5 root growth phenotype at 27°C (Figure 2D). Surprisingly, we found that CAB3p:DOF-HY5 could also rescue the hy5 root growth defects (Figure 2D), indicating that HY5 mobility is not required for HY5-dependent shoot-to-root communication. These results are consistent with the finding that HY5 can trigger an unknown signal in the shoot to control root thermomorphogenesis (Gaillochet et al., 2020). HY5 expression in the shoot and the root was weaker in CER6p:DOF-HY5 line number 9 than that in other lines derived from this construct (Figure 1D and Supplemental Figure 3A). We could only detect a few YFP-positive spots in the upper portions of the root, and this line showed markedly weaker rescue of the root phenotype than the higher-expressing lines, indicating that we could successfully observe different levels of HY5 activity (Figure 2D and Supplemental Figures 1B and 2B). Therefore, the rescue of the hy5 root phenotype, which was observed in CAB3p:DOF-HY5 seedlings, most likely cannot be explained by the undetectable level of active HY5 in roots, as the weak rescue of the phenotype in weakly-expressing CER6p:DOF-HY5 line number 9 suggests that rescue of hy5 root phenotype depends on HY5 level. As roots are not usually exposed to light in natural conditions, we examined whether HY5 activity in the shoot could rescue the reduced growth phenotype of the hy5 primary root when the roots were in the dark. Although the root growth of CER6p:DOF-HY5 and HY5p:HY5-GFP plants was similar to that of wild-type plants, we found that CAB3p:DOF-HY5 only partially rescued the hy5 root phenotype (Figure 2E and 2F). As this experiment was performed in the presence of sucrose, we repeated it in the absence of sucrose (Figure 2G). We observed similar results, suggesting that this phenotype was not sucrose-dependent. Therefore, the partial rescue of the hy5 primary root growth phenotype was probably not due to the lack of HY5 mobility, as the mobile form of HY5 had a similar primary root growth phenotype when the roots were covered (Chen et al., 2016).
Our results showed that HY5 mobility was not required for HY5-dependent shoot-to-root signaling. However, when roots were in the dark, local HY5 expression in roots was required for fully functional roots. Auxin and GA are known to travel from the shoot to the root to regulate root architecture, and HY5 has been shown to regulate the levels of both hormones (Bhalerao et al., 2002, Sibout et al., 2006, Weller et al., 2009, Regnault et al., 2015, Gangappa and Kumar, 2017). Therefore, they likely represent the mobile signals downstream of HY5 for root-to-shoot communication.
Tissue Autonomous Expression of HY5 Regulates Lateral Root Development
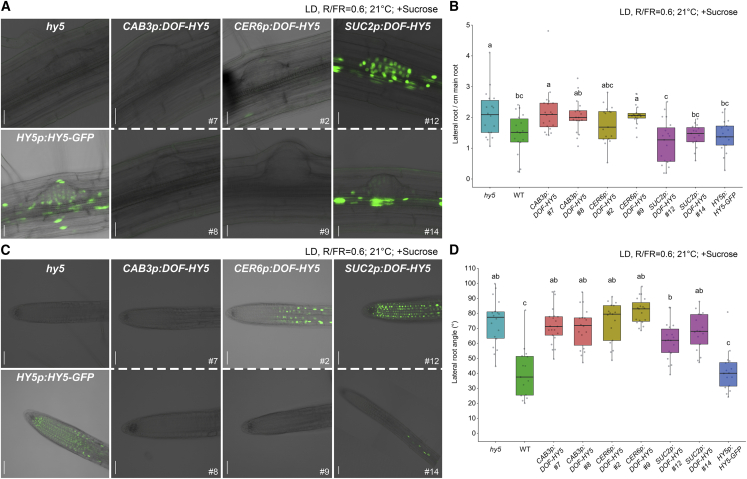
In addition to root elongation, HY5 regulates root branching by reducing LR density in white and far-red light conditions (WL + FR). HY5 does this by causing a reduction in auxin transport into the LR primordium, thus repressing LR formation (van Gelderen et al., 2018a). Given that DOF-HY5 is most likely not mobile, this prompted us to ask whether the local activity of HY5 in the LR primordium is needed to rescue the hy5 LR phenotype in low R/FR light conditions (WL + FR, R:FR = 0.6). We did not observe rescue of the LR density phenotype in the CAB3p:DOF-HY5 lines. Furthermore, we found that CER6p:DOF-HY5, which is expressed in the root but not in or close to the LR primordium, failed to rescue the LR density phenotype of hy5 (Figure 3A and 3B). By contrast, HY5 expressed in the LR formation zone using the SUC2 or HY5 native promoter did rescue the LR density phenotype, suggesting that HY5 is needed in the LR formation zone to rescue the hy5 mutant phenotype (Figure 3A and 3B).
Figure 3.
HY5 That is Restricted to the Hypocotyl Cannot Rescue the Lateral Roots Phenotypes of hy5 in Shade
(A and C) Representative fluorescence images and bright-field images of lateral root (LR) primordia (A) and LR tips (C) from seedlings expressing HY5 (green) under the control of different tissue-specific promoters or HY5p:HY5-GFP in the hy5 mutant background. As these constructs contained different fluorophores that were expressed at different levels, image capture settings were different for each image. Scale bars, A, 20 μm; C, 50 μm.
(B and D) LR density (the number of all emerged lateral roots divided by the total length of the main root) (B) and LR angle (D). In (D), the values of the angle between the longest lateral root tip and the main root are shown (see also Supplemental Figure 5). All images and measurements in (A)–(D) were taken of 10-day-old seedlings grown in white and far-red light (127 μE, R:FR = 0.6) LD at 21°C on media with sucrose. Different letters denote statistical differences (p < 0.05) among samples, as assessed by one-way ANOVA and Tukey HSD (n > 15).
HY5 also regulates LR gravitropism (Lee et al., 2016). In contrast to the LR density phenotype, the LR gravitropism phenotype of the hy5 mutant could not be rescued by the expression of HY5 with non-native promotors (Figure 3C and 3D and Supplemental Figure 5). Partial rescue of this phenotype was conferred by SUC2p:DOF-HY5 line number 12, which was expressed in the LR tip (Figure 3C and 3D); however, SUC2p:DOF-HY5 line number 14, which lacked expression in the LR tip, did not rescue the phenotype. Although SUC2 and HY5 promoters both drive expression in the same region of the LR tip, differences in their expression levels in the root may account for the different levels of rescue (Figure 2B and Supplemental Figure 3B). These results indicate that HY5 needs to be expressed locally in the LR tip during formation and elongation, which is consistent with previous findings (Lee et al., 2016, van Gelderen et al., 2018a).
Together with previous studies, these results suggest that HY5 regulates root growth via at least two mechanisms. Firstly, the HY5 protein can activate a signal in the shoot, which travels to the root where it directly regulates root growth or activates a root-specific signal, possibly including local transcription of HY5. Secondly, the HY5 protein can move from the shoot to the root where it will regulate target genes, along with its own promoter (Chen et al., 2016). In both cases, local HY5 expression is needed in the root to achieve the full rescue of the hy5 phenotype (Chen et al., 2016, Zhang et al., 2017, van Gelderen et al., 2018a). Further studies are needed to understand the differences between CAB3p:HY5-GFP and CAB3p:DOF-HY5, which affected HY5 mobility. We conclude that these differences were due to the different tags used. However, the mechanism of HY5 mobility at physiological levels in wild-type plants needs further investigation. Clarifying these issues will allow us to better understand the roles of mobile signals that coordinate growth between tissues.
Methods
Plant Materials, Growth Conditions, and Seedling Measurements
All Arabidopsis thaliana plants were of the Columbia (Col-0) background and grown in long-day conditions (16 h light, 8 h dark), unless otherwise indicated. The hy5 mutant, which carried an early stop codon (Q2∗), as well as HY5p:HY5-GFP hy5 and CAB3p:HY5-GFP hy5 lines, were prepared as previously described (Lian et al., 2011, Chen et al., 2016). Seeds were sterilized, stratified in the dark at 4°C, and germinated in the indicated light and temperature conditions on plates without sucrose: 0.5× Linsmaier and Skoog medium (Caisson Laboratories, Smithfield, UT, USA) with 0.8% phyta-agar (Caisson Laboratories), or with sucrose: 0.5× Murashige and Skoog medium (Caisson Laboratories) with 2.5 mM MES (Acros Organic, Hampton, NH, USA), 1% sucrose (Fisher Bioreagents, Hampton, NH, USA), and 0.8% agar (Caisson Laboratories), as indicated in the figures and in Supplemental Tables 1 and 2.
Assays to determine hypocotyl length, root length, and LR number were performed on seedlings grown vertically. Root images were acquired using a multiplex scanning system as described by Slovak et al. (2014). The root elongation rate was calculated by subtracting the length at 3 days after germination (3 DAG) from that at 6 DAG (Figure 2C and 2D) or subtracting the length at 4 DAG from that at 9 DAG (Figure 2G and 2F). Hypocotyl and root measurements were performed using NIH ImageJ software (Schindelin et al., 2012).
DNA Constructs and Plant Transformation
Vectors for transformation into plants were constructed using the multisite Gateway system (Invitrogen). HY5 was PCR amplified from cDNA using B2r-HY5_F and B3-HY5stop_R primers (Supplemental Table 2) and cloned into pDONR-P2RP3. CER6 and SUC2 promoters were prepared in pDONR-P4P1R as previously described (Procko et al., 2016). The CAB3 promoter was PCR amplified from genomic DNA using B4-CAB3p_F and B1r-CAB3p_R primers (Supplemental Table 2) (Procko et al., 2016). The DOF (HA-YFP-HA) tag in pDONR-P221 was prepared as previously described (Burger et al., 2017). CAB3p:DOF-HY5, CER6p:DOF-HY5, and SUC2p:DOF-HY5 were recombined into the destination vector pK7m34GW. hy5 mutant plants were transformed with Agrobacterium tumefaciens GV3101 using the floral dip method (Clough and Bent, 1998), and segregation analysis of antibiotic resistance was used to isolate single-insertion homozygous lines.
Western Blot Analysis
Western blots were performed on protein extracts from 25 roots and 20 shoots harvested at 6 DAG. Tissues were homogenized, and 2× loading buffer (18 μl of β-ME to 1 ml of 2× NuPAGE LDS sample buffer; NP0008, Thermo Fisher Scientific, Waltham, MA, USA) was added to roots (70 μl) and shoots (140 μl) and then boiled for 5 min. Proteins were separated on 4%–12% Bis-tris gels (Invitrogen, Carlsbad, CA, USA) and transferred onto membranes under semi-dry conditions (Pierce G2 Fast Blotter, Thermo Fisher Scientific). Membranes were incubated with antibodies against αHA-HRP (1:2000, 12013819001, Roche), αHY5(N) (1:5000, R1245-1b, ABiocode), and α-actin (1:70000, A0408, Sigma), followed by a secondary goat anti-mouse antibody (1:70000, 1706516, Bio-Rad) for α-actin and a goat anti-rabbit antibody (1:5000, 1706515, Bio-Rad) for αHY5(N). Actin was visualized by reprobing membranes.
Expression Analysis
For qRT–PCR, RNA was extracted using the RNeasy Micro kit (QIAGEN). cDNA was prepared using the Maxima First-Strand cDNA Synthesis kit (Thermo Fisher Scientific) with 1 μg of RNA. qRT–PCR analysis was carried out using the CFX384 Real-Time PCR Detection system (Bio-Rad), with Premix Ex Taq II (TaKaRa, #RR820A). mRNA levels were calculated relative to the internal control IPP2 as described by Shleizer-Burko et al. (2011). Details of the primers used for qRT–PCR are provided in Supplemental Table 3.
Microscopy and Imaging
Fluorescence images were acquired with a Zeiss LSM 710 confocal microscope. The YFP signal was detected at wavelengths of 514 nmEX and 519–562 nmEM. The GFP signal was detected at wavelengths of 488 nmEX and 502–554 nmEM. The chlorophyll signal was detected at wavelengths of 633 nmEX and 647–721 nmEM. Hypocotyl sections were prepared by embedding seedlings in 2% low-melting agarose (V2111, Promega). After 30–60 min, one section was generated with a razor blade (270100, Thermo Fisher Scientific), and the dissected block was placed on the coverslip.
Funding
The study was supported by grants from the National Institute of General Medical Sciences of the National Institutes of Health (5R35GM122604-02_05) and the Howard Hughes Medical Institute to J.C., a grant from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM127759) to W.B., and start-up funds from the Salk Institute for Biological Studies to W.B.. Y.B. was funded by an EMBO Fellowship (ALTF 785-2013) and BARD (FI-488-13).
Author Contributions
Conceptualization, Y.B. and C.G.; Methodology, Y.B., C.G., A.S., J.C., and W.B.; Investigation, Y.B. and C.G.; Writing – Original Draft, Y.B. and A.S.; Writing – Review and Editing, Y.B., C.G., A.S., J.C., and W.B.; Funding Acquisition, J.C. and W.B.; Supervision, J.C. and W.B..
Acknowledgments
We thank Dr. Xiangdong Fu for kindly providing the HY5p:HY5-GFP and CAB3p:HY5-GFP seeds. No conflicts of interest are declared.
Published: May 16, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Contributor Information
Joanne Chory, Email: chory@salk.edu.
Wolfgang Busch, Email: wbusch@salk.edu.
Supplemental Information
References
- Bellstaedt J., Trenner J., Lippmann R., Poeschl Y., Zhang X., Friml J., Quint M., Delker C. A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 2019;180:757–766. doi: 10.1104/pp.18.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao R.P., Eklöf J., Ljung K., Marchant A., Bennett M., Sandberg G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 2002;29:325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- Burger M., Willige B.C., Chory J. A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nat. Commun. 2017;8:2201. doi: 10.1038/s41467-017-02347-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y., Seluzicki A., Zander M., Pedmale U.V., Ecker J.R., Chory J. Chimeric activators and repressors define HY5 activity and reveal a light-regulated feedback mechanism. Plant Cell. 2020;32:967–983. doi: 10.1105/tpc.19.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yao Q., Gao X., Jiang C., Harberd N.P., Fu X. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 2016;26:640–646. doi: 10.1016/j.cub.2015.12.066. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coruzzi G., Bush D.R. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 2001;125:61–64. doi: 10.1104/pp.125.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Nakamura S., Araki T., Mochizuki N., Nagatani A. Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell. 2005;17:1941–1952. doi: 10.1105/tpc.105.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C., Burko Y., Platre M.P., Zhang L., Simura J., Kumar V., Ljung K., Chory J., Busch W. Shoot and root thermomorphogenesis are linked by a developmental trade-off. bioRxiv. 2020 doi: 10.1101/2020.05.07.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant. 2016;9:1353–1365. doi: 10.1016/j.molp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Kumar S.V. DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 2017;18:344–351. doi: 10.1016/j.celrep.2016.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyomarc’h S., Leran S., Auzon-Cape M., Perrine-Walker F., Lucas M., Laplaze L. Early development and gravitropic response of lateral roots in Arabidopsis thaliana. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:1509–1516. doi: 10.1098/rstb.2011.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J.H., Kim J.H., Kim S.G., Sim H.J., Lee G., Halitschke R., Baldwin I.T., Kim J.I., Park C.M. Shoot phytochrome B modulates reactive oxygen species homeostasis in roots via abscisic acid signaling in Arabidopsis. Plant J. 2018;94:790–798. doi: 10.1111/tpj.13902. [DOI] [PubMed] [Google Scholar]
- Hooker T.S., Millar A.A., Kunst L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 2002;129:1568–1580. doi: 10.1104/pp.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Song K., Park E., Kim K., Bae G., Choi G. Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously. Plant Cell. 2016;28:2770–2785. doi: 10.1105/tpc.16.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Schopfer P. Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2012;109:11217–11221. doi: 10.1073/pnas.1203746109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Ohnishi A., Sasaki D., Fujii S., Iwase A., Sugimoto K., Masuda T., Wada H. Shoot removal induces chloroplast development in roots via cytokinin signaling. Plant Physiol. 2017;173:2340–2355. doi: 10.1104/pp.16.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U., Briggs W.R. Growth, in vivo extensibility, and tissue tension in developing pea internodes. Plant Physiol. 1988;86:306–311. doi: 10.1104/pp.86.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera U., Niklas K.J. The epidermal-growth-control theory of stem elongation: an old and a new perspective. J. Plant Physiol. 2007;164:1395–1409. doi: 10.1016/j.jplph.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Ha J.H., Kim S.G., Choi H.K., Kim Z.H., Han Y.J., Kim J.I., Oh Y., Fragoso V., Shin K. Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci. Signal. 2016;9:ra106. doi: 10.1126/scisignal.aaf6530. [DOI] [PubMed] [Google Scholar]
- Li Q.F., He J.X. BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant. 2016;9:113–125. doi: 10.1016/j.molp.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.D., Zhou C.M., Xu P.B., Luo Q., Lian H.L., Yang H.Q. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant. 2015;8:467–478. doi: 10.1016/j.molp.2014.11.025. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paultre D.S.G., Gustin M.P., Molnar A., Oparka K.J. Lost in transit: long-distance trafficking and phloem unloading of protein signals in Arabidopsis homografts. Plant Cell. 2016;28:2016–2025. doi: 10.1105/tpc.16.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C., Burko Y., Jaillais Y., Ljung K., Long J.A., Chory J. The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 2016;30:1529–1541. doi: 10.1101/gad.283234.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C., Crenshaw C.M., Ljung K., Noel J.P., Chory J. Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 2014;165:1285–1301. doi: 10.1104/pp.114.241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A., Fiene G., Fackendahl P., Hoecker U. The Arabidopsis repressor of light signaling SPA1 acts in the phloem to regulate seedling de-etiolation, leaf expansion and flowering time. Development. 2011;138:1851–1862. doi: 10.1242/dev.061036. [DOI] [PubMed] [Google Scholar]
- Reed R.C., Brady S.R., Muday G.K. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 1998;118:1369–1378. doi: 10.1104/pp.118.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnault T., Daviere J.M., Wild M., Sakvarelidze-Achard L., Heintz D., Carrera Bergua E., Lopez Diaz I., Gong F., Hedden P., Achard P. The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat. Plants. 2015;1:15073. doi: 10.1038/nplants.2015.73. [DOI] [PubMed] [Google Scholar]
- Salisbury F.J., Hall A., Grierson C.S., Halliday K.J. Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 2007;50:429–438. doi: 10.1111/j.1365-313X.2007.03059.x. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199–202. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q.M., Yang X., Song L., Xue H.W. Arabidopsis MSBP1 is activated by HY5 and HYH and is involved in photomorphogenesis and brassinosteroid sensitivity regulation. Mol. Plant. 2011;4:1092–1104. doi: 10.1093/mp/ssr049. [DOI] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N. Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development. 2011;138:695–704. doi: 10.1242/dev.056770. [DOI] [PubMed] [Google Scholar]
- Sibout R., Sukumar P., Hettiarachchi C., Holm M., Muday G.K., Hardtke C.S. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2006;2:e202. doi: 10.1371/journal.pgen.0020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovak R., Goschl C., Su X., Shimotani K., Shiina T., Busch W. A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell. 2014;26:2390–2403. doi: 10.1105/tpc.114.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek R.E., Ausubel F.M., Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Tao Y., Ferrer J.L., Ljung K., Pojer F., Hong F., Long J.A., Li L., Moreno J.E., Bowman M.E., Ivans L.J. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Sauer N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- van Gelderen K., Kang C., Paalman R., Keuskamp D., Hayes S., Pierik R. Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell. 2018;30:101–116. doi: 10.1105/tpc.17.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K., Kang C., Pierik R. Light signaling, root development, and plasticity. Plant Physiol. 2018;176:1049–1060. doi: 10.1104/pp.17.01079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J.L., Hecht V., Vander Schoor J.K., Davidson S.E., Ross J.J. Light regulation of gibberellin biosynthesis in pea is mediated through the COP1/HY5 pathway. Plant Cell. 2009;21:800–813. doi: 10.1105/tpc.108.063628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li C., Zhang J., Wang J., Yang J., Lv Y., Yang N., Liu J., Wang X., Palfalvi G. Dissection of HY5/HYH expression in Arabidopsis reveals a root-autonomous HY5-mediated photomorphogenic pathway. PLoS One. 2017;12:e0180449. doi: 10.1371/journal.pone.0180449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.