TO THE EDITOR
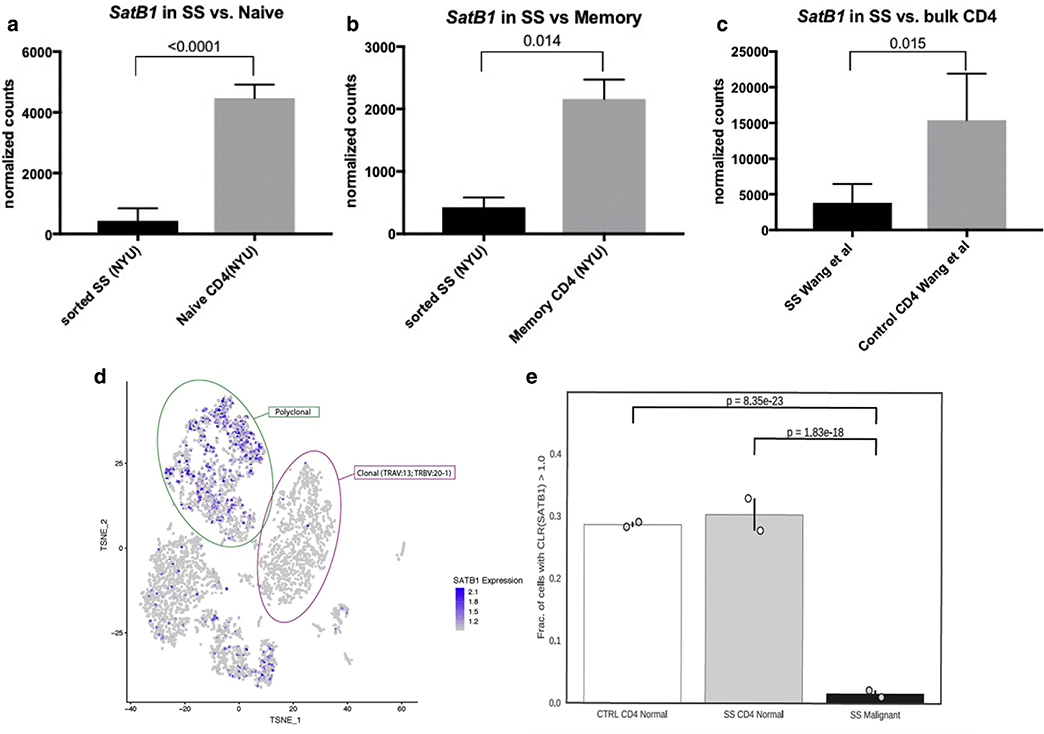
We read with interest the Letter to the Editor by Gagat et al.(2019) on the ambiguous role of special AT-rich sequence-binding protein-1 (SATB1) expression in malignant tumors. The authors make a strong point that miR-155/SATB1 axis contributes to apoptosis resistance not only in malignant T cells but is also critical in regulating cell death in a wide range of cell lines from hematologic malignancies to solid tumors. Among other findings, the authors reported that low SATB1 expression is associated with resistance to apoptosis and poor responses to UV treatment in mycosis fungoides (MF). Along with the observation that SATB1 repression by miR-155 promotes aberrant expression of cytokines and malignant growth factors (Fredholm et al., 2018), these new findings further suggest that SATB1 should be viewed as a tumor suppressor in the context of cutaneous T-cell lymphoma (CTCL) and may indeed prove to be a valuable prognostic marker in this malignancy (Fredholm et al., 2018; Grzanka et al., 2015; Poglio and Merlio, 2018). Indeed, lymphomatoid papulosis and other forms of indolent CTCL are associated with an increased expression of SATB1 (Sun et al., 2018), whereas Sézary syndrome (SS), an aggressive leukemic form of CTCL, is associated with low SATB1 expression (Wang et al., 2011). Our analysis of previously published data on SS (Fanok et al., 2018) showed low SATB1 expression in malignant T cells when compared with either naive or memory CD4+ T cells from healthy controls (Figure 1a and b). Malignant T cells from patients with SS express significantly lower levels of SATB1 when compared with both naive and memory T cells, indicating that this SATB1 downregulation is not due to intrinsic differences between memory and naive T-cell phenotypes. Analysis of another RNA sequencing dataset from a previously published large cohort of patients further validated our observation that malignant SS cells are characterized by low SATB1 expression (Wang et al., 2015) (Figure 1c). As the malignant T-cell population is known to display a large degree of heterogeneity at a single-cell level (Buus et al., 2018), we performed Expanded CRISPR-compatible Cellular Indexing of Transcriptomes and Epitopes by sequencing on peripheral blood mononuclear cells from 2 patients with SS. This expanded Cellular Indexing of Transcriptomes and Epitopes by sequencing approach enabled us to simultaneously examine the transcriptome, surface phenotype, and T-cell receptor (TCR) rearrangement of individual cells without any presorting (Mimitou et al., 2019). Using this approach, we were able to interrogate the transcriptome of individual malignant T-cell clones, defined by the singular clonality of their TCR (complementarity-determining region 3 sequence of TCRα and TCRβ), and compare their transcriptional level of SATB1 with that of the polyclonal non-malignant T cells from the blood of the same patient and healthy controls. Zeroing in on clonal T cells also allowed us to study the gene expression specific to malignant cells at a far greater degree of confidence than what is possible when relying on surface markers. In these particular patients with SS, the clonal malignant CD4+ T cells appeared in distinct clusters from polyclonal non-malignant CD4+ T cells on the t-distributed stochastic neighbor embedding dimensional reduction plots (Figure 1d). Notably, malignant T cells in these 2 patients were almost exclusively negative for SATB1, whereas SATB1 was robustly expressed in polyclonal, non-malignant CD4+ T cells from the same individuals and in T-helper lymphocytes from the blood of 2 healthy individuals (Figure 1e).
Figure 1. Malignant T cells display low SATB1 expression.
(a, b) Bar graphs: Normalized counts from bulk RNA sequencing analysis comparing sorted (a) naive Th versus malignant SS cells and (b) memory T-helper versus malignant SS cells. T-helper naive cells (n = 3) and memory T cells (n = 4) were sorted from healthy individuals, and malignant cells (n = 7) were sorted based on expression of CD3 and CD4 and lack of or low expression of CD7 and CD26 in malignant T cells as described in Fanok et al., 2018. (c) Bar graphs showing relative expression from bulk sorted and sequenced CD4+ Th cells and SS cells from Wang et al., 2015.(d) Representative t-SNE plot showing transcriptome-based clustering and levels of SATB1 expression (blue) in PBMCs from a patient with SS. Clonal malignant CD4+ T cells (1352) expressing TRAV 35: CAGQLRNAGGTSYGKLTF and TRBV 20–1: CSARFLRGGYNEQFF are encased in pink, whereas polyclonal non-malignant CD4+ T cells (1025) are encased in green. (e) Scatter plot showing frequency of clonal malignant (black) and polyclonal non-malignant (grey) CD4+ T cells from 2 patients with SS and CD4+ T cells (white) from 2 healthy individuals with normalized SATB1 expression > 1.0. A total of 29 of 1352 and 18 of 1730 total malignant cells had SATB1 expression > 1.0, whereas 337 of 1025 and 357 of 1288 non-malignant cells and 308 of 1089 and 387 of 1331 total CD4+ T cells from 2 healthy individuals were above that threshold. CTRL, control; PBMC, peripheral blood mononuclear cells; SATB1, special AT-rich sequence-binding protein-1; SS, Sézary syndrome; Th, T helper; t-SNE, t-distributed stochastic neighbor embedding.
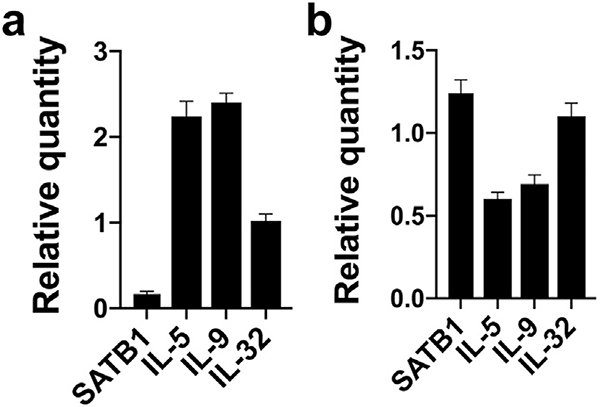
In addition, we found that a malignant SS T-cell line SeAx also displays low but detectable levels of SATB1 when compared with non-malignant CD4+ T-cell lines (unpublished data). Because Sézary syndrome is associated with increased Th2 cytokines and SATB1 inhibits expression of IL-5 and IL-9 in MF cells, we examined whether even a low level of SATB1 had an impact on the expression of cytokines in SeAx cells. Small interfering RNA-mediated knock down of SATB1 triggered a selective increase in IL-5 and IL-9 expression without affecting IL-32 expression (Figure 2a), indicating that even a residual, low expression of SATB1 modulates cytokine expression. This is consistent with previous findings that SATB1 can act as a brake on cytokine expression in both healthy and malignant T cells. Because SATB1 is downregulated by miR-155, a micro-RNA highly expressed in malignant T cells, we examined whether silencing of miR-155 enhanced SATB1 expression and decreased cytokine expression in SeAx cells. Indeed, inhibition of miR-155 triggered an increased expression of SATB1 and a concomitant decrease in IL-5 and IL-9 expression, whereas the expression of IL-32 remained unchanged (Figure 2b). These findings indicate that miR-155, via inhibition of SATB1, is involved in the regulation of cytokine expression in SS cells, as previously reported in MF (Fredholm et al., 2018). However, since miR-155 inhibition had only a modest effect on SATB1 expression, it is likely that other mechanisms are also involved in the repression of SATB1. Li et al. (2018) reported a deficient expression in malignant T cells of SNF5, a core member of the SWI/SNF chromatin-remodeling complex, which is an upstream regulator of SATB1. Notably, forced expression of SNF5 triggered SATB1 expression and apoptosis in SS cells (Li et al., 2018) supporting the hypothesis that malignant transformation of T cells involves multiple pathways that target and silence SATB1.
Figure 2. Inhibition of SATB1 promotes expression of IL-5 and IL-9.
The malignant SS cell line SeAx was treated with (a) siRNA against SATB1 or non-target control or (b) antagomir-155 or antagomir control. RNA was purified 48 hours after treatment, and gene expression was analyzed by qPCR with GAPDH as control. Results are shown as relative values to expression in non-target controls arbitrarily defined as 1. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qPCR, quantitative reverse transcriptase in real time; SATB1, special AT-rich sequence-binding protein-1; SS, Sézary syndrome; siRNA, small interfering RNA.
It is worth noting that in their recent Letter to the Editor Gao et al. (2019) highlight the “multifaceted” role of SATB1 in discrete entities and stages of CTCL emphasizing that it is found to be highly expressed in some subsets (MF, primary anaplastic large T-cell lymphoma and breast implant-associated anaplastic large-cell lymphoma) while being downregulated in SS. We agree that the expression and possible downstream effects of SATB1 in hematologic malignancies and solid tumors are dependent on the context of the tumor, the cell of origin, and epigenetic landscapes. Furthermore, since TCR activation can affect SATB1 expression (Stephen et al., 2017), and TCR signaling capacity and TCR expression are known to be variable in CTCL malignancies, this may be an additional important factor in the modulation of SATB1 expression and its downstream effects. We propose that SATB1 expression in malignant T cells impacts multiple aspects of the malignant transformation and disease progression through direct effects on the malignant T cell per se (such as resistance to apoptosis and enhanced proliferation) and indirect effects involving cytokine release and modulation of the tumor environment. Several lines of evidence indicate that malignant T cells—via crosstalk with non-malignant T cells and stromal cells—orchestrate a special inflammatory microenvironment (Krejsgaard et al., 2017). Since repression of SATB1 promotes survival, proliferation, and “malignant inflammation,” this pathway represents an intriguing target and should be further investigated as a possible biomarker of disease progression in CTCL.
ACKNOWLEDGMENTS
Work in Dr. Ødum’s laboratory was supported by the LEO Foundation and grants from The Novo Nordisk Research Foundation (NNF14OC0012345), the Danish Cancer Society (Kræftens Bekæmpelse), the Fight Cancer Program (Knæk Cancer), and The Lundbeck Foundation.
Work in Dr. Koralov’s laboratory was supported by the National Institutes of Health R01 grant (HL-125816), the Judith and Stewart Colton Center for Autoimmunity Pilot grant, and a grant from the Drs. Martin and Dorothy Spatz Foundation. Angelina Seffens is supported by Howard Hughes Medical Institute Medical Scholar Fellowship.
Abbreviations
- CTCL
cutaneous T-cell lymphoma
- MF
mycosis fungoides
- SATB1
special AT-rich sequence-binding protein-1
- SS
Sézary syndrome
- TCR
T-Cell receptor
Footnotes
CONFLICT OF INTEREST
Peter Smibert is an author on a patent disclosing the Cellular Indexing of Transcriptomes and Epitopes by sequencing methods used in the study. The rest of the authors state no conflict of interest.
Data availability statement
Datasets related to this article have been deposited in the Gene Expression Omnibus with accession number GSE126310 with further data submission pending. Additional sample information can be obtained by contacting Koralov laboratory.
REFERENCES
- Buus TB, Willerslev-Olsen A, Fredholm S, Blümel E, Nastasi C, Gluud M, et al. Single-cell heterogeneity in Sezary syndrome. Blood Adv 2018;2:2115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanok MH, Sun A, Fogli LK, Narendran V, Eckstein M, Kannan K, et al. Role of dysregulated cytokine signaling and bacterial triggers in the pathogenesis of cutaneous T-cell lymphoma. J Invest Dermatol 2018;138:1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredholm S, Willerslev-Olsen A, Met Ö, Kubat L, Gluud M, Mathiasen SL, et al. SATB1 in malignant T cells. J Invest Dermatol 2018;138:1805–15. [DOI] [PubMed] [Google Scholar]
- Gagat M, Grzanka D, Krajewski A. Ambiguous role of SATB1 expression in malignant tumors. J Invest Dermatol 2019;139:1608–10. [DOI] [PubMed] [Google Scholar]
- Gao Y, Sun J, Yi S, Tu P, Wang Y. Response to the commentary “Ambiguous role of SATB1 expression in malignant tumors”. J Invest Dermatol 2019;139:1611–2.30876801 [Google Scholar]
- Grzanka D, Gagat M, Izdebska M, Marszalek A. Expression of special AT-rich sequence-binding protein 1 is an independent prognostic factor in cutaneous T-cell lymphoma. Oncol Rep 2015;33:250–66. [DOI] [PubMed] [Google Scholar]
- Krejsgaard T, Lindahl LM, Mongan NP, Wasik MA, Litvinov IV, Iversen L, et al. Malignant inflammation in cutaneous T-cell lymphoma-a hostile takeover. Semin Immunopathol 2017;39:269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang J, Yu M, Wang Y, Zhang H, Yin J, et al. SNF5 deficiency induces apoptosis resistance by repressing SATB1 expression in Sezary syndrome. Leuk Lymphoma 2018;59:2405–13. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Cheng A, Montalbano A, Hao S, Stoeckius M, Legut M, et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 2019;16:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poglio S, Merlio JP. SATB1 is a pivotal epigenetic biomarker in cutaneous T-cell lymphomas. J Invest Dermatol 2018;138:1694–6. [DOI] [PubMed] [Google Scholar]
- Stephen TL, Payne KK, Chaurio RA, Allegrezza MJ, Zhu H, Perez-Sanz J, et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity 2017;46:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yi S, Qiu L, Fu W, Wang A, Liu F, et al. SATB1 defines a subtype of cutaneous CD30(+) lymphoproliferative disorders associated with a T-helper 17 cytokine profile. J Invest Dermatol 2018;138:1795–804. [DOI] [PubMed] [Google Scholar]
- Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet 2015;47:1426–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su M, Zhou LL, Tu P, Zhang X, Jiang X, et al. Deficiency of SATB1 expression in Sezary cells causes apoptosis resistance by regulating FASL/CD95L transcription. Blood 2011;117:3826–35. [DOI] [PubMed] [Google Scholar]