Abstract
Cajal bodies (CBs) are subnuclear domains involved in the formation of ribonucleoproteins (RNPs) including small nuclear RNPs (snRNPs). CBs associate with specific gene loci, which impacts expression and provides a platform for the biogenesis of the nascent transcripts emanating from these genes. Here we report that CBs can associate with the C19MC microRNA (miRNA) gene cluster, which suggests a role for CBs in the biogenesis of animal miRNAs. The machinery involved in the formation of miRNAs includes the Drosha/DGCR8 complex, which processes primary-miRNA to precursor miRNA. Further processing of precursor miRNA by Dicer and other components generates mature miRNA. To test if CBs influence the expression and formation of miRNAs, we examined two representative miRNAs (miR-520 h and let-7a) in conditions that disrupt CBs. CB disruption correlates with alterations in the level of primary and mature miRNA and the let-7a mRNA target, HMGA2. We have also found that the processing of some small CB-specific RNAs (scaRNAs) is directly mediated by the Drosha/DGCR8 complex. ScaRNAs form scaRNPs, which play an important role in snRNP formation. Collectively, our results demonstrate that CBs and the miRNA processing machinery functionally interact and together contribute to the biogenesis of miRNAs and snRNPs.
INTRODUCTION
Drosha and DGCR8 are well-characterized components of the microRNA (miRNA) processing pathway that produce precursor-mRNA from primary-miRNA (Bernstein et al., 2001; Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001; Lee et al., 2003; Denli et al., 2004; Zeng et al., 2005). Interestingly, stem/loop fragments derived from certain scaRNAs (small Cajal body [CB]-specific RNAs) are approximately the same size (70–80 nt) (Tycowski et al., 2004) as precursor-miRNAs generated by Drosha/DGCR8, suggesting that some scaRNAs may be unorthodox substrates for the miRNA processing machinery. We have published data in support of this hypothesis (Logan et al., 2018). CBs are subnuclear domains involved in small nuclear ribonucleoprotein (snRNP) biogenesis (Darzacq et al., 2002; Kiss, 2004). Modifications to the snRNA component of snRNPs are required for proper snRNP and spliceosomal function (Yu et al., 1998). These modifcations are guided by scaRNAs as part of scaRNPs in CBs (Darzacq et al., 2002; Kiss, 2004). In cell types lacking CBs, modification of snRNA takes place in the nucleoplasm (Liu et al., 2009; Makarova and Kramerov, 2011; Deryusheva et al., 2012; Marnef et al., 2014). In addition to RNP formation, CBs physically associate with many different genes and gene clusters (Frey et al., 1999). This CB/gene association impacts expression and biogenesis of the nascent transcripts (Frey et al., 1999; Sawyer et al., 2016; Wang et al., 2016). No studies have shown that CBs associate with miRNA gene clusters. In the human genome, there are 153 gene clusters encoding 468 miRNAs (Kabekkodu et al., 2018). One of these miRNA gene clusters is C19MC (Bortolin-Cavaille et al., 2009). C19MC is expressed primarily in placental tissue and is maternally imprinted (Noguer-Dance et al., 2010; Bellemer et al., 2012). Using a placental line (JEG-3), RNA fluorescence in situ hybridization (FISH) showed that Drosha and DGCR8 colocalize with the primary-miRNA transcript at the paternally expressed C19MC locus (Bellemer et al., 2012). We hypothesize that CBs associate with miRNA gene clusters including C19MC, and this association influences miRNA biogenesis. We further hypothesize that CBs and the miRNA processing machinery functionally interact, with each of these entities contributing to the activity of the other.
We report here that CBs associate with the C19MC miRNA gene cluster and disruption of CBs alters the amount of a representative C19MC miRNA, miR-520 h. Another miRNA encoded in clusters is let-7a, and we have found that CB alteration influences the level of primary-let-7a, mature let-7a, and a major let-7a target mRNA, HMGA2. These findings support a role for CBs in miRNA expression and biogenesis. Disruption of CBs was accomplished by reduction of coilin, the CB marker protein (Tucker et al., 2001; Strzelecka et al., 2010); SMN, the survival of motor neuron protein (Lorson and Androphy, 1998; Bertrandy et al., 1999; Hebert et al., 2001; Jones et al., 2001; Pellizzoni et al., 2001; Mahmoudi et al., 2010; Broome and Hebert, 2012; Enwerem et al., 2014; Machyna et al., 2014); TDP-43, an RNA-binding protein that targets certain scaRNAs to CBs (Izumikawa et al., 2019); and WRAP53, which plays a role in scaRNP and telomerase formation (Richard et al., 2003; Tycowski et al., 2009; Venteicher et al., 2009). Importantly, alteration of miRNA levels is differentially impacted depending on which protein enriched in the CB is reduced. We have also found that the Drosha/DGCR8 complex can directly process a specific scaRNA, providing evidence in support of a regulatory pathway in which snRNP formation/activity is governed by miRNA component processing of scaRNAs. In summary, our findings indicate that CBs and the miRNA processing machinery synergistically interact and together contribute to miRNA and snRNP biogenesis.
RESULTS AND DISCUSSION
CB association with the expressed C19MC locus
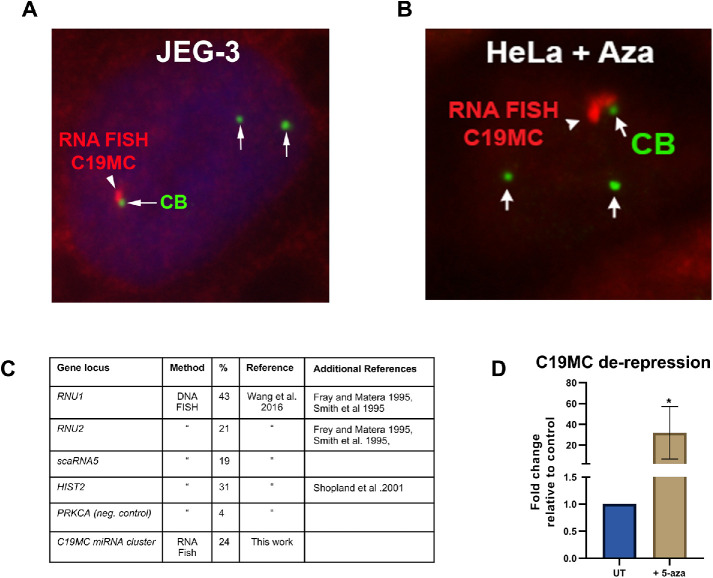
To examine if CBs associate with miRNA gene clusters, we used JEG-3 cells and conducted RNA FISH to detect primary-miRNA from the C19MC locus and immunofluorescence (IF) to detect CBs. The association of CBs with the transcriptionally active C19MC cluster was observed in 24% of cells, a value equivalent to known CB association with other gene loci (Figure 1). Since previous work has shown that both Drosha and DGCR8 colocalize with the primary-miRNA transcript at the expressed C19MC locus (Bellemer et al., 2012), we costained JEG-3 cells for DGCR8 and coilin (or a coilin-GFP construct). As shown in Supplemental Figure S1, associations of CBs with the expressed C19MC locus (as demarcated by DGCR8) are detected. Prior work has shown that expression of the C19MC cluster in nonplacental cells can be induced epigenetically with 5-aza-2′-deoxycytidine (5-aza, a DNA methylation inhibitor; Bellemer et al., 2012). We treated HeLa cells with 5-aza and verified that the C19MC cluster was de-repressed using qRT-PCR (Figure 1). We then conducted RNA FISH for the C19MC primary-miRNA transcript and IF to detect CBs. We observe associations of CBs with the C19MC cluster in 5–aza-treated HeLa cells as found in JEG-3 cells (Figure 1). These findings demonstrate that CBs can associate with a miRNA gene cluster and thus may play a role in the global expression of all miRNAs encoded in gene clusters (Kabekkodu et al., 2018).
FIGURE 1:
CBs associate with the transcriptionally active C19MC miRNA gene cluster. (A) RNA FISH and IF were used to detect the C19MC primary-miRNA transcript (red) and CBs (green, anti-SMN) in JEG-3 cells. (B) De-repression of the C19MC locus in HeLa cells by 5-aza). RNA FISH and IF were used to detect the C19MC primary-miRNA transcript (red) and CBs (green, anti-coilin). (C) Table of CB association frequency with selected gene loci (Frey and Matera, 1995; Smith et al., 1995; Shopland et al., 2001; Wang et al., 2016). (D) Histogram of C19MC de-repression in HeLa cells treated with 2.5 μM 5-aza for 72 h detected by qRT-PCR: n = 6 biological repeats; *p < 0.05, error bar represents SD. The C19MC primary transcript was amplified using primers that bind exons. GAPDH was used as the normalizer and data is shown with untreated set to 1.
Dysregulation of miR-520 h levels when CBs are disrupted
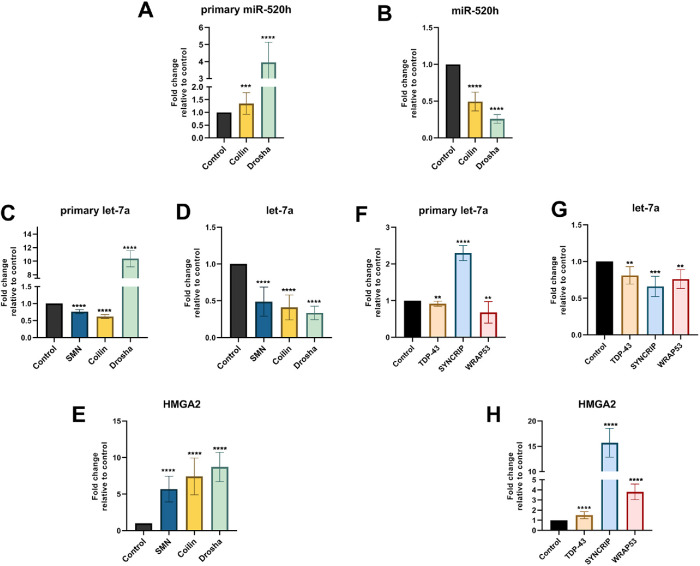
To generate additional evidence supporting a functional relationship between CBs and miRNA gene clusters, we investigated if CB disruption impacts the level of representative miRNAs. We first examined miR-520 h, which is one of the 46 miRNAs encoded by the C19MC miRNA gene cluster. For this analysis, JEG-3 cells were treated with control, coilin, or Drosha siRNA for 5 d followed by detection of primary and mature miR-520 h. Drosha knockdown does not impact CBs, but is a positive control for altered miRNA processing. Coilin knockdown abolishes canonical CBs (Lemm et al., 2006). As shown in Figure 2A, the relative level of primary-miR-520 h was slightly, but significantly, increased by coilin reduction compared with control. As expected, Drosha reduction results in a more dramatic increase in primary-miR-520 h. Analysis of mature miR-520 h shows that both coilin and Drosha knockdown result in the decrease of mature-miR-520 h (Figure 2B). These findings show that disruption of CBs as a consequence of coilin knockdown can impact the levels of a miRNA.
FIGURE 2:
Knockdown of CB-enriched proteins disrupts miRNA biogenesis. JEG-3 (A, B) and HeLa (C–H) cells were transfected with siRNA for 120 h followed by RNA isolation and qRT-PCR to detect primary-miR-520 h (A), mature-miR-520 h (B), primary-let-7a (C, F), mature-let7a (D, G), and HMGA2 mRNA (E, H). The relative amount of a given amplicon in the control siRNA condition was set to 1. Data were derived from multiple biological (at least three) and technical (at least two) repeats, and error bars denote SD. ****p < 0.0001, ***p < 0.001, **p < 0.005.
The level of miRNA let-7a and its target mRNA, HMGA2, are altered on CB protein reduction
Let-7a is part of the well-characterized let-7 miRNA family and can be encoded in clusters (Reinhart et al., 2000; Lee et al., 2016). A major target for let-7a is HMGA2 mRNA (Lee and Dutta, 2007). To examine if CB disruption impacts let-7a and HMGA2 levels, HeLa cells were treated for 5 d with control, coilin, or SMN siRNA. SMN knockdown has been shown to alter CBs (Lemm et al., 2006). Drosha knockdown served as a positive control for the detection of 1) increased primary-let-7a, 2) decreased mature-let-7a, and 3) increased HGMA2 mRNA. This was observed (Figure 2, C–E). Unlike Drosha knockdown, reduction of SMN or coilin results in a decrease of primary-let-7a (Figure 2C), suggesting that the disruption of CBs in HeLa cells affects miRNA formation upstream of the cropping step performed by Drosha/DGCR8. Similar to Drosha knockdown, however, reduction of SMN or coilin decreases the levels of mature let-7a (Figure 2D). Hence disruption of CBs, accomplished by the reduction of SMN or coilin, is correlated with decreased levels of let-7a. We next examined the level of HMGA2. As expected, Drosha knockdown results in increased levels of HMGA2 mRNA (Figure 2E). Significant increases in the level of HMGA2 mRNA were also observed in cells with reduced SMN or coilin (Figure 2E). The reduction of SMN and coilin (CB proteins) therefore impacts a well-characterized miRNA (let-7a) and alters the level of the main target for this miRNA, HMGA2 mRNA. We note that the 5-d knockdown conditions utilized here in HeLa cells did not significantly decrease snRNP resources, as evidenced by no significant changes in the amount of endogenous β-actin pre-mRNA (Supplemental Figure S2). We also did not detect a uniform increase in the amount of 5 different pre-mRNAs encoding intronic miRNAs (Supplemental Figure S2). In fact, coilin reduction results in a decrease in amplicons for three of the five pre-mRNAs investigated. One exception is the miR-15b-16-2 amplicon, which shows that SMN KD slightly increases the amount of this pre-mRNA (Supplemental Figure S2). Collectively, therefore, this analysis shows that the CB KD conditions used here do not drastically reduce splicing resources such that the splicing of β-actin pre-mRNA and the pre-mRNA of five host genes encoding miRNAs is uniformly reduced. Very interestingly, the release of intron-encoded miRNAs by Drosha/DGCR8 does not require intron excision by splicing before cropping takes place (Kim and Kim, 2007). Thus it is possible that even if the conditions we use here reduce the splicing of introns containing miRNAs, the release of these miRNAs by Drosha/DGCR8 would not be affected. Disrupted splicing, however, would be expected to decrease the production of intron-containing mirtrons which are generated by a Drosha-independent pathway. To further support a role for the CB in miRNA biogenesis, we monitored the level of primary-let-7a and mature-let-7a after 72 h of SMN or coilin KD. It is not expected that splicing resources would be significantly reduced with three days of disrupted CBs. As shown in Supplemental Figure S2 (G and H), although primary-let-7a levels are not significantly altered, we do detect a decrease in the amount of mature-let-7a with 72 h coilin KD. Our studies of miR-520 h and let-7a thus support the hypothesis that CBs regulate miRNA expression.
Although SMN and coilin are well-characterized proteins that are enriched in the CB, other components present in CBs, TDP-43, and WRAP53, may also impact miRNA biogenesis. To test this, HeLa cells were treated for 5 d with control, TDP-43, or WRAP53 siRNA, followed by assessment of primary-let7a, mature-let-7a, and HMGA2 mRNA levels. Typical knockdown efficiencies are shown in Supplemental Figures S3 and S4. To serve as a positive control, cells were also treated with siRNAs to SYNCRIP (Synaptotagmin Binding Cytoplasmic RNA Interacting Protein), a participant in primary-miRNA processing (Chen et al., 2020). Although previous work observed that SYNCRIP reduction does not alter primary let-7a levels (Chen et al., 2020), we have found that SYNCRIP knockdown increases primary-let7a (Figure 2F). One possible explanation for this discrepancy is that we conducted knockdowns for 5 d whereas the previous report reduced SYNCRIP for 2 d (Chen et al., 2020). However, in agreement with previous work, we observe that mature-let-7a levels are decreased on SYNCRIP knockdown (Figure 2G) and HMGA2 mRNA is correspondingly increased (Figure 2H). Unlike SYNCRIP and Drosha, reduction of TDP-43 and WRAP53 results in a similarly altered profile for let-7a and HMGA2 RNA as observed for SMN and coilin knockdown. Specifically, knockdown of TDP-43 and WRAP53 decreases primary let-7a and mature let-7a levels and increases HMGA2 mRNA levels (Figure 2, F–H). Importantly, however, the magnitude of these changes varies depending on which protein enriched in the CB is reduced. In summary, we have identified that components of the CB contribute to the biogenesis of two different miRNAs (miR-520 h and let-7a) encoded by two different miRNA gene clusters. It will be of interest to determine the points of activity along the miRNA biogenesis pathway for SMN, coilin, TDP-43, and WRAP53 and assess how these components interact with and/or impact the function of the Drosha/DGCR8 complex and the SYNCRIP/Drosha/DGCR8 complex.
Altered processing of ectopic scaRNA9 on TDP-43 and SYNCRIP reduction
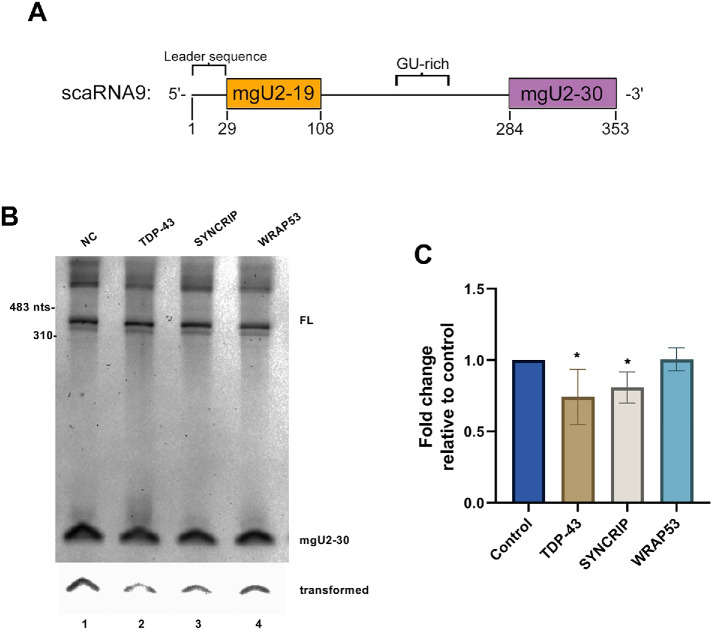
Three scaRNAs, scaRNA2, scaRNA9, and scaRNA17, have been shown to be processed (Tycowski et al., 2004), resulting in the generation of fragments that are approximately the same size as precursor-miRNA generated by Drosha/DGCR8. One of these scaRNAs, scaRNA9, generates two different fragments: mgU2-19 and mgU2-30 (Tycowski et al., 2004) (Figure 3A). The nomenclature of these fragments reflects their target site of ribose methylation. Namely, mgU2-19 serves as a methylation guide for nucleotide 19 in U2 snRNA and mgU2-30 guides the ribose methylation of U2 snRNA at nucleotide 30 (Tycowski et al., 2004). We have published that the processing of primary-scaRNA9 is reduced on SMN or Drosha knockdown (Logan et al., 2018), which suggests that components of the CB and the miRNA processing machinery together contribute toward the processing of specific primary-scaRNAs. To extend these studies, we evaluated the level of the mgU2-30 fragment generated from ectopically expressed scaRNA9 by Northern blotting in conditions with reduced levels of TDP-43, SYNCRIP, or WRAP53 (Figure 3B). Quantification of the data in Figure 3B, and other experiments, is shown in Figure 3C. For this quantification, the mg-U2 signal for a given condition was divided by the full-length scaRNA9 signal in that exact lane. This value is then normalized to that obtained with control siRNA. From this analysis, we have found that TDP-43 knockdown reduces the mgU2-30 fragment/full-length scaRNA9 ratio relative to that obtained with control siRNA by approximately 25%. Additionally, knockdown of SYNCRIP, which has not previously been shown to impact CB function, likewise results in a slight decrease in the relative amount of the mgU2-30 fragment derived from primary-scaRNA9. In contrast, WRAP53 knockdown does not result in a significant change in the amount of the mgU2-30 fragment derived from ectopically expressed primary-scaRNA9. These findings demonstrate that CB proteins differentially impact the processing of a specific primary-scaRNA, and SYNCRIP, which is part of the miRNA biogenesis machinery, contributes toward scaRNA fragment generation as does Drosha (Logan et al., 2018).
FIGURE 3:
Altered processing of ectopic scaRNA9 on TDP-43 or SYNCRIP knockdown. (A) Schematic of full-length scaRNA9 annotating the mgU2-19 and mgU2-30 domains and other sequence elements (Enwerem et al., 2015; Poole et al., 2016, 2017). (B) Detection of the mgU2-30 fragment and full-length (FL) scaRNA9 by Northern blotting. RNA was isolated from cells transfected with siRNA then subsequently transfected with plasmid encoding scaRNA9. The bottom panel is a transformed section of the top panel containing the mgU2-30 signal. (C) Histogram generated from the quantification of the data shown in B and other experiments. For each condition, the mgU2-30 signal was divided by the FL scaRNA9 signal in that lane and normalized to that obtained with control siRNA. N = 3 biological repeats, *p < 0.05, error bars represent SD.
ScaRNAs as noncanonical substrates for Drosha/DGCR8
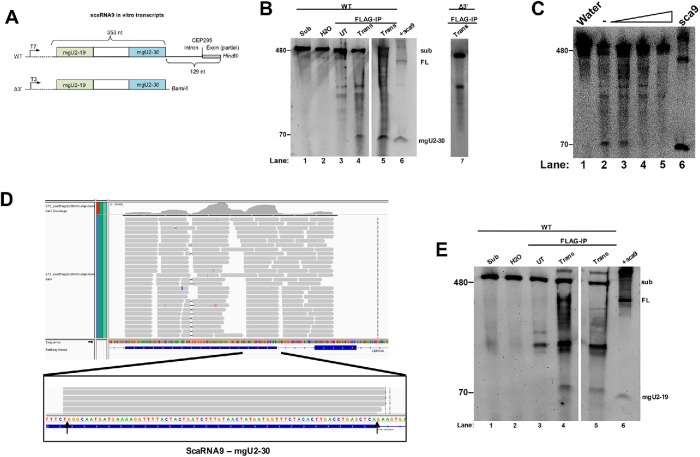
Our previously published data (Logan et al., 2018), and that shown in Figure 3, support a role for Drosha and SYNCRIP in the processing of primary-scaRNA. To directly show that the Drosha/DGCR8 complex can process scaRNAs, we conducted in vitro processing assays using two different in vitro transcribed scaRNA9 transcripts. The wild-type (WT) scaRNA9 transcript contains the entire scaRNA9 353 nt sequence, plus an additional 129 nt downstream of the mgU2-30 domain. The scaRNA9 Δ3′ transcript does not contain this downstream sequence (Figure 4A). These two transcripts served as substrates in the Drosha in vitro processing assay, as previously described (Lee and Kim, 2007). Briefly, cells were cotransfected with myc-Drosha and FLAG-DGCR8 plasmids, followed by immunoprecipitation and complex capture with anti-FLAG beads. The beads were then incubated with transcripts followed by RNA isolation and processing was detected by Northern blotting using a probe that hybridizes to the mgU2-30 fragment of scaRNA9. If the Drosha/DGCR8 complex can process scaRNA9, we expect that a fragment corresponding in size to the mgU2-30 fragment (69 nt) would be detected after incubation with the Drosha/DGCR8 beads. This is what we have observed (Figure 4B). Incubation of the WT scaRNA9 substrate with water or FLAG beads from untransfected cells does not generate an appropriately sized fragment (lanes 2 and 3). However, WT scaRNA9 substrate incubated with FLAG-beads from Drosha/DGCR8 transfected-lysate generates a fragment approximately the same size as the mgU2-30 fragment (compare the 70 nt band in lanes 4 and 5 to that in lane 6). Lane 6 contains RNA isolated from cells transfected with scaRNA9 plasmid. Hence, in vitro processing of WT scaRNA9 transcript by Drosha/DGCR8 generates a fragment that is approximately the same size as the mgU2-30 fragment detected in cellular RNA. Since we are detecting these fragments by Northern blotting, the sequence of the fragment generated in vitro by Drosha/DGCR8 must be highly similar to the mgU2-30 fragment generated by cells. A fragment similar in size to mgU2-30 was not detected when reactions were conducted using Drosha/DGCR8 FLAG beads with the scaRNA9 Δ3′ substrate (lane 7). Thus sequences downstream of the end of scaRNA9 contain important cis element determinants for Drosha/DGCR8-mediated processing of scaRNA9. Previous reports have also emphasized the requirement for upstream and downstream cis elements in Drosha/DGCR8 processing (Lee and Kim, 2007), and we note that the scaRNA9 Δ3′ substrate lacks the CNNC element present in WT primary-scaRNA9. As an additional control, we set up processing reactions with Drosha/DGCR8 FLAG beads using the WT scaRNA9 transcript as described above, but with increasing salt concentration. Salt addition decreased the amount of processing, demonstrating that the mgU2-30 fragment is enzymatically generated by Drosha/DGCR8 (Figure 4C, compare the amount of 70 nt fragment present in lanes 4 and 5 to that in lane 2. Lane 6 contains RNA from cells transfected with scaRNA9 DNA).
FIGURE 4:
In vitro processing of primary-scaRNA9 by Drosha/DGCR8. (A) WT and Δ3’ scaRNA in vitro transcribed substrates. (B) Northern blot of RNA isolated from the in vitro Drosha/DGCR8 processing assays. FL scaRNA9 and the mgU2-30 fragment were detected with a probe that hybridizes to the mgU2-30 region. Substrate was incubated with FLAG beads from Drosha/FLAG-DGCR8 transfected lysate (Trans, lanes 4, 5, and 7). Control reactions include substrate (lane 1), substrate incubated with water alone (lane 2), substrate incubated with FLAG beads from nontransfected cell lysate (lane 3), and a positive control of ectopically expressed scaRNA9 RNA (lane 6). (C) Processing assay with Drosha/DGCR8 FLAG beads using WT scaRNA9 substrate and increasing amounts of salt (lanes 3–5). Ectopically expressed scaRNA9 RNA is shown in lane 6. (D) RNA sequencing reads of in vitro Drosha processed scaRNA9. Arrows indicate the 5′ and 3′ ends of mgU2-30 as described previously (Tycowski et al., 2004). (E) Northern blot of RNA generated from in vitro Drosha processing assay and probed with a scaRNA9 mgU2-19 probe. A band around 79 nts appears in lanes 4 and 5, consistent with the mgU2-19 fragment. This gel was organized as described in B.
To unambiguously demonstrate that Drosha/DGCR8 can generate the scaRNA9 mgU2-30 fragment in vitro, we conducted a Drosha/DGCR8 processing reaction using the WT scaRNA9 substrate followed by gel electrophoresis, band excision of the 70 nt region, RNA isolation, and RNA sequencing. As shown in Figure 4D, numerous reads spanning scaRNA9 were obtained, but many reads demarcate the mgU2-30 region. In fact, RNA sequencing shows that the 5′ and 3′ ends of the mgU2-30 fragment generated by in vitro Drosha/DGCR8 processing closely align with that described previously (Figure 4D, bottom panel, arrows) (Tycowski et al., 2004). These findings prove that Drosha/DGCR8 can process scaRNA9, and recapitulate in vitro that observed in cells. Moreover, we have also found by in vitro processing assays that Drosha/DGCR8 can generate the mgU2-19 fragment of scaRNA9 (Figure 4E). Thus Drosha/DGCR8 can, in vitro, generate the mgU2-30 and mgU2-19 fragments from primary-scaRNA9.
Functional interaction between CBs and the miRNA processing machinery
The data presented in this report describe, for the first time, that components of the miRNA processing machinery can utilize scaRNA as a substrate, and CBs (or CB proteins) impact miRNA biogenesis. We hypothesize that regulated processing of primary-scaRNA is a mechanism to control snRNA modifications and snRNP activity. Drosha/DGCR8 processing of certain scaRNAs (i.e., scaRNA2, scaRNA9 and scaRNA17) will control how much primary-scaRNA goes toward scaRNP formation (needed for snRNA modification) versus fragment generation. Previous reports have shown that scaRNA 2, 9, and 17 fragments are enriched in the nucleolus as well as the nucleoplasm (CB) (Tycowski et al., 2004), and we have suggested that these fragments form regulatory RNPs that influence snoRNP activity (Poole et al., 2017; Burke et al., 2018, 2019). Drosha/DGCR8 processing of specific scaRNAs, therefore, could also impact rRNA modification. In regard to the contribution of CBs to miRNA biogenesis, recent work has implicated miRNA dysregulation as a contributor to spinal muscular atrophy (SMA), the leading genetic cause of infant mortality (Goncalves et al., 2018). Most cases of SMA are caused by insufficient levels of SMN, the survival of motor neuron protein (Lefebvre et al., 1995), and reduced amounts of Drosha are observed in SMA (Goncalves et al., 2018). The involvement of CBs in the formation of miRNAs encoded in gene clusters more clearly links SMN with the miRNA processing machinery. The elucidation of these pathways will thus provide insight into fundamental cellular functions and clarify the SMA disease state. A model of the interconnections between primary-miRNA and primary-scaRNA processing is shown (Figure 5). In cell types that lack CBs, we posit that the nucleoplasmic pool of SMN, coilin, and other proteins enriched in the CB still participate in a miRNA biogenesis pathway, but the impact of these proteins on the efficiency of this process will likely be reduced. To test this hypothesis, we conducted experiments in the WI-38 primary cell line, which does not have a high percentage of cells with CBs. We have found that coilin reduction in WI-38 cells for 5 d does not significantly decrease the levels of primary-let7a or mature-let7a as observed in HeLa cells (Supplemental Figure S5). However, as observed in the HeLa line, Drosha reduction for 5 d in the WI-38 line greatly increases primary-let7a and correspondingly decreases mature let-7a. These findings suggest that altered CB function is responsible for the dysregulation of miRNA biogenesis we have observed.
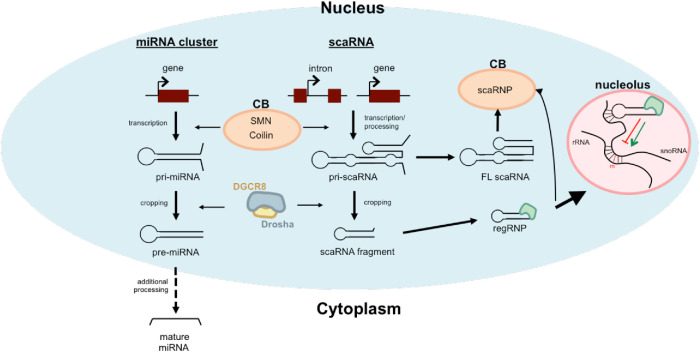
FIGURE 5:
Model of the interconnections between CBs and the miRNA processing machinery. Although miRNAs were not identified in the coilin interactome (Machyna et al., 2014), we hypothesize that proteins enriched in the CB, such as SMN and coilin, transiently participate in early steps of miRNA and scaRNA biogenesis while Drosha/DGCR8 perform cropping reactions on both primary-miRNAs and certain primary-scaRNAs. We further hypothesize that the activity of Drosha/DGCR8 on primary-scaRNA will indirectly influence snRNP activity by regulating scaRNP levels.
Interestingly, in plants a subnuclear structure known as the Dicing body contains the miRNA processing machinery (Fang and Spector, 2007). However, some components of the RNAi pathway are present in plant CBs, such as argonaute 4 (Li et al., 2006; Pontes and Pikaard, 2008). Additionally, coilin negative CBs have been identified in plants. These structures contain some components normally found in CBs, such as SmB and SmD3, and the plant miRNA processing components DCL1, HYL1, and SE (Pontes and Pikaard, 2008). These findings demonstrate the variety of methods employed to efficiently generate miRNAs in plants, and our findings suggest that connections between CBs and miRNA processing components also exist in animals.
MATERIALS AND METHODS
Cell lines, plasmids, transfections, and treatments
HeLa, JEG-3, and WI-38 cell lines were obtained from the American Type Culture Collection. Cells were cultured as previously described (Enwerem et al., 2014). All siRNAs were obtained from Integrated DNA Technologies (Coralville, IA) and utilized with RNAiMax (Invitrogen, Carlsbad, CA) per the manufacturer’s protocol. TDP-43: forward, 5-CUAAUUCUAAGCAAAGCCAAGAUGA-3 reverse: 5-UCAUCUUGGCUUUGCUUAGAAUUAGGA-3; SYNCRIP, forward: 5-GCUCUAUUCCUAAGAGUAAAACCAA-3 reverse, 5-UUGGUUUUACUCUUAGGAAUAGAGCCC-3. Negative control, SMN, Drosha, coilin, and WRAP53 siRNAs were previously described (Poole et al., 2016; Logan et al., 2018). For 120-h transfections, cells were transfected at day 0, transfected again at day 3, and then harvested at day 5, resulting in 120 h of transfection. For siRNA/ DNA transfection combination, HeLa cells were treated with negative control, TDP-43, SYNCRIP, or WRAP53 siRNA for 48 h and then transfected with scaRNA9 pcDNA3.1+ (as previously described in Poole et al., 2016) for 24 h resulting in 72 h knockdown and 24 h scaRNA9 expression. Note that endogenous scaRNA9 is only faintly visible using this method (Poole et al., 2016), thus the majority of the detected full-length scaRNA9 and mgU2-30 fragment is from the processing of ectopically expressed primary-scaRNA9. DNA transfections were conducted using FuGene HD (Promega, Madison, WI) according to the manufacturer’s protocol.
IF with RNA FISH
The C19MC cluster is present on chromosome 19. JEG-3 cells contain three chromosome 19s (two maternal and one paternal). Since the C19MC locus is maternally imprinted, the RNA FISH signal for the expressed C19MC locus comes from the paternal chromosome 19. HeLa and JEG-3 cells were grown on 2-well glass slides. HeLa cells were treated with 5-aza for 48 h. JEG-3 and HeLa cells were first prepared using the turboFISH protocol from the Raj lab at the University of Pennsylvania. Cells were washed with 1× phosphate-buffered saline (PBS), permeabilized with –20°C methanol, and incubated in –20°C for 10 min. Cells were then hybridized in 2× SSC with 10% dextran sulfate, 10% formamide, 3 mM Alexa Fluor 594 tagged probe (5′-Alex594 N/ATTTTCCTTGACCAGGTTAAAATGGACACAAAAATAAAGATGCATTTA-3), and 4′,6-diamidino-2-phenylindole (DAPI) for 20 min at 37°C. This probe binds intronic sequences in the C19MC primary transcript (Noguer-Dance et al., 2010). Cells were then washed in prewarmed 2× SSC with 10% formamide wash buffer 3× during a 37°C incubation. Finally, cells were washed with 2× SSC. For IF, cells were washed in 1× PBS and then fixed in 4% paraformaldehyde, permeabilized in 0.5% Triton, and blocked in 10% normal goat serum as previously described (Logan et al., 2018). Cells were then probed with 1:200 anti-SMN mouse monoclonal antibody (BD Biosciences, San Jose, CA) or 1:200 anti-coilin mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in 10% NGS at 37°C for 30 min. Slides were then washed with 1× PBS and incubated with 1:600 Alexa Fluor 488 (A11012, Invitrogen, Carlsbad, CA) goat anti-mouse (green) in 10% NGS at 37°C for 30 min. Slides were then washed in 1× PBS and DAPI stained to detect the nucleus followed by coverslip mounting with Antifade (Invitrogen, Carlsbad, CA). Where noted, the C19MC locus in JEG-3 cells was demarcated by staining with anti-DGCR8 (Abcam, Cambridge, MA) and CBs was detected using anti-coilin antibodies described above or detection of coilin-GFP. Images were captured on a Nikon Eclipse E600 epiflourescence microscope, and digital images were taken using Photometics CoolSnap HQ2 CCD camera and processed using MetaView software. PowerPoint and Adobe Photoshop Elements 7 were used in the preparation of images, as previously described (Poole et al., 2016).
Quantitative real-time PCR
RNA was extracted with TRI-REAGENT (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s suggested protocol. Reactions were set up with 50 ng total RNA in Brilliant II SYBR Green qRT-PCR master mix (Agilent, Santa Clara, CA) using an Agilent MX3000P qRT-PCR system. Amplification rates, Ct values, and dissociation curve analyses of products were determined using MxPro (version 4.01) software. Relative expression was determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Microsoft Excel was used for post-hoc statistical analysis using the Student’s t test with p < 0.05 considered significant. Histograms were prepared using GraphPad Prism. Oligonucleotides used were obtained from Integrated DNA Technologies (Coralville, Iowa) and are as follows: GAPDH, previously described (Burke et al., 2018);
C19MC primary (exons) transcript, forward 5-ACTGTGTGTCCCTGTGCTGGAACTCAAGTGAAC-3, reverse 5-CCAAGAGAATTGGGCTGGATTTCTAGAGAA-3 (Bortolin-Cavaille et al., 2009);
primary-miR-520 h, forward 5-CCAGAGTGCTGGAGCAAGAAG-3, reverse 5-CAGCATCAACTTCAACACTGC-3.
Primary let-7, forward 5-GATTCCTTTTCACCATTCACCCTGGATGTT-3, reverse 5-TTTCTATCAGACCGCCTGGATGCAGACTTT-3 (Heo et al., 2008);
HMGA2, forward 5-CACTCCAAGTCTCTTCCCTTTCCAAGC-3 (Lee and Dutta, 2007), reverse 5′-CCTCTTCGGCAGACTCTTGTGAGGA-3 (Agostini et al., 2015).
To assess if splicing is disrupted in HeLa cells after 5 d of CB component knockdown, isolated RNA was subjected to qRT-PCR to detect β-actin unspliced and spliced RNA. The primers used are as follows:
β-actin, exonic forward 5-GGGCCGTCTTCCCCTCCATCGTGG-3, exonic reverse 5-GTACTTCAGGGTGAGGATGCCTCTCTTG-3, intronic reverse 5-CACCCCGGAAACCGGGAGGCTCCTG-3.
Spliced β-actin mRNA was detected using exonic forward and exonic reverse primers. Unspliced β-actin pre-mRNA was detected using exonic forward and intronic reverse primers. We also examined the splicing of five different messages that encode intronic miRNAs by qRT-PCR. Primers described previously were used (Kim and Kim, 2007). These primers bind exonic and intronic regions such that the encoded miRNA is flanked by the primers. The following amplicons were analyzed: MCM7 (miR-106b region), CTDSP1 (miR-26b region), PANK1 (miR-107 region), EGFL7 (miR-126 region), and SMC4 (miR-15b-16-2 region).
For RT-PCR detection of miRNAs, the miRCURY LNA RT and PCR kits (Qiagen, Germantown, MD) were used according to the manufacturer’s protocol. Primers for 5S rRNA, mature let-7, and mature miR-520 h designed for use in the miRCURY LNA system were obtained from the manufacturer (Qiagen, Germantown, MD).
Northern blotting
Total RNA was isolated using TRI-Reagent (Molecular Research Center, Cincinnati, OH). For total RNA, typically 10–15 μg was run on a 6% denaturing polyacrylamide gel (Invitrogen, Carlsbad, CA) in 1× Tris-Borate-EDTA (TBE) at 200V. The gel was then washed in 1× TBE and then transferred onto a positively charged nylon membrane (Invitrogen, Carlsbad, CA) with the iBlot Gel Transfer device (Life Technologies, Grant Island, NY) using program 5 for 5 min. After transfer, the membrane was rinsed in ultrapure water, allowed to dry, and then subjected to a UV cross-linker (UVP, Upland, CA) at a setting of 120,000 μJ/cm2. The membrane was then placed in a hybridization bottle and prehybridized using Ultrahyb Ultrasensitive Hybridization buffer (Ambion Life Technologies, Grand Island, NY) for 30 min at 42°C in a hybridization oven. The DNA oligo probe used for scaRNA 9 full length and mgU2-30 fragment detection was 5′ DIG labeled and previously described (Poole et al., 2017). Membranes were then prepared for detection using the DIG Wash and Block kit (Invitrogen, Carlsbad, CA) following the manufacturer’s suggested protocol with the Anti-DIG antibody used at 1:10,000. Detection was carried out using CSPD (Roche, Mannheim, Germany) following the manufacturer’s suggested protocol. Blots were imaged using a Chemidoc imager (Bio-Rad, Hercules, CA). Where noted, adjustments to images were made using the transformation settings on QuantityOne software and applied across the entire image.
In vitro Drosha/DGCR8 processing assay
Myc-tagged Drosha and FLAG-tagged DGCR8 were obtained from Addgene (Watertown, MA). HeLa cells were cotransfected with myc-Drosha and FLAG-DGCR8 DNA for 24 h. After 24 h, cells were harvested using a KCl lysis buffer (20 mM Tris-HCl, 180 mM KCl, 0.2 mM EDTA) followed by sonication with a Fisher Scientific sonic dismembrator (model 100) for 6× for 5 s each using the output setting of 1. The FLAG-DGCR8/Drosha complex was immunoprecipitated using anti-FLAG-M2 affinity agarose beads (Sigma Aldrich, St. Louis, MO) overnight and washed 3× with KCl lysis buffer. The substrate for the processing assay was generated by digesting pBluescript KS+ containing scaRNA9 with a 3′ extension (Enwerem et al., 2014) with HindIII and linear DNA was gel purified using QIAquick Gel Extraction Kit (Qiagen, Germantown, MD) following the manufacturer’s protocol. In vitro transcription of linear DNA with T7 polymerase was accomplished using the T7 Megascript Kit (Invitrogen, Carlsbad, CA). Processing assays were also conducted with a scaRNA9 transcript lacking the 3′ extension (Δ3′). To generate the scaRNA9 Δ3′ transcript, pBluescript KS+ scaRNA9 Δ3′ DNA (Enwerem et al., 2014) was linearized with BamHI, gel purified, and subjected to in vitro transcription with T3 polymerase using the T3 Megascript Kit (Invitrogen, Carlsbad, CA). Transcripts were gel purified using ZR small-RNA Page Recovery Kit (Zymo Research, Irvine, CA) and added to the washed anti-FLAG beads along with 80 mM MgCl2 and RNAse inhibitor (ThermoFisher, Waltham, MA). The reaction was allowed to incubate for 90 min at 37°C. Control reactions include substrate incubated with water alone and substrate incubated with FLAG beads from nontransfected cell lysate. The RNA was then isolated using Tri-Reagent (Molecular Research Center, Cincinnati, OH) following the manufacturer’s protocol and subjected to Northern blotting using 5′ DIG probes specific for full-length scaRNA9 and the mgU2-30 fragment (described above) or the mgU2-19 fragment (5-GTAGACTGGAAAGACTTCTGATGCTCAGATTTGGCTAGTTTCATCATTGA-3). Where indicated in Figure 4C, NaCl was added to the reactions at a final concentration of 10, 50, and 100 mM.
RNA sequencing
An in vitro Drosha/DGCR8 processing assay using scaRNA9 substrate was conducted as described above. RNA isolated from the reaction was run on a denaturing polyacrylamide and a region of the gel corresponding to 70 nts (the size of mgU2-30 and mgU2-19) was excised and the RNA in this gel fragment was purified with the ZR small-RNA Page Recovery Kit (Zymo Research, Irvine, CA). The isolated RNA was then provided to the University of Mississippi Medical Center (UMMC) Molecular and Genomics Core Facility (www.umc.edu/genomicscore). Samples underwent an initial quality control step to determine RNA concentration (Qubit Fluorimeter) and integrity (QIAxcel Advanced System). Subsequently, samples (along with control samples) were used to develop a small RNA library using Illumina TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA) with modification to excise appropriate band in the 160–200 bp range (as opposed to miRNA in the 140–160 bp range). The library was sequenced using iSeq Reagent Kit on Illumina iSeq platform. The sequencing reads (1,590,886) were uploaded, evaluated for quality, and small RNA adaptor trimmed using Illumina BaseSpace Cloud Computing platform. The trimmed FASTQ sequence was aligned to the human reference genome (hg19 [RefSeq]) using RNA-Seq Alignment Application. The aligned reads (BAM files) were visualized using Integrative Genomics Viewer.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Intramural Research Support Program of The University of Mississippi Medical Center. The work performed through the UMMC Molecular and Genomics Facility is supported, in part, by funds from the National Institute of General Medical Sciences, including Mississippi INBRE (P20GM103476), Center for Psychiatric Neuroscience–Center for Biomedical Research Excellence (COBRE) (P30GM103328), Obesity, Cardiorenal and Metabolic Diseases–COBRE (P20GM104357), and Mississippi Center of Excellence in Perinatal Research–COBRE (P20GM121334).
Abbreviations used:
- 5-aza
5-aza-2′-deoxycytidine
- CB
Cajal body
- DAPI
4′,6-diamidino-2-phenylindole
- FISH
fluorescence in situ hybridization
- IF
immunofluorescence
- miRNA
microRNA
- PBS
phosphate-buffered saline
- scaRNA
small Cajal body-specific RNA
- SMA
spinal muscular atrophy
- SMN
survival of motor neuron protein
- snRNP
small nuclear ribonucleoprotein
- SYNCRIP
Synaptotagmin Binding Cytoplasmic RNA Interacting Protein
- TBE
Tris-Borate-EDTA
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-02-0144) on May 20, 2020.
REFERENCES
- Agostini A, Panagopoulos I, Andersen HK, Johannesen LE, Davidson B, Trope CG, Heim S, Micci F. (2015). HMGA2 expression pattern and TERT mutations in tumors of the vulva. Oncol Rep , 2675–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemer C, Bortolin-Cavaille ML, Schmidt U, Jensen SM, Kjems J, Bertrand E, Cavaille J. (2012). Microprocessor dynamics and interactions at endogenous imprinted C19MC microRNA genes. J Cell Sci , 2709–2720. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature , 363–366. [DOI] [PubMed] [Google Scholar]
- Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S. (1999). The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum Mol Genet , 775–782. [DOI] [PubMed] [Google Scholar]
- Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. (2009). C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res , 3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome HJ, Hebert MD. (2012). In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS One , e36300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MF, Logan MK, Hebert MD. (2018). Identification of additional regulatory RNPs that impact rRNA and U6 snRNA methylation. Biol Open , bio036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke MF, McLaurin DM, Logan MK, Hebert MD. (2019). Alteration of 28S rRNA 2’-O-methylation by etoposide correlates with decreased SMN phosphorylation and reduced Drosha levels. Biol Open , bio041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chan J, Chen W, Li J, Sun M, Kannan GS, Mok YK, Yuan YA, Jobichen C. (2020). SYNCRIP, a new player in pri-let-7a processing. RNA , 290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. (2002). Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. EMBO J , 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature , 231–235. [DOI] [PubMed] [Google Scholar]
- Deryusheva S, Choleza M, Barbarossa A, Gall JG, Bordonne R. (2012). Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells. RNA , 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwerem II, Velma V, Broome HJ, Kuna M, Begum RA, Hebert MD. (2014). Coilin association with Box C/D scaRNA suggests a direct role for the Cajal body marker protein in scaRNP biogenesis. Biol Open , 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwerem II, Wu G, Yu YT, Hebert MD. (2015). Cajal body proteins differentially affect the processing of box C/D scaRNPs. PLoS One , e0122348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol , 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Bailey AD, Weiner AM, Matera AG. (1999). Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol , 126–135. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. (1995). Coiled Bodies Contain U7 Small Nuclear RNA and Associate with Specific DNA Sequences in Interphase Cells. Proc Natl Acad Sci USA , 5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves I, Brecht J, Thelen MP, Rehorst WA, Peters M, Lee HJ, Motameny S, Torres-Benito L, Ebrahimi-Fakhari D, Kononenko NL, et al (2018). Neuronal activity regulates DROSHA via autophagy in spinal muscular atrophy. Sci Rep , 7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell , 23–34. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. (2001). Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev , 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. (2008). Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell , 276–284. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science , 834–838. [DOI] [PubMed] [Google Scholar]
- Izumikawa K, Nobe Y, Ishikawa H, Yamauchi Y, Taoka M, Sato K, Nakayama H, Simpson RJ, Isobe T, Takahashi N. (2019). TDP-43 regulates site-specific 2’-O-methylation of U1 and U2 snRNAs via controlling the Cajal body localization of a subset of C/D scaRNAs. Nucleic Acids Res , 2487–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. (2001). Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J Biol Chem , 38645–38651. [DOI] [PubMed] [Google Scholar]
- Kabekkodu SP, Shukla V, Varghese VK, D’Souza J, Chakrabarty S, Satyamoorthy K. (2018). Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc , 1955–1986. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev , 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. (2007). Processing of intronic microRNAs. EMBO J , 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2004). Biogenesis of small nuclear RNPs. J Cell Sci , 5949–5951. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. (2001). A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science , 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature , 415–419. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. (2007). The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev , 1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Han S, Kwon CS, Lee D. (2016). Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell , 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim VN. (2007). In vitro and in vivo assays for the activity of Drosha complex. Methods Enzymol , 89–106. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell , 155–165. [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. (2006). Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell , 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell , 93–106. [DOI] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. (2009). Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell , 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KL, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods , 402–408. [DOI] [PubMed] [Google Scholar]
- Logan MK, Burke MF, Hebert MD. (2018). Altered dynamics of scaRNA2 and scaRNA9 in response to stress correlates with disrupted nuclear organization. Biology Open , bio037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ. (1998). The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum Mol Genet , 1269–1275. [DOI] [PubMed] [Google Scholar]
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. (2014). The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell , 389–399. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. (2010). WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol , e1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova JA, Kramerov DA. (2011). SNOntology: Myriads of novel snoRNAs or just a mirage? BMC Genomics , 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Richard P, Pinzon N, Kiss T. (2014). Targeting vertebrate intron-encoded box C/D 2’-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res , 6616–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. (2010). The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet , 3566–3582. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. (2001). The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr Biol , 1079–1088. [DOI] [PubMed] [Google Scholar]
- Pontes O, Pikaard CS. (2008). siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev , 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AR, Enwerem II, Vicino IA, Coole JB, Smith SV, Hebert MD. (2016). Identification of processing elements and interactors implicate SMN, coilin and the pseudogene-encoded coilp1 in telomerase and box C/D scaRNP biogenesis. RNA Biol , 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AR, Vicino I, Adachi H, Yu YT, Hebert MD. (2017). Regulatory RNPs: a novel class of ribonucleoproteins that potentially contribute to ribosome heterogeneity. Biol Open , 1342–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature , 901–906. [DOI] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. (2003). A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J , 4283–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer IA, Sturgill D, Sung MH, Hager GL, Dundr M. (2016). Cajal body function in genome organization and transcriptome diversity. Bioessays , 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. (2001). Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell , 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Carter K, Johnson C, Lawrence J. (1995). U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem , 473–485. [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. (2010). Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol , 403–409. [DOI] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. (2001). Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol , 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Aab A, Steitz JA. (2004). Guide RNAs with 5’ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol , 1985–1995. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. (2009). A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell , 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science , 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. (2016). Cajal bodies are linked to genome conformation. Nat Commun , 10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. (1998). Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J , 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. (2005). Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J , 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.