Abstract
Membrane trafficking during cytokinesis is essential for the delivery of membrane lipids and cargoes to the division site. However, the molecular mechanisms are still incompletely understood. In this study, we demonstrate the importance of uncharacterized fission yeast proteins Mso1 and Sec1 in membrane trafficking during cytokinesis. Fission yeast Mso1 shares homology with budding yeast Mso1 and human Mint1, proteins that interact with Sec1/Munc18 family proteins during vesicle fusion. Sec1/Munc18 proteins and their interactors are important regulators of SNARE complex formation during vesicle fusion. The roles of these proteins in vesicle trafficking during cytokinesis have been barely studied. Here, we show that fission yeast Mso1 is also a Sec1-binding protein and Mso1 and Sec1 localize to the division site interdependently during cytokinesis. The loss of Sec1 localization in mso1Δ cells results in a decrease in vesicle fusion and cytokinesis defects such as slow ring constriction, defective ring disassembly, and delayed plasma membrane closure. We also find that Mso1 and Sec1 may have functions independent of the exocyst tethering complex on the plasma membrane at the division site. Together, Mso1 and Sec1 play essential roles in regulating vesicle fusion and cargo delivery at the division site during cytokinesis.
INTRODUCTION
Cytokinesis is the final stage of the cell division cycle, which is required for a cell to separate after mitosis and well conserved from yeast to animal cells (Balasubramanian et al., 2004; Pollard and Wu, 2010; Meitinger and Palani, 2016; Gerien and Wu, 2018). Cytokinesis begins with the selection of the division site followed by the formation of an actomyosin ring. The ring then constricts, which is coupled with plasma membrane deposition and extracellular matrix formation or remodeling. Finally, the daughter cells separate to form two new cells. During fission yeast cytokinesis, cells select the cell equator as the division site to assemble the ring (Barr and Gruneberg, 2007; Pollard and Wu, 2010). The ring constricts as the septum is constructed to separate the two daughter cells. Targeted membrane deposition at the division site is important for the delivery of machinery that drives cytokinesis, as well as the membrane itself that is required to close the new end of the daughter cell (Albertson et al., 2005). Thus, it is important to understand vesicle trafficking and how vesicles are tethered and fused at the division site.
Vesicle fusion is a multi-step process (Cucu et al., 2017; Wickner and Rizo, 2017; Gerien and Wu, 2018). First, a vesicle must approach the target membrane, either by transportation on actin cables or microtubules or by random walk (Chang and Martin, 2009; Lo Presti and Martin, 2011; Snaith et al., 2011). Tethering occurs next, involving interactions between the incoming vesicle and the target membrane, and is mediated in many cases by multi-subunit tethering complexes, or MTCs (Dubuke and Munson, 2016). MTCs help guide the vesicle to the membrane for fusion. The following step is docking, which takes place when SNARE proteins on the vesicle and target membrane interact with each other to form a trans-SNARE complex. Finally, the vesicle fuses with the target membrane when the two membranes merge and the now cis-SNARE complexes are disassembled (Whyte and Munro, 2002). The SNARE proteins are essential for the fusion process (McNew et al., 2000), usually with three SNAREs on the target membrane (t-SNAREs) and one on the vesicle membrane (v-SNARE). Each SNARE contains a domain called a SNARE helix. The SNARE helixes from the four SNARE motifs interact to form a helix bundle that drives fusion (Sutton et al., 1998). Previous studies have shown that while the formation of cognate SNARE complexes can be specific, most of these complexes depend on other factors to become fusogenic in vivo (Furukawa and Mima, 2014).
Tethering proteins are important for the efficiency and specificity of vesicle fusion (Yu and Hughson, 2010). The tethers interact with the vesicle first and are believed to help guide the vesicle to the SNAREs on the target membrane (Heider and Munson, 2012; Dubuke and Munson, 2016). The tethers have also been shown to increase the efficiency of vesicle fusion (Zick and Wickner, 2014). The exocyst complex, the best known tether on the plasma membrane, is an octameric MTC composed of subunits Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84 (TerBush et al., 1996). The six Sec subunits were identified in the original landmark screen for mutants that were defective in secretion (Novick et al., 1980), while the Exo70 and Exo84 subunits were identified in later studies (TerBush et al., 1996; Guo et al., 1999). The exocyst is required for Golgi-derived vesicles to fuse at the plasma membrane and is used for polarized exocytosis (He and Guo, 2009; Yu and Hughson, 2010). In fission yeast, the exocyst localizes to the cell tips and the division site and is essential for cell separation (Wang et al., 2002). Sec3 and Exo70 can localize the exocyst complex to the sites of polarized growth (Bendezú et al., 2012; Jourdain et al., 2012). sec8 mutant cells are defective in secretion, with lower acid phosphatase secretion and vesicle accumulation near the plasma membrane (Wang et al., 2002).
Besides tethers, another set of proteins involved in vesicle fusion is the Sec1/Munc18 (SM) family proteins. They are conserved proteins that interact with SNAREs (Carr and Rizo, 2010; Jiao et al., 2018). Both budding yeast and fission yeast contain four SM proteins: Sec1, Vps45, Sly1, and Vps33 (Rizo and Sudhof, 2012). In budding yeast, Sec1 is essential and involved in exocytosis and vesicle fusion at the plasma membrane (Carr et al., 1999). It localizes to the plasma membrane and is concentrated at the bud neck (Scott et al., 2004). Sec1 is a homologue of mammalian Munc18a and has been shown to interact with the fully assembled SNARE complex (Carr et al., 1999; Jiao et al., 2018). However, Sec1 has not been studied in fission yeast.
SM binding partners have also emerged to play a role in vesicle fusion. In mammals, the protein Mint1 interacts with Munc18-1 in neuronal cells (Okamoto and Südhof, 1997). Mso1 in budding yeast is homologous to the PTB domain of Mint1 (Knop et al., 2005). Mso1 localizes to the plasma membrane and interacts with Sec1 at the bud tip and bud neck (Aalto et al., 1997; Castillo-Flores et al., 2005; Weber et al., 2010). Deletion of Mso1 is not lethal, and growth of mso1Δ cells on both synthetic and rich media at different temperatures is not significantly affected, but vesicles were seen to accumulate in the bud in mso1Δ cells (Aalto et al., 1997). Mso1 has also been shown to interact with the membrane and is suggested to interact with both the target membrane and the vesicle membrane (Weber-Boyvat et al., 2013). These interactions suggest that Mso1 has multiple roles in vesicle fusion, and its functions seem to lie at crossroads between Sec1, SNARE proteins, and the exocyst complex. Mso1 was shown to be essential for membrane fusion during prospore formation (Knop et al., 2005). Although the interactions of Mso1 and Sec1 have been well studied, the roles of these proteins during cytokinesis have not been studied in fungi or in animals.
In the fission yeast Schizosaccharomyces pombe, SM proteins and the proteins that interact with them have rarely been studied (Snaith et al., 2011). Additionally, the roles of these proteins during cytokinesis have not been investigated. Here we show that loss of Mso1 leads to decreased vesicle fusion at the division site and defects in ring constriction and disassembly during cytokinesis. These defects are due to loss of Sec1 localization at the division site, as similar defects are observed in sec1 mutant cells. Our results also suggest that Sec1 and Mso1 have functions in exocytosis independent of the exocyst complex at the plasma membrane.
RESULTS
Mso1 localizes to the division site during cytokinesis
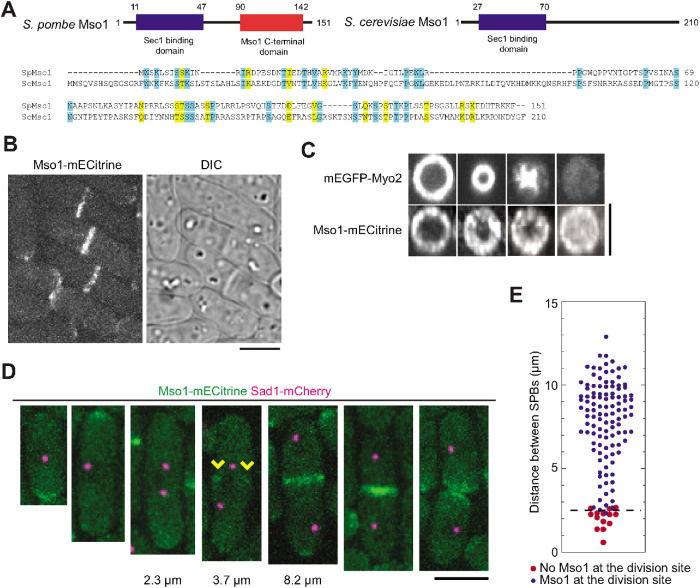
S. pombe Mso1 (SPAC1F5.05c, 16.66 kDa) was reported to localize to the division site and cell tips in a genome-wide protein localization study (Matsuyama et al., 2006). Analysis of the Mso1 sequence revealed similarity to two regions of Mso1 from Saccharomyces cerevisiae, a Sec1 binding domain at the N-terminus and an Mso1 C-terminal domain (Pfam PF14475 and PF14477; Figure 1A, top). Sequence alignment corroborated low similarities between the two proteins with 12% identity and 19% similarity in amino acids (Figure 1A, bottom). We created a homology model for S. pombe Mso1 based on the human homologue, the Mint1 PTB domain (Supplemental Figure S1A). This model showed a core of beta sheets with several helices. Both of the predicted domains were exposed to the outer surface in the model.
FIGURE 1:
Mso1 localizes to the division site in fission yeast. (A) Domain organization and sequence alignment of Mso1 from S. pombe and S. cerevisiae. Identical and similar (V/I/L, D/E, K/R, N/Q, and S/T) residues are shaded in blue and yellow, respectively. (B) Mso1 localizes to cell tips during interphase and to the division site during cytokinesis. DIC: differential interference contrast. (C) End-on projections of division site showing the localization of Myo2 and Mso1 in cells at similar stages of septation. Mso1 localizes across the division plane as the Myo2-labeled contractile ring constricts inward. (D, E) Micrographs (D) and quantification (E) of Mso1 appearance at the division site relative to the distance between SPBs (before spindle breakdown). (D) The arrowheads indicate Mso1 appearance at the division site. Distances between SPBs before ring constriction are indicated below the micrographs. (E) The dash line marks the boundary between cells with (blue circles) and without (red circles) Mso1-mECitrine signal at the division site. Bars, 5 µm.
We confirmed that Mso1 was present at cell tips during interphase and at the division site during cytokinesis (Figure 1B). As the actomyosin ring (marked by Myo2) constricted, Mso1 was added to the leading edge of the cleavage furrow and localized across the entire division plane by the end of ring constriction (Figure 1C). To determine when Mso1 appears at the cell division site, we imaged Mso1 together with spindle pole body (SPB) protein Sad1 as a cell-cycle marker. Mso1 started to accumulate at the division site during early anaphase when two SPBs were ∼2.5 µm apart (Figure 1, D and E). Consistently, Mso1 appeared at the division site when Rlc1 nodes coalesced into a compact contractile ring (Supplemental Figure S1B), which happens at the start of anaphase (Wu et al., 2003). The localization of Mso1 was not altered by disruption of the actin or microtubule cytoskeleton (Supplemental Figure S1, C and D). Taken together, the timing and pattern of Mso1 localization suggest that it plays a role in cytokinesis.
Loss of Mso1 leads to defects in late stages of cytokinesis
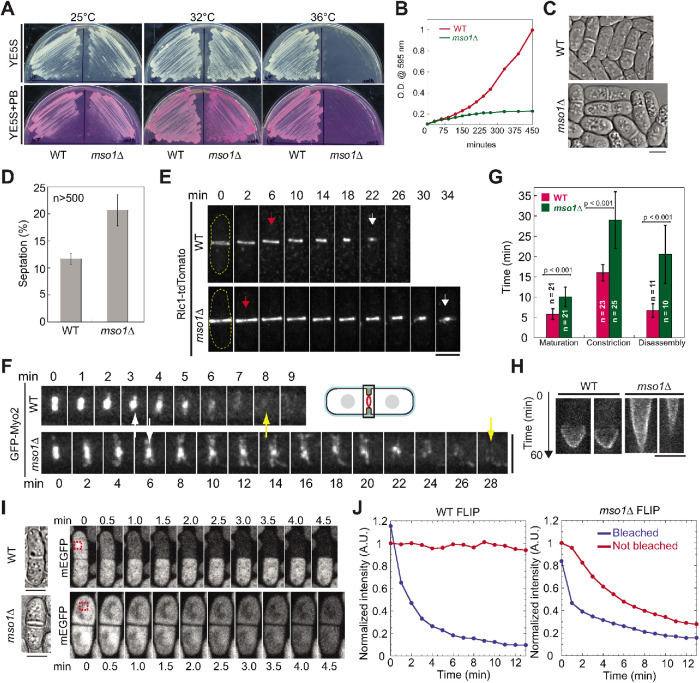
To determine the function of Mso1 during cytokinesis, we analyzed the phenotype when Mso1 was deleted. When cultured on rich media at 25°C (Figure 2A), mso1∆ cells grew similarly to wild-type (WT) cells. At 36°C, however, mso1∆ cells could not form colonies (Figure 2A), which is consistent with the genome-wide deletion-phenotype study (Hayles et al., 2013). At the intermediate temperature of 32°C, mso1∆ cells could grow but were stained dark red on YE5S + phloxin B (PB) plates, indicating more dead cells (Figure 2A). Growing mso1∆ cells in liquid medium at 36°C revealed that the cells were slow growing, sick, and vacuolated, and had more visible septa compared with WT (Figure 2, B–D). The higher septation index indicates that cells spend longer during septum formation and/or have a delay in cell separation.
FIGURE 2:
Mso1 is important for cytokinesis. (A) Wild-type (WT) and mso1∆ cells grown on YE5S and YE5S + PB plates at 25°C, 32°C, and 36°C. (B) Growth curves of WT and mso1∆ cells in liquid cultures at 36°C. (C, D) DIC images (C) and septation index (D; mean ± SD) of WT and mso1∆ cells grown in YE5S liquid media at 36°C for 4 h. (D) For each strain, three sets of data were measured, with all sets having >500 cells. (E–G) The contractile ring (marked with Rlc1 or Myo2) is defective in mso1∆ cells after growth at 36°C for 2 h. (E, F) Time course of contractile-ring constriction (E) and disassembly (F) in WT and mso1∆ cells. The arrows indicate the start (red) and end (white) of ring constriction and disappearance (yellow) of Myo2 signal. (G) Quantification of ring maturation (Rlc1), constriction (Rlc1), and disassembly (Myo2). See Materials and Methods for the definition of each stage. Error bars are SDs. (H) Kymographs of different Ain1-mEGFP expressing cells showing ring constriction in WT and mso1Δ cells. Kymographs are 60 min from top to bottom with each line representing 1 min. (I, J) Exchange of free GFP between daughter cells is observed in mso1∆ but not in WT cells after ring constriction. Time courses (I) and intensity measurements (J) of fluorescence loss in photobleaching (FLIP) assays in WT and mso1∆ cells after growth at 36°C for 2 h. The box with red dash lines in I marks the bleached region. Bars, 5 µm.
To determine the exact cytokinesis defects in mso1∆ cells, we quantified ring kinetics using either tagged myosin light chain Rlc1 or myosin heavy chain Myo2. Time-lapse microscopy revealed that the actomyosin ring was defective in several stages of cytokinesis (Figure 2, E–G). The maturation, constriction, and disassembly of the ring were significantly delayed in mso1∆ compared with WT cells (Figure 2, E–G). The last two stages took ∼2x and ∼3x longer than in WT cells, respectively (Figure 2G). During ring disassembly, we detected abnormal myosin linear structures across the division plane that were not observed in WT cells (Figure 2F).
These ring disassembly defects suggested some problems in contractile-ring constriction and/or plasma-membrane closure in mso1∆ cells. We observed ring constriction again using actinin Ain1- monomeric enhanced green fluorescent protein (mEGFP), whose concentration in the ring is constant during constriction in contrast to the increased concentrations of Myo2 and Rlc1 (Wu and Pollard, 2005), to see whether the ring stalled during constriction. We confirmed that the constriction took much longer in mso1∆ than WT cells, but the ring did not appear to pause or stall at any point (Figure 2H). Then we performed fluorescence loss in photobleaching (FLIP) to determine whether there was an exchange of free cytoplasmic GFP between the daughter cells after ring constriction, as determined by Rlc1-tdTomato. In WT, when GFP was bleached in one daughter cell, no signal was lost in the paired other daughter cell. In contrast, 53% mso1∆ cells (n = 22) exchange GFP across the division site as evidenced by the loss of GFP signal in the unbleached daughter cell (Figure 2, I and J; Supplemental Movies 1 and 2). The exchange of materials across the division site indicates a failure to properly close the plasma membrane after the ring constriction.
Movie S1.
A WT cell in a FLIP experiment shows loss of GFP signal on one side of the septum but not the other. The red square indicates where the cell was bleached. The cell was bleached every 30 seconds, with an image captured just before and after the bleaching event. Display rate is 7 frames per second.
Movie S2.
An mso1Δ cells in a FLIP experiment shows loss of GFP signal on both sides of the septum. The red square indicates where the cell was bleached. The cell was bleached every 30 seconds, with an image captured just before and after the bleaching event. Display rate is 7 frames per second.
Loss of Mso1 compromises fusion of vesicles at the division site
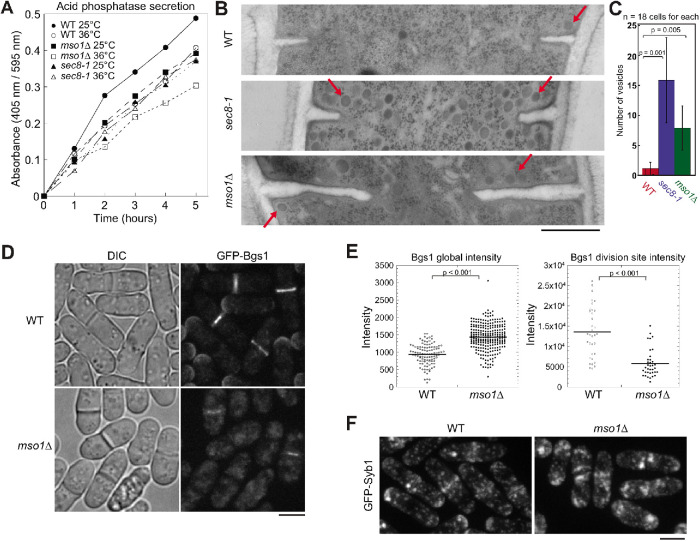
We next investigated what causes the cytokinetic defects in mso1∆ cells. Previous studies found that exocytosis is essential for cytokinesis in fission yeast (Wang et al., 2002, 2016; Zhu et al., 2018). An acid phosphatase secretion assay was utilized to measure the amount of secretion in mso1∆ cells. We used exocyst mutant sec8-1 as a control because it is known to have reduced exocytosis and secretion (Wang et al., 2002). As expected, sec8-1 had lower secretion of acid phosphatase than WT (Figure 3A). Similarly, mso1∆ cells secreted less acid phosphatase at both 25 and 36°C (Figure 3A). This suggests that exocytosis may be compromised in mso1∆ cells.
FIGURE 3:
Loss of Mso1 leads to exocytosis defects. (A) Acid phosphatase secretion assays of WT, mso1∆, and sec8-1 cells grown at 25°C or at 36°C. The absorbance at 405 nm was divided by the OD595 nm of cells. (B, C) Electron microscopy images (B) and quantification (C) of vesicle (examples marked by arrows) accumulation in WT, sec8-1, and mso1∆ cells grown at 36°C for 2 h. (C) Vesicles within 250 nm from the division plane in a thin EM section were counted. Error bars shown are SDs. (D, E) Micrographs (D) and quantification (E) of β-glucan synthase Bgs1 intensity in WT and mso1∆ cells after growth at 36°C for 2 h. (F) Localization of v-SNARE Syb1 in WT and mso1∆ cells after growth at 36°C for 2 h. Bars, 500 nm (B) and 5 µm (D, F).
We next used electron microscopy and electron tomography to examine the exocytic vesicles in WT and mso1∆ cells. Secretory vesicles (examples marked by arrows) accumulated near the ingressing septa and cell tips in mso1∆ cells (Figure 3B; Supplemental Figure S2, A and B; Supplemental Movies 3 and 4). While each thin section of WT cells had on average 1 ± 1 vesicle near the division site, mso1∆ cells had 8 ± 4 vesicles nearby (Figure 3, B and C). In comparison, sec8-1 cells had 16 ± 7 vesicles at the division site (Figure 3, B and C). The accumulation of vesicles suggested that vesicles are delivered to the division site but cannot fuse with the plasma membrane in mso1∆ cells. Consistently, the level of β-glucan synthase Bgs1, one of the vesicle cargoes, was significantly reduced at the division site even though the global Bgs1 level increased in mso1∆ cells (Figure 3, D and E). We also observed an accumulation of vesicle marker v-SNARE Syb1 at the division site and cell tips in mso1∆ cells grown at 36°C (Figure 3F). Both Syb1 and t-SNARE Psy1 protein levels were significantly increased in mso1∆ cells when quantified using fluorescence intensity (Supplemental Figure S3, A and B). Taken together, mso1∆ cells have reduced vesicle fusion, which compromises the delivery of membrane and cargoes to the plasma membrane.
Movie S3.
Electron tomography reconstruction of a WT cell during cytokinesis. A few vesicles can be seen near the ingressing septum. Bar, 100 nm.
Movie S4.
Electron tomography reconstruction of an mso1Δ cell during cytokinesis. Many vesicles can be seen accumulated near the ingressing septum. Bar, 100 nm.
Mso1 interacts with the SM protein Sec1 and is important for Sec1 localization
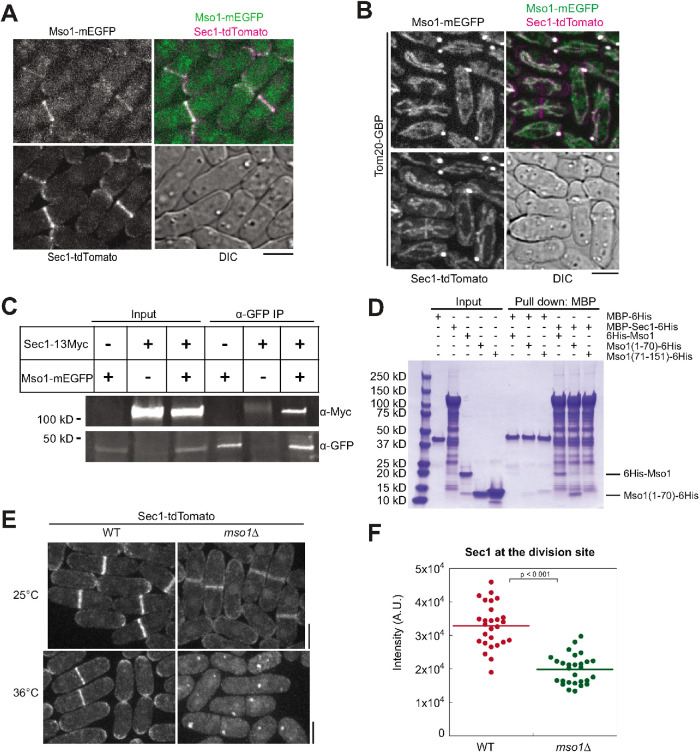
Next we investigated how Mso1 affects vesicle fusion. Budding yeast Mso1 interacts with the SM protein Sec1 for SNARE complex assembly during exocytosis (Carr et al., 1999; Weber et al., 2010). We therefore tested whether fission yeast Mso1 is also an SM protein interactor. Mso1 and Sec1 colocalized at the division site and cell tips (Figure 4A). To determine whether the two proteins interact, Mso1-mEGFP was ectopically targeted to mitochondria using Tom20-GFP-binding protein (GBP) (Rothbauer et al., 2008; Luo et al., 2014; Wang et al., 2016). The ectopically targeted Mso1 was able to recruit Sec1-tdTomato to the mitochondria efficiently (Figure 4B). Mso1 did not recruit all Sec1, as some Sec1 still localized to the division site and cell tips, possibly due to the higher cellular level of Sec1. As controls, we confirmed that Tom20-GBP could not recruit tdTomato-tagged Sec1 to mitochondria and no signals bled through between the red and green channels (Supplemental Figure S4A). The mistargeting of Sec1 by Mso1 suggests that the two proteins physically interact directly or in a complex in vivo. Indeed, Sec1-13Myc coimmunoprecipitated with Mso1-mEGFP from fission yeast cell lysate (Figure 4C). To verify whether the Sec1 binding occurs via the N-terminal portion of Mso1 (Figure 1A), we purified recombinant fragments of Mso1 comprising the N-terminal residues 1–70 and C-terminal residues 71–151. We found that the full-length Mso1 and the Mso1(1–70), but not Mso1(71–151), were pulled down by MBP-Sec1 in in vitro binding assays (Figure 4D). This further supports the physical interaction between Sec1 and Mso1 and the location of the Sec1 binding region in Mso1.
FIGURE 4:
Mso1 colocalizes and interacts with the SM protein Sec1. (A) Mso1 and Sec1 colocalize at the division site and at cell tips. (B) Mso1-mEGFP ectopically targeted to mitochondria by Tom20-GBP can recruit Sec1-tdTomato. (C) Mso1-mEGFP is able to pull down Sec1-13Myc from S. pombe cell lysates in co-IP assay. (D) SDS–PAGE gel showing in vitro binding assays between Sec1 and Mso1 using purified proteins. MBP-Sec1 can pull down full-length Mso1 and Mso1(1-70), but not Mso1(71-151). Bead bound MBP-Sec1 or an MBP control was incubated with Mso1, Mso1(1-70), or Mso1(71-151). (E) Sec1 localization in mso1∆ cells grown at 25°C or 36°C for 2 h. (F) Sec1-tdTomato intensity at the division site at 25°C. WT, n = 27 cells; mso1Δ, n = 29 cells. Bars, 5 µm.
To determine the functional importance of the Mso1-Sec1 interaction, we analyzed the effect of loss of Mso1 on the localization of Sec1. While Sec1 still localized to the cell tips and division site in mso1∆ cells at 25°C, its intensity was significantly reduced (Figure 4, E and F). Moreover, Sec1 was abolished from the plasma membrane and aggregated into cytoplasmic puncta when mso1∆ cells were shifted to the restrictive temperature of 36°C for 2 h (Figure 4E). These puncta did not colocalize with Syb1 marked secretory vesicles or Atg8 marked autophagosomes (Supplemental Figure S4B). Thus, Mso1 is crucial for Sec1 localization, especially at 36°C.
Sec1 is crucial for Mso1 localization and cytokinesis
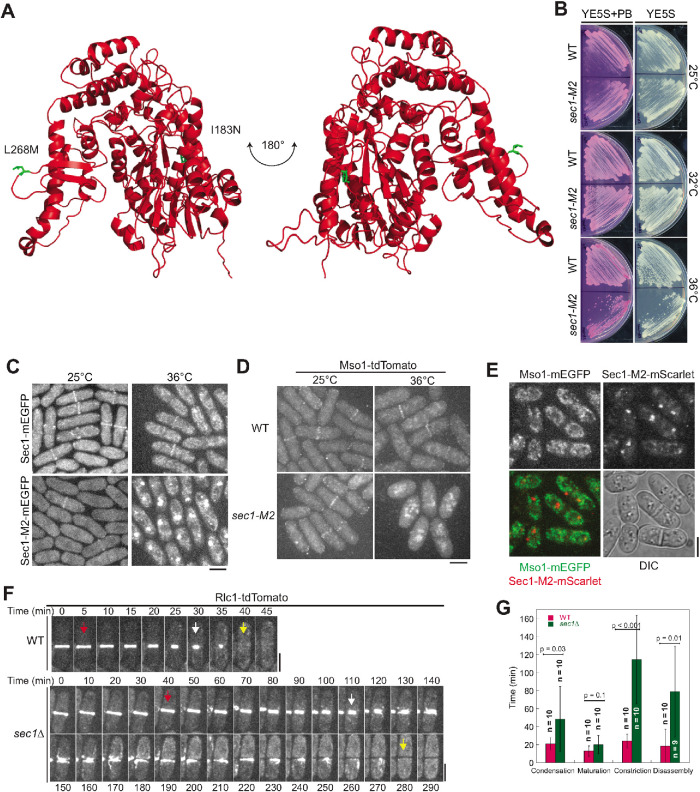
To further investigate the relationship between Mso1 and Sec1, we created a sec1 temperature-sensitive mutant, sec1-M2, using the marker (his5+) reconstitution mutagenesis method (Tang et al., 2011; Lee et al., 2014; Zhu et al., 2018). sec1-M2 contains two mutations in the ORF: I183N and L268M. We performed homology modeling for Sec1 using the human Munc18 crystal structure (PDB: 4cca) as a template (Hackmann et al., 2013). The mutated residues in sec1-M2 (highlighted green) are on the surface of Sec1 (Figure 5A) and may be important for its interactions with other proteins. sec1-M2 cells grew slowly and showed increased cell death at 36°C (Figure 5B). Sec1-M2-mEGFP was lost from the plasma membrane and gradually accumulated into aggregates at the restrictive temperature of 36°C, similar to Sec1 observed in mso1∆ cells (Figure 5C). This loss of Sec1 localization also resulted in a loss of Mso1 localization from the plasma membrane and Mso1 accumulation in the cytoplasmic puncta at the restrictive temperature (Figure 5D). However, these Mso1 puncta were distinct from those formed by Sec1-M2-mEGFP (Figure 5E). Therefore, Sec1 and Mso1 are interdependent for localization on the plasma membrane. The Psy1 level was significantly higher in sec1-M2 than in WT cells (Supplemental Figure S3, C and D), similar to those in mso1Δ cells (Supplemental Figure S3, A and B); however, the Syb1 level was not changed (Supplemental Figure S3, C and D).
FIGURE 5:
Sec1 is important for cytokinesis. (A) Homology modeling of Sec1 using the crystal structure of human Munc18-2 (PDB: 4cca) as a template. Residues mutated in the sec1-M2 mutant are highlighted in green. (B) The sec1-M2 mutant is temperature sensitive and defective in cell growth at 36°C. Cells were grown on YE5S and YE5S + PB plates for 2 d. (C) Sec1-M2 loses localization on the plasma membrane and accumulates to cytoplasmic puncta after grown at 36°C for 2 h. (D) Mso1 loses localization on the plasma membrane in sec1-M2 cells grown at 36°C for 4 h. (E) Cytoplasmic puncta of Sec1-M2 and Mso1 do not colocalize in sec1-M2 cells grown at 36°C for 2 h. (F, G) Time courses (F) and quantification (G) of Rlc1-tdTomato labeled contractile ring in WT and sec1∆ cells grown at 25°C. The arrows indicate the start (red) and end (white) of ring constriction and the end (yellow) of ring disassembly. Error bars shown are SDs. Bars, 5 µm.
To test whether Sec1 and Mso1 have a similar function in cytokinesis, we examined the kinetics of the contractile ring in the sec1∆ mutant because sec1-M2 may still retain some Sec1 functions. Sec1 is an essential gene (Kim et al., 2010; Hayles et al., 2013), so we performed tetrad fluorescence microscopy to examine the ring before cell death (Coffman et al., 2013; Lee et al., 2014; Davidson et al., 2016a). Each dissected tetrad from the sec1+/sec1∆ rlc1-tdTomato-natMX6/rlc1-tdTomato-natMX6 diploid strain contains two sec1+ spores and two sec1∆ spores. As expected, sec1+ spores germinated and grew normally. sec1∆ spores germinated normally, but the cells divided only two to four times before stopping growing and dying (Supplemental Figure S4C). The sec1∆ cells displayed cytokinesis defects similar to those observed in mso1∆ cells, taking dramatically longer to complete constriction and to disassemble the contractile ring (Figure 5, F and G; Supplemental Movies 5 and 6). Ring constriction took 115 ± 49 min in sec1∆ compared with 24 ± 8 min in WT. Disassembly of the ring took 79 ± 50 min in sec1∆ as opposed to 19 ± 19 min in WT. We also observed abnormal myosin structures during ring disassembly, with strand-like structures emanating from the division plane. Thus, loss of Mso1 or Sec1 has similar but not identical effects on the contractile ring and cytokinesis.
Movie S5.
Contractile-ring kinetics in WT cells. Tetrad Fluorescence Microscopy of Rlc1-tdTomato in a WT colony shows ring formation, constriction, and disassembly. Time stamp is hour : minute.
Movie S6.
Contractile-ring kinetics in sec1Δ cells. Tetrad Fluorescence Microscopy of Rlc1-tdTomato in a sec1Δ colony shows ring formation, constriction and disassembly. Time stamp is hour : minute
Mso1 and Sec1 have functions independent of the exocyst complex
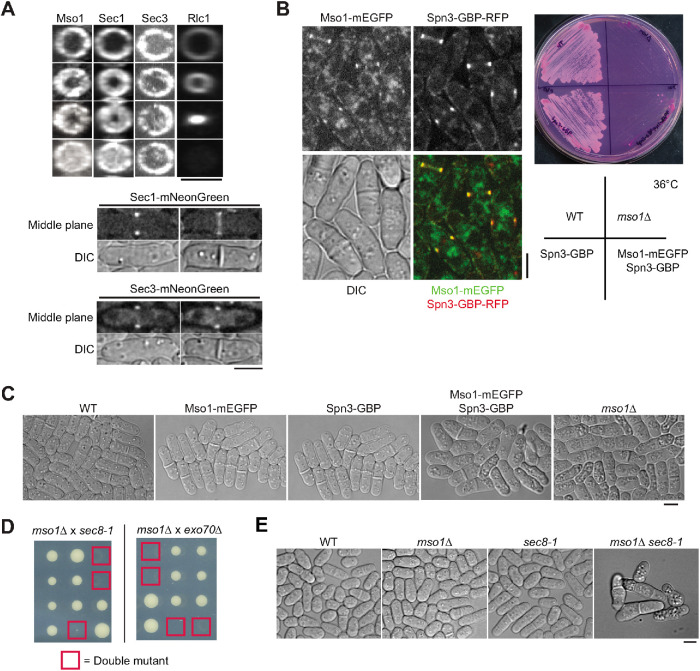
Both Mso1 and Sec1 interact with the exocyst complex in budding yeast (Knop et al., 2005; Morgera et al., 2012). Thus, it has been assumed that Mso1/Sec1 and the exocyst work together for vesicle fusion on the plasma membrane during exocytosis. However, no colocalization between them has been reported. After ring constriction, both Sec1 and Mso1 localized across the division plane (Figure 6A). However, the exocyst complex (marked with the Sec3 subunit) concentrated on the outer rim of the division site with little signal at the interior (Figure 6A). Their nonoverlapping localization in fission yeast suggests that Sec1 and Mso1 have functions independent of the exocyst complex.
FIGURE 6:
Mso1 has a function independent of the exocyst complex. (A) Mso1 and Sec1 do not have identical localization with the exocyst. (Top) End-on views of division site localization of Mso1, Sec1, exocyst subunit Sec3, and Rlc1 in cells at similar stages of septation. (Bottom) Middle focal planes and DIC images of Sec1 and Sec3 before (left) and after (right) septum formation. (B) Ectopic targeting of Mso1 to septin rings. (Left) Fluorescence images shown are the middle focal plane. (Right) Growth of the indicated strains at 36°C for 2 d. (C) WT, Mso1-mEGFP, Spn3-GBP, Mso1-mEGFP Spn3-GBP, and mso1Δ strains imaged after 8.5 h growth in liquid culture at 36°C. (D, E) Mutations in mso1 and exocyst are synthetic lethal at 25°C. (D) Tetrad analysis of crosses between mso1∆ and sec8-1 or exo70∆. Red squares mark the predicted double mutants. (E) Images of representative colonies from the cross between mso1∆ and sec8-1. Bars, 5 µm.
To test the importance of the interior fraction of Mso1 on the division plane, we ectopically targeted Mso1 to the septin ring (Figure 6B), which colocalizes with the exocyst complex at the rim of the division plane (Martín-Cuadrado et al., 2005; Pérez et al., 2015). One of the septin functions is acting as a scaffold to recruit other proteins to the division site (Addi et al., 2018; Zheng et al., 2018; Marquardt et al., 2019). Once the contractile-ring constriction begins, the septins separate into two rings and stay at the rim of the cleavage plane. Mso1 was efficiently concentrated at the outer rim of the division plane by septin Spn3-GBP (Figure 6B). When grown at 36°C, cells of strain mso1-mEGFP spn3-GBP, but not other control strains, grew slowly similar to mso1Δ (Figure 6B). While the WT and control strains had normal morphology, the cells of mso1-mEGFP spn3-GBP and mso1Δ showed increased cell death and visible vacuolation in differential interference contrast (DIC) images (Figure 6C). This suggests that the ability of Mso1 and Sec1 to help vesicle fusion at the interior of the division plane is important for proper cytokinesis.
To further investigate the genetic interaction between mso1 and the exocyst complex, mso1∆ was crossed to exocyst mutants sec8-1 or exo70∆. These mutations were synthetic lethal as no double mutant colonies could be recovered even at 25°C (Figure 6D). While the single mutants grew well, double mutant cells often died after a few cell divisions (Figure 6E). The synthetic lethal interactions suggest that Mso1 and the exocyst are also on parallel pathways during exocytosis. Thus, the exocyst complex may tether vesicles for Sec1 and Mso1 mediated vesicle fusion at the rim of the division site, but another tether may cooperate with Sec1 and Mso1 at the interior of the division plane.
DISCUSSION
Our work highlights the importance of cellular trafficking and vesicle fusion during cytokinesis. Mso1 plays an important role in localizing the SM protein Sec1 in fission yeast. Mso1 and Sec1 work together for the proper delivery of both membranes and cargoes at the division site. In their absence, cytokinesis becomes delayed and defective, which leads to severe defects in cell division and cell death.
The roles of Mso1 and Sec1 in vesicle fusion during cytokinesis
The role of Mso1 in cytokinesis had not been studied previously, and its function in regulating Sec1 is incompletely understood. Here we observed that cytokinesis is dramatically affected by the loss of Mso1 (at the restrictive temperature) or Sec1. Ring constriction and ring disassembly in the mutants take significantly longer than in WT cells, with abnormal myosin structures appearing during ring disassembly. These myosin structures may be a response to the failure of properly sealing the barrier between the two daughter cells that we observed in FLIP experiments (Figure 2, I and J), which likely arises from a lack of plasma membrane, but not from stalled ring constriction (Figure 2H). The loss of Mso1 reduces vesicle fusion at the division site and prevents the efficient delivery of essential cargoes, such as β-glucan synthase Bgs1 (Figure 3). Without Mso1, Sec1 can still localize to the plasma membrane at low temperature, although less efficiently. When the temperature is increased, cellular traffic speeds up and the membrane may become more fluid, putting stress on the exocytic pathway and the plasma membrane. This stress together with the absence of Mso1 may lead to the complete loss of Sec1 localization and its aggregation into puncta. Without Sec1 present at the plasma membrane to promote vesicle fusion, vesicles accumulate near the plasma membrane, and cytokinesis is impaired. These results suggest that Mso1 functions to maintain the proper localization of Sec1 at the division site for vesicle fusion. Our data highlight the important roles of Mso1 and Sec1 in membrane insertion in yeast cytokinesis. While Sec1 had not been studied during cytokinesis in animal and fungal cells, SM family protein KEULE has been shown to stabilize the fusion competent conformation of syntaxin KNOLLE during Arabidopsis cytokinesis (Park et al., 2012). Our study provides another example to support the theory that targeted membrane addition is the most conserved aspect of cytokinesis (Hales et al., 1999).
Mso1 and Sec1 may work with vesicle tethers other than the exocyst at the division site
Previous works in budding yeast revealed that Mso1 and Sec1 cooperate with the exocyst tethering complex to aid vesicle fusion at the plasma membrane. The N-terminus of Mso1 is found in complex with exocyst components as well as Sec1 and SNARE components in coimmunoprecipitation (co-IP) experiments (Knop et al., 2005). Purified Sec1 interacts with the exocyst subunit Sec6 during in vitro binding experiments (Morgera et al., 2012). The Mso1 C-terminus, specifically the last 22 amino acids, is required for polarized membrane targeting and may aid in the targeting of Sec1 (Weber-Boyvat et al., 2011). In mammalian cells, an N-terminal peptide sequence in t-SNARE syntaxin interacts with the hydrophobic binding pocket of the SM protein Munc18 (Hu et al., 2007). The cognate SNARE for Sec1, Sso1 in budding yeast, does not contain the N-terminal peptide sequence, and Sec1 lacks the hydrophobic binding pocket, which suggest Sec1’s interaction with the SNARE complex is different from that of Munc18a (Togneri et al., 2006). Nevertheless, both Sec1 and Munc18a appear to promote vesicle fusion. Studies have shown that the Sec1 C-terminal tail interacts with SNARE complexes and promotes complex formation (Weber-Boyvat et al., 2011). Sec1 likely plays a role in fusion before and after SNARE complex formation in cooperation with the exocyst complex (Hashizume et al., 2009).
In fission yeast, we also observe colocalization and maybe cooperation among Mso1, Sec1, and the exocyst at cell tips and at the outer rim of the division site. However, our microscopy data reveal that Mso1 and Sec1 also have a very distinct localization from the exocyst at the interior of the division plane. This and other experiments suggest that Mso1 and Sec1 may also drive the fusion of vesicles tethered independently from the exocyst complex. Indeed, the mislocalization of Mso1 to the rim of the division site in spn3-GBP mso1-mEGFP cells suggests that Mso1 plays important roles at the interior of the division site (Figure 6, B and C), although we cannot rule out the possibility that the mistargeted Mso1 at the rim led to the defects in the spn3-GBP mso1-mEGFP cells. Our previous work showed that the TRAPP-II complex can tether vesicles at the interior of the division site (Wang et al., 2016). This TRAPP-II mediated exocytosis depends in part on the UNC-13/Munc13 protein Ync13 (Zhu et al., 2018). Interestingly, plant SM protein KEULE’s phragmoplast localization has been shown to be regulated by the TRAPP-II complex (Steiner et al., 2016). Together, these data suggest that Mso1 and Sec1 may have functions independent of the exocyst tethering complex and may cooperate with other vesicle tethers at the division site. Such an interaction would suggest as yet unexplored functions for Mso1 and Sec1. Further experiments are needed to determine whether the TRAPPII complex and Mso1/Sec1 work together at the interior of the division plane for vesicle fusion.
In the future, it will be important to understand how Sec1 and Mso1 coordinate exocytosis and endocytosis, as both are essential for successful cytokinesis (Gerald et al., 2001; Feng et al., 2002; Montagnac et al., 2008; Giansanti et al., 2015). Ync13 has been shown to play roles in the regulation of both exocytosis and endocytosis in fission yeast (Zhu et al., 2018), and Munc13 has been shown to cooperate with the SM protein Munc18 to regulate SNARE protein conformation (Ma et al., 2011; Yang et al., 2015). Thus, Ync13 is a candidate for additional regulation of the fusion complex containing SNAREs, Sec1, Mso1, Rab GTPase Sec4, and tethering proteins.
As the role of fission yeast Mso1 in exocytosis appears to be very similar to that of Mso1 in budding yeast and Mint1 in humans, we expect these homologues and the SM proteins to play roles during cytokinesis similar to those we have observed here. Thus, it will be interesting to investigate the roles of Mso1, Mint1, and the SM proteins in cytokinesis to confirm whether these functions are conserved in future studies.
MATERIALS AND METHODS
Strain construction and genetic and molecular methods
The strains used in this study are listed in Table 1. Genes were deleted or tagged at the C-terminal or N-terminal end at the endogenous locations using PCR-based gene targeting methods (Bähler et al., 1998). All tagged proteins use their native promotors except the free GFP. To observe sec1∆ cells, we deleted one copy of sec1 in a diploid strain rlc1-tdTomato-natMX6/rlc1-tdTomato-natMX6 ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 to obtain the strain sec1+/sec1∆. The diploid cells were then sporulated on a SPA5S plate, and the spores were dissected on rich medium YE5S (yeast extract plus five supplements) plate. Cells from the germinated spores were grown at 25°C and imaged using tetrad fluorescence microscopy (Coffman et al., 2013; Lee et al., 2014; Davidson et al., 2016a).
TABLE 1:
S. pombe strains used in this study.
| Strain | Genotype | Figure/Movie/Reference |
|---|---|---|
| JW5663 | h– mso1-mECitrine-kanMX6 ade6-210 ura4-D18 leu1-32 | Figures 1, B and C, and 6A; Supplemental Figure S1, C and D |
| JW1109 | h+ kanMX6-Pmyo2-mEGFP-myo2 ade6-M210 leu1-32 ura4-D18 | Figures 1C and 2, F and G |
| JW5813 | h+ mso1-mECitrine-kanMX6 sad1-mCherry-natMX6 ade6-M210 leu1-32 ura4-D18 | Figure 1, D and E |
| JW5665 | h– mso1∆::kanMX6 ade6-210 ura4-D18 leu1-32 | Figures 2, A, B, C, and D, 3, A, B, and C, and 6, B, C, D, and E; Supplemental Figure S2B; Supplemental Movie 4 |
| JW81 | h– ade6-210 ura4-D18 leu1-32 | Figures 2, A, B, C, and D, 3, A, B, and C, 5B, and 6, B and C; Supplemental Figure S2A; Supplemental Movie 3 |
| JW5673 | h+ mso1Δ::kanMX6 rlc1-tdTomato-natMX6 sad1-mEGFP-kanMX6 ade6-210 ura4-D18 leu1-32 | Figure 2, E and G |
| JW2178 | h+ rlc1-tdTomato-natMX6 sad1-mEGFP-kanMX6 ade6-M210 ura4-D18 leu1-32 | Figures 2, E and G, and 6A |
| JW6802 | mso1∆::kanMX6 kanMX6-Pmyo2-mEGFP-myo2 ade6-210 ura4-D18 leu1-32 | Figure 2, F and G |
| JW1144 | h– ain1-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | Figure 2H |
| JW9555 | mso1Δ::kanMX6 ain1-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | Figure 2H |
| JW3313 | h– kanMX6-3nmt1-mEGFP rlc1-tdTomato-natMX6 ade6-M210 leu1-32 ura4-D18 | Figure 2, I and J; Supplemental Movie 1 |
| JW6158 | mso1∆::kanMX6 kanMX6-3nmt1-mEGFP rlc1-tdTomato-natMX6 ade6-210 leu1-32 ura4-D18 | Figure 2, I and J; Supplemental Movie 2 |
| MBY887 | h+ sec8-1 ura4-D18 leu1-32 | Figures 3, A, B, and C, and 6, D and E; Wang et al., 2002 |
| JW5249 | GFP-bgs1-leu1+ bgs1Δ::ura4+ rlc1-tdTomato-natMX6 ade6-M210 leu1-32 ura4-D18 | Figure 3, D and E |
| JW5840 | mso1∆::kanMX6 GFP-bgs1-leu1+ bgs1Δ::ura4+ rlc1-tdTomato-natMX6 ade6-M210 leu1-32 ura4-D18 | Figure 3, D and E |
| IJ253 | h– GFP-syb1-kanMX6 ade6 leu1 ura+ | Figure 3F; Supplemental Figure S3, A, B, C, and D; Jourdain et al., 2012 |
| JW6199 | mso1∆::kanMX6 GFP-syb1-kanMX6 ade6 leu1 ura4-D18 | Figure 3F; Supplemental Figure S3, A and B |
| JW7361 | sec1-tdTomato-natMX6 mso1-mEGFP-kanMX6 ade6-210 leu1-32 ura4-D18 | Figure 4A |
| JW7068 | sec1-tdTomato-natMX6 mso1-mEGFP-kanMX6 tom20-GBP-hphMX6 ade6-210 leu1-32 ura4-D18 | Figure 4B |
| JW7628 | sec1-13Myc-kanMX6 mso1-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | Figure 4C |
| JW5723 | h– mso1-mEGFP-kanMX6 ade6-210 leu1-32 ura4-D18 | Figures 4C and 6C |
| JW7614 | h– sec1-13Myc-kanMX6 ade6-210 leu1-32 ura4-D18 | Figure 4C |
| JW6115 | h– sec1-tdTomato-natMX6 ade6-210 leu1-32 ura4-D18 | Figure 4, E and F |
| JW6631 | sec1-tdTomato-natMX6 mso1∆::kanMX6 ade6-210 leu1-32 ura4-D18 | Figure 4, E and F |
| JW9103 | h– sec1-M2-his5+-kanMX6 his5Δ ade6-210 leu1-32 ura4 | Figure 5B |
| JW7021 | h– sec1-mEGFP-kanMX6 ade6-210 leu1-32 ura4-D18 | Figure 5C |
| JW9105 | h+ sec1-M2-mEGFP-hphMX6-his5+-kanMX6 his5Δ? ade6-210 leu1-32 ura4 | Figure 5C |
| JW5724 | h– mso1-tdTomato-natMX6 ade6-210 leu1-32 ura4-D18 | Figure 5D |
| JW9109 | sec1-M2-his5+-kanMX6 mso1-tdTomato-natMX6 his5Δ? ade6-210 leu1-32 ura4 | Figure 5D |
| JW9169 | mso1-mEGFP-kanMX6 sec1-M2-mScarlet-I-hphMX6-his5+-kanMX6 his5Δ? ade6-210 leu1-32 ura4-D18 | Figure 5E |
| JW6114 | h+/h– sec1∆::kanMX6/sec1+ rlc1-tdTomato-natMX6/rlc1-tdTomato-natMX6 ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | Figure 5, F and G; Supplemental Figure S4C; Supplemental Movies 5 and 6 |
| JW7568 | h– sec1-mNeonGreen-kanMX6 ade6-M210 leu1-32 ura4-D18 | Figure 6A |
| JW7580 | h– sec3-mNeonGreen-kanMX6 ade6-M210 leu1-32 ura4-D18 | Figure 6A |
| JW7392 | spn3-GBP-RFP-hphMX6 mso1-mEGFP-kanMX6 ade6-210 leu1-32 ura4-D18 | Figure 6B |
| JW7230 | h– spn3-GBP-hphMX6 ade6-M210 leu1-32 ura4-D18 | Figure 6, B and C |
| JW7393 | spn3-GBP-hphMX6 mso1-mEGFP-kanMX6 ade6-210 leu1-32 ura4-D18 | Figure 6, B and C |
| JW2716 | h+ exo70Δ::kanMX4 ade6 leu1-32 ura4-D18 | Figure 6D |
| JW7575 | mso1-mNeonGreen-kanMX6 rlc1-tdTomato-natMX6 ade6-M210 leu1-32 ura4-D18 | Supplemental Figure S1B |
| JW2180 | psy1Δ::kanMX6 leu1+::GFP-psy1 ade6-M210 ura4 | Supplemental Figure S3, A, B, C, and D |
| JW7605 | mso1∆::kanMX6 psy1Δ::kanMX6 leu1+::GFP-psy1 ade6-M210 ura4 | Supplemental Figure S3, A and B |
| JW9554 | sec1-M2-his5+-kanMX6 psy1Δ::kanMX6 leu1+::GFP-psy1 his5Δ? ade6-M210 ura4 | Supplemental Figure S3, C and D |
| JW9553 | sec1-M2-his5+-kanMX6 GFP-syb1-kanMX6 his5Δ? ade6 leu1 ura+ | Supplemental Figure S3, C and D |
| JW6957 | h– mso1-mEGFP-kanMX6 tom20-GBP-hphMX6 ade6-210 leu1-32 ura4-D18 | Supplemental Figure S4A |
| JW7075 | sec1-tdTomato-natMX6 tom20-GBP-hphMX6 ade6-210 leu1-32 ura4-D18 | Supplemental Figure S4A |
| JW9570 | kanMX6-Patg8-mEGFP-atg8 sec1-tdTomato-natMX6 mso1∆::kanMX6 ade6-M210 leu1-32 ura4-D18 | Supplemental Figure S4B |
| JW9558 | sec1-tdTomato-natMX6 mso1∆::kanMX6 GFP-syb1-kanMX6 ade6-M210 leu1-32 ura4-D18 | Supplemental Figure S4B |
Mso1(1-70) and Mso1(71-151) were cloned into pET21a vector between the T7 tag and the 6His tag by Gibson assembly (Gibson et al., 2009). Full-length Mso1 was cloned into the pCOLADuet vector using BamHI and SalI restriction sites after 6His. Sec1 was cloned into the pMAL-c5x vector after MBP and a 6His tag was added to the C-terminal of Sec1 by Gibson assembly. pMAL-c5x was modified by adding the TEV site and 6His tag to the C-terminal of MBP to express MBP-6His. The constructs were confirmed by sequencing.
The temperature-sensitive sec1-M2 mutant was created using marker reconstitution mutagenesis (Tang et al., 2011; Lee et al., 2014). Briefly, sec1 was cloned into a pHis5C plasmid with the C-terminal portion of his5 after the 3′UTR of sec1. Error prone PCR was performed using a mutagenic cocktail (8 mM dTTP, 8 mM dCTP, 48 mM MgCl2, and 5 mM MnCl2) in addition to the normal PCR mix containing 0.2 mM dNTPs, NEB Taq polymerase, and Taq Pol buffer. The PCR product was then transformed into strain JW7670 (h– sec1-his5∆C-kanMX6 his5∆ ade6-210 leu1-32 ura4) and positive transformants were selected on medium without histidine. The positive colonies were then grown on YE5S + PB plates at a range of temperatures from 25°C to 36°C and screened by temperature sensitivity. Finally, we sequenced the mutants to identify mutations located in the ORF but not in the UTRs.
For latrunculin A (Lat-A) treatment, 1 ml of cells was concentrated to 100 µl and incubated with 100 µM Lat-A or equal volume of dimethyl sulfoxide (DMSO) for 10 min. Cells were then imaged on bare slides (Wu et al., 2001; Zhu et al., 2018). For methyl benzimidazole-2-yl carbamate (MBC) treatment, 1 ml for cells were incubated with 5 µl 5 mg/ml MBC or DMSO for 15 min. Cells were imaged on gelatin pads containing the same concentration of MBC or DMSO (Wu et al., 2011; Zhu et al., 2018).
For the acid phosphatase secretion assay (Wang et al., 2002), cells were grown in EMM5S (Edinburgh minimal medium plus five supplements) to log phase at 25°C or shifted to 36°C from 25°C at time zero. To measure acid phosphatase secretion, cell cultures (1.5 ml) were centrifuged and 500 µl of the supernatant was mixed with 500 µl of substrate (2 mM p-nitrophenyl phosphate, 0.1 M sodium acetate, pH 4.0). The reaction was incubated at 25°C for 10 min before quenching with 500 µl of 1 M sodium hydroxide and measuring the absorbance at 405 nm. The secretion and cell density (OD at 595 nm) were measured for each sample every hour. The 405 nm absorbance at time zero was subtracted for each culture.
Microscopy
Strains stored at –80°C were streaked onto YE5S and grown for 2 d. Fresh cells were inoculated into YE5S liquid cultures and grown in exponential phase for ∼2 d at 25°C. Cells were collected at 3000 rpm for 30 s and then washed with EMM5S to reduce autofluorescence. n-Propyl gallate was added to a final concentration of 50 nM from a 10X stock solution (in EMM5S) during the second wash to reduce phototoxicity and photobleaching. Cells were then imaged on glass slides with a gelatin pad (20% gelatin in EMM5S + 50 nM n-propyl gallate) at ∼23°C. For cells imaged at 36°C, the cells were grown in liquid culture for ∼2 d and then shifted to 36°C and grown for the time indicated before imaging. These cells were then collected at 3000 rpm for 30 s and placed on a prewarmed coverglass-bottom dish (0420041500C; Bioptechs, Butler, PA). The cells were then covered with a piece of prewarmed YE5S agar and imaged on a temperature-controlled stage (Stage Top Incubator INUB-PPZI2-F1 with UNIV2-D35 dish holder; Tokai Hit, Shizuoka-ken, Japan) set to 36°C.
Microscopy was performed as previously described (Davidson et al., 2016a). We used a spinning-disk confocal system (UltraVIEW Vox CSUX1 system; PerkinElmer, Waltham, MA) with 440-, 488-, 515-, and 561-nm solid-state lasers and back thinned electron-multiplying charge-coupled device (EMCCD) cameras (C9100-13 or C9100-23B; Hamamatsu Photonics, Bridgewater, NJ) on a Nikon Ti-E microscope with a 100x/1.4 numerical aperture (NA) Plan-Apo objective lens (Nikon, Melville, NY). DIC images for septation quantification were taken on a Nikon Eclipse Ti inverted microscopy equipped with a DS-QI1 Nikon cooled digital camera (Nikon, Melville, NY).
Images were analyzed using Volocity (PerkinElmer) and ImageJ (National Institutes of Health, Bethesda, MD). Fluorescence images shown are maximum-intensity projections of image stacks with 0.4–0.6 µm spacing except where noted.
For quantification of the kinetics of the contractile ring, stages were defined as follows: ring formation lasted from the appearance of myosin nodes until the appearance of a single, compact, and cohesive ring; maturation lasted from the end of node condensation until before the ring could be seen constricting at the fast phase (Ramos et al., 2019); constriction lasted from the end of maturation until the myosin (Myo2 or Rlc1) reached peak pixel intensity when the ring constricted to a spot; and disassembly lasted from the end of constriction until the myosin signal disappeared from the division site. Two tailed Student’s t test was used to determine whether values were significantly different in this study.
FLIP assay
FLIP was performed on the Photokinesis unit of the Ultraview Vox CSUX1 system using cells expressing Rlc1-tdTomato and diffusible cytoplasmic GFP. A single image was taken to identify appropriate cells to bleach. We did FLIP on cells that had finished ring constriction (Rlc1 had reached peak pixel intensity when the ring constricted to a spot or during ring disassembly). Then movies were captured by taking a single image at the central focal plane, immediately bleaching selected regions of interest, and then taking another image. After a 30 s delay, this process was repeated. To analyze the data, the mean intensity of each daughter cell was measured and corrected for background fluorescence (from regions without cells) and photobleaching from imaging. These values were then normalized to the starting intensity of the unbleached daughter cell.
Electron microscopy
Electron microscopy and electron tomography were performed at the Boulder Electron Microscopy Services at the University of Colorado at Boulder. Cells were grown in YE5S at 25°C for ∼48 h and then shifted to 36°C for 2 h. Cells were then harvested and frozen using a Wohlwend Compact 02 High Pressure Freezer, freeze-substituted in the presence of 2% osmium tetroxide and 0.1% uranyl acetate in acetone, and embedded in Epon-Araldite epoxy resin. These frozen pellets were then sectioned and stained with uranyl acetate and lead citrate and imaged on a Philips CM100 transmission electron microscope (FEI, Hillsboro, OR) (Giddings et al., 2001; Lee et al., 2014; Davidson et al., 2016b; Liu et al., 2016; Zhu et al., 2018). To quantify the number of vesicles at the division site, we drew a rectangle approximately 500 nm wide (along the cell long axis) centered on the septum and counted vesicles inside that area in a single thin section. For electron tomography, multiple serial sections were imaged using tilt series and then aligned using gold fiducial markers. The aligned projections were then reconstructed to show depth in the cell.
IP and Western blotting
IP and Western blotting assays were carried out as described previously (Lee and Wu, 2012; Liu et al., 2016). Using S. pombe cell extracts, Mso1-mEGFP was pulled down with protein G covalently coupled magnetic Dynabeads (100.04D; Invitrogen, Carlsbad, CA) using polyclonal anti-GFP antibodies (NB600-308; Novis Biologicals, Littleton, CO). These beads were washed five times before the proteins were eluted by boiling in sample buffer. The samples were then run on SDS–PAGE gels and transferred to nitrocellulose membrane. Proteins were detected with monoclonal anti-GFP antibody (1:2000 dilution; 11814460001; Roche, Mannheim, Germany) or monoclonal anti-Myc antibody (1:5000 dilution; 9E10; Santa Cruz Biotechnology, Dallas, TX). Secondary antibody was anti-mouse IRDye 680RD (1:10,000; 925-68070; LiCOR, Lincoln, NE). Blots were imaged on a LiCOR Odyssey (LiCOR).
Protein purification and in vitro binding assays
We purified recombinant proteins by transforming the plasmids into Rosetta (DE3) pLysS Escherichia coli cells (Novagen). Mso1-6His, Mso1(1-70)-6His, and Mso1(71-151)-6His were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 37°C for 4 h. MBP-Sec1-6His and MBP-6His were expressed with 0.5 mM IPTG at 25°C for 15 h. Purifications of 6His-tagged proteins were carried out as previously described (Zhu et al., 2013). The proteins were purified with Talon metal affinity resin (635501; Clontech, Mountain View, CA) in extraction buffer (50 mM sodium phosphate, pH 8.0, 400 mM NaCl, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 20 mM imidazole) with EDTA-free protease inhibitor tablet (Roche) and eluted with elution buffer (50 mM sodium phosphate, pH 8.0, 400 mM NaCl, 10 mM β-mercaptoethanol, 1 mM PMSF, and 300 mM imidazole). The purified proteins were then dialyzed into the binding buffer (137 mM NaCl, 2 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.5 mM dithiothreitol, and 10% glycerol, pH 7.4).
For in vitro binding assays between MBP-Sec1-6His and 6His-Mso1, Mso1(1-70)-6His, and Mso1(71-151)-6His, purified proteins were dialyzed into the binding buffer. We incubated 1 ml MBP-Sec1-6His (5 µM) or 90 µl MBP-6His (27 µM) control with 500 µl Amylose beads for 1 h at 4°C and washed the beads eight times with 1 ml of the binding buffer each time to remove unbound proteins. Then 1 ml 6His-Mso1, Mso1(1-70)-6His, or Mso1(71-151)-6His (10 µM) was incubated with the 100 µl beads with MBP-Sec1-6His or MBP-6His for 1 h at 4°C. After eight washes with 1 ml of the binding buffer each time, the beads were boiled with sample buffer for 5 min. Then the samples were run on SDS–PAGE gel and detected with Coomassie Blue staining.
Homology modeling
Homology modeling was performed using Modeller 9.14 (Sali and Blundell, 1993). The structure of human Munc18-2 (Protein Data Bank (PDB): 4cca [ Hackmann et al., 2013]) was used to build the model for Sec1. S. pombe Sec1 and the Munc18-2 sequences were aligned. A model was then built based on this alignment using Modellers’ automodel. The best one out of the five models generated was selected to use. A model of Mso1 was constructed in a similar manner, using the structure of human Mint1 (PDB: 4DBB [ Matos et al., 2012]) as template.
Supplementary Material
Acknowledgments
We thank Mohan Balasubramanian, Kathy Gould, Sophie Martin, Thomas Pollard, John Pringle, and Takashi Toda for strains and plasmids; the Anita Hopper, James Hopper, Stephen Osmani, and Zhengrong Wu laboratories for equipment, reagents, and technical support; Mary Morphew and Thomas Giddings, Jr., at the University of Colorado for EM; and current and past members of the Wu laboratory for helpful discussions and suggestions. Research reported in this publication was supported by a Pelotonia Postdoctoral Fellowship to S.Z. and by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under Award Number R01GM118746 to J.-Q.W. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations used:
- DIC
differential interference contrast
- EMM5S
Edinburgh minimal medium plus five supplements
- FLIP
fluorescence loss in photobleaching
- GBP
GFP-binding protein
- MBC
methyl benzimidazole-2-yl carbamate
- MTC
multi-subunit tethering complex
- PB
phloxin B
- SM
Sec1/Munc18
- SPB
spindle pole body
- WT
wild type
- YE5S
yeast extract plus five supplements.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0067) on May 20, 2020.
REFERENCES
- Aalto MK, Jäntti J, Ostling J, Keränen S, Ronne H. (1997). Mso1p: a yeast protein that functions in secretion and interacts physically and genetically with Sec1p. Proc Natl Acad Sci USA , 7331–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addi C, Bai J, Echard A. (2018). Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr Opin Cell Biol , 27–34. [DOI] [PubMed] [Google Scholar]
- Albertson R, Riggs B, Sullivan W. (2005). Membrane traffic: a driving force in cytokinesis. Trends Cell Biol , 92–101. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast , 943–951. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. (2004). Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol , R806–R818. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. (2007). Cytokinesis: placing and making the final cut. Cell , 847–860. [DOI] [PubMed] [Google Scholar]
- Bendezú FO, Vincenzetti V, Martin SG. (2012). Fission yeast Sec3 and Exo70 are transported on actin cables and localize the exocyst complex to cell poles. PLoS One , e40248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. (1999). Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol , 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Rizo J. (2010). At the junction of SNARE and SM protein function. Curr Opin Cell Biol , 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Flores A, Weinberger A, Robinson M, Gerst JE. (2005). Mso1 is a novel component of the yeast exocytic SNARE complex. J Biol Chem , 34033–34041. [DOI] [PubMed] [Google Scholar]
- Chang F, Martin SG. (2009). Shaping fission yeast with microtubules. Cold Spring Harb Perspect Biol , a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Sees JA, Kovar DR, Wu J-Q. (2013). The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J Cell Biol , 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucu B, Degreif D, Bertl A, Thiel G. (2017). Vesicle fusion and fission in plants and yeast. Cell Calcium , 40–45. [DOI] [PubMed] [Google Scholar]
- Davidson R, Liu Y, Gerien KS, Wu J-Q. (2016a). Real-time visualization and quantification of contractile ring proteins in single living cells. Methods Mol Biol , 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R, Pontasch JA, Wu J-Q. (2016b). Sbg1 is a novel regulator for the localization of the β-glucan synthase Bgs1 in fission yeast. PLoS One , e0167043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuke ML, Munson M. (2016). The secret life of tethers: the role of tethering factors in SNARE complex regulation. Front Cell Dev Biol , 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Schwarz H, Jesuthasan S. (2002). Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp Cell Res , 14–20. [DOI] [PubMed] [Google Scholar]
- Furukawa N, Mima J. (2014). Multiple and distinct strategies of yeast SNAREs to confer the specificity of membrane fusion. Sci Rep , 4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald NJ, Damer CK, O’Halloran TJ, De Lozanne A. (2001). Cytokinesis failure in clathrin-minus cells is caused by cleavage furrow instability. Cytoskeleton , 213–223. [DOI] [PubMed] [Google Scholar]
- Gerien KS, Wu J-Q. (2018). Molecular mechanisms of contractile-ring constriction and membrane trafficking in cytokinesis. Biophys Rev , 1649–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Vanderleest TE, Jewett CE, Sechi S, Frappaolo A, Fabian L, Robinett CC, Brill JA, Loerke D, Fuller MT, et al. (2015). Exocyst-dependent membrane addition is required for anaphase cell elongation and cytokinesis in Drosophila . PLoS Genet , e1005632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods , 343–345. [DOI] [PubMed] [Google Scholar]
- Giddings TH, O’Toole ET, Morphew M, Mastronarde DN, McIntosh JR, Winey M. (2001). Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol , 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Grant A, Novick P. (1999). Exo84p is an exocyst protein essential for secretion. J Biol Chem , 23558–23564. [DOI] [PubMed] [Google Scholar]
- Hackmann Y, Graham SC, Ehl S, Honing S, Lehmberg K, Arico M, Owen DJ, Griffiths GM. (2013). Syntaxin binding mechanism and disease-causing mutations in Munc18–2. Proc Natl Acad Sci USA , E4482–E4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales KG, Bi E, Wu J-Q, Adam JC, Yu I-C, Pringle JR. (1999). Cytokinesis: an emerging unified theory for eukaryotes? Curr Opin Cell Biol , 717–725. [DOI] [PubMed] [Google Scholar]
- Hashizume K, Cheng Y-S, Hutton JL, Chiu C-H, Carr CM. (2009). Yeast Sec1p functions before and after vesicle docking. Mol Biol Cell , 4673–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P. (2013). A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol , 130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Guo W. (2009). The exocyst complex in polarized exocytosis. Curr Opin Cell Biol , 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider MR, Munson M. (2012). Exorcising the exocyst complex. Traffic , 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SH, Latham CF, Gee CL, James DE, Martin JL. (2007). Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci USA , 8773–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, He M, Port SA, Baker RW, Xu Y, Qu H, Xiong Y, Wang Y, Jin H, Eisemann TJ, et al. (2018). Munc18–1 catalyzes neuronal SNARE assembly by templating SNARE association. Elife , e41771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain I, Dooley HC, Toda T. (2012). Fission yeast Sec3 bridges the exocyst complex to the actin cytoskeleton. Traffic , 1481–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. (2010). Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol , 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Miller KJ, Mazza M, Feng D, Weber M, Keränen S, Jäntti J. (2005). Molecular interactions position Mso1p, a novel PTB domain homologue, in the interface of the exocyst complex and the exocytic SNARE machinery in yeast. Mol Biol Cell , 4543–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wang N, Hu W, Schott K, Bähler J, Giddings TH, Pringle JR, Du L-L, Wu J-Q. (2014). Regulation of spindle pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol Biol Cell , 2735–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wu J-Q. (2012). Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J Cell Sci , 2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lee I-J, Sun M, Lower CA, Runge KW, Ma J, Wu J-Q. (2016). Roles of the novel coiled-coil protein Rng10 in septum formation during fission yeast cytokinesis. Mol Biol Cell , 2528–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, Martin SG. (2011). Shaping fission yeast cells by rerouting actin-based transport on microtubules. Curr Biol , 2064–2069. [DOI] [PubMed] [Google Scholar]
- Luo G, Zhang J, Guo W. (2014). The role of Sec3p in secretory vesicle targeting and exocyst complex assembly. Mol Biol Cell , 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Li W, Xu Y, Rizo J. (2011). Munc13 mediates the transition from the closed syntaxin– Munc18 complex to the SNARE complex. Nat Struct Mol Biol , 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt J, Chen X, Bi E. (2019). Architecture, remodeling, and functions of the septin cytoskeleton. Cytoskeleton , 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Cuadrado AB, Morrell JL, Konomi M, An H, Petit C, Osumi M, Balasubramanian M, Gould KL, Del Rey F, de Aldana CRV. (2005). Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol Biol Cell , 4867–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos MF, Xu Y, Dulubova I, Otwinowski Z, Richardson JM, Tomchick DR, Rizo J, Ho A. (2012). Autoinhibition of Mint1 adaptor protein regulates amyloid precursor protein binding and processing. Proc Natl Acad Sci USA , 3802–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. (2006). ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe . Nat Biotechnol , 841–847. [DOI] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Sollner TH, Rothman JE. (2000). Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature , 153–159. [DOI] [PubMed] [Google Scholar]
- Meitinger F, Palani S. (2016). Actomyosin ring driven cytokinesis in budding yeast. Semin Cell Dev Biol , 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G, Echard A, Chavrier P. (2008). Endocytic traffic in animal cell cytokinesis. Curr Opin Cell Biol , 454–461. [DOI] [PubMed] [Google Scholar]
- Morgera F, Sallah MR, Dubuke ML, Gandhi P, Brewer DN, Carr CM, Munson M. (2012). Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Mol Biol Cell , 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell , 205–215. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Südhof TC. (1997). Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem , 31459–31464. [DOI] [PubMed] [Google Scholar]
- Park M, Touihri S, Muller I, Mayer U, Jurgens G. (2012). Sec1/Munc18 protein stabilizes fusion-competent syntaxin for membrane fusion in Arabidopsis cytokinesis. Dev Cell , 989–1000. [DOI] [PubMed] [Google Scholar]
- Pérez P, Portales E, Santos B. (2015). Rho4 interaction with exocyst and septins regulates cell separation in fission yeast. Microbiology , 948–959. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. (2010). Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol , 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M, Cortes JCG, Sato M, Rincon SA, Moreno MB, Clemente-Ramos JA, Osumi M, Perez P, Ribas JC. (2019). Two S. pombe septation phases differ in ingression rate, septum structure, and response to F-actin loss. J Cell Biol , 4171–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. (2012). The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices-guilty as charged? Annu Rev Cell Dev Biol , 279–308. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics , 282–289. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. (1993). Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol , 779–815. [DOI] [PubMed] [Google Scholar]
- Scott BL, Van Komen JS, Irshad H, Liu S, Wilson KA, McNew JA. (2004). Sec1p directly stimulates SNARE-mediated membrane fusion in vitro . J Cell Biol , 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Thompson J, Yates JR, Sawin KE. (2011). Characterization of Mug33 reveals complementary roles for actin cable-dependent transport and exocyst regulators in fission yeast exocytosis. J Cell Sci , 2187–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner A, Müller L, Rybak K, Vodermaier V, Facher E, Thellmann M, Ravikumar R, Wanner G, Hauser M-T, Assaad Farhah F. (2016). The membrane-associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis . Mol Plant , 528–540. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature , 347–353. [DOI] [PubMed] [Google Scholar]
- Tang X, Huang J, Padmanabhan A, Bakka K, Bao Y, Tan BY, Cande WZ, Balasubramanian MK. (2011). Marker reconstitution mutagenesis: a simple and efficient reverse genetic approach. Yeast , 205–212. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. (1996). The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae . EMBO J , 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Togneri J, Cheng Y-S, Munson M, Hughson FM, Carr CM. (2006). Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p. Proc Natl Acad Sci USA , 17730–17735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang X, Liu J, Trautmann S, Balasundaram D, McCollum D, Balasubramanian MK. (2002). The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe . Mol Biol Cell , 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lee I-J, Rask G, Wu J-Q. (2016). Roles of the TRAPP-II complex and the exocyst in membrane deposition during fission yeast cytokinesis. PLoS Biol , e1002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Chernov K, Turakainen H, Wohlfahrt G, Pajunen M, Savilahti H, Jäntti J. (2010). Mso1p regulates membrane fusion through interactions with the putative N-peptide-binding area in Sec1p domain 1. Mol Biol Cell , 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Boyvat M, Aro N, Chernov KG, Nyman T, Jäntti J. (2011). Sec1p and Mso1p C-terminal tails cooperate with the SNAREs and Sec4p in polarized exocytosis. Mol Biol Cell , 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Boyvat M, Zhao H, Aro N, Yuan Q, Chernov K, Peränen J, Lappalainen P, Jäntti J. (2013). A conserved regulatory mode in exocytic membrane fusion revealed by Mso1p membrane interactions. Mol Biol Cell , 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JRC, Munro S. (2002). Vesicle tethering complexes in membrane traffic. J Cell Sci , 2627–2637. [DOI] [PubMed] [Google Scholar]
- Wickner W, Rizo J. (2017). A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol Biol Cell , 707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Bähler J, Pringle JR. (2001). Roles of a fimbrin and an α-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell , 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Kuhn JR, Kovar DR, Pollard TD. (2003). Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell , 723–734. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science , 310–314. [DOI] [PubMed] [Google Scholar]
- Wu P, Zhao R, Ye Y, Wu J-Q. (2011). Roles of the DYRK kinase Pom2 in cytokinesis, mitochondrial morphology, and sporulation in fission yeast. PLoS One , e28000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wang S, Sheng Y, Zhang M, Zou W, Wu L, Kang L, Rizo J, Zhang R, Xu T, et al. (2015). Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol , 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I-M, Hughson FM. (2010). Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol , 137–156. [DOI] [PubMed] [Google Scholar]
- Zheng S, Dong F, Rasul F, Yao X, Jin Q-W, Zheng F, Fu C. (2018). Septins regulate the equatorial dynamics of the separation initiation network kinase Sid2p and glucan synthases to ensure proper cytokinesis. FEBS J , 2468–2480. [DOI] [PubMed] [Google Scholar]
- Zhu Y-H, Hyun J, Pan Y-Z, Hopper JE, Rizo J, Wu J-Q. (2018). Roles of the fission yeast UNC-13/Munc13 protein Ync13 in late stages of cytokinesis. Mol Biol Cell , 2259–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-H, Ye Y, Wu Z, Wu J-Q. (2013). Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol Biol Cell , 3187–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Wickner WT. (2014). A distinct tethering step is vital for vacuole membrane fusion. Elife , e03251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.