Abstract
Objective:
The objectives of this study were to determine the biofilm-forming capability and antimicrobial susceptibility of Escherichia coli recovered from bovine endometritis samples.
Materials and Methods:
A total of 120 uterine specimens were collected from cows suffering from endometritis for bacteriological examination. Antimicrobial susceptibility testing was carried out for all isolated E. coli by using the disc diffusion method. The isolates were phenotypically studied for biofilm-forming ability by cultivation on yeast extract -casamino acids Congo red agar (CRA). Some randomly selected isolates were chosen for the molecular identification of some virulence and resistance genes.
Results:
A total of 58(48.3%) E. coli isolates could be isolated from the 120 samples. Antimicrobial susceptibility testing exhibited that 91.4%, 79.3%, 79.3%, 74.1%, and 58.6% of the isolates were sensitive to gentamicin, amoxicillin-clavulanic acid, ciprofloxacin, cephalexin, and sulfamethoxazole- trimethoprim, respectively. On the other hand, 91.4% and 70.7% isolates were resistant to cefotaxime and doxycycline, respectively. Cultivation on CRA revealed that 46.6% of isolates were biofilm producers. The molecular detection of resistance and virulence genes declared that all isolates harbored blaTEM, sul1, tetA, qnrS, blaCTX-M, and fimH with a percentage of 100%, papC (40%), and hlyA (10%). FimH was the most prevalent biofilm-associated gene.
Conclusion:
The present study highlights the high prevalence of multi-drug- resistant E. coli associated with bovine endometritis. The detection of the fimH gene is circumstantial evidenced that this gene has a crucial role in biofilm formation in intrauterine pathogenic E. coli.
Keywords: Biofilm E. coli, endometritis, resistance, virulence
Introduction
Endometritis is known to be one of the major diseases, which upset the reproductive performance of cattle and reduce livestock productivity [1]. Following parturition, the invasion of the endometrium with different bacterial species (more than 200) occurs, but not all these bacteria considered as pathogens [2]. The initial step in developing bovine endometritis is the infection of the endometrium with Escherichia coli preceded by further bacteria such as Arcanobacterium pyogenes [3]. In a study, E. coli was regarded as the main associated bacteria in clinical and subclinical endometritis samples [4].
Moreover, cows with positive uterine E. coli cultures did not become pregnant to the same degree as cows without E. coli in their uteri [5]. The crucial pathogenicity characters of E. coli include epithelial cell adhesion, flagella-mediated motility, exotoxins, and lipopolysaccharides. Endometrial pathogenic E. coli strains were more adherent and invasive for the endometrial cells in vitro than that isolated from the uteri of clinically healthy animals and triggered the ultimate inflammatory response [3]. Carniello et al. [6] stated that the means of bacterial protection other than the expression of resistance genes include the production of a large quantity of extracellular polymeric substance (EPS) throughout the process of biofilm formation. This EPS is composed mainly of exopolysaccharides that form the main structure of biofilm and serve in bacterial resistance to antibiotics and host immunity [7].
Three major components, including surface, microbes, and slime EPS, constitute the output of biofilm so that it can be discarded by removing either of these components [8]. The process of biofilm formation occurs by cell-to-cell communication that is known as quorum sensing, where the accumulation of signaling molecules in the extracellular environment occurs, leading to the regulation of specific gene expression [9]. Ahmadi et al. [10] have observed the opaque liquid or some particles in uterine lavage fluid by normal saline of repeat breeder cows at estrus phase that leads to a question about the nature of these particles and the possibility of the presence of bacterial biofilm. Biofilm development is a multistep process, where it begins with the bacteria preliminary adherence to the substratum and permanent attachment followed by their colonization, in which gene alteration and protein expression occur, subsequently the exponential growth phase. The formation of EPS and water channels promotes the supply of nutrients, which results in the maturation of biofilms [9].
For biofilm formation, a set of genes is required for the initial bacterial adhesion, maturation, and production of EPS [11]. Some recent studies recognized that genes encoding certain E. coli virulence factors, such as fimH, papC, and hlyA, are responsible for bacterial adhesion and associated with bovine endometritis [12–14], where FimH (a type 1 pilus component) is E. coli specific gene and considerably associated with metritis and endometritis in cattle [13]. Consequently, the ability to produce biofilm in endometrial pathogenic E. coli hinders antimicrobial therapy. Hence, the current work aimed to investigate biofilm-forming capability and antimicrobial resistance of E. coli recovered from bovine endometritis in Egypt at Beni-Suef and Fayum governorates.
Materials and Methods
Ethical approval
The study was approved from Beni-Suef University, Institutional Animal Care and Use Committee (BSU-IACU/http://www.bsu.edu.eg).
Samples
A total of 120 uterine samples, including uterine discharges, vaginal swabs, and uterine lavages, were collected for bacteriological examination under complete aseptic conditions. They were collected from various dairy farms in Beni-Suef and Fayoum governorates in the period from February to June 2019. The samples were sent to the laboratory with a minimum of delay to avoid the dryness of samples.
Isolation and biochemical identification of E. coli
A loopful from each sample was inoculated into the tryptone soya broth (TSB) and incubated at 37°C for 16–18 h. After the incubation period, one loopful from the TSB culture was inoculated onto MacConkey agar to be incubated at 37°C for 24–48 h. Pink colonies were picked up for morphological and biochemical identification using oxidase, indole production, methyl red, Voges Proskauer, citrate utilization, and urease tests as well as growth on triple sugar iron agar as described by Quinn et al. [15].
Antimicrobial susceptibility testing of E. coli isolates
The standard disk diffusion technique was used against seven different antimicrobial disks, according to Clinical and Laboratory Standards Institute (CLSI) [16]. The suspensions of the isolates equivalent to 0.5 McFarland standards turbidity were prepared, and Mueller Hinton agar plates were inoculated. Antimicrobial disks [amoxicillin-clavulanic acid (30 μg), cephalexin (30 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), gentamicin (10 μg) and sulfamethoxazole-trimethoprim (25 μg)], representing the antimicrobials mostly used in the treatment of uterine affections under field conditions, were applied on the plates. The tested isolates were categorized as sensitive, intermediate sensitive, or resistant, according to CLSI [16].
Biofilm formation of identified E. coli isolates
Congo red (CR) assay for bacteria, as described by Zhou et al. [17], was used for the detection of biofilm formation on yeast extract-casamino acids (YESCA) CR agar plates after pre-enrichment of the isolates on Luria–Bertani agar medium. For good induction of curli production, the isolates were grown on YESCA CR agar plates at 26°C for 48 h; after that, the color of the bacterial colonies was checked, where the red-stained colonies considered as positive for curli production, and on the other hand, pink or white colonies considered as negative.
Detection of resistance and virulence genes of E. coli isolates
Ten E. coli isolates were selected for genotypic characterization by polymerase chain reaction (PCR) to detect the presence of several virulence and resistance-associated genes such as fimH, papC, hlyA, blaTEM, sul1, tetA, qnrS, and blaCTX-M using their specific forward and reverse primers as shown in Table 1. The selected isolates exhibited a multidrug resistance pattern, which was resistance to at least one agent in three or more antimicrobial classes [18]. As well, they were representing different resistance patterns and positive for phenotypic biofilm formation. The positive control DNA was obtained from confirmed positive E. coli field isolate in RLQP (Reference laboratory for veterinary quality control on poultry production, Dokki, Giza, Egypt). On the contrary, a negative control is a PCR mixture free from the DNA template.
Table 1. Oligonucleotide primers used for amplification of virulence and resistance-associated genes.
| Annealing temp. | Product | Reference | Primer sequence(5′-3′) | Target Gene |
|---|---|---|---|---|
| 50°C | 508-bp | [40] | TGCAGAACGGATAAGCCGTGG | fimH |
| GCAGTCACCTGCCCTCCGGTA | ||||
| 60°C | 1,177-bp | [41] | AACAAGGATAAGCACTGTTCTGGCT | hlyA |
| ACCATATAAGCGGTCATTCCCGTCA | ||||
| 58°C | 501-bp | [42] | TGATATCACGCAGTCAGTAGC | papC |
| CCGGCCATATTCACATAA | ||||
| 54°C | 516-bp | [43] | ATCAGCAATAAACCAGC | blaTEM |
| CCCCGAAGAACGTTTTC | ||||
| 60°C | 433-bp | [44] | CGGCGTGGGCTACCTGAACG | sul1 |
| GCCGATCGCGTGAAGTTCCG | ||||
| 50°C | 576-bp | [45] | GGTTCACTCGAACGACGTCA | tetA |
| CTGTCCGACAAGTTGCATGA | ||||
| 55°C | 417-bp | [46] | ACGACATTCGTCAACTGCAA | qnrS |
| TAAATTGGCACCCTGTAGGC | ||||
| 54°C | 593-bp | [47] | ATGTGCAGYACCAGTAARGTKATGGC | blaCTX-M |
| TGGGTRAARTARGTSACCAGAAYCAGCGG |
Results
Escherichia coli prevalence
A total number of 58 E.coli isolates were recovered from 120 uterine samples by a ratio of 48.3%.
Antimicrobial susceptibility of recovered E. coli
The antimicrobial susceptibility testing of E. coli isolates (n = 58) showed that 91.4%, 79.3%, 79.3%, 74.1%, and 58.6% of them were sensitive to gentamicin, amoxicillin-clavulanic acid, ciprofloxacin, cephalexin, and sulfamethoxazole-trimethoprim, respectively. On the contrary, 91.4% and 70.7% were resistant to cefotaxime and doxycycline, respectively. The detailed results of each antimicrobial are shown in Table 2. Of the 58 isolates, 36 (62.07%) were classified as multidrug resistant (MDR).
Table 2. Antimicrobial susceptibility of different E. coli isolates (n = 58).
| Antimicrobial class | Antimicrobial disk | Resistant | Intermediate | Sensitive | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||
| β-lactamase stable | Amoxicillin-clavulanic acid | 7 | 12.1 | 5 | 8.6 | 46 | 79.3 |
| β-lactams | Cephalexin | 15 | 25.9 | – | – | 43 | 74.1 |
| Cephalosporins | Cefotaxime | 53 | 91.4 | – | – | 5 | 8.6 |
| Fluoroquinolones | Ciprofloxacin | 7 | 12.1 | 5 | 8.6 | 46 | 79.3 |
| Tetracyclines | Doxycycline | 41 | 70.7 | 5 | 8.6 | 12 | 20.7 |
| Aminoglycosides | Gentamicin | – | – | 5 | 8.6 | 53 | 91.4 |
| Potentiated sulfonamide | Sulfamethoxazole-trimethoprim | 24 | 41.4 | – | – | 34 | 58.6 |
Biofilm formation on YESCA CR agar
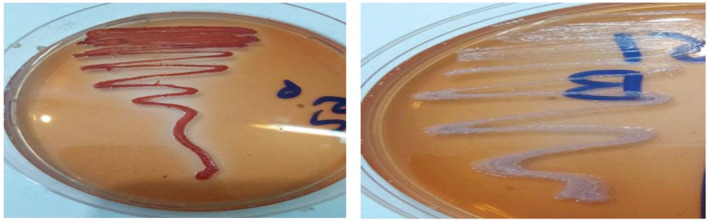
Of the total tested isolates (n = 58), 27 E. coli isolates (46.6%) were grown as red colonies on YESCA CR agar and described as biofilm positive. In comparison, 31 isolates (53.4%) were grown as white colonies and described as negative for biofilm formation, as shown in Figure 1.
Figure 1. Cultivation of E. coli on YESCA CR agar. Left side = E. coli colonies appeared red (biofilm positive). Right side = E. coli colonies seemed to be white on YESCA CR agar (biofilm negative).
Association between antimicrobial resistance and biofilm formation
Of 27 biofilm-producing E. coli isolates, 22(81.5%) were recorded as MDR.
Detection of antimicrobial resistance and virulence genes of E. coli
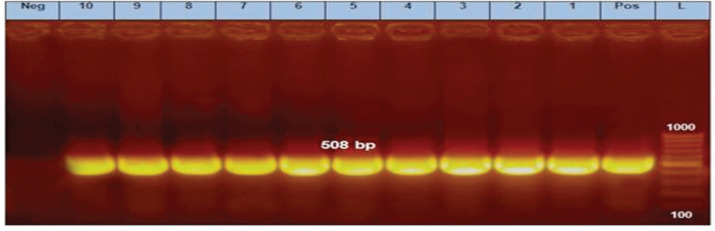
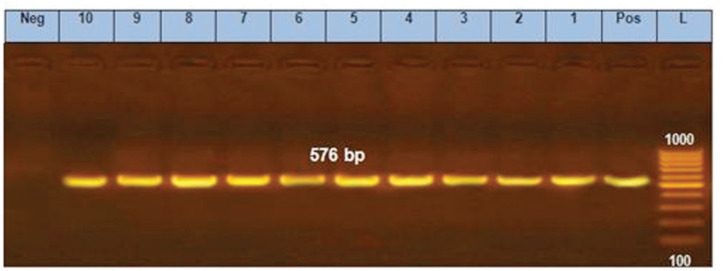
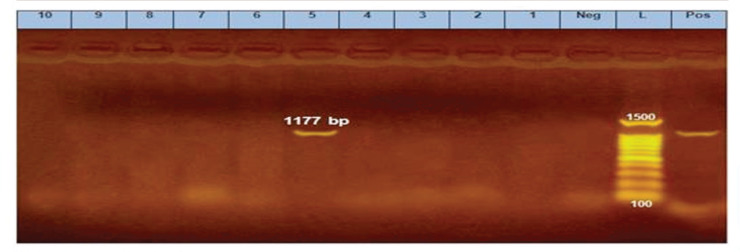
Ten E. coli isolates were tested using PCR for the detection of fimH, papC, hlyA, blaTEM, sul1, tetA, qnrS, and blaCTX-M. All of them harbored fimH gene, four of them (40%) contain papC gene, only one isolate (10%) exhibited hlyA gene, and all of them carried all the tested antimicrobial resistance genes (blaTEM, sul1, tetA, qnrS, and blaCTX-M) (Figs. 2–9).
Figure 2. PCR amplification of the fimH gene at 508-bp fragment. Lanes 1–10 showed positive amplification of the fimH gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
Figure 9. PCR amplification of tetA gene at 576-bp fragment. Lanes 1–10 showed positive amplification of tetA gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
Discussion
The present study revealed that E. coli is one of the most significant bacteriological risk factors of bovine endometritis. It was isolated by a percentage of 48.3%, where many other studies confirmed by Kasimanickam et al. [19], who isolated E. coli by 45%. The high prevalence of E. coli in bovine endometritis may be connected to the existence of these bacteria in enteric microflora, in addition to the proximity of the rectum and external genital tract, which donate to uterine contamination by these enteric bacteria [20].
The antimicrobial sensitivity and resistance patterns of E. coli isolates by the disk diffusion method against seven diverse antimicrobial agents of five different classes were studied.
Cefotaxime was the most antimicrobial agent showing resistance by the percentage of 91.4%, followed by doxycycline (70.7%).
On the other hand, E. coli found to be 91.4% sensitive to gentamicin, 79.3% to both amoxicillin-clavulanic acid and ciprofloxacin, 74.1% to cephalexin, and 58.6% to sulfamethoxazole-trimethoprim.
The percentage of cefotaxime resistance of intrauterine pathogenic E. coli was 70.8% in a study that was performed by Ma et al. [14]. The high percentage of cefotaxime resistance in this study may be related to the extensive use of third-generation cephalosporin ceftiofur in the treatment of endometritis.
In the present study, E. coli showed high resistance to doxycycline, whereas Zhao et al. [21] recorded a high resistance, to a certain degree, (46%) against this antibiotic. This high resistance in this study may be related to the widespread use of the broad-spectrum antibiotic oxytetracycline in uterine irrigation as one of the methods for the treatment of endometritis, either clinical or subclinical.
Figure 3. PCR amplification of the hlyA gene at 1177-bp fragment. Lane 5 showed positive amplification of the hlyA gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
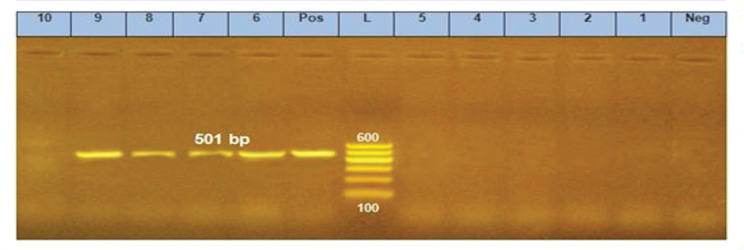
Figure 4. PCR amplification of papC gene at 501 bp fragment. Lanes 6–9 showed positive amplification of papC gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
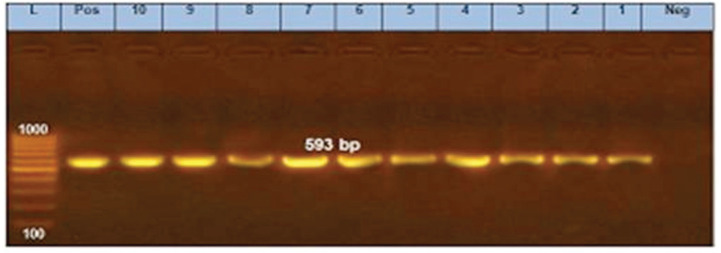
Figure 5. PCR amplification of the blaCTX-M gene at 593-bp fragment. Lanes 1–10 showed positive amplification of the blaCTX-M gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
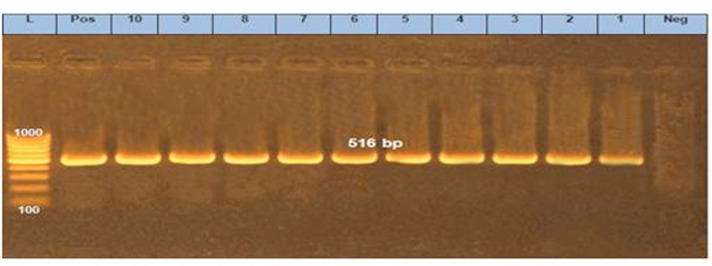
Figure 6. PCR amplification of the blaTEM gene at 516bp fragment. Lanes 1–10 showed positive amplification of the blaTEM gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
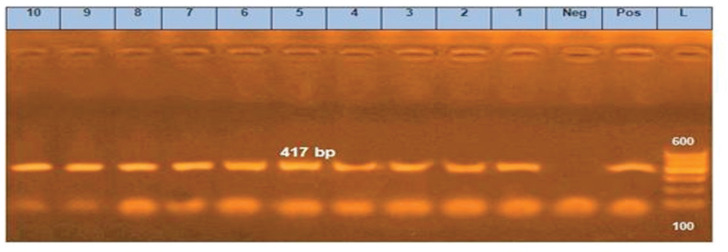
Figure 7. PCR amplification of a gene at 417-bp fragment. Lanes 1–10 showed positive amplification of a gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
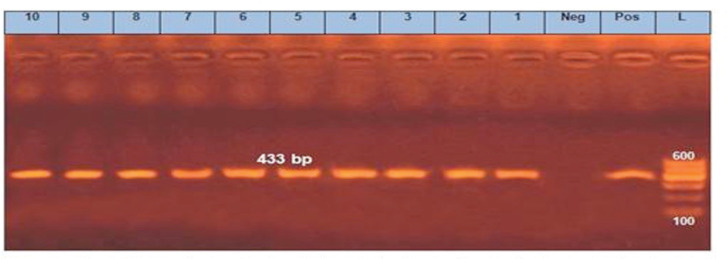
Figure 8. PCR amplification of the sul1 gene at 433-bp fragment. Lanes 1–10 showed positive amplification of the sul1 gene. Pos = Positive control; Neg = negative control; L = 100-bp DNA molecular size ladder.
The same results of high sensitivity to gentamicin and ciprofloxacin were obtained by Zhao et al. [21], where they recorded 60.1% and 77.1% sensitivity against them, respectively.
Brodzki et al. [22] found that E. coli isolated from bovine uteri was sensitive to amoxicillin-clavulanic acid by the percentage of 100% supporting the results of high sensitivity to this antibiotic. On the contrary, Li-ming et al. [23] found that E. coli was highly resistant to amoxicillin.
The present study declared that intrauterine pathogenic E. coli is highly sensitive to cephalexin, but Dutt et al. [24] found that E. coli isolates were 100% resistant to it.
In this study, the sensitivity against sulfamethoxazole-trimethoprim was 58.6%, whereas Zhao et al. [25] reported 100% resistance against it. Based on the finding of this study, 62.07% of E. coli isolates were resistant to 3–5 categories of antimicrobials. Ma et al. [14] reported that all intrauterine pathogenic E. coli were MDR. Furthermore, Zhao et al. [21] isolated 148 E. coli isolates from the cases of bovine endometritis, and 132 (89.2%) out of them were MDR.
Biofilm formation is a mechanism for bacterial resistance and also for bacterial virulence [26], where it increases the antimicrobial resistance up to 1,000 folds to inactivate organisms developing inside a biofilm, and high antimicrobial concentrations are required [27]. This resistance may be due to the inadequate concentration of antimicrobials that reach certain parts of the biofilms and metabolic inactivity, in addition to the existence of active antibiotic degradation mechanisms that contribute to the cessation of drug accumulation to a sufficient concentration [10].
For the detection of biofilm in E. coli isolates, Reichhardt et al. [28] concluded that CR dye can bind to curled whole cells, without inhibition of growth, and can be used to comparatively measure the whole-cell curliation, where E. coli accumulate extracellular adhesive amyloid fibers termed curli which enable the bacterial adhesion and encourage the biofilm formation.
The current study reported that 46.6% of the recovered E. coli isolates were phenotypically positive for biofilm formation. Moori Bakhtiari et al. [29] reported that 53.3% and 16.6% of E. coli isolates were moderately and strongly biofilm producers, respectively.
Cephalosporin resistance is linked to the genes that encode for β-lactamases such as blaTEM, blaCTX-M, and blaCMY [30]. Moreover, blaCTX-M genes are the most common type of extended-spectrum β-lactamases with high clinical significance [14]. In this study, blaTEM and blaCTX-M were identified in all selected E. coli isolates, whereas Zhao et al. [21] detected them by a percentage of 30.4%, as well they concluded that blaTEM gene was predominant in E. coli isolates that were resistant to quinolones, whereas, in this study, qnrS gene also detected in all selected isolates.
Sul1 gene is a plasmid-borne sulfonamide resistance gene that is linked to the universal and long-known sulfonamide resistance in Gram-negative bacteria [31]. This gene determined in all selected E. coli isolates. Similarly, tetA gene detected in all the studied isolates that encodes the synthesis of the protein responsible for the efflux pump process, which is the most common resistance mechanism for tetracycline and its analogs [32].
In this study, the virulence-associated fimH gene identified in all tested isolates that were phenotypically positive for biofilm formation. The same high gene prevalence was also mentioned by Bicudo et al. [20], where it was detected in more than 90% of uterine isolates of cows, which reinforces the effect of this adhesion in early uterine contamination. FimH is a Type 1 pili correlated to adherence, invasion, and biofilm formation in the epithelial cells of host tissues [33]. Moreover, Bicudo et al. [20] clarified that fimH has an essential role in the establishing of E. coli in the endometrium, increasing the risk of endometritis and the failure in the consequent pregnancy when detected in cows at 1–3 days postpartum. In addition, the treatment of endometrial pathogenic E. coli with mannose resulted in a reduction of their ability to adhere to the endometrial cells, which confirms the expression of the fimH gene [34].
The papC gene which also encodes for bacterial adhesion detected in 4 out of the 10 selected isolates (40%). In a study conducted by Kassé et al. [35], papC gene was detected by 9% in E. coli isolates associated with postpartum metritis in cattle.
Alpha-hemolysin (hlyA) gene identified only in one of the selected isolates (n = 10) by a percentage of 10%, where it is a pore-forming cytotoxin that is responsible for lysis of the cell wall of the host cells including leukocytes, erythrocytes, and endothelial cells [36].
Silva et al. [12] did not find any relation between hlyA and fimH genes in the occurrence of bovine metritis, and on the other hand, Bicalho et al. [37] proposed a relationship between the presence of hlyA gene and the presence of fimH gene in the occurrence of bovine metritis, but the expression of hemolysin must be considered an extra mechanism of E. coli pathogenicity, favoring the development of extra-intestinal infections, included in bovine endometritis [35].
In the present study, of 27 biofilm-producing E. coli isolates, 22 (81.5%) were recorded as MDR that declares the correlation between the antimicrobial resistance and the biofilm formation, and similar results were also obtained by Neupane et al. [38] and Karigoudar et al. [39].
Conclusion
The present study highlights the high incidence of MDR E. coli associated with bovine endometritis. The detection of fimH gene is circumstantial evidence that this gene has a significant role in biofilm formation in intrauterine pathogenic E. coli. Moreover, there was a high antimicrobial resistance of E. coli isolates in addition to its correlation with biofilm formation.
Acknowledgments
The authors are thankful to the Mrs. Soma Gamal for her excellent technical assistance
Conflict of interest
All the authors contributed equally. Other than this, there was no conflict of interest among the authors.
References
- [1].Adnane M, Kaidi R, Hanzen C, England GCW. Risk factors of clinical and subclinical endometritis in cattle: a review. Turk J Vet Anim Sci. 2017;4:1–11. https://doi.org.10.3906/vet-1603-63. [Google Scholar]
- [2].Drillich M, Wagener K. Pathogenesis of uterine diseases in dairy cattle and implications for fertility. Anim Reprod. 2018;15:879–85. doi: 10.21451/1984-3143-AR2018-0023. https://doi.org.10.21451/1984-3143-AR2018-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sheldon IM, Rycroft AN, Dogan B, Craven M, Bromfield JJ, Chandler A, et al. Specific strains of Escherichia coli are pathogenic for the endometrium of cattle and cause pelvic inflammatory disease in cattle and mice. PLoS One. 2010;5(2):e9192. doi: 10.1371/journal.pone.0009192. https://doi.org.10.1371/journal.pone.0009192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Salah N, Yimer N, Wahid H, Rosnina Y, Khairani B, Omar MA. Agreement among bacteriological findings, vaginal discharges, and endometrial cytology for endometritis detection in postpartum beef cows. Emirates J Food Agric. 2017;29(5):396–403. https://doi.org.10.9755/ejfa.2016-11-1561. [Google Scholar]
- [5].Ordell A, Unnerstad HE, Nyman A, Gustafsson H, Båge R. A longitudinal cohort study of acute puerperal metritis cases in Swedish dairy cows. Acta Vet Scand. 2016;58:79. doi: 10.1186/s13028-016-0257-9. https://doi.org.10.1186/s13028-016-0257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Carniello V, Peterson BW, van der Mei HC, Busscher HJ. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv Colloid Interface Sci. 2018;261:1–14. doi: 10.1016/j.cis.2018.10.005. https://doi.org.10.1016/j.cis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- [7].Ostapska H, Howell PL, Sheppard DC. Deacetylated microbial biofilm exopolysaccharides: it pays to be positive. PLoS Pathog. 2018;14(12):e1007411. doi: 10.1371/journal.ppat.1007411. https://doi.org.10.1371/ journal.ppat.1007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Murugan K, Usha M, Malathi P, Al-Sohaibani AS, Chandrasekaran M. Biofilm forming multi drug resistant staphylococcus spp. among patients with conjunctivitis. Pol J Microbiol. 2010;59(4):233–9. [PubMed: 21466040] [PubMed] [Google Scholar]
- [9].Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8(76):1–10. doi: 10.1186/s13756-019-0533-3. https://doi.org.10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ahmadi MR, Derakhshandeh A, Shirian S, Daneshbod Y, Ansari-Lari M, Nazifi S. Detection of bacterial biofilm in uterine of repeat breeder dairy cows. Asian Pacific J Reprod. 2017;6:136–9. https://doi:10.12980/apjr.6.20170308. [Google Scholar]
- [11].Schiebel J, Böhm A, Nitschke J, Burdukiewicz M, Weinreich J, Ali A, et al. Genotypic and phenotypic characteristics in association with biofilm formation in different pathotypes of human clinical Escherichia coli isolates. Appl Environ Microbiol. 2017;83(24):e01660–17. doi: 10.1128/AEM.01660-17. https://doi.org.10.1128/AEM.01660-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Silva E, Leitão S, Tenreiro T, Pomba C, Nunes T, da Costa LL, et al. Genomic and phenotypic characterization of Escherichia coli isolates recovered from the uterus of puerperal dairy cows. J Dairy Sci. 2009;92(12):6000–10. doi: 10.3168/jds.2009-2358. https://doi.org.10.3168/jds.2009-2358. [DOI] [PubMed] [Google Scholar]
- [13].Bicalho MLS, Machado VS, Oikonomou G, Gilbert RO, Bicalho RC. Association between virulence factors of Escherichia coli, fusobacterium necrophorum and arcanobacterium pyogenes and uterine diseases of dairy cows. Vet Microbiol. 2012;157(12):125–31. doi: 10.1016/j.vetmic.2011.11.034. https://doi.org.10.1016/j.vetmic.11.034. [DOI] [PubMed] [Google Scholar]
- [14].Ma Z, Ginn A, Kang M, Galvão KN, Jeong KC. Genomic and virulence characterization of intrauterine pathogenic Escherichia coli with multi-drug resistance isolated from cow uteri with metritis. Front Microbiol. 2018;9:3137. doi: 10.3389/fmicb.2018.03137. https://doi.org.10.3389/fmicb.2018.03137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Quinn PJ, Markey BK, Carter ME, Donnelly WJC, Leonard FC, Maguire D. Veterinary microbiology and microbial diseases. 1st. Published Blackwell Science; Hoboken, NJ: 2002. [Google Scholar]
- [16].Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. Twenty-Seven Informational Supplement. 2018;37(1):M100–S27. [Google Scholar]
- [17].Zhou Y, Smith DR, Hufnagel DA, Chapman MR. Experimental manipulation of the microbial functional amyloid called curli. Methods Mol Biol. 2013;966:53–75. doi: 10.1007/978-1-62703-245-2_4. https://doi.org.10.1007/978-1-62703-245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. doi: 10.1111/j.1469-0691.2011.03570.x. https://doi.org.10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- [19].Kasimanickam VR, Owen K, Kasimanickam RK. Detection of genes encoding multi-drug resistance and biofilm virulence factor in uterine pathogenic bacteria in postpartum dairy cows. Theriogenology. 2016;85(2):173–9. doi: 10.1016/j.theriogenology.2015.10.014. https://doi.org.10.1016/j.theriogenology.2015.10.014. [DOI] [PubMed] [Google Scholar]
- [20].Bicudo LC, Oba E, Bicudo SD, Leite DS, Siqueira AK, Monobe MMS, et al. Virulence factors and phylogenetic group profile of uterine Escherichia coli in early postpartum of high-producing dairy cows. Anim Prod Sci. 2019;59:1898–1905. https://doi.org.10.1071/AN17729. [Google Scholar]
- [21].Zhao HX, Zhao JL, Shen JZ, Fan HL, Guan H, An XP, et al. Prevalence and molecular characterization of fluoroquinolone resistance in Escherichia coli isolates from dairy cattle with endometritis in China. Microb Drug Resist. 2014;20(2):162–9. doi: 10.1089/mdr.2013.0073. https://doi.org.doi:10.1089/mdr.2013.0073. [DOI] [PubMed] [Google Scholar]
- [22].Brodzki P, Bochniarz M, Brodzki A, Wrona Z, Wawron W. Trueperella pyogenes and Escherichia coli as an etiological factor of endometritis in cows and the susceptibility of these bacteria to selected antibiotics. Pol J Vet Sci. 2014;17(4):657–64. doi: 10.2478/pjvs-2014-0096. https://doi.org.10.2478/pjvs-2014-0096. [DOI] [PubMed] [Google Scholar]
- [23].Li-ming Y, Yi-hao W, Yu P, Jiang-tao MIN, Su-qin H, Wei-yun ZHU. Genomic characterization and antimicrobial susceptibility of bovine intrauterine Escherichia coli and its relationship with postpartum uterine infections. J Integr Agric. 2016;15(6):1345–54. https://doi.org.10.1016/S2095-3119(15)61170-4. [Google Scholar]
- [24].Dutt R, Singh G, Singh M, Sharma M, Dalal J, Chandolia RK. Diagnosis of subclinical endometritis in murrah buffaloes through cytobrush technique. Int J Curr Microbiol Appl Sci. 2017;6(11):494–9. https://doi.org.10.20546/ijcmas.2017.611.059. [Google Scholar]
- [25].Zhao HX, Shen JZ, An XP, Fan HL, Cao JS, Li PF. Characterization of integrons in multiple antimicrobial resistant Escherichia coli isolates from bovine endometritis. Res Vet Sci. 2011;91(3):412–4. doi: 10.1016/j.rvsc.2010.09.004. https://doi.org.10.1016/j.rvsc.2010.09.004. [DOI] [PubMed] [Google Scholar]
- [26].Cepas V, López Y, Muñoz E, Rolo D, Ardanuy C, Martí S, et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb Drug Resist. 2019;25(1):72–9. doi: 10.1089/mdr.2018.0027. https://doi:10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- [27].Hall CW, Mah T. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. https://doi:10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- [28].Reichhardt C, Jacobson AN, Maher MC, Uang J, McCrate OA, Eckart M, et al. Congo red interactions with curli-producing E. coli and native curli amyloid fibers. PLoS One. 2015;10(10):e0140388. doi: 10.1371/journal.pone.0140388. https://doi.org.10.1371/journal.pone.0140388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Moori Bakhtiari N, Gooraninezhad S, Karami M. Biofilm-producing ability of bovine extraintestinal pathogenic Escherichia coli and its correlation with attachment factors. Jundishapur J Heal Sci. 2018;10(3):e77130. https://doi.org.10.5812/jjhs.77130. [Google Scholar]
- [30].Li XZ, Mehrotra M, Ghimire S, Adewoye L. Beta lactam resistance and beta-lactamases in bacteria of animal origin. Vet Microbiol. 2007;121(3–4):197–214. doi: 10.1016/j.vetmic.2007.01.015. https://doi.org.10.1016/j.vetmic.2007.01.015. [DOI] [PubMed] [Google Scholar]
- [31].Jiang H, Cheng H, Liang Y, Yu S, Yu T, Fang J, et al. Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from penaeus vannamei and pork from large markets in Zhejiang, China. Front Microbiol. 2019;10:1787. doi: 10.3389/fmicb.2019.01787. https://doi:10.3389/fmicb.2019.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ozgumus OB, Celik-Sevim E, Alpay-Karaoglu S, Sandalli C, Sevim A. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. J Microbiol. 2007;45(5):379–87. [PubMed: 17978796] [PubMed] [Google Scholar]
- [33].Bien J, Sokolova O, Bozko P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol. 2012;2012:681473. doi: 10.1155/2012/681473. https://doi.org.10.1155/2012/ 681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sheldon IM, James GC, John JB. Tolerance and innate immunity shape the development of postpartum uterine disease and the impact of endometritis in dairy cattle. Annu Rev Anim Biosci. 2019;15:361–84. doi: 10.1146/annurev-animal-020518-115227. https://doi:10.1146/annurev-animal-020518-115227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kassé FN, Fairbrother JM, Dubuc J. Relationship between Escherichia coli virulence factors and postpartum metritis in dairy cows. J Dairy Sci. 2016;99:1–12. 4656–67. doi: 10.3168/jds.2015-10094. https://doi.org.10.3168/jds.2015-10094. [DOI] [PubMed] [Google Scholar]
- [36].Henriques S, Silva E, Silva MF, Carvalho S, Diniz P, Lopes L, et al. Immunomodulation in the canine endometrium by uteropathogenic Escherichia coli. Vet Res. 2016;47(114):1–17. doi: 10.1186/s13567-016-0396-z. https://doi:10.1186/s13567-016-0396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bicalho RC, Machado VS, Bicalho MLS, Gilbert RO, Teixeira AGV, et al. Molecular and epidemiological characterization of bovine intrauterine Escherichia coli. J Dairy Sci. 2010;93(1–2):5818–30. doi: 10.3168/jds.2010-3550. https://doi.org.10.3168/jds.2010-3550. [DOI] [PubMed] [Google Scholar]
- [38].Neupane S, Pant ND, Khatiwada S, Chaudhary R, Banjara MR. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting shree Birendra hospital, Chhauni, Kathmandu. Antimicrob. Resist Infect Control. 2016;5(5):1–5. doi: 10.1186/s13756-016-0104-9. https://doi.org.10.1186/s13756-016-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Karigoudar RM, Karigoudar MH, Wavare SM, Mangalgi SS. Detection of biofilm among uropathogenic Escherichia coli and its correlation with antibiotic resistance pattern. J Lab Physicians. 2019;11:17–22. doi: 10.4103/JLP.JLP_98_18. https://doi.org.10.4103/JLP.JLP_98_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ghanbarpour R, Salehi M. Determination of adhesin encoding genes in Escherichia coli isolates from omphalitis of chicks. Am J Anim Vet Sci. 2010;5(2):91–6. https://doi.org.10.3844/ajavsp.2010.91.96. [Google Scholar]
- [41].Piva IC, Pereira AL, Ferraz LR, Silva RSN, Vieira AC, Blanco JE, et al. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasília, Brazil. J Clin Microbiol. 2003;41(5):1827–32. doi: 10.1128/JCM.41.5.1827-1832.2003. https://doi.org.10.1128/jcm.41.5.1827-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wen-Jie J, Zhi-Ming Z, Yong-Zhi Z, Ai-Jian Q, Hong-Xia S, Yue-Long L, et al. Distribution of virulence-associated genes of avian pathogenic Escherichia coli isolates in China. Agric Sci China. 2008;7(12):1511–5. https://doi.org.10.1016/S1671-2927(08)60410-1. [Google Scholar]
- [43].Colom K, Pèrez J, Alonso R, Fernández-Aranguiz A, Lariňo E, Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223(2):147–51. doi: 10.1016/S0378-1097(03)00306-9. https://doi.org.10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- [44].Ibekwe AM, Murinda SE, Graves AK. Genetic diversity and antimicrobial resistance of Escherichia coli from human and animal sources uncovers multiple resistances from human sources. PLoS One. 2011;6(6):e20819. doi: 10.1371/journal.pone.0020819. https://doi.org.10.1371/journal.pone.0020819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Randall LP, Cooles SW, Osborn MK, Piddock LJV, Woodward MJ. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother. 2004;53(2):208–16. doi: 10.1093/jac/dkh070. https://doi.org.10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- [46].Robicsek A, Strahilevitz J, Sahm DF, Jacoby GA, Hooper DC. qnr prevalence in ceftazidime-resistant enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother. 2006;50(8):2872–4. doi: 10.1128/AAC.01647-05. https://doi.org.10.1128/AAC.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Archambault M, Petrov P, Hendriksen RS, Asseva G, Bangtrakulnonth A, Hasman H, et al. Molecular characterization and occurrence of extended-spectrum beta-lactamase resistance genes among Salmonella enterica serovar Corvallis from Thailand, Bulgaria, and Denmark. Microb Drug Resist. 2006;12(3):192–8. doi: 10.1089/mdr.2006.12.192. https://doi.org.10.1089/mdr.2006.12.192. [DOI] [PubMed] [Google Scholar]