Abstract
Extracellular vesicles (EVs) are membrane-enclosed particles that are released by virtually all cells from all living organisms. EVs shuttle biologically active cargo including protein, RNA, and DNA between cells. When shed by cancer cells, they function as potent intercellular messangers with important functional consequences. Cells produce a diverse spectrum of EVs, spanning from small vesicles of 40–150 nm in diameter, to large vesicles up to 10 μm in diameter. While this diversity was initially considered to be purely based on size, it is becoming evident that different classes of EVs, and different populations within one EV class may harbor distinct molecular cargo and play specific functions. Furthermore, there are considerable cell type-dependent differences in the cargo and function of shed EVs. This review focuses on the most recent proteomic studies that have attempted to capture the EV heterogeneity by directly comparing the protein composition of different EV classes and EV populations derived from the same cell source. Recent studies comparing protein composition of the same EV class(es) derived from different cell types are also summarized. Emerging approaches to study EV heterogeneity and their important implications for future studies are also discussed.
Keywords: biomarkers, cancer, extracellular vesicles, heterogeneity, protein profiling
1. Introduction
Extracellular vesicles (EVs) are membrane-enclosed structures of variable size (40 nm–10 μm) that are released by all cells into the extracellular space and can be detected in all body fluids.[1–7] EVs function as mediators of communication between adjacent or distant cells and have been recognized as promoters of cancer progression.[8–12] EVs contain and likely transfer to the recipient cells functional biomolecules,[9,13–20] including proteins,[9,16,17] lipids,[21–23] RNA,[18,19] and DNA.[20,24,25] This EV cargo is typically representative of the molecular composition of the cells they originate from, making EVs a good source of circulating biomarkers.[4,19,26] Additionally, recent studies have demonstrated enrichment of certain molecules in EVs in comparison with their cells of origin, suggesting that EV analysis provides an information that tissue specimen evaluation alone would not.[14,27]
Based on their size, EVs can be broadly divided into two classes: small EVs (s-EVs), and large EVs (l-EVs). Although the size ranges for the two classes are not precisely defined and can vary depending on the model system and measurement approach, we refer to the EVs in the 40– 150 nm size range as s-EVs and to the EVs in the 200 nm–10 μm size range as l-EVs (Figure 1). Exosomes are one of the most studied types of s-EVs, which originate from multi-vesicular bodies (MVB). MVB are formed in the cell cytoplasm by inward invagination of limiting membrane of late endosomes.[28] When the membrane of MVB fuses with the plasma membrane of the cell, exosomes are released into the extracellular space. Most studies of s-EVs refer to their EVs as exosomes; however, the endosomal origin of these EVs is difficult to prove and the common methods for isolating s-EVs do not separate endosome-derived s-EVs from non-endosomal s-EVs, such as small vesicles originating at the plasma membrane.[26,29–32] Therefore, here we will refer to all EVs within the size range of 40–150 nm as s-EVs. Ectosomes are EVs of nonendosomal origin ranging from 100 nm and up to a few micrometers in diameter that form at the plasma membrane and are released into the extracellular space by budding off of their cell of origin.[33–35] Due to their large size range, ectosomes can be found in both s-EV and l-EV preparations.[30,36–38] l-EVs are also commonly referred to as microvesicles.
Figure 1.
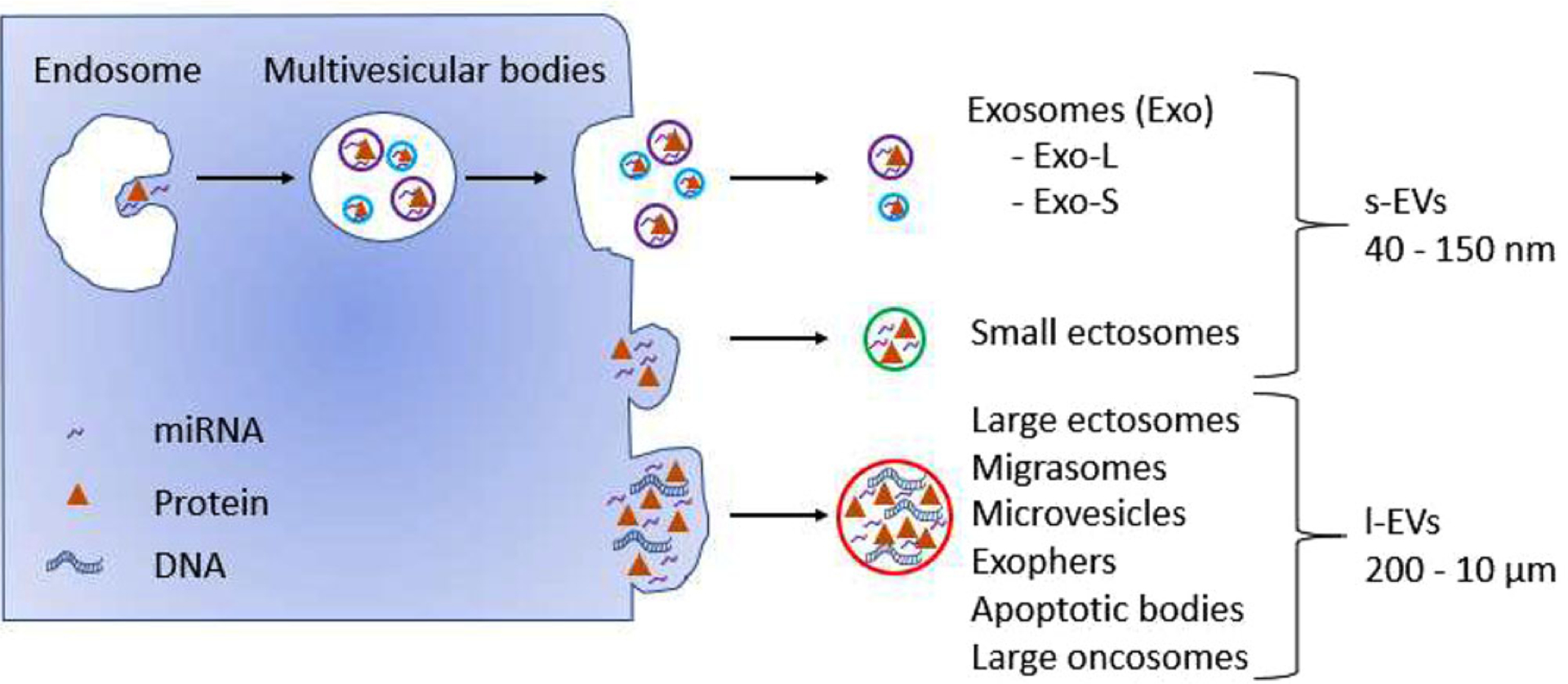
Heterogeneity of EV classes released by cells. Cells release EVs that can be classified into either s-EVs (40–150 nm) or l-EVs (200 nm–10 μm) according to their size. s-EVs encompass endosome-derived exosomes, which can be further subdivided into small (Exo-S) and large (Exo-L) exosomes, as well as nonendosomal small ectosomes. l-EVs include large ectosomes, microvesicles, migrasomes, exophers, apoptotic bodies, and large oncosomes.
Some EVs are linked to a defined physiological state of the parental cell or/and a particular cell type. Among them, migrasomes are l-EVs that are released from the tips and intersections of retraction fibers, which are long tubular strands left behind migrating cells.[38,39] These connections eventually break, leading to vesicle release.[39] Migrasomes are up to 3 μm in diameter and contain numerous smaller vesicles (50–100 nm), giving them a “pomegranate”-like appearance. Another example of l-EVs are exophers, which are l-EVs of ≈4 μm in diameter containing protein aggregates and organelles that are produced by adult neurons from Caenorhabditis elegans.[40] Exophers can form under native physiological cellular conditions; however, their formation is enhanced in proteotoxically stressed neurons, which seem to function better than similarly stressed neurons without exophers.[40] A common type of EVs linked to a physiological state of the parental cells is apoptotic bodies. Apoptotic bodies are l-EVs (800 nm–5 μm) that are released into the extracellular space as the apoptotic cell disassembles.[41–43] Large oncosomes (LO)[14,44–47] are atypically large l-EV of 1–10 μm in diameter, which are released by membrane blebbing of amoeboid cancer cells exhibiting highly migratory and metastatic properties. This process is modulated by loss of the Diaphanous-related formin 3 (DIAPH3), which results in activation of RhoA-ROCK kinase pathway, a major mechanism regulating the transition of cancer cells motility from mesenchymal to amoeboid.[14,44–47]
Although distinct EV classes seem to be well-defined, in practice EV pellets comprise different EV classes and populations (Figure 2), which are heterogeneous with respect to size and molecular cargo, as well as, potentially, to function. For example, within the class of s-EVs obtained from the same cell source, different populations of s-EVs, based on size, density, and protein expression, have been described.[48,49] Moreover, recently a novel class of the smallest extracellular particles reported so far (35 nm in diameter), termed exomeres, was identified by Zhang et al.[29] While still poorly characterized, it is evident that exomeres present distinct properties when compared to s-EVs.
Figure 2.
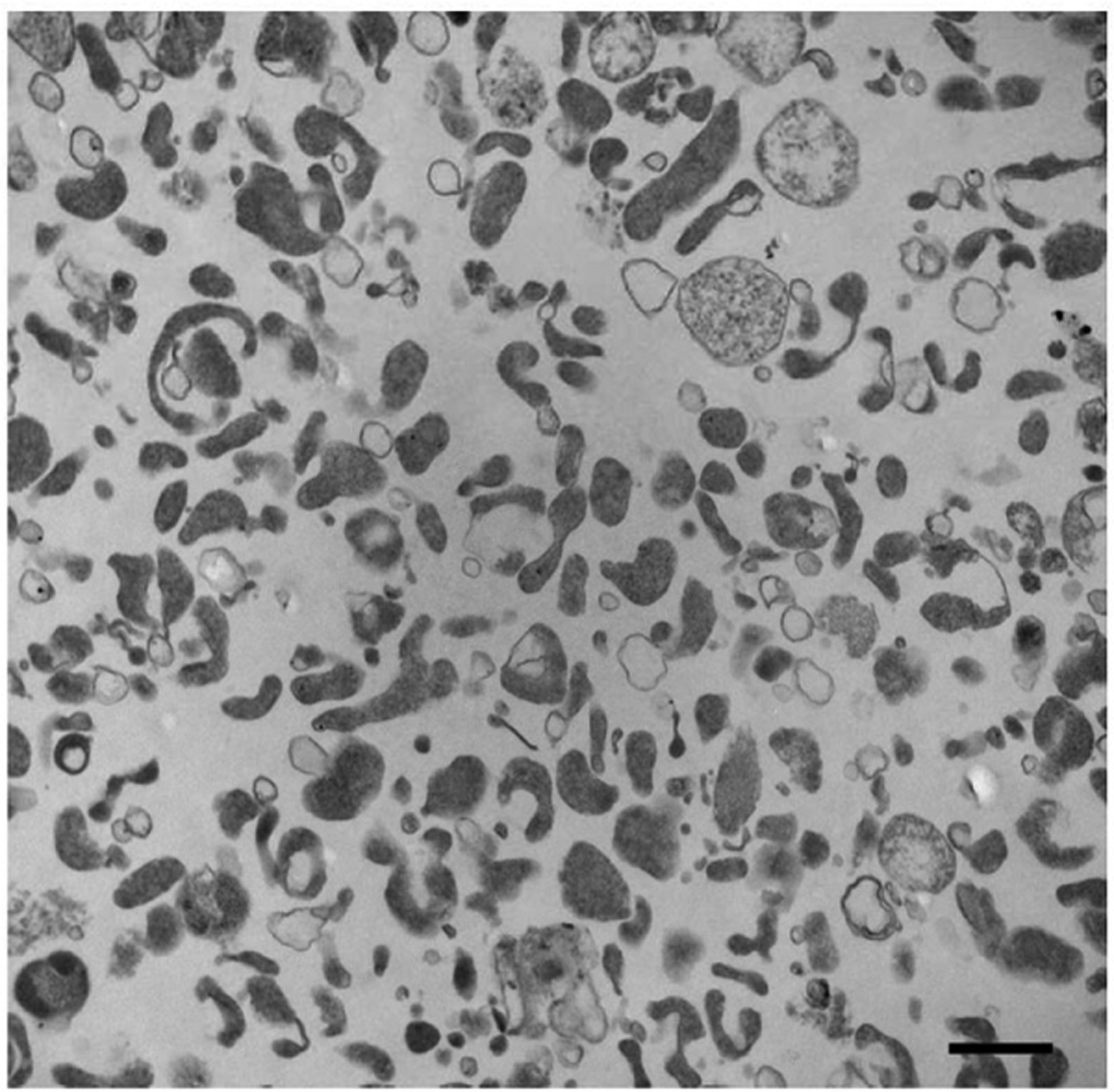
Heterogeneity of EVs recovered from prostate cancer cell media by ultracentrifugation at 10 000 × g. Transmission electron microscopy image showing particles of different sizes and densities in the EV pellet stained with osmium tetroxide. Prior to pelleting EVs at 10 000 × g for 30 min, conditioned cell media was cleared by two low-speed centrifugation steps: 300 × g for 10 min, followed by 2800 × g for 10 min. Scale bar: 600 nm
The discovery of all these EV types has recently brought significant attention to the molecular mechanisms underlying EV heterogeneity.[28,50] However, separating and defining discreet EV populations has been a challenging task due to limitations of the standard methodologies, the disparate and often poorly described isolation strategies, as well as the lack of systematical biochemical and biophysical characterization in published reports. These issues have stimulated an immense interest in improving methodologies for EV isolation and in identifying specific markers that allow discrimination between different classes/populations of EVs.
Given that proteins are critical bioactive constituents of EV cargo, proteomic approaches are appealing to compare molecular profiles of different EV types and provide invaluable insights into the nature of EV heterogeneity. Moreover, EV cargo has been demonstrated to include cancer-specific proteins[51–55] and EV potential to serve as a source of circulating biomarkers has been under vigorous investigation. Therefore, defining discrete EV populations enriched in tumor-derived proteins, as well as development of novel technologies that would allow EV isolation from clinical samples more efficiently, is of utter importance.
2. Heterogeneity of EVs
Different biogenesis and subcellular origin of EVs (endosomal vs nonendosome-derived) could be one of the major sources of EV heterogeneity. Therefore, in the past few years, there have been several attempts to comparatively analyze by MS the protein cargo of different EV populations after separating EVs by size and density (using differential centrifugation/filtration/density gradient)[47,56–58] (summarized in Table 1). These initial studies focused mainly on characterizing the protein cargo of s-EVs versus l-EVs and they identified several common proteins but also an abundance of unique proteins in each EV class. For example, in one of the early studies from the Simpson group, the authors compared the proteome of s-EVs and l-EVs from the same cell source (colon cancer cell line LIM1863).[56] They employed sequential centrifugal ultrafiltration to separate the two EV classes and reported isolation of morphologically homogenous population of s-EVs, and a heterogeneous population of l-EVs. Western blot characterization demonstrated that s-EVs contained proteins that had been frequently shown to be enriched in endosome-derived EVs (Alix, TSG101, CD81, and CD63), while l-EVs were significantly depleted of these proteins. Comparative protein profiling of s-EVs and l-EVs using gel-based LC–MS/MS (GeLC–MS/MS) revealed 354 and 606 unambiguous protein identifications, respectively, with 256 proteins in common. A total of 350 proteins were uniquely identified in l-EVs for the first time, the most abundant of which were members of the septin family, KIF23, CSE1L, and RACGAP1. Functionally, l-EVs promoted invasion in the EV-treated recipient cells to a significantly greater extent (threefold) than s-EVs.[56]
Table 1.
Summary of the findings reported in the most recent studies that performed a side-by-side comparison of the proteomes of different EV classes.
| Proteins enriched in | |||||
|---|---|---|---|---|---|
| Cell line(s) | Isolation strategy | Proteomics Approach | l-EVs | s-EVs | Reference |
| Primary monocyte-derived dendritic cells | Differential ultracentrifugation (l-EVs: 10 000 × g, 40 min; s-EVs: 100000 × g, 90 min) followed by density gradient | GeLC-MS/MS | Actinin-4, Mitofilin, RACGAP1 | TSG101.ADAM10, Syntenin-1, EHD4 (CD9, CD63, CD81) | [57] |
| LI M1863 (colon cancer) | Sequential centrifugal ultrafiltration (l-EVs: >0.65 μm, 3000 × g; s-EVs: <0.1 μm, 3000 × g) | CeLC-MS/MS | KIF23, CSETL, RACGAP1 | Alix, TSG101,CD81, CD63 | [56] |
| NCI-60 | Polyethylene glycol-based precipitation (ExtraPEC) followed by ultracentrifugation (100 000 × g, 70 min) | LC-MS/MS | CD81, Alix, HSC70 | [61] | |
| U87 (glioblastoma), Huh7 (hepatocellular carcinoma), bone-marrow-derived MSC | Differential ultracentrifugation (l-EVs: 10 000 × g, 30 min; s-EVs: 100 000 × g, 90 min) | LC-MS/MS | MSC: PLP2 | U87and Huh7: CD63; | [58] |
| U87:Tsg101, Alix, CD82 | |||||
| DU145-shDIAPH3 (prostate cancer) | Differential ultracentrifugation (l-EVs: 10 000 × g, 30 min; s-EVs: 100 000 × g, 60 min) followed by density gradient | SILAC | GAPDH, CPI, LDHB, HSPA5, MDH, GOT1, CK18 | CD81.CD9, ITGA3, ITGAV, ICAM, CD44 | [47] |
Another study, from the Théry laboratory,[57] employed the same MS approach (GeLC–MS/MS) to perform a comprehensive comparative proteomic analysis of different classes of EVs released by dendritic cells. s- and l-EVs were separated by their sedimentation speed and density. The study identified syntenin-1, TSG101, ADAM10, and EHD4 as proteins enriched in s-EVs. It also identified common proteins present in all classes of EVs, which included some proteins that had previously been described as canonical s-EV markers (MHC class I and class II molecules, heat-shock proteins 70, flotillins, and actin). Immuno-isolation using various anti-tetraspanin antibodies (CD9, CD63, or CD81) enabled identification of different populations of s-EVs corresponding to endosome-derived and plasma membrane-or early endosome-derived EVs. These data suggested that not all s-EVs expressing tetraspannins are derived from the endosomal machinery, and those that do not always express the same tetraspannin profile. These findings are in agreement with previous study(ies) from the Gould group demonstrating that certain cells (e.g., Jurkat T cells) exhibit discrete domains of the plasma membrane that are enriched for endosomal proteins and serve as sites of immediate s-EV biogenesis.[31]
Using SILAC quantitative proteomics analysis, our group compared protein cargo of s-EVs and l-EVs LO.[47] The study identified a number of proteins that were common to s-EVs and LO from prostate cancer cells. However, 25% of the proteins were differentially represented in the two EV classes. Functional pathway analysis demonstrated that enzymes involved in glucose, glutamine, and amino acid metabolism, such as GAPDH, GPI, LDHB, HSPA5, MDH, and GOT family members, are enriched in LO. Conversely, proteins enriched in s-EVs belonged to adhesion molecules and integrin family proteins, in line with data from other groups.[59,60] In addition, among the proteins enriched in LO, cytokeratin 18 (CK18) was found to be one of the most abundant (within the top 5%) and was used to develop an assay to detect LO in the circulation and tissues of mice and patients with prostate cancer.
Attempts to compare protein sets identified in discrete EV classes among different studies are still limited. Additionally, the differing protein profiles identified in those studies might result from the use of disparate cell sources of EVs or the differences in the proteome of the producer cells (e.g., in cancer cells vs benign cells, or in cancer cell lines with different metastatic potential). However, the identified differences might also reflect the disparate EV isolation methods used by the researchers, which yield EV pellets of varying composition. For example, proteomic profile of s-EVs isolated using differential centrifugation followed by density gradient purification might be distinct from proteomic profile of s-EVs isolated using PEG precipitation, which also contain larger particles and/or protein contaminants, even when those s-EVs are derived from the same cell source. Finally, apart from different cell source and isolation methods, treatment also can change the composition of s-EVs.[61] Therefore, analysis of proteome profiles of discrete EV classes isolated from different cell sources using the same method could provide valuable information on the extent of the heterogeneity of EVs. This is important as it would add to the increasing evidence that different proteomic profiles might reflect the biological variability among the various EV sources.
One such study was performed by the Khvorova group.[58] The study compared protein and lipid composition of l-EVs and s-EVs isolated by differential ultracentrifugation from glioblastoma, hepatocellular carcinoma, or bone marrow mesenchymal stem cells (MSC). LC–MS/MS identified 3532 proteins and MS/MSALL identified 1961 lipid species. An interesting finding of the study is that the s-EV proteome, rather than the l-EV proteome, closely resembled the proteome of the donor cells thus allowing to distinguish between cancer cells and MSC. In contrast, this resemblance was not preserved at the level of lipids. In fact, lipidomic profiles did not distinguish the MSC and hepatocellular carcinoma cells. Functionally, extracellular matrix, receptors, heparin-binding, phospholipid-binding, integrin, immune response, and cell adhesion functions were characteristic for s-EVs, whereas mitochondrial, endoplasmic reticulum, and proteasomal functions were enriched in l-EVs. This result is in line with the findings of the above mentioned study on LO that also reported the enrichment of proteins involved in adhesion, motility, and response to hypoxia in s-EVs, and the enrichment of proteins involved in metabolic processes in LO.[47] The authors concluded that although s-EVs and l-EVs can be distinguished at the proteome level based on both their size and the cellular origin, they did not display s-EV- or l-EV-specific protein markers that could be universally found across all cell types examined.
The Meckes Jr. laboratory[62] approached the issue of EV heterogeneity from a different angle. In their study, they did not separate s- and l-EVs, instead they used a PEG-based method for enrichment of EVs (ExtraPEG) followed by ultracentrifugation. They compared protein profiles of EV preparations representing 60 different cancer cell lines from nine distinct tissue types in an attempt to identify common EV protein cargo, as well as cancer type-specific biomarkers. Across the 60 cell lines examined, LC–MS/MS identified a total of 6071 proteins, 213 of which were common among all cell lines, likely reflecting common intracellular EV machinery. When enrichment of traditional EV proteins was analyzed, only CD81, Alix, and HSC70 were found to be present across all samples. Other proteins that had been previously reported to be frequently enriched in EVs, such as CD63, CD9, TSG101, Syntenin-1, and flotillin-1, were present in at least two-thirds of the samples. The expression of some of the canonical EV proteins varied greatly in EVs of different cellular origin. Interestingly, clustering of EV protein profiles revealed that some of them clustered together based on the metastatic potential of their donor cell or growth type (adherent/suspension) rather than based on the tissue type. For example, the protein profiles of EVs derived from highly metastatic breast cancer cell lines clustered together away from the EV protein profiles derived from nonmetastatic cell lines. Similarly, the basement membrane glycoprotein agrin (AGRN) was absent only in EVs secreted from leukemia cancer lines grown in suspension, while the adhesion molecule ICAM3 was present predominately in the leukemia-derived EVs. More studies like this–that allow direct comparison of protein profiles of EVs isolated from multiple cell sources using the same method–would contribute enormously to the field. However, methods that ensure isolation of high-purity EV populations should be used.[8,63]
The main findings, as well as isolation strategies and biological sources used in the comparative studies discussed in this section, are summarized in Table 1. Taken together, these studies demonstrate that both biological variability and differences in isolation techniques contribute substantially to the observed interclass heterogeneity of EVs, and highlight the importance of standardization of isolation methods in order to be able to reliably identify the biological component. Further proteomic comparisons of different high-purity EV classes are needed to shed light on the common markers present in each EV class in healthy and disease models. These studies will allow for a greater understanding of the composition of the diverse populations of EVs that are isolated from biofluids, such as blood, and may aid in the identification of EV class-specific disease-associated markers.
3. Heterogeneity Within One EV Class
On top of the functional and compositional diversity between EV classes, there is an additional layer of heterogeneity within each EV class as well. One of the most striking examples of this heterogeneity is represented by differences in protein composition of s-EVs isolated from the apical and basolateral sides of polarized epithelial cells. Nearly two decades ago, the Heyman and Simpson groups independently showed that apical and basolateral s-EVs released from the same cells contain different protein cargo[64,65] The Heyman laboratory isolated and characterized apical and basolateral s-EVs from electrically tight polarized colonic adenocarcinoma cells by ultracentrifugation. They showed difference in protein composition by MALDI–TOF-MS,[64] despite both populations having similar levels of MHC class I and II proteins. A few years later, the Simpson lab used sequential immunoaffinity capture with an A33 antibody, followed by immuno-capture with an EpCAM antibody to isolate basolateral (A33-Exo) and apical (EPCAM-Exo) s-EVs, respectively, from human colon carcinoma cells. The two populations of s-EVs were profiled using GeLC-MS/MS.[65] The fact that EpCAM-Exo were enriched in apical sorting proteins, while A33-Exo were enriched in basolateral proteins is in line with subcellular localizations of EpCAM to the apical surface and A33 to the basolateral surface of LIM1863 cells. The two s-EV populations exhibited distinct protein expression patterns with A33-Exo containing MHC class I molecules, which were not identified in EpCAM-Exo. Additionally, protein profiling of LIM1863 shed l-EVs demonstrated that protein expression in l-EVs was distinct from both A33- and EpCAM-Exo. These early studies suggest that polarized cells release different populations of s-EVs that are shed into different biological compartments.
Recently three independent labs have compared the protein cargo of apical and basolateral s-EVs isolated by ultracentrifugation from polarized epithelial cells grown on transwell inserts[66–68] All three studies demonstrated, together with a 70% overlap between apical and basolateral s-EVs, some qualitative and quantitative significant differences confirming the earlier experiments in apical and basolateral EV proteins. However, there appears to be significant tissue- or cell line-dependent differences in the composition of apical and basolateral s-EVs. Syntenin-1 has been proposed as a reference gene for apical and basolateral s-EVs[67] and it is present in apical and basolateral s-EVs from porcine retinal epithelial cells and MCF10A human mammary epithelial cells.[66] However, Syntenin-1 was only barely detected in basolateral s-EVs and was not detected at all in apical EVs from HuMEC human mammary epithelial cells and MCFDCIS human breast (ductal carcinoma in situ) cells.[66] This could be due to differences in the donor cells, which differed in terms of species and tissue: human breast[66] and porcine retina.[67]
The traditional EV proteins CD63[65–68] and CD81[65,67,68] were enriched in apical s-EVs in comparison to the basolateral s-EVs, indicating that they may have potential as markers for at least a subset of apically shed EVs. In contrast, traditional EV proteins were not commonly enriched in basolateral in comparison to apical EVs across these different studies. Interestingly, Alix (PDCD6IP) was found in similar proportions in both apical and basolateral s-EVs in several studies[65–67] Conversely, conflicting results were found for TSG101. In s-EVs isolated from polarized MDCK cells, TSG101 was found to be enriched in basolateral s-EVs,[68] while in s-EVs isolated from polarized human breast epithelial cells and polarized porcine retinal epithelial cells, TSG101 was enriched in apical s-EVs. In contrast, A33 and EPCAM immunoaffinity isolation of s-EV populations did not enrich for TSG101 in either population.[65] In all the three studies showing enrichment of TSG101 in apical or basolateral s-EVs the producer cells were grown on transwell inserts and s-EVs were isolated from conditioned media by ultracentrifugation followed by density gradient separation, thus it is unlikely that the difference in TSG101 is due to methodology differences[66–68] It is possible that the differential secretion of TSG101 in apical or basolateral s-EVs is due to its specific localization in different cell types. Since TSG101 is a key component of the ESCRT-1 complex, this could indicate that ESCRT-mediated EV cargo sorting is largely apical or basolateral depending on the physiological context. However, additional studies are necessary to draw this conclusion.
Aside from differences in apical versus basolateral secretion, there are also different s-EV populations that can be distinguished based upon size. Using the novel asymmetric flow fieldflow fractionation (AF4) technology, the Lyden’s team recently reported that s-EVs can be subdivided into small and large populations, Exo-S (60–80 nm) and Exo-L (90–120 nm), as well as very small non-vesicular extracellular particles termed exomeres (≈35 nm), which lack an external membrane.[29] MS from these subpopulations indicated that the Exo-S class was enriched in proteins associated with endosomes, MVBs, vacuoles, and phagocytic vesicles, while the Exo-L was enriched in proteins associated with plasma membrane, late endosomes, and Golgi suggesting differences in biogenesis. In contrast, exomeres were depleted of membrane-associated proteins and enriched in ECM, ER, and mitochondrial proteins. Although CD81, CD9, CD63, and TSG101 were enriched in both Exo-S and Exo-L in comparison to exomeres, there were large differences in the relative abundance within each population when compared between five different cell lines. This result highlights the variability of expression for these proteins across different sources, which impairs their use as universal markers. In vivo all three nanoparticle subpopulations were taken up mostly by hemopoietic organs, with some uptake observed in other organs including lungs, lymph nodes, and kidneys. Exo-L displayed notably larger uptake in the lymph nodes and bone than Exo-S and exomeres, suggesting that EV size could be used to separate populations of s-EVs with different organotropism.
In parallel work, the Wood’s laboratory reported that B16F10 melanoma cells released two discrete populations of s-EVs termed low density exosomes (LD-Exo) and high density exosomes (HD-Exo), which had different equilibration times in density gradients, protein/RNA cargo, as well as functional effects on target cells.[69] When s-EV pellets were loaded on top of a sucrose density gradient and spun for 16 h, LD-Exo and HD-Exo separated into distinct fractions. However, when the density gradient was spun for 72 h, EV markers and particles were mainly detected in the LD-Exo fractions, suggesting that HD-Exo have delayed equilibration. Both populations expressed EV-enriched proteins Alix and TSG101. Notably, the ceramide inhibitor GW4869 resulted in loss of both Alix and TSG101 and decreased s-EV particle counts in both populations, suggesting that these populations are dependent on ceramide biosynthesis. Electron microscopy and Nanoparticle Tracking Analysis (NTA) indicated that HD-Exo were smaller (mode size 66 nm) and more homogenous in size compared to LD-Exo (mode size 117 nm), and it was hypothesized that this difference in size contributes to the delayed equilibration time. Similar LD-Exo and HD-Exo populations were found from several mouse and human cell lines as well as human plasma, suggesting that they are not cell line specific. NanoLC–MS/MS proteomic analysis revealed high overlap between HD-Exo and LD-Exo protein content, with ≈8.5% proteins unique to HD-Exo and ≈39.5% proteins unique to LD-Exo. Bioanalyzer profiles of the RNA content showed that HD-Exo lacked rRNA and had a broader small RNA distribution profile compared to LD-Exo. Treatment of H5V endothelial cells with HD-Exo or LD-Exo resulted in subtle differences in expression profiles; however, physiological phenotypes were not tested/validated in this study.
Together these findings suggest that even a single class of EVs can be further subdivided into different populations based upon properties such as physiological compartment and size. Importantly these divisions are not mutually exclusive as the Takada laboratory found HD-Exo and LD-Exo populations (which may differ by size) in apical but not basolateral s-EVs isolated from MDCK cells.[68] The Heyman laboratory found opposing results showing that apical s-EVs from colonic adenocarcinoma cells were more homogenous in size than basolateral s-EVs, which may contain both Exo-S and Exo-L populations,[64] hinting that these populations are dynamic in respects to each other and may differ between cell lines and possibly culture conditions. EVs secreted from the apical versus basolateral compartments of the cell are exposed to different physiological environments (and thus may have different recipients) and Exo-L have greater bone and lymph node tropism than Exo-S and exomeres, indicating that individual EV populations may communicate with different recipient cells using specific mechanisms (Figure 3).[69] If care is not taken to separate these EV populations, study conclusions will be limited and not physiologically relevant.
Figure 3.
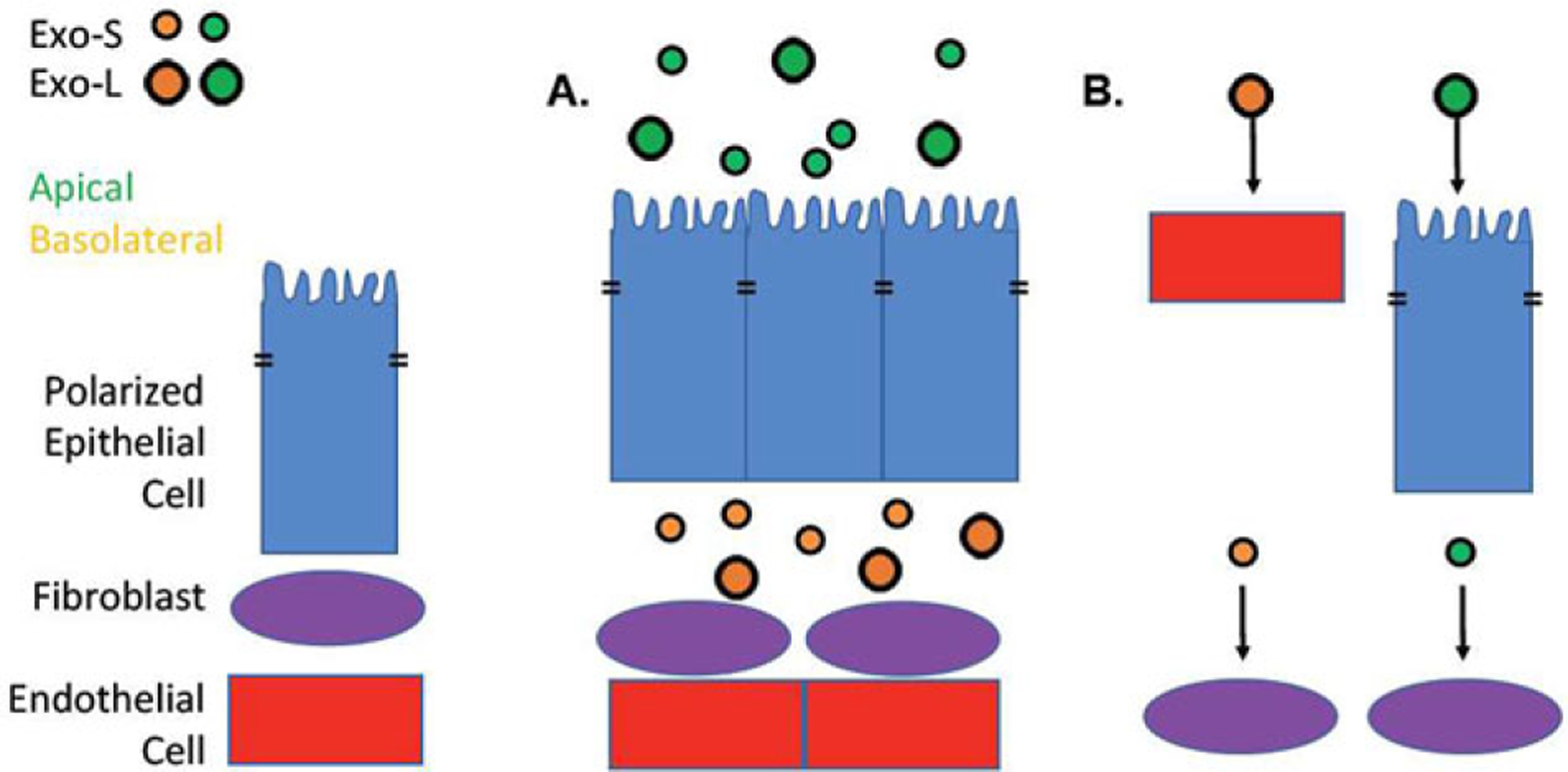
Complexity of exosomes secreted by polarized epithelia. A) Polarized epithelial cells secrete s-EVs with different protein cargo from the apical (green) versus basolateral (yellow) sides of the cell, potentially exposing them to different recipient cells. These s-EVs can be further subdivided into small (Exo-S, 60–80 nm) and large (Exo-L, 90–120 nm). B) Differences in tropism as well as available recipient cells may result in different EV populations signaling with different recipient cells. For instance, the basolateral secreted EVs have a greater chance to come into contact with endothelial cells and fibroblasts than apical secreted EVs.
4. Emerging Approaches to Study EV Heterogeneity
In order to study EV heterogeneity and to reliably identify protein biomarkers specific to different EV populations, it is crucial to improve our ability to separate discrete homogenous EV populations, which has been one of the main challenges in the field. Novel techniques employ commonly identified EV markers to further characterize the diversity of protein cargo in EV populations differentially expressing those markers.
In one such recent study, the Weissleder group developed a single EV analysis (SEA) technology and used it for multiplexed profiling of EVs.[70] s-EVs from glioblastoma cells (Gli36) were immobilized on a functionalized surface through biotin/neutravidin interaction and sequentially imaged for a selection of EV markers (CD9, CD81, and CD63) or putative glioblastoma tumor markers (EGFR, EGFRvIII, IDH1, IDH1-R32, PDPN, PDGFRa, PDL1, and PDL2). The analysis showed a nonhomogeneous distribution of classical endosomal EV markers among s-EVs: 54% of EVs expressed CD63, 26% CD81, and only 4.8% of EVs expressed CD9. This result is in line with other reports[57,65–68] showing that the proteins considered as general s-EV markers are not present in all s-EVs. Among the selected tumor markers, EGFR was found to be the most prevalent (71%), followed by IDH1, PDGFRa, and EGFRvIII (28–6.5%). Overall, for a given cell type, cumulative distribution showed that >90% EV expressed four or less markers. When clustered by the method for dimensionality reduction tSNE, the most clustered population was positive for CD9/EGFRvIII. Some markers were expressed in many clustered populations (e.g., CD63 in eight populations), whereas others were present in much fewer (e.g., PDGFRa, only in 1 population). These data demonstrate that the choice of a ‘common’ vesicular or/and a cancer-specific marker for EV capture can lead to isolation of different EV populations, which in turn can result in very different findings.
To examine EV heterogeneity at a population level, the Boilard lab took advantage of FACS followed by[71] spanning-tree progression analysis of density-normalized events (SPADE) analysis, which clusters events based on similarity and allows for better identification of rare subpopulations. EVs isolated from stimulated platelets and erythrocytes were organized based on the expression of CD41a, CD235a, phosphatidylserine, mitochondrial content, size, and complexity (granularity). Based on the default settings, SPADE auto-suggested ten EV populations, some of which were identified by the authors as originating from erythrocytes, platelets, or mitochondria. The use of SPADE would allow analysis of FACS data from multiple channels at once; however, in its current state, more work needs to be done on the sets of defined, highly purified EV populations to confirm the results obtained using this tool. It also remains questionable whether these are physiologically relevant populations or a by-product of this innovative analysis.
The Yan laboratory employed high-sensitivity flow cytometry (HSFCM), which allows detection of single EVs (down to 40 nm size), to compare and characterize protein profiles of EVs isolated from normal colorectal cells and colorectal cancer cells, as well as plasma EVs from colorectal cancer patients.[51] HSFCM identified 85–90% of particles isolated from cultured cells as EVs, while only 70% of particles isolated from platelet-free patient plasma corresponded to vesicles (probably due to large number of contaminating lipoprotein particles in plasma). EV analysis by HSFCM showed similar size distributions to the cryo-TEM analysis, suggesting that HSFCM is accurately measuring EV size. Authors reported that 55.2% of colorectal cancer-derived EVs (HCT15) were positive for CD9, 46.0% positive for CD63, and 55.0% were positive for CD81, providing additional evidence that these proteins are not present on all EVs. Double staining analysis indicated that 31.8% EVs were CD9+/CD81+, 31.1% were CD63+/CD81+, and 22.3% were CD9+/CD63+, highlighting the heterogeneity of these traditional EV markers within individual EVs. The expression of CD147 had been previously shown to be increased in colorectal cancer EVs. In this study, CD147 was present on 10% of EVs from normal colorectal cells (CCD-18Co) and increased to 51.4% and 47.5% in HCT15 and HCT116 colorectal cells, respectively. Importantly, there were significantly more CD147+ EVs in plasma of colorectal cancer patients of all stages (including stage 1) compared to healthy controls, suggesting that CD147+ plasma EVs may be useful for colorectal cancer diagnosis. In comparison there was no significant difference in EV numbers in plasma of healthy donors vs. colorectal cancer patients due to large (40–50-fold) variability between individuals, indicating that analysis of EV populations is necessary for diagnostic purposes. Importantly, HSFCM only requires 50 μL of platelet-free plasma to analyze plasma EV in comparison to more traditional approaches that usually require much larger volumes.
A large interindividual variability has emerged as one of main obstacles in the proteomic analysis of clinical samples and has been reported in a number of studies that interrogated the potential of EV protein cargo as a source of biomarkers in cancer diagnostics/prognostics.
5. Proteomic Profiling of EVs in Clinical Samples
As carriers of disease-associated molecules that can be found in biofluids, EVs have great potential as a noninvasive source of biomarkers for diseases such as cancer. Recently three groups (Hwang, Buzás, and Xiao) have performed comparative proteomic analysis on EVs from disease-associated biofluids.[52,53,72] Proteomic analysis of EVs from biofluids has been shown to decrease the background signal derived from highly abundant proteins, such as creatine in urine, resulting in greater sensitivity.[72] This allows identification of additional proteins that would not be detected in the source biofluid. For example, a large number of EV proteins isolated from urine specimens of bladder cancer patients were not identified by proteomics perfomed on straight urine without EV isolation.[72] In line with this, a large number of EV proteins isolated from pancreatic juice of patients with chronic pancreatitis or pancreatic cancer were not identified when proteomics was performed on pancreatic juice without EV isolation.[53] In both cases, the number of proteins that were identified only in the EV samples were ≥40%. These data suggest that EV proteins can potentially achieve better separation of patients and healthy individuals. In addition, different EV classes seem to contain distinct cargo. For instance, in the study from Buzás group, mucin-5AC and mucin-6 were uniquely identified in l-EVs, while mucin-16 and dihydropyrimidinase-related protein 2 were exclusive to s-EVs isolated from the pancreatic juice of pancreatic cancer patients.[53] Similarly, the study from Xiao lab showed that BPIFA1 and CRNN were significantly enriched in l-EVs, while MUC5B and IQGAP were enriched in s-EVs from the saliva of lung cancer patients compared to healthy donors. Importantly, these differences were not observed when saliva was analyzed without EV isolation.[52]
Altogether, these findings suggest not only that EVs containing cancer-specific proteins can be successfully isolated from clinical samples, but also that enriching for EV proteins is important to increase the S/N ratio of cancer circulating biomarkers. However, the number of studies that focus on proteomic analysis of EVs, and in particular of different EV classes, in clinical samples is still limited to make universal conclusions that can be translated into clinical practice (reviewed in refs. 73 and 74). Additionally, these studies require larger numbers of patients for an adequate power analysis, and standardization of the collection procedures to minimize interstudy variability.
6. Conclusions
MS is the most powerful approach for identification and quantification of protein components of EVs. This characterization is essential for understanding EV biogenesis and function, as well as for distinguishing among different EV classes and subtypes, and identification of disease-specific markers. However, in the literature published to date, there is a lack of agreement among the reports on the protein markers specific to different EV classes/populations. And often this lack of agreement is due to a remarkable EV heterogeneity that has been largely ignored in the past and is emerging as functionally important.
Heterogeneity of EVs has been one of the main limitations for the use of proteomics in EV analysis. Most of the currently available EV isolation methods yield mixed EV populations and, therefore, impede differential analysis of discrete EV types. In addition to the differences in isolation techniques, biological variability contributes significantly to EV heterogeneity. Relative enrichment of a protein in a given EV class/population can be cell type-dependent. Moreover, it can depend on the subcellular compartment from which these EVs have originated, as illustrated by different populations of s-EVs generated by the same cell. Together, biological and technical variability would inevitably lead to identification of differing EV protein markers by different studies. This highlights the importance of standardization of isolation methods in order to be able to reliably identify EV protein components.
EV protein cargo represents a valuable source of prognostic or/and diagnostic biomarkers in a number of pathological conditions, including cancer, and it has been demonstrated that circulating EVs could be successfully isolated from clinical samples. However, the application of proteomic analysis to patient-derived EVs is further complicated by sometimes striking interindividual variability, as well as often very limited amount of material available for analysis. The rapid progress in the development of proteomic technologies would likely address the latter issue in the near future and allow accurate analysis of scant amounts of EV protein. And a steadily increasing number of studies on patient-derived EVs would hopefully help overcome the interindividual variability issue by providing more data and, thus, increasing its reliability/statistical power.
Tune in to:
Explore the LC-MS portfolio and meet the expanded Orbitrap Exploris MS systems in our Exhibit Hall
Learn from mass spectrometry experts, such as Proffesor Alexander Makarov himself, about Orbitrap mass spectrometry technology and the applications it enables
Browse posters and short presentations in your application area.
Biographies

Tatyana Vagner is a project scientist at Cedars-Sinai Medical Center in Los Angeles (USA). She obtained her Ph.D. in molecular medicine from the University of Auckland, New Zealand. Her current research focuses on development of EV-based liquid biopsy platforms and discovery of circulating EV biomarkers in cancer. She has also been investigating the origin of the DNA associated with cancer-specific EVs, termed large oncosomes, and the potential of the EV-derived DNA for early detection of metastasis, prediction of the aggressive features of the disease, and monitoring cancer patients’ response to treatment.

Andrew Chin is a postdoctoral scientist at Cedars-Sinai Medical Center in Los Angeles (USA). He obtained his Ph.D. at the Irell and Manella Graduate School of Biological Sciences (City of Hope) in Duarte (USA) studying the effects of the EV-encapsulated plant-derived miRNA miR-159 on breast cancer. His current interest is in understanding the heterogeneity of EV and how they influence diseases such as prostate cancer. He is a Chesapeake Urology Associates Sanford J. Siegel, MD, Prostate Cancer research scholar and his current work focuses on the functional role of miRNA in large oncosomes in prostate cancer.

Dolores Di Vizio is a Professor at Cedars-Sinai Medical Center. Her background is in cancer pathology and molecular and cell biology. More recently, her expertise has been extended to Extracellular Biology. She is an Executive Board Member of the International Society of Extracellular Vesicles (ISEV). Her group studies the molecular mechanisms of progression to advanced disease in human tumors, with a particular emphasis on large oncosomes, EVs shed by fast migrating and metastatic amoeboid cancer cells. Her lab is currently profiling the large oncosomes and other EV populations by NGS and proteomics for functional and molecular characterization.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Javier Mariscal, Department of Surgery, Division of Cancer Biology and Therapeutics, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA.
David M. Engman, Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA
Dolores Di Vizio, Department of Surgery, Division of Cancer Biology and Therapeutics, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA; Department of Pathology and Laboratory Medicine, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA; Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA; Department of Medicine, University of California, Los Angeles, CA, 90095, USA.
References
- [1].Figueroa JM, Skog J, Akers J, Li H, Komotar R, Jensen R, Ringel F, Yang I, Kalkanis S, Thompson R, Loguidice L, Berghoff E, Parsa A, Liau L, Curry W, Cahill D, Bettegowda C, Lang FF, Chiocca EA, Henson J, Kim R, Breakefield X, Chen C, Messer K, Hochberg F, Carter BS, Neuro Oncol 2017, 19, 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fais S, O’Driscoll L, Borras FE, Buzas E, Camussi G, Cappello F, Carvalho J, Cordeiro Da Silva A, Del Portillo H, El Andaloussi S, Ficko Trček T, Furlan R, Hendrix A, Gursel I, Kralj-Iglic V, Kaeffer B, Kosanovic M, Lekka ME, Lipps G, Logozzi M, Marcilla A, Sammar M, Llorente A, Nazarenko I, Oliveira C, Pocsfalvi G, Rajendran L, Raposo G, Rohde E, Siljander P, Van Niel G, Vasconcelos MH, Yáñez-Mó M, Yliperttula ML, Zarovni N, Zavec AB, Giebel B, ACS Nano 2016, 10, 3886. [DOI] [PubMed] [Google Scholar]
- [3].Witwer KW, Buzás EI, Bemis LT, Bora A, Lässer C, Lötvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Théry C, Wauben MH, Hochberg F, J. Extracell. Vesicles 2013, 2, 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C, Curr. Biol 2009, 19, 1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lässer C, Seyed Alikhani V, Ekström K, Eldh M, Torregrosa Pare-des P, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H, J. Transl. Med 2011, 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van Eijndhoven MAJ, Zijlstra JM, Groenewegen NJ, Drees EEE, van Niele S, Baglio SR, Koppers-Lalic D, van der Voorn H, Libregts SFWM, Wauben MHM, de Menezes RX, van Weering JRT, Nieuwland R, Visser L, van den Berg A, de Jong D, Pegtel DM, JCI Insight 2016, 1, 89631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van Den Berg MS, Willemsen R, Luider T, Paša-Tolić L, Jenster G, PLoS One 2013, 8, 82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach J-M, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, et al. J. Extracell. Vesicles 2018, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar CM, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D, Nat. Med 2012, 18, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO, Nat. Cell Biol 2007, 9, 654. [DOI] [PubMed] [Google Scholar]
- [11].Segura E, Amigorena S, Théry C, Blood Cells Mol. Dis 2005, 35, 89. [DOI] [PubMed] [Google Scholar]
- [12].Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, De Wit E, Berenguer J, Ellenbroek SIJ, Wurdinger T, Pegtel DM, Van Rheenen J, Cell 2015, 161, 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sidhu SS, Mengistab AT, Tauscher AN, LaVail J, Basbaum C, Oncogene 2004, 23, 956. [DOI] [PubMed] [Google Scholar]
- [14].Morello M, Minciacchi VR, De Candia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chung LWK, Gandellini P, Freeman MR, Demichelis F, Di Vizio D, De Candia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chung LWK, Gandellini P, Freeman MR, Demichelis F, Di Vizio D, Cell Cycle 2013, 12, 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Minciacchi VR, Spinelli C, Reis-Sobreiro M, Cavallini L, You S, Zandian M, Li X, Chiarugi P, Cocucci E, Bhowmick NA, Di Vizio D, Adam RM, Posadas EM, Viglietto G, Freeman MR, Cocucci E, Bhowmick NA, Di Vizio D, Cancer Res 2017, 77, 2306. [DOI] [PubMed] [Google Scholar]
- [16].Demory Beckler M, Higginbotham JN, Franklin JL, Ham A-J, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ, Mol. Cell. Proteomics 2013, 12, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J, Nat. Cell Biol 2008, 10, 619. [DOI] [PubMed] [Google Scholar]
- [18].Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F, Nat. Commun 2011, 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO, Nat. Cell Biol 2008, 10, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D, Cell Res 2014, 24, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Skotland T, Hessvik NP, Sandvig K, Llorente A, J. Lipid Res 2018, jlr. R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Skotland T, Ekroos K, Kauhanen D, Simolin H, Seierstad T, Berge V, Sandvig K, Llorente A, Eur. J. Cancer 2017, 70, 122. [DOI] [PubMed] [Google Scholar]
- [23].Lydic TA, Townsend S, Adda CG, Collins C, Mathivanan S, Reid GE, Methods 2015, 87, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch O, Zhang J, Weitz J, Chin L, Futreal A, Kalluri R, J. Biol. Chem 2014, 289, 3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J, Nat. Commun 2011, 2, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li B, Antonyak MA, Zhang J, Cerione RA, Oncogene 2012, 31, 4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Conley A, Minciacchi VR, Lee DH, Knudsen BS, Karlan BY, Citrigno L, Viglietto G, Tewari M, Freeman MR, Demichelis F, Di Vizio D, RNA Biol 2017, 14, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Colombo M, Raposo G, Théry C, Annu. Rev. Cell Dev. Biol 2014, 30, 255. [DOI] [PubMed] [Google Scholar]
- [29].Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhães A, Ferreira JA, Osório H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, Lyden D, Nat. Cell Biol 2018, 20, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].D’Souza-Schorey Crislyn C, Clancy JW, Genes Dev 2012, 26, 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JEK, Gould SJ, Sandefur S, Varthakavi V, Cell Biol J. 2006, 172, 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, Holowka DA, Cerione RA, Proc. Natl. Acad. Sci. USA 2011, 108, 4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stein JM, Luzio JP, Biochem. J 1991, 274, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cocucci E, Meldolesi J, Curr. Biol 2011, 21, 940. [DOI] [PubMed] [Google Scholar]
- [35].Keerthikumar S, Gangoda L, Liem M, Fonseka P, Atukorala I, Ozcitti C, Mechler A, Adda CG, Ang C-S, Mathivanan S, Oncotarget 2015, 6, 15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Larson MC, Woodliff JE, Hillery CA, Kearl TJ, Zhao M, Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2012, 1821, 1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lima LG, Chammas R, Monteiro RQ, Moreira MEC, Barcinski MA, Cancer Lett 2009, 283, 168. [DOI] [PubMed] [Google Scholar]
- [38].Whiteside TL, Br. J. Cancer 2005, 92, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y, Chen L, Yan X, Du Y, Yu L, Cell Res 2015, 25, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, Taub D, Parker JA, Neri C, Gabel CV, Hall DH, Driscoll M, Nature 2017, 542, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Elmore S, Toxicol. Pathol 2007, 35, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Turiák L, Misják P, Szabó TG, Aradi B, Pálóczi K, Ozohanics O, Drahos L, Kittel Á, Falus A, Buzás EI, Vékey K, J. Proteomics 2011, 74, 2025. [DOI] [PubMed] [Google Scholar]
- [43].Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J, Extracell J. Vesicles 2013, 2, 20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, Demichelis F, Freeman MR, Cancer Res 2009, 69, 5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, Mulholland D, Rotinen M, Hager MH, Insabato L, Moses MA, Demichelis F, Lisanti MP, Wu H, Klagsbrun M, Bhowmick NA, Rubin MA, D’Souza-Schorey C, Freeman MR, Am. J. Pathol 2012, 181, 1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hager MH, Morley S, Bielenberg DR, Gao S, Morello M, Holcomb IN, Liu W, Mouneimne G, Demichelis F, Kim J, Solomon KR, Adam RM, Isaacs WB, Higgs HN, Vessella RL, Di Vizio D, Freeman MR, EMBO Mol. Med 2012, 4, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Minciacchi VR, You S, Spinelli C, Morley S, Zandian M, Aspuria P, Cavallini L, Ciardiello C, Sobreiro MR, Morello M, Kharmate G, Jang SC, Kim D, Hosseini-Beheshti E, Tomlinson Guns E, Gleave M, Gho YS, Mathivanan S, Yang W, Freeman MR, Di Vizio D, Reis Sobreiro M, Morello M, Kharmate G, Jang SC, Kim D, Hosseini-Beheshti E, Tomlinson Guns E, Gleave M, Gho YS, Mathivanan S, Yang W, Freeman MR, Di Vizio D, Guns ET, Gleave M, Gho YS, Mathivanan S, Yang W, Freeman MR, Di Vizio D, Oncotarget 2015, 6, 11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zaborowski MP, Balaj L, Breakefield XO, Lai CP, Bioscience 2015, 65, 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xu R, Greening DW, Zhu H, Takahashi N, Simpson RJ, J. Clin. Invest 2016, 126, 1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kowal J, Tkach M, Théry C, Curr. Opin. Cell Biol 2014, 29, 116. [DOI] [PubMed] [Google Scholar]
- [51].Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W, Wang S, Li Z, Chen C, Li L, Wu L, Yan X, ACS Nano 2018, 12, 671. [DOI] [PubMed] [Google Scholar]
- [52].Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S, Jiang X, Fan LY, Ji L, Guan X, Cao CX, Xiao H, J. Proteome Res 2018, 17, 1101. [DOI] [PubMed] [Google Scholar]
- [53].Osteikoetxea X, Benke M, Rodriguez M, Pálóczi K, Sódar BW, Szvicsek Z, Szabó-Taylor K, Vukman KV, Kittel Á, Wiener Z, Vékey K, Harsányi L, Szűcs Á, Turiák L, Buzás EI, Biochem. Biophys. Res. Commun 2018, 499, 37. [DOI] [PubMed] [Google Scholar]
- [54].Lane RE, Korbie D, Hill MM, Trau M, Clin. Transl. Med 2018, 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Choi D, Spinelli C, Montermini L, Rak J, Proteomics 2018, 1800169. [DOI] [PubMed] [Google Scholar]
- [56].Xu R, Greening DW, Rai A, Ji H, Simpson RJ, Methods 2015, 87, 11. [DOI] [PubMed] [Google Scholar]
- [57].Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C, Proc. Natl. Acad. Sci. USA 2016, 113, E968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Haraszti RA, Didiot MC, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, Gao F, Narain NR, DiFiglia M, Kiebish MA, Aronin N, Khvorova A, Extracell J. Vesicles 2016, 5, 32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hoshino A, Costa-Silva B, Shen T-LL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Jørgen Labori K, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, Nature 2015, 527, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Krishn SR, Singh A, Bowler N, Duffy AN, Friedman A, Fedele C, Kurtoglu S, Tripathi SK, Wang K, Hawkins A, Sayeed A, Goswami CP, Thakur ML, Iozzo RV, Peiper SC, Kelly WK, Languino LR, Matrix Biol 2018. 10.1016/j.matbio.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Soekmadji C, Riches JD, Russell PJ, Ruelcke JE, McPherson S, Wang C, Hovens CM, Corcoran NM, BioResource TAPCC, Hill MM, Nelson CC, Oncotarget 2017, 8, 52237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, Meckes DG Jr., Oncotarget 2016, 7, 86999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, Gho YS, V Kurochkin I, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Théry C, Extracell J. Vesicles 2014, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M, Gastroenterology 2001, 121, 337. [DOI] [PubMed] [Google Scholar]
- [65].Tauro BJ, Greening DW, Mathias RA, Mathivanan S, Ji H, Simpson RJ, Mol. Cell. Proteomics 2013, 12, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chin AR, Yan W, Cao M, Liu X, Wang SE, Mammary Gland Biol J. Neoplasia 2018, 23, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Klingeborn M, DIsmuke WM, Skiba NP, Kelly U, Stamer WD, Bowes Rickman C, Sci. Rep 2017, 7, 4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen Q, Takada R, Noda C, Kobayashi S, Takada S, Sci. Rep 2016, 6, 35562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Willms E, Johansson HJ, Mäger I, Lee Y, Blomberg KEM, Sadik M, Alaarg A, Smith CIIEE, Lehtiö J, El Andaloussi S, Wood MJA, Vader P, Sci. Rep 2016, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, Chiocca EA, Breakefield XO, Lee H, Weissleder R, ACS Nano 2018, 12, 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Marcoux G, Duchez AC, Cloutier N, Provost P, Nigrovic PA, Boilard E, Sci. Rep 2016, 6, 35928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lee J, McKinney KQ, Pavlopoulos AJ, Niu M, Kang JW, Oh JW, Kim KP, Hwang S, Mol. Cells 2018, 41, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Fendl B, Weiss R, Fischer MB, Spittler A, Weber V, Biochem. Biophys. Res. Commun 2016, 478, 168. [DOI] [PubMed] [Google Scholar]
- [74].Yuana Y, Böing AN, Grootemaat AE, van der Pol E, Hau CM, Cizmar P, Buhr E, Sturk A, Nieuwland R, J. Extracell. Vesicles 2015, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]


