Abstract
Cancer progression is influenced by changes in the tumor microenvironment, such as the stiffening of the extracellular matrix. Yet our understanding of how cancer cells sense and convert mechanical stimuli into biochemical signals and physiological responses is still limited. The long noncoding RNA nuclear paraspeckle assembly transcript 1 (NEAT1), which forms the backbone of subnuclear “paraspeckle” bodies, has been identified as a key genetic regulator in numerous cancers. Here, we investigated whether paraspeckles, as defined by NEAT1 localization, are mechanosensitive. Using tunable polyacrylamide hydrogels of extreme stiffnesses, we measured paraspeckle parameters in several cancer cell lines and observed an increase in paraspeckles in cells cultured on soft (3 kPa) hydrogels compared with stiffer (40 kPa) hydrogels. This response to soft substrate is erased when cells are first conditioned on stiff substrate, and then transferred onto soft hydrogels, suggestive of mechanomemory upstream of paraspeckle regulation. We also examined some well-characterized mechanosensitive markers, but found that lamin A expression, as well as YAP and MRTF-A nuclear translocation did not show consistent trends between stiffnesses, despite all cell types having increased migration, nuclear, and cell area on stiffer hydrogels. We thus propose that paraspeckles may prove of use as mechanosensors in cancer mechanobiology.
INTRODUCTION
Tissue mechanics change gradually during development and aging, and even more dynamically with disease progression. In cancer, the stiffness of tumor tissue increases primarily due to the excessive deposition and reorganization of extracellular matrix (ECM) proteins, such as collagen, fibronectin, and laminin (Cox and Erler, 2011; An et al., 2019). These aberrant changes in stiffness and ECM are well associated with invasion, metastasis, and poor survival in breast cancer patients (Acerbi et al., 2015; Zhou et al., 2015). However, how biomechanical changes within the tumor microenvironment alter mechanosensation of cancer cells needs further investigation.
The ability of cells to convert mechanical stimuli into biochemical signals, through a process known as mechanotransduction, has been well studied using adult stem cells such as bone marrow–derived stem cells (BMSCs) and adipose-derived stem cells (Engler et al., 2006; Choi et al., 2012; Wen et al., 2014). Stem cell mechanosensation ultimately leads to changes in cell fate such as cell morphology, proliferation, migration, and differentiation (Sheetz et al., 1998; Lo et al., 2000; Engler et al., 2006; Dupont et al., 2011; Wei et al., 2015). Several markers including YAP/TAZ and MRTF-A, which shuffle between the nucleus and cytoplasm, and lamin A, have been described as being “mechanosensitive.” These markers respond differently depending on the stiffness of substrate that cells are cultured on (Hadden et al., 2017; Kim et al., 2019; Major et al., 2019). Furthermore, BMSCs also showed “mechanomemory” responses by retaining the memory of YAP/TAZ nuclear translocation experienced on one stiffness, even after cells were transferred to a new stiffness environment (Yang et al., 2014).
Mechanotransduction can also induce structural changes within the nucleus, such as changes in chromatin organization and nuclear envelope composition (Dahl et al., 2008; Alam et al., 2016; Le et al., 2016). “Paraspeckles” are stress-induced subnuclear bodies found within the interchromatin space of mammalian nuclei (Fox et al., 2002; An et al., 2019). Paraspeckles are RNA–protein granules marked by the long noncoding RNA (lncRNA) nuclear paraspeckle assembly transcript 1 (NEAT1), bound by several paraspeckle proteins including paraspeckle component 1 (PSPC1), non-POU domain-containing octamer binding (NONO), and splicing factor proline/glutamine-rich (SFPQ) (Fox et al., 2002, 2005; Clemson et al., 2009; Naganuma and Hirose, 2013). lncRNAs in general are emerging as important gene regulators in cancer, and NEAT1 is strongly correlated with poor prognosis and metastasis in many cancer subtypes, such as breast, lung, and prostate cancers (Chakravarty et al., 2014; Sun et al., 2016; Shin et al., 2019).
Here, we have sought to investigate whether paraspeckles are mechanosensitive to substrate stiffness and subject to mechanomemory. Furthermore, we investigated whether cancer cells were susceptible to changes in morphology, migration, and expression, and translocation of mechanomarkers, upon culture on two extreme stiffness substrates. This study expands our understanding of cancer mechanobiology by discovering a role for ECM stiffness in subnuclear organization via mechanotransduction.
RESULTS AND DISCUSSION
Paraspeckles in cancer cells have an inverse relationship to substrate stiffness
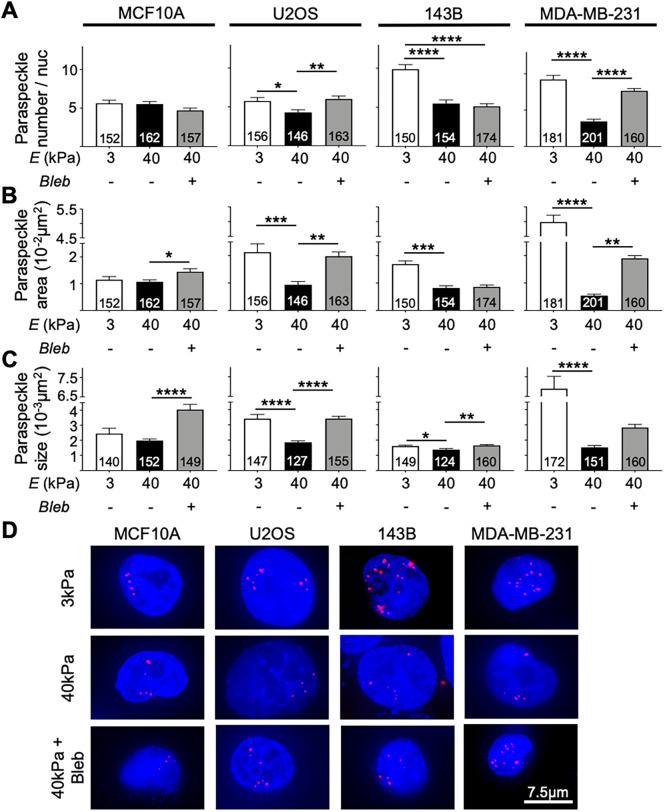
As paraspeckles are stress-responsive nuclear bodies that change in abundance depending on cellular homeostasis, we sought to investigate paraspeckle responsiveness in cells, when grown on soft or stiff substrates. We used a variety of cancer cell lines, as well as a normal epithelial cell line. We used fluorescence in situ hybridization (FISH) against the paraspeckle marker, the lncRNA, NEAT1, to measure differences in paraspeckle parameters following 48 h of cell growth on hydrogel substrates tuned to either 3 kPa or 40 kPa stiffness. Figure 1 shows that all cancer cell lines examined, including U2OS, 143B, and MDA-MB-231, displayed a clear difference in paraspeckles when grown on soft 3 kPa substrates compared with cells grown on stiff 40 kPa hydrogels (Figure 1, A–C). All cancer cell lines displayed an increased paraspeckle number per nucleus, total paraspeckle area (a measure of how much nuclear area was represented by paraspeckle-associated fluorescence), and average paraspeckle size on soft substrates, compared with stiff (Figure 1, A–C). In contrast, the noncancer MCF10A breast epithelial cell line showed no differences in paraspeckle parameters between the two stiffnesses, suggesting that paraspeckle responses to stiffness may be cancer specific (Figure 1, A–C). For the cancer cells, the difference in paraspeckle number for cells grown on soft compared with stiff substrates was more pronounced for the cell lines of metastatic origin, including the metastatic osteosarcoma-derived 143B cells (1.8-fold increase in paraspeckles on soft vs. stiff substrates) and the metastatic breast cancer MDA-MB-231 cells (2.7-fold increase in paraspeckles on soft vs. stiff substrates), compared with the nonmetastatic U2OS osteosarcoma cell line (1.3-fold). Thus, paraspeckles in metastatic cell lines may be more susceptible to changes in substrate stiffness than in nonmetastatic lines. These observations may be linked to the preestablished correlation between NEAT1/paraspeckles and cancer progression and metastasis, and to the role for paraspeckles in cell plasticity (Li and Cheng, 2018; Modic et al., 2019).
FIGURE 1:
Paraspeckle expression on 3 kPa and 40 kPa in MCF10A, U2OS, 143B, and MDA-MB-231 cells. (A) The average number of paraspeckles per nucleus was higher in cancer cell lines cultured on 3 kPa hydrogels compared with 40 kPa hydrogels in U2OS (5.75 vs. 4.28), 143B (9.88 vs. 5.47), and MDA-MB-231 (9.22 vs. 3.44) cell lines. No difference in paraspeckle number was observed in the MCF10A cell line. Treatment of cells cultured on 40 kPa hydrogels with blebbistatin revealed an increase in paraspeckle number in the U2OS (4.28 to 6.00) and MDA-MB-231 (3.44 to 7.65) cell lines. (B) Paraspeckle total area showed the same trend in U2OS (0.021 µm2 vs. 0.0094 µm2), 143B (0.017 µm2 vs. 0.0082 µm2), and MDA-MB-231 (0.055 µm2 to 0.0054 µm2) cell lines but not in the MCF10A cell line. Blebbistatin treatment also resulted in increased paraspeckle area in MCF10A, 143B, and MDA-MB-231 cell lines. (C) Analysis of paraspeckle size revealed that paraspeckles appeared larger in size in cancer cells cultured on 3 kPa hydrogels vs. 40 kPa hydrogels in U2OS (0.0034 µm2 vs. 0.0019 µm2), 143B (0.0016 µm2 vs. 0.0014 µm2), and MDA-MB-231 (0.0069 µm2 vs. 0.0015 µm2) cell lines. Blebbistatin treatment further increased paraspeckle size in all cell lines. (D) Representative images showing paraspeckles (red) in nuclei (blue) taken at 60× magnification. Scale bar = 7.5 µm. Data are shown as mean ± SEM. Numbers of nuclei used in analyses were indicated per bar graph. *, p < 0.05; **, p < 0.01; ***, p < 0.001, and ****, p < 0.0001.
Myosin-II traction forces suppress paraspeckle expression
We reasoned that mechanotransduction may be responsible, at least in part, for the suppression of paraspeckle abundance when cells were grown on the stiff substrate. When cultured on stiff substrates, cells exhibit greater traction forces compared with cells cultured on soft substrates (Lo et al., 2000). Therefore, our data suggests an inverse relationship between paraspeckles and traction force, with cancer cells cultured on soft substrates exhibiting a greater number, total area, and average size of paraspeckles, compared with cells cultured on stiff substrates. To test paraspeckle responsiveness to mechanotransduction, we treated cells with blebbistatin, a nonmuscle myosin-2 inhibitor in order to inhibit cytoskeletal signal propagation on cells cultured on the 40 kPa stiffness, then again measured paraspeckle parameters. The outline of cells as depicted by actin immunofluorescence staining shows the disruption of cell structure following blebbistatin treatment in cells cultured on 40 kPa hydrogels (Supplemental Figure 1). We observed that blebbistatin treatment led to an increased number of paraspeckles/nuclei in U2OS and MDA-MB-231 cells (Figure 1A), increased paraspeckle area in MCF10A, U2OS, and MDA-MB-231 cells (Figure 1B), and increased paraspeckle size in all cell lines (Figure 1C). Thus, our data indicates that traction force–mediated mechanotransduction is playing a role in the suppression of paraspeckles in cancer cells cultured on stiff substrates.
Paraspeckles in cancer cells cultured on soft substrates are more heterogeneous in distribution
Further analysis of our data revealed that paraspeckles in cancer cell lines appear more heterogeneous in distribution on soft substrates compared with stiff. Of the MDA-MB-231 cells grown on 40 kPa substrates, 80% contain between zero and five paraspeckles, compared with only 30% of cells when grown on 3 kPa substrate (Supplemental Figure 1). The other cancer cell lines show the same pattern. Again, the noncancer MCF10A cell line showed no obvious trend in distribution with 58% and 61% of cells displaying between zero and five paraspeckles per nucleus, cultured on 3 kPa and 40 kPa hydrogels, respectively (Supplemental Figure 2). It is well known that phenotypic and functional heterogeneity is characteristic of cancer cells, and in particular, differential expression of lncRNAs between cells has been observed in cancer (Meacham and Morrison, 2013). Our data suggests that substrate stiffness may mediate the variation in the distribution of paraspeckles in cancer cell lines.
Paraspeckles show signs of mechanomemory when cultured on stiff substrates in MDA-MB-231 metastatic breast cancer cells
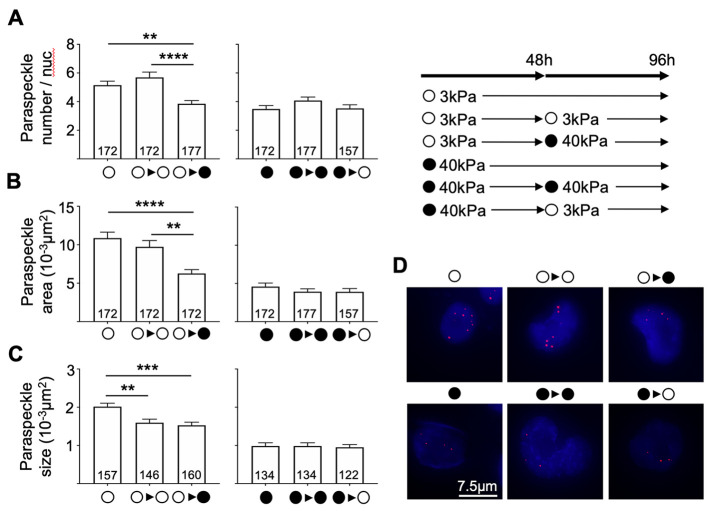
We next explored the possibility that paraspeckle suppression on stiff substrates might be subject to mechanomemory, and used the MDA-MB-231 breast cancer cell line for these experiments, as they displayed the most obvious paraspeckle differences when cultured on different substrates. We cultured cells on one substrate for 48 h, and then transferred the cells to a different stiffness substrate, as well as the same stiffness substrate as a control. Following a further 48 h culture, we fixed the cells and analyzed paraspeckle parameters. As shown in Figure 2, MDA-MB-231 cells showed increased paraspeckle number, total area, and paraspeckle size when cultured on 3 kPa hydrogels compared with 40 kPa hydrogels, and these levels remained elevated when cells were transferred from 3 kPa to 3 kPa (5.15 to 5.69 average number of paraspeckles/nuclei and 0.011 to 0.010 µm2 total paraspeckle area); however, they significantly decreased when MDA-MB-231 cells were transferred from 3 kPa to 40 kPa hydrogels (5.15 to 3.84 average number of paraspeckles/nuclei and 0.011 µm2 to 0.0063 µm2 for the total paraspeckle area; Figure 2A). Thus, the stiffer substrate was able to change the cellular paraspeckle program that had been established on the soft substrate. In contrast, when MDA-MB-231 cells were cultured on 40 kPa hydrogels (where paraspeckle levels were initially lower than that of the 3 kPa hydrogel condition) and transferred onto 3 kPa hydrogels, we observed that paraspeckle parameters did not change, suggestive of paraspeckle mechanomemory (Figure 2B). Of note, paraspeckle size decreased when cells were collected from 3 kPa hydrogels (0.002 µm2) and transferred onto new 3 kPa (0.0016 µm2) and 40 kPa hydrogels (0.0015 µm2), indicating that this parameter may be sensitive to cell passaging. Overall, these data suggest a dominant paraspeckle-suppressive signal that occurs in cancer cells when they are grown on stiff substrates, with a memory of this suppression that persists even when switched to a softer substrate. Consistent with our findings, previous work reported that when human mesenchymal stem cells (hMSCs) were cultured on stiff substrates, YAP/TAZ localized in the nucleus and that this persisted upon culture on soft substrates over a few days (Yang et al., 2014). Further work found that global histone acetylation and chromatin condensation were higher in hMSCs cultured on stiff substrates compared with soft, and that this persisted following culture on soft substrates for a prolonged period, suggesting that epigenetic modifications may provide a means of remembering mechanical input (Killaars et al., 2019). We therefore speculate that the mechanomemory responses in paraspeckles may be explained by epigenetic changes, likely laid down as chromatin marks at the NEAT1 promoter; however, this is yet to be tested.
FIGURE 2:
Paraspeckles in MDA-MB-231 cells following conditioning of cells on hydrogels of one stiffness, then transferring cells onto hydrogels of the opposite stiffness, as well as a hydrogel of the same stiffness as a control. (A) The average number of paraspeckles per nucleus and paraspeckle total area remained elevated when cells were conditioned on 3 kPa hydrogels and transferred onto new 3 kPa hydrogels; however, paraspeckle size decreased. When MDA-MB-231 cells were conditioned on 3 kPa hydrogels and transferred onto hydrogels of 40 kPa stiffness, reduced levels of paraspeckle number (5.15 to 3.84), total area (0.011 µm2 to 0.0063 µm2), and size (0.002 µm2 to 0.001 µm2) were observed. (B) Cells that were conditioned on hydrogels of 40 kPa stiffness and transferred onto hydrogels of the same 40 kPa stiffness, as well as the opposite 3 kPa stiffness, showed no changes in paraspeckle parameters. (C) Representative images showing paraspeckles (red) in nuclei (blue) taken at 60× magnification. Scale bar = 7.5 µm. Data are shown as mean ± SEM. **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001.
Cells appear larger and morphologically different when cultured on stiff substrates
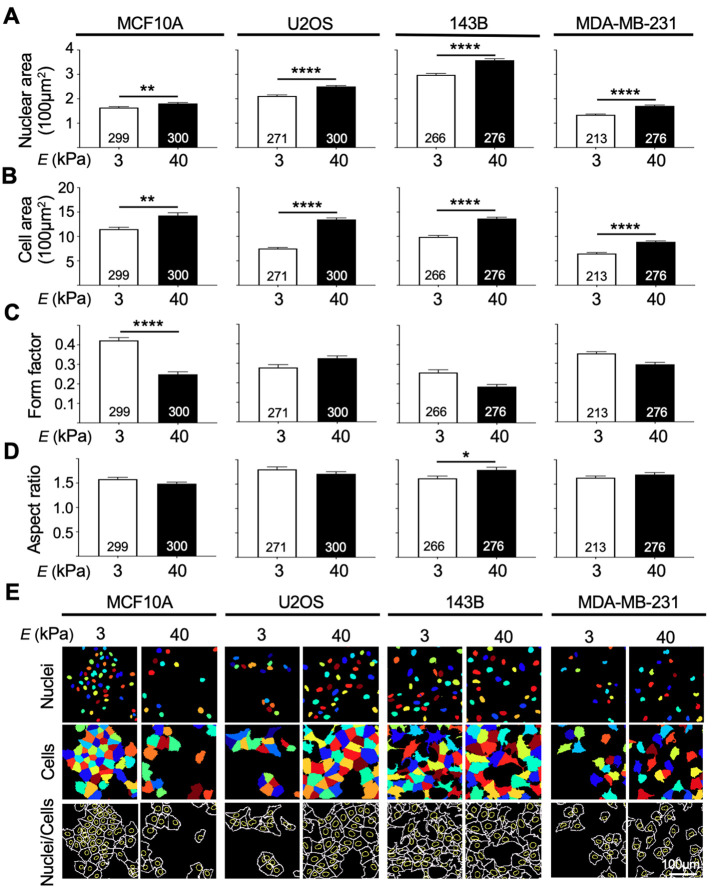
To further assess mechanotransduction, we investigated cell morphological parameters that have previously been linked to cell growth on different stiffness substrates. All cancer cell lines, as well as the normal breast epithelial MCF10A cells, had greater nuclear and cell areas when grown on stiff 40 kPa hydrogels compared with cells grown on soft 3 kPa hydrogels (Figure 3, A and B). This positive correlation between ECM stiffness and nuclear and cell area has been well described in several studies, including in breast cancer, and may be explained by the increase in traction forces exerted to the ECM via integrins (Hynes, 1987; Yeung et al., 2005; Kass et al., 2007; Califano et al., 2008; Mouw et al., 2014; Hadden et al., 2017). Comparing different cell sizes, we observed that MDA-MB-231 cells were smaller overall than the MCF10A cells, with an average size of 650.03 µm2 and 887.26 µm2 when grown on 3 kPa and 40 kPa, respectively, compared with MCF10A cells (3 kPa, 1150.34 µm2, and 40 kPa, 1428.4 µm2). Previous literature has described both breast and osteosarcoma metastatic cells to be smaller than primary nonmetastatic cancer cells (Bell and Waizbard, 1986; Lyons et al., 2016). Because we saw this trend in our breast cancer cell lines, but not in the osteosarcoma cell lines, cancer cell morphology may be cell-type specific. Next, form factor was used to investigate the degree of circularity of cells, with a value closer to one indicating a perfect circle. All cell lines, with the exception of U2OS, had a higher form factor when cells were cultured on 3 kPa hydrogels, compared with 40 kPa hydrogels indicating that the 3 kPa substrate led to a more circular cell shape (Figure 3C). Analysis of the aspect ratio (major axis/minor axis) revealed no changes in this parameter when cells were grown on different stiffness, with the exception being a slight increase in the aspect ratio for the two metastatic cell lines 143B and MDA-MB-231, when cultured on stiff 40 kPa hydrogels compared with soft 3 kPa hydrogels (Figure 3D). Consistent with our findings, previous work reported that breast cancer cells grown on stiff substrates may appear more elongated in shape, potentially assisting epithelial-to-mesenchymal transition and thus promoting metastasis (Syed et al., 2017). Thus, these data show that in terms of morphology, cancer cells respond to stiff substrates by increasing cell size and becoming less round, in a similar manner to what has been reported thus far in the field of mechanobiology.
FIGURE 3:
Nuclear area, cell area, and shape of cells revealed that all cell lines appeared morphologically different when cultured on 3 kPa and 40 kPa hydrogels. (A, B) All four cell lines had larger nuclei and cell area when cultured on 40 kPa hydrogels compared with 3 kPa hydrogels. (C) Analysis of form factor, representing the circulatory of cells (1 = a perfect circle) showed that MCF10A, 143B, and MDA-MB-231 cells appeared more circular when cultured on 3 kPa hydrogels. (D) Aspect ratio (X/Y) revealed that 143B cells had a larger X/Y ratio when cultured on stiff substrates. (E) Outlines of nuclear and cell images showing differences in size and morphology of cell cultured on both conditions. Outlines were obtained from images taken at 20× magnification and visualized in multicolor images by CellProfiler using F-actin for cytoplasmic and DAPI for nuclear boundary recognition. Scale bar = 100 µm. Data are shown as mean ± SEM. Numbers of cells used in analyses were indicated per bar graph. *, p < 0.05; **, p < 0.01; and ****, p < 0.0001.
Cells have increased migration on stiffer substrates
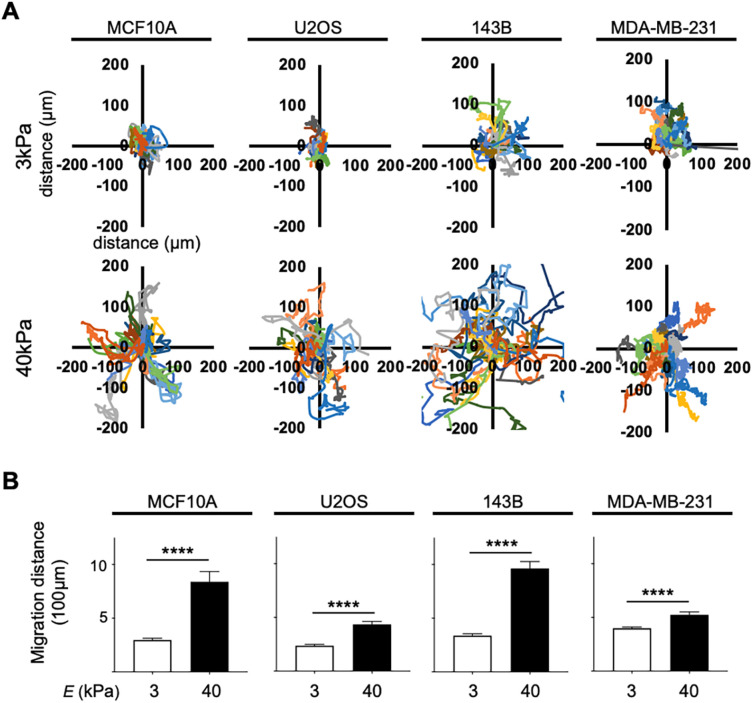
Migration tracking was next used to examine how readily cancer and normal epithelial cells moved when cultured on different stiffness hydrogels over 24 h. Migration tracking showed that all four cell lines displayed increased migration on stiff 40 kPa hydrogels compared with soft 3 kPa hydrogels (Figure 4A). This follows the same patterns as reported in several other cell types including 3T3 mouse fibroblasts and SaI/N transformed fibroblastic cells (Pelham and Wang, 1997; Tzvetkova-Chevolleau et al., 2008). Examining cancer subtypes, we found that the average distance traveled in the metastatic osteosarcoma cell line 143B was significantly greater compared with the average total distance traveled by the nonmetastatic osteosarcoma cell line U2OS in cells cultured on both 3 kPa (337 µm for 143B, and 236 µm for U2OS) and 40 kPa hydrogels (963 µm for 143B, and 434 µm for U2OS; Figure 4B). This is consistent with several previous studies that have reported that increased tissue rigidity can increase migration in breast cancer, pancreatic cancer, glioma, and colorectal cancer (Ulrich et al., 2009; Tilghman et al., 2010; Baker et al., 2012; Kraning-Rush and Reinhart-King, 2012; Haage and Schneider, 2014; Lin et al., 2018). Overall, these results indicate that cancer and epithelial cells respond to stiff substrates by increasing motility, and that overall metastatic osteosarcoma cells display increased motility, compared with nonmetastatic osteosarcoma cells.
FIGURE 4:
Migration tracking of MCF10A, U2OS, 143B, and MDA-MB-231 cells on 3 kPa vs. 40 kPa hydrogels showed increased migratory trends on stiff substrates. (A) Rose plots corrected to 0,0 showing representative tracks taken by cells cultured on 3 kPa and 40 kPa (n = 20). (B) Total distance traveled confirmed that cells from all cell lines cultured on 40 kPa hydrogels migrated a greater distance (n = 80) compared with cells cultured on 3 kPa hydrogels (n = 80). Data are shown as mean ± SEM. ****, p < 0.0001.
Lamin A expression, and YAP and MRTF-A nuclear translocation do not show consistent changes in cells grown on different stiffness
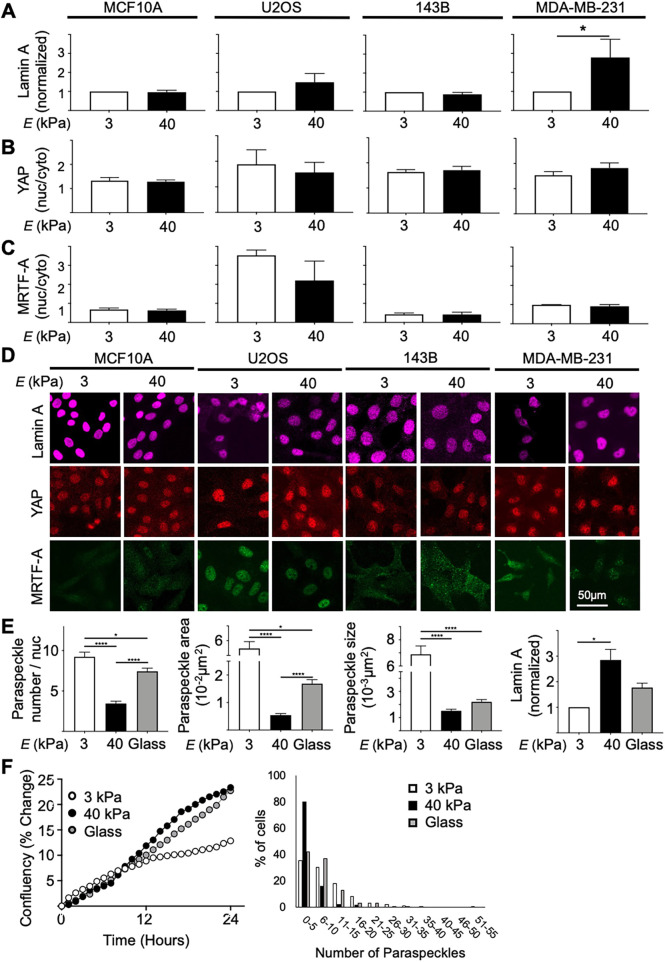
We next examined the degree of lamin A expression, as well as the nuclear/cytoplasmic YAP and MRTF-A ratios, which have previously had clear trends with respect to substrate stiffness described in the stem cell field (Dupont et al., 2011; Swift et al., 2013; Hadden et al., 2017; Kim et al., 2019; Major et al., 2019). The expression of lamin A in all cell lines measured showed no clear differences when cells were grown on substrates of different stiffness, with the exception of MDA-MB-231 cells, that had normalized lamin A expression approximately threefold higher in cells cultured on 40 kPa hydrogels compared with 3 kPa hydrogels, consistent with observations made by Swift et al. (2013) (Figure 5A). We also considered the origins of each line, from different tissues of the body with different innate stiffness. We found that U2OS and 143B cells, originating from bone tissue, had significantly higher levels of lamin A as determined by fluorescent intensity, compared with MCF10A and MDA-MB-231 cells, which originate from breast tissue, irrespective of being cultured on 3 kPa or 40 kPa hydrogels (Supplemental Figure 3).
FIGURE 5:
(A) Normalized lamin A expression (n = 3, 100 cells/repeat) revealed that lamin A levels did not change pending on stiffness in MCF10A, U2OS, and 143B cell lines; however, the MDA-MB-231 cell line showed increased normalized lamin A expression in cells cultured on 40 kPa hydrogels compared with soft 3 kPa hydrogels. (B, C) Quantification of YAP and MRTF-A nuclear/cytoplasmic ratio showed no trend between stiffness for both markers in MCF10A, U2OS, 143B, and MDA-MB-231 cell lines (n = 3, 100 cells/repeat). The ratios between markers followed similar trends between cell lines and appeared significantly elevated in the U2OS cell line compared with all other cell lines. (D) Representative images of lamin A, YAP, and MRTF-A staining in all four cell lines at 3 kPa and 40 kPa stiffness taken using confocal microscopy at 20× magnification. (E) Paraspeckle parameters (paraspeckle number, total area, and average size) in MDA-MB-231 cells cultured on 3 kPa, 40 kPa, and glass (E = GPa range) revealed a nonlinear trend between stiffness and paraspeckles. This trend was inverse to the relationship between lamin A and the same stiffness conditions. (F) Investigation of cell proliferation (determined by % confluency changes over time) and paraspeckles in MDA-MB-231 revealed that although cells cultured on 40 kPa and glass exhibited similar proliferation rates, paraspeckle abundance in these two conditions differed. Scale bar = 50 µm. Data are shown as mean ± SEM. *, p < 0.05.
For YAP and MRTF-A nuclear/cytoplasmic ratios, we observed no consistent trend in subcellular distribution when cells were grown on either stiffness in any of the cell lines tested (Figure 5, B and C). Although trends between increasing nuclear translocation of YAP when cells are grown on increasing stiffness substrates have been well accepted in the stem cell field, the role of YAP translocation in the context of mechanotransduction and cancer remains controversial (Dupont et al., 2011; Tan et al., 2018; Qin et al., 2019). A study using colorectal cancer cells showed increased YAP translocation into the nucleus when cells were cultured on substrates with increased stiffness (Tan et al., 2018). However, in a separate study on the same MDA-MB-231 used here, there was no change observed in YAP translocation when cells were grown on substrates of varying stiffness, suggesting that YAP nuclear translocation as a result of altered substrate stiffness may be cell line specific (Qin et al., 2019). Although MRTF-A nuclear translocation in the context of substrate stiffness has not been as extensively studied as YAP translocation, our MRTF-A staining, as with the YAP translocation data, showed no clear trend in terms of different behavior in cells grown on different stiffness. Furthermore, vinculin staining, which is typically enhanced in cells cultured on stiff substrates and used to show focal adhesions at the cell–cell and cell–matrix junctions, revealed more focal adhesions in cells grown on 40 kPa hydrogels compared with soft 3 kPa hydrogels (Supplemental Figure 4; Yamashita et al., 2014; Omachi et al., 2017). Collectively, the lamin A expression and YAP and MRTF-A nuclear translocation in our cell lines, did not follow typical trends between stiffness as previously reported in the stem cell field, suggesting that epithelial and cancer cell lines may be influenced by mechanotransduction differently.
Mechanosensation of MDA-MB-231 on nonphysiological stiffness
Interestingly, paraspeckle parameters (number, total area, and average size) in MDA-MB-231 cells cultured on plastic plates or glass coverslips showed levels in between what was observed in MDA-MB-231 cells cultured on 3 kPa and 40 kPa conditions (Figure 5E). Although inverted, the trend was very consistent with lamin A levels in the same stiffness conditions. As paraspeckle parameters on glass did not match either of 3 kPa or 40 kPa, previous culture on plastic or coverslip had minimum effect on mechanomemory experiments. This comparison between hydrogels and coverslips clearly highlights the importance of the physiological environment in cellular and molecular biology.
Furthermore, substrate stiffness can lead to differences in proliferation rates in various cell lines, including MDA-MB-231; however, no correlation between proliferation and paraspeckle expression was observed in our data (Figure 5, E and F). Cells on glass had a similar proliferation rate to cells on 40 kPa; however, cells on glass had more paraspeckles with an increased heterogeneous distribution compared with cells on 40 kPa. Tilghman et al. (2010), showed no significant differences in cell cycle (G1, S, and G2 phases) between MDA-MB-231 cells on 0.15 versus 4.8 kPa. Moreover, Fox et al. showed loss of paraspeckles only during telophase (Fox et al., 2005), a cell cycle stage that would typically only take up a very small proportion of the overall cell cycle. If there was a subpopulation of cells arrested in telophase on the 40 kPa hydrogels, we would expect to have two distinct populations in paraspeckle numbers. However, because we had a relatively normal distribution of paraspeckles in the 40 kPa condition (Figure 5F and Supplemental Figure 2), our data suggests a very limited representation of cells in telophase in our experiment.
Proposed mechanisms for paraspeckle mechanosensation
Paraspeckle abundance and size is determined by the level of the lncRNA NEAT1 (Hirose et al., 2014; Wang et al., 2018). Although our study did not characterize the mechanisms underlying paraspeckle mechanosensation to substrate stiffness, we speculate that the reduced levels of paraspeckles in response to stiffness are the result of suppression in NEAT1 transcription.
Our data presents an opposing view on what is considered “traditional” in the field of mechanobiology, given most mechanosensitive markers have shown positive correlations with stiffness (e.g., increased lamin A expression, increased nuclear localization of YAP and MRTF-A, faster migration, and larger nuclei). Although counterintuitive, several other studies have reported similar “nontraditional” trends that are in line with our study. A study investigating the nuclear translocation of protein four-and-a-half LIM domains 2 (FHL2) using human foreskin fibroblasts, showed an increase in nuclear localization of FHL2 when cells were cultured on soft (8.78 kPa) substrates compared with mid (20.2 kPa) and stiff (75.3 kPa) hydrogels (Nakazawa et al., 2016). Furthermore, research investigating matrix models of scars revealed that in mesenchymal stem cells (MSCs), a strong smooth muscle actin repressor, NKX2.5, slowly exited the nucleus on rigid matrices (Dingal et al., 2015). The overexpression of NKX2.5 overrode rigid phenotypes, inhibiting smooth muscle actin and cell spreading, whereas cytoplasm-localized NKX2.5 mutants degraded in well-spread cells.
Ongoing work in understanding the role of mechanotransduction in altering gene expression in response to force, have highlighted the role of chromatin remodeling and subsequent epigenetic regulation in gene expression. A recent study described that chromatin reorganization in response to force, can induce polycomb repressive complex 2 (PRC-2) mediated global transcription silencing. This study reported a concomitant increase in H3K27me3, a gene-silencing marker, and decrease in active RNA polymerase II, upon prolonged strain in epithelial stem cells (ESCs; Le et al., 2016). Publicly available RNA-seq data from this study revealed that NEAT1 levels decreased approximately twofold in response to strain in ESCs in this context (unpublished data). We therefore speculate that decreased paraspeckle expression in response to stiffness is due to the suppression in NEAT1 transcription, which may be the result of PRC2-H3K27me3–linked epigenetic silencing. This suppression of transcription may counteract the more usual induction of NEAT1/paraspeckles that is observed with many other cell stresses (An et al., 2019).
Future experiments testing a role for such epigenetic regulation of NEAT1 will be important to determine factors upstream of NEAT1 in this context. The field of mechanobiology is expanding, and alternative mechanisms inducing gene expression, aside from traditional activation of mechanotransduction signaling pathways, are becoming apparent. Although further research should investigate this, we believe that our study along with the studies mentioned above, all showing nonclassic mechanoresponses, may be of benefit one day. We believe these “counterintuitive” findings, although currently posing as a challenge, will potentially assist in identifying other mechanotransduction pathways that may currently be unknown.
Summary
In summary, here we showed that paraspeckles are mechanosensitive in U2OS, 143B, and MDA-MB-231 cancer cell lines, with greater fold changes observed in metastatic cancer cell lines. The paraspeckle trend showed a consistent inverse relationship with morphological and migratory properties, confirming their mechanosensitivity. We propose that paraspeckles may be a better marker of mechanotransduction for cancer cells, in contrast to lamin A expression and the nuclear translocation of YAP and MRTF-A that showed no obvious changes in cells cultured on different substrate stiffnesses.
MATERIALS AND METHODS
Cell culture
Cell lines including MCF10A breast epithelial, U2OS osteosarcoma, 143B metastatic osteosarcoma, and MDA-MB-231 metastatic breast cancer were used for this study. U2OS, 143B, and MDA-MB-231 cells were cultured in DMEM (Invitrogen) + 10% fetal bovine serum + 1% penicillin-streptomycin. MCF10A cells were cultured in DMEM/F12 (Invitrogen) + 2% horse serum, 1% penicillin-streptomycin, 0.05% hydrocortisone, 0.01% cholera toxin, and 0.1% insulin. Cells were maintained in an ESCO CelCulture CO2 incubator at 5% CO2 and 37°C. Inhibition experiments (inhibition of myosin-II forces) were performed by allowing cells to adhere for 24 h and then treating cells with 50 µM blebbistatin (Sigma-Aldrich) for a further 24 h. Cells were washed with phosphate-buffered saline (PBS) three times for 5 min and immediately fixed using 4% paraformaldehyde (PFA; sc-281692; Santa Cruz) in preparation for FISH and immunofluorescence, as described below.
Polyacrylamide gel fabrication and functionalization
Polyacrylamide hydrogels of 3 kPa and 40 kPa (E: Young’s modulus) were fabricated by creating polymer solutions containing 10% acrylamide monomers (Bio-Rad) with 0.03% and 0.3% (vol/vol) N,N′-methylene-bis-acrylamide cross-linker (Bio-Rad), respectively (Tse and Engler, 2010). Petri dishes containing 20 ml 100% ethanol + 600 µl acetic acid + 100 µl 3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich) and 20 ml 100% ethanol were prepared. Coverslips (Menzel Glasser; 25 mm) were subject to UV radiation on both sides for 1 min on each side and were soaked in the methacrylate solution for 5 min, followed by ethanol for 3 min. Coverslips were then removed and allowed to air dry. Glass microscope slides (HURST Scientific) were prepared for the fabrication of hydrogels by coating with dichlorodimethylsilane (DCDMS; Sigma-Aldrich) and spreading using a kimwipe. Aliquots of polymer solutions of 1 ml for each 3 kPa and 40 kPa were acquired and 10 μl of 10% (wt/vol) ammonium persulfate (APS; Sigma-Aldrich) was added and mixed using a vortex for 1 s. Then, 1 μl of N,N,N′,N′- tetramethylethylenediamine (TEMED; Bio-Rad) was added to each aliquot and mixed for 1 s using a vortex. Working quickly, 250 µl of the polymer solution + APS + TEMED was added onto the pre prepared/DCDMS-coated glass microscope slide for each hydrogel, and the ready methacrylated coverslips were transferred on top. Gels were allowed to polymerize for 15 min and then were stored in six-well plates in PBS. Hydrogels were functionalized using 0.2 mg/ml sulfosuccinimidyl 6-(4-azido-2-nitrophenylamino) hexanoate (sulfo-SANPAH) diluted in 50 mM HEPES, pH 8.5, and placed under a UVP Benchtop transilluminator (365 nm) for 10 min. Hydrogels were washed twice using (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES; Sigma-Aldrich) and protein coated with 25 µg/ml fibronectin (Sigma-Aldrich) overnight at 37°C. Prior to seeding, hydrogels were placed under the UV transilluminator (305 nm) for 20 min.
Mechanomemory experimental setup
MDA-MB-231 cells were cultured for 48 h on 2 × 3 kPa hydrogels and 2 × 40 kPa hydrogels prepared as described above. One hydrogel of each stiffness was used as a control (control sample) and the other hydrogel of each stiffness was used to condition cells and then split onto new hydrogels (conditioning sample). Cells for the control sample were seeded at a density of 5000 cells/µm2 and cells for the conditioning sample were seeded at a density of 10,000 cells/µm2. At 48 h, the control samples for each stiffness were allowed to grow for a further 48 h before fixing, while the conditioning samples were subject to 0.2 ml of TrypLE Express (Life Technologies) to allow for cell detachment. Each cell suspension was split in two and cells were seeded on new 3 kPa and 40 kPa hydrogels and allowed to grow for a further 48 h. All cells for the mechanomemory experiment were grown for a total of 96 h.
FISH
Cells on hydrogels were fixed with 4% PFA (sc-281692; Santa Cruz) at 48 h following seeding for nonmechanomemory experiments and 96 h following seeding for mechanomemory experiments. Cells were permeabilized with 70% ethanol overnight. Stellaris RNA-FISH was performed as per the manufacturer’s instructions using probes targeting 5′ NEAT1 (SMF-2036-1) labeled with Quaser 570 Dye (1:100; Biosearch Technologies). Cells were stained with DAPI (4′,6-diamidino-2-phenylindol; 1:15,000) in diethylpyrocarbonate (DEPC) water for 1 min at room temperature. VectaShield (Vector Laboratories) was used to mount hydrogels onto microscope slides and coverslips were sealed using nail polish. Fluorescence signals were imaged at 60× magnification using the DeltaVision Elite imaging system and SoftWoRx software. Images were acquired as Z-stacks of 0.2 μm increments and were subjected to deconvolution and quick projection. NIS-Elements Advanced (4.0) software (Nikon) was used to quantify paraspeckles by identifying the region of interest (ROI; nuclei) and detecting binary thresholds representing paraspeckles, within the ROI. Paraspeckle data was presented as (i) average number of paraspeckles per nuclei, (ii) average total paraspeckle area, and (iii) average paraspeckle size. The pixel to µm conversion factor was 0.011.
Immunofluorescence
Cells on hydrogels were fixed as previously described and were permeabilized with 1% Triton-X (Sigma-Aldrich) for 15 min at room temperature. Cells were costained using primary antibodies: YAP (sc-101199; Santa Cruz; 1:100), lamin A (sc-20681; Santa Cruz), MRTF-A (sc-390324; Santa Cruz), and vinculin (ab18058; Abcam; 1:100) diluted in 2% bovine serum albumin (Sigma-Aldrich) in PBS for 1 h at 37°C. Cells were then washed using PBS three times for 5 min. Secondary antibodies including Alexa Fluor 594 (ab150116; Thermo Fisher Scientific Company; 1:200) and Alexa Fluor 647 (ab150095; Thermo Fisher Scientific Company; 1:200) were conjugated against primary antibodies along with rhodamine-conjugated phalloidin (r415; Invitrogen; 1:100) for 1 h at 37°C. DAPI (1:15,000; 4′,6-diamidino-2-phenylindole) in DEPC water was counterstained for 1 min at room temperature and coverslips were washed with PBS. Coverslips were mounted onto microscope slides using VectaShield (Vector Laboratories) mounting media and sealed with nail polish. Imaging was done using a Nikon C2+ confocal microscope and NIS-Elements Advanced (4.0) software (Nikon). Images were processed using CellProfiler using a custom made pipeline to measure protein expression levels and determine nuclear area, cellular area, and form factor (4 × π × area/perimeter2).
Cell migration tracking and proliferation analysis
Following 4 h post seeding once cells had adhered, hydrogels were placed in an IncuCyte S3 live-cell imaging system (Essen Biosciences). The IncucCyte was set to acquire images of each gel at 10× magnification every 15 min for 24 h for both 3 kPa and 40 kPa hydrogels. For the cell migration tracking analysis, images were exported and videos were created using Fiji software. Four videos of randomly selected regions within the gel were analyzed and one reference video to normalize for shaking of hydrogels that occurred during imaging was obtained. Cells were numbered using a random number generator and 20 cells per video were selected to be manually tracked. A total of 80 cells per condition were analyzed (20 per video). Cells were manually tracked using the manual tracking plugin function on Fiji, ensuring to exclude any cells that had left the frame. Cell proliferation data was acquired using the IncuCyte proliferation assay to determine cell proliferation as occupied area (% confluence) at each time point. Proliferation data was presented as % change in confluency over time.
Statistical analysis
All data has been presented as mean ± SEM and all experiments were performed in triplicate. Data in Figures 1 and 2 were analyzed using one-way ANOVA, while all other data was analyzed using the Students t test. Multiple comparisons of Tukey post tests were used where necessary. Data presented was interpreted as *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001.
Supplementary Material
Acknowledgments
This work was supported by an ARC Future Fellowship FT180100204 (to A.H.F.), the Cancer Council of Western Australia (to A.H.F.), National Health and Medical Research Council PG1098449 (to Y.S.C.), Heart Foundation Future Leader Fellowship 101173 (to Y.S.C.), and the Australian Government Research Training Program Scholarship (to V.T.).
Abbreviations used:
- BMSCs
bone marrow-derived stem cells
- ECM
extracellular matrix
- FHL2
four-and-a-half LIM domains 2
- hMSCs
human mesenchymal stem cells
- lncRNA
long noncoding RNA
- MRTF-A
Myocardin-related transcript factor A
- NEAT1
nuclear paraspeckle assembly transcript 1
- NONO
non-POU domain-contain octamer-binding
- PRC-2
Polycomb repressive complex 2
- PSPC1
paraspeckle component 1
- SFPQ
splicing factor proline/glutamine-rich
- YAP/TAZ
Yes-associated protein/PDZ-binding protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-02-0097) on April 15, 2020.
REFERENCES
- Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM. (2015). Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb) , 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SG, Zhang Q, Prasad N, Li Y, Chamala S, Kuchibhotla R, Kc B, Aggarwal V, Shrestha S, Jones AL, et al. (2016). The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep , 38063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Tan JT, Shelkovnikova TA. (2019). Stress granules regulate stress-induced paraspeckle assembly. J Cell Biol , 4127–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AM, Bird D, Lang G, Cox TR, Erler JT. (2012). Lysyl oxidase enzymatic function increases stiffness to drive colorectal cancer progression through FAK. Oncogene , 1863. [DOI] [PubMed] [Google Scholar]
- Bell CD, Waizbard E. (1986). Variability of cell size in primary and metastatic human breast carcinoma. Invasion Metastasis , 11–20. [PubMed] [Google Scholar]
- Califano JP, Reinhart-King CAJC, Bioengineering M. (2008). A balance of substrate mechanics and matrix chemistry regulates endothelial cell network assembly. , 122. [Google Scholar]
- Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. (2014). The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun , 5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Vincent LG, Lee AR, Dobke MK, Engler AJ. (2012). Mechanical derivation of functional myotubes from adipose-derived stem cells. Biomaterials , 2482–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell , 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TR, Erler JT. (2011). Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech , 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJS, Lammerding J. (2008). Nuclear shape, mechanics, and mechanotransduction. Circ Res , 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingal PCDP, Bradshaw AM, Cho S, Raab M, Buxboim A, Swift J, Discher DE. (2015). Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nat Mater , 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature , 179–183. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. (2006). Matrix elasticity directs stem cell lineage specification. Cell , 677–689. [DOI] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. (2005). P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell , 5304–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. (2002). Paraspeckles: a novel nuclear domain. Curr Biol , 13–25. [DOI] [PubMed] [Google Scholar]
- Haage A, Schneider IC. (2014). Cellular contractility and extracellular matrix stiffness regulate matrix metalloproteinase activity in pancreatic cancer cells. , 3589–3599. [DOI] [PubMed] [Google Scholar]
- Hadden WJ, Young JL, Holle AW, McFetridge ML, Kim DY, Wijesinghe P, Taylor-Weiner H, Wen JH, Lee AR, Bieback K, et al. (2017). Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc Natl Acad Sci USA , 5647–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G. (2014). NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell , 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. (1987). Integrins: A family of cell surface receptors. Cell , 549–554. [DOI] [PubMed] [Google Scholar]
- Kass L, Erler JT, Dembo M, Weaver VM. (2007). Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. Int J Biochem Cell Biol , 1987–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killaars AR, Grim JC, Walker CJ, Hushka EA, Brown TE, Anseth KS. (2019). Extended Exposure to Stiff Microenvironments Leads to Persistent Chromatin Remodeling in Human Mesenchymal Stem Cells. Adv Sci , 1801483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Young JL, Holle AW, Jeong K, Major LG, Jeong JH, Aman ZM, Han D-W, Hwang Y, Spatz JP, Choi YS. (2019). Stem cell mechanosensation on gelatin methacryloyl (GelMA) stiffness gradient hydrogels. Ann Biomed Eng , 893–902. [DOI] [PubMed] [Google Scholar]
- Kraning-Rush CM, Reinhart-King CA. (2012). Controlling matrix stiffness and topography for the study of tumor cell migration. Cell Adh Migr , 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le HQ, Ghatak S, Yeung CY, Tellkamp F, Gunschmann C, Dieterich C, Yeroslaviz A, Habermann B, Pombo A, Niessen CM, Wickstrom SA. (2016). Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol , 864–875. [DOI] [PubMed] [Google Scholar]
- Li Y, Cheng C. (2018). Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am J Cancer Res , 81–90. [PMC free article] [PubMed] [Google Scholar]
- Lin F, Zhang H, Huang J, Xiong C. (2018). Substrate stiffness coupling TGF-β1 modulates migration and traction force of MDA-MB-231 human breast cancer cells in vitro. ACS Biomater Sci Eng , 1337–1345. [DOI] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. (2000). Cell movement is guided by the rigidity of the substrate. Biophys J , 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Alizadeh E, Mannheimer J, Schuamberg K, Castle J, Schroder B, Turk P, Thamm D, Prasad A. (2016). Changes in cell shape are correlated with metastatic potential in murine and human osteosarcomas. Biol Open , 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major LG, Holle AW, Young JL, Hepburn MS, Jeong K, Chin IL, Sanderson RW, Jeong JH, Aman ZM, Kennedy BF, et al. (2019). Volume adaptation controls stem cell mechanotransduction. ACS Appl Mater Interfaces , 45520–45530. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. (2013). Tumour heterogeneity and cancer cell plasticity. Nature , 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modic M, Grosch M, Rot G, Schirge S, Lepko T, Yamazaki T, Lee FCY, Rusha E, Shaposhnikov D, Palo M, et al. (2019). Cross-regulation between TDP-43 and paraspeckles promotes pluripotency-differentiation transition. Mol Cell , 951–965.e913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, et al. (2014). Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med , 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T, Hirose T. (2013). Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol , 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Sathe AR, Shivashankar GV, Sheetz MP. (2016). Matrix mechanics controls FHL2 movement to the nucleus to activate p21 expression. Proc Natl Acad Sci USA , E6813–E6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omachi T, Ichikawa T, Kimura Y, Ueda K, Kioka N. (2017). Vinculin association with actin cytoskeleton is necessary for stiffness-dependent regulation of vinculin behavior. PLoS One , e0175324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham RJ, Wang Y-l. (1997). Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA , 13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Lv X, Li P, Yang R, Xia Q, Chen Y, Peng Y, Li L, Li S, Li T, et al. (2019). Matrix stiffness modulates ILK-mediated YAP activation to control the drug resistance of breast cancer cells. Biochim Biophys Acta, Mol Basis Dis , 165625. [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Felsenfeld DP, Galbraith CG. (1998). Cell migration: regulation of force on extracellular-matrix-integrin complexes. Trends Cell Biol , 51–54. [DOI] [PubMed] [Google Scholar]
- Shin VY, Chen J, Cheuk IWY, Siu M-T, Ho C-W, Wang X, Jin H, Kwong A. (2019). Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis , 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D. (2016). Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget , 51784–51814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. (2013). Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science , 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed S, Schober J, Blanco A, Zustiak SP. (2017). Morphological adaptations in breast cancer cells as a function of prolonged passaging on compliant substrates. PLoS One , e0187853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Huang Y, Pei Q, Liu H, Pei H, Zhu H. (2018). Matrix stiffness mediates stemness characteristics via activating the Yes-associated protein in colorectal cancer cells. J Cell Biochem, 10.1002/jcb.27532. [DOI] [PubMed] [Google Scholar]
- Tilghman RW, Cowan CR, Mih JD, Koryakina Y, Gioeli D, Slack-Davis JK, Blackman BR, Tschumperlin DJ, Parsons JT. (2010). Matrix rigidity regulates cancer cell growth and cellular phenotype. PLoS One , e12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. (2010). Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol Chapter 10, Unit 10.16. [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Stephanou A, Fuard D, Ohayon J, Schiavone P, Tracqui P. (2008). The motility of normal and cancer cells in response to the combined influence of substrate rigidity and anisotropic nanostructure. Biomaterials , 1541–1551. [DOI] [PubMed] [Google Scholar]
- Ulrich TA, de Juan Pardo EM, Kumar S. (2009). The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res , 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu SB, Wang MR, Yao RW, Wu D, Yang L, Chen LL. (2018). Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol , 1145–1158. [DOI] [PubMed] [Google Scholar]
- Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, Yang J. (2015). Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol , 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. (2014). Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater , 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Ichikawa T, Matsuyama D, Kimura Y, Ueda K, Craig SW, Harada I, Kioka N. (2014). The role of the interaction of the vinculin proline-rich linker region with vinexin α in sensing the stiffness of the extracellular matrix. J Cell Sci , 1875–1886. [DOI] [PubMed] [Google Scholar]
- Yang C, Tibbitt MW, Basta L, Anseth KS. (2014). Mechanical memory and dosing influence stem cell fate. Nat Mater , 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. (2005). Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion , 24–34. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Qutaish M, Han Z, Schur RM, Liu Y, Wilson DL, Lu ZR. (2015). MRI detection of breast cancer micrometastases with a fibronectin-targeting contrast agent. Nat Commun , 7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.