Abstract
During the initial stages of cell division, the cytoskeleton is extensively reorganized so that a bipolar mitotic spindle can be correctly assembled. This process occurs through the action of molecular motors, cytoskeletal networks, and the nucleus. How the combined activity of these different components is spatiotemporally regulated to ensure efficient spindle assembly remains unclear. To investigate how cell shape, cytoskeletal organization, and molecular motors cross-talk to regulate initial spindle assembly, we use a combination of micropatterning with high-resolution imaging and 3D cellular reconstruction. We show that during prophase, centrosomes and nucleus reorient so that centrosomes are positioned on the shortest nuclear axis at nuclear envelope (NE) breakdown. We also find that this orientation depends on a combination of centrosome movement controlled by Arp2/3-mediated regulation of microtubule dynamics and Dynein-generated forces on the NE that regulate nuclear reorientation. Finally, we observe this centrosome configuration favors the establishment of an initial bipolar spindle scaffold, facilitating chromosome capture and accurate segregation, without compromising division plane orientation.
INTRODUCTION
Chromosome segregation requires the assembly of a bipolar mitotic spindle. While multiple pathways contribute to spindle assembly (Prosser and Pelletier, 2017), in human somatic cells centrosomes play a dominant role. During prophase, centrosome separation occurs independently of nuclear envelope breakdown (NEB) in a kinesin–5-dependent manner (Whitehead et al., 1996). Accordingly, depletion or inhibition of kinesin-5 prevents centrosome separation, generating monopolar spindles and mitotic arrest (Kapoor et al., 2000). Other players have been involved in this process, including Myosin II (Rosenblatt et al., 2004), actin (Cao et al., 2010), microtubule pushing forces (Cytrynbaum et al., 2003; Tanenbaum and Medema, 2010), and Dynein, both at the nucleus (Bolhy et al., 2011; Raaijmakers et al., 2012; Baffet et al., 2015; De Simone et al., 2016; Boudreau et al., 2019) and at the cell cortex (Cytrynbaum et al., 2003; De Simone et al., 2016). How the forces generated by these components are functionally coordinated to ensure efficient spindle assembly remains unclear.
Entry into mitosis involves loss of interphase adhesion complexes (Dao et al., 2009; Dix et al., 2018; Lock et al., 2018), changes in interphase microtubule organization (Rusan et al., 2002; McHedlishvili et al., 2018), and dynamics (Zhai et al., 1996) and retraction of cell margins (Cramer and Mitchison, 1997; Maddox and Burridge, 2003). This enables mitotic cells to round up and assemble a stiff actin cortex (Maddox and Burridge, 2003; Carreno et al., 2008; Kunda et al., 2008). Although the changes observed in the microtubule and actin cytoskeletons during early mitosis occur simultaneously, they are thought to be independently regulated (Matthews et al., 2010; McHedlishvili et al., 2018). However, several lines of evidence suggest a functional connection between the two cytoskeletal systems at this stage. Actin helps in initial centrosome separation (Cao et al., 2010) and spindle assembly (Chan et al., 2014; Plessner et al., 2019), and its accumulation on the nuclear envelope (NE) facilitates chromosome congression (Booth et al., 2019). Moreover, actin localizes to centrosomes in an Arp2/3-dependent manner (Farina et al., 2016, 2019) to assist in microtubule network remodeling (Farina et al., 2019; Inoue et al., 2019).
Once the mitotic spindle is assembled, cortical force generators determine metaphase spindle orientation (Thery et al., 2005, 2007; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012). These are activated by external cues (Fink et al., 2011) and generate pulling forces on astral microtubules (Grill et al., 2003; Grill and Hyman, 2005; Bosveld et al., 2016) to align the spindle with the long cell axis (Hertwig, 1884), ultimately defining the division plane. However, whether centrosome separation and early spindle assembly follow the same cortical cues remains unknown. Defining how external and internal signals are integrated during mitosis to ensure efficient spindle assembly and robust division plane orientation is essential, since prophase centrosome positioning is required for accurate chromosome segregation (Kaseda et al., 2012; Silkworth et al., 2012).
Here, we performed a high-resolution analysis of centrosome, cytoskeleton, and nuclear behavior during mitotic entry in human cells, followed by 3D cellular reconstruction and centrosome tracking. We show that during mitotic entry, the centrosomes axis does not align according to external cues. Instead, the centrosomes and nucleus reorient so that centrosomes are positioned on the shortest nuclear axis at NEB. This depends on a combination of Dynein-dependent nuclear movement and Arp2/3-mediated centrosome positioning. As a result, the formation of an initial bipolar spindle scaffold is facilitated, ensuring maximum exposure of kinetochores to microtubules and improving chromosome segregation fidelity.
RESULTS
Centrosomes position on the shortest nuclear axis at NEB
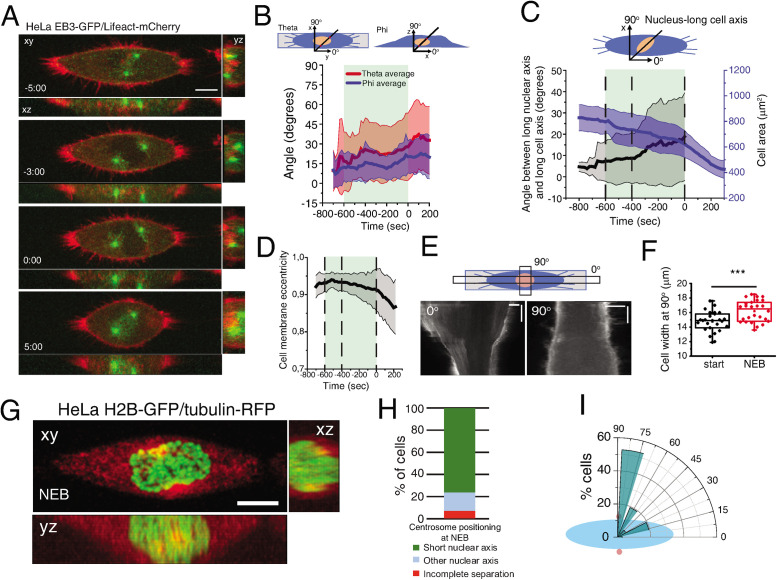
To characterize mitotic spindle assembly at high spatiotemporal resolution, we performed 4D imaging in HeLa cells. We observed that when cells were seeded on a substrate that does not activate integrin signaling (poly-l-lysine; PLL), centrosomes separated independently of NEB (Supplemental Figure S1A), as reported previously (Whitehead et al., 1996; Kaseda et al., 2012). However, when seeded on integrin-activating fibronectin (FBN), ∼82% of the cells separated their centrosomes to opposite sides of the nucleus before NEB. Moreover, cells that had an increased spreading area at NEB showed longer intercentrosome distances (Supplemental Figure S1B), suggesting that centrosome separation prior to NEB is a function of the adhesion area. To normalize cell area and shape in 2D, we seeded cells on defined FBN micropatterns and monitored centrosome dynamics, cell membrane, and nuclear shape (Figure 1A), which were subsequently reconstructed using specifically developed computational algorithms (Supplemental Figure S2). Centrosome dynamics relative to the micropattern was defined by two angles, theta and phi, reflecting movements in xy (azimuth) and xz (inclination), respectively (Figure 1B). These varied between 0o (aligned with the long axis of the pattern) and 90o (perpendicular to the pattern). We anticipated that separated centrosomes should align with the long axis of the micropattern due to the distribution of retraction fibers imposed by extracellular matrix organization (Thery et al., 2005; Fink et al., 2011). However, during mitotic entry, centrosomes deviated from the underlying micropattern, as observed by the high variability of theta and phi (Figure 1B). This was accompanied by a movement of the nucleus relative to the long axis of the pattern, as well as a decrease in cell area (Figure 1C). Due to the shape asymmetry of the line micropattern, we could calculate cell membrane eccentricity, which varied between 1 (completely elongated cell) and 0 (spherical cell). As cells progressed toward NEB, membrane eccentricity decreased (Figure 1D), which was due to a retraction of the long cell axis (Figure 1E, 0o) and a simultaneous increase in cell width, perpendicularly to the pattern (Figure 1, E and F, 90o; ***p < 0.001). Interestingly, during the rounding process, the centrosomes and nucleus reoriented so that centrosomes were positioned on the shortest nuclear axis at NEB (∼80% of cells; Figure 1, G–I; Supplemental Movie S1).
FIGURE 1:
Characterization of early spindle assembly. (A) Frames from a movie of a cell seeded on a line micropattern showing movement of the centrosomes toward the shortest nuclear axis. Time is in minutes:seconds. Time zero corresponds to NEB. (B) Characterization of centrosome orientation vector in xy (theta; red) and z (phi; blue) for cells seeded on line micropatterns (n = 30). Line corresponds to average and shaded area to SD. (C) Quantification of cell area (μm2; blue) and angle between nucleus-long cell axis (black) for cells on line micropatterns (n = 37). Lines correspond to average and shaded areas represent SD. (D) Cell membrane eccentricity during mitotic entry for cells on line micropatterns. Line represents average value and shaded area represents SD. (E) Kymograph from cell expressing Lifeact-mCherry seeded on a line micropattern, during mitotic cell rounding. Zero degrees corresponds to the long cell axis and 90° to the perpendicular orientation. (F) Cell width (μm) perpendicular to the pattern (n = 16; ***p < 0.001). (G) Representative frame from a movie of a cell expressing H2B-GFP/tubulin-RFP showing centrosome and nucleus orientation at NEB. (H) Quantification of centrosome separation behavior at NEB for cells seeded on line micropatterns. (I) Polar plot quantifying centrosome positioning (red circles) relative to the longest nuclear axis (blue ellipse) at NEB for cells seeded on line micropatterns. All experiments were replicated at least three times.
Movie S1.
HeLa cell expressing EB3‐GFP/Lifeact‐mCherry seeded on a line micropattern during mitotic entry, related to Figure 1
Centrosome positioning requires nuclear and centrosome movement
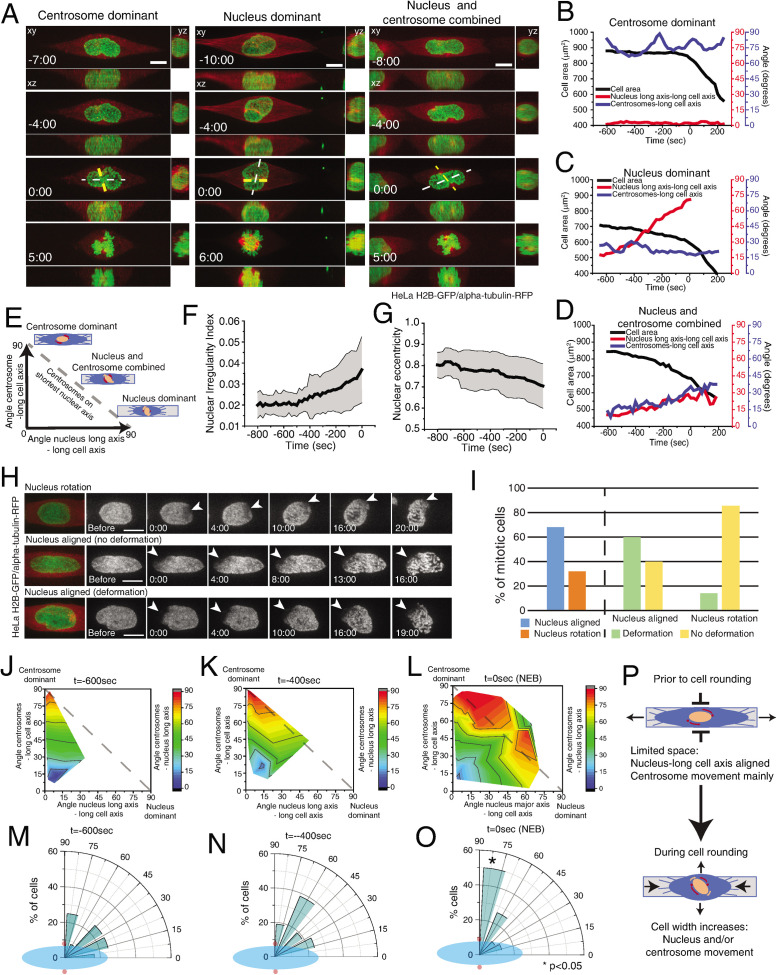
Our observations suggest that prophase centrosome positioning on the shortest nuclear axis is a result of the combined motion of centrosomes and nuclear reorganization. To directly test this, we analyzed the relative contribution of each component for the positioning of centrosomes on the shortest nuclear axis. We reasoned that if positioning on the shortest nuclear axis depended exclusively on centrosome movement (centrosome dominant), the centrosomes-long nuclear axis angle would tend to 90o and the nucleus would remain aligned with the micropattern. On the other hand, if this mechanism required only nuclear reorientation, then the nucleus long axis–long cell axis angle would tend to 90o, and the centrosomes movement would be residual. If both components were involved, then we would observe a combined contribution of both nucleus and centrosomes (Figure 2, A–E). Accordingly, when cell rounding was limited, the nucleus aligned with the long cell axis and centrosomes deviated from the pattern (“centrosome dominant;” Figure 2, A and B). When cell rounding is more pronounced, the long nuclear axis tends to deviate from the long cell axis (“nucleus dominant;” Figure 2, A and C). In intermediate cases, both centrosomes and nucleus deviated from the long cell axis (Figure 2, A and D).
FIGURE 2:
Centrosome positioning requires centrosome and nucleus movement. (A) Selected frames from movies of HeLa cells expressing H2B-GFP/alpha-tubulin-RFP seeded on line micropatterns showing different pathways for centrosomes–nucleus axis orientation (n = 38). White line shows the long nuclear axis and yellow lines show centrosomes axis. Time lapse is 20 s. Time is in minutes:seconds. Time zero corresponds to NEB. Scale bars, 10 μm. Representative plots showing the correlation between centrosome-long cell axis (blue), long nuclear axis-long cell axis (red), and cell area (black) for centrosome-dominant (B), nucleus-dominant (C), and nucleus–centrosome-combined (D) pathways. (E) Positioning of centrosomes on the shortest nuclear axis can be achieved by a combination of centrosome movement and nuclear rotation. Quantification of nuclear irregularity index (F) and nuclear eccentricity (G) for cells entering mitosis. (H) Time-lapse imaging of photobleached H2B-GFP during mitotic entry (n = 22). Time lapse is 20 s. Scale bars, 10 μm. Time is in minutes:seconds. (I) Quantification of the percentage of nuclei that rotate or deform during mitotic entry. Quantification of the contribution of centrosome displacement (angle between centrosomes-long cell axis) and nucleus displacement (angle nucleus long axis-long cell axis) for centrosome positioning on the shortest nuclear axis (angle centrosomes-long nuclear axis) at –600 s (J), –400 s (K), and NEB (L). Distribution of centrosome positioning (red circles) relative to the longest nuclear axis (blue ellipse) at –600 s (M), –400 s (N), and NEB (O). (P) Before cell rounding, centrosome–nucleus axis orientation depends mainly on centrosome movement due to the limitation in space. During mitotic rounding, cell width increases, allowing nuclear rotation.
Since the nucleus undergoes extensive changes during prophase, we decided to clarify if the nucleus-dominant behavior is due to nuclear rotation or a change in nuclear shape. As cells progressed toward NEB, nuclear irregularity increased (Figure 2F) and eccentricity decreased (Figure 2G), suggesting that nuclear shape is indeed changing. Next, to evaluate if nuclei also rotated during this stage, we performed photobleaching of H2B-GFP (Figure 2H). Under these conditions, 32% of the nuclei rotated, whereas 68% remained aligned with the long cell axis (Figure 2, H and I). Moreover, a significant percentage of aligned nuclei showed significant deformation (Figure 2I), leading to the generation of a new short axis. We conclude that nuclear orientation during prophase is determined by a combination of nuclear rotation and deformation.
Taken together, our results reveal that multiple components contribute for centrosome positioning on the shortest nuclear axis. Accordingly, at earlier time points (600 s before NEB) when cells had not rounded up significantly, nuclear movement was limited and correct positioning depended mainly on centrosome motion and nuclear deformation (Figure 2, J and M). However, as rounding progressed, the nucleus was now able to reorient (Figure 2, K, L, and N), contributing to correct centrosome positioning (Figure 2O, *p < 0.05). Overall, these observations suggest that cell rounding enables nuclear reorientation to facilitate the positioning of centrosomes on the shortest nuclear axis at NEB (Figure 2P).
Centrosome positioning on the shortest nuclear axis depends on cell adhesion area but not cell shape
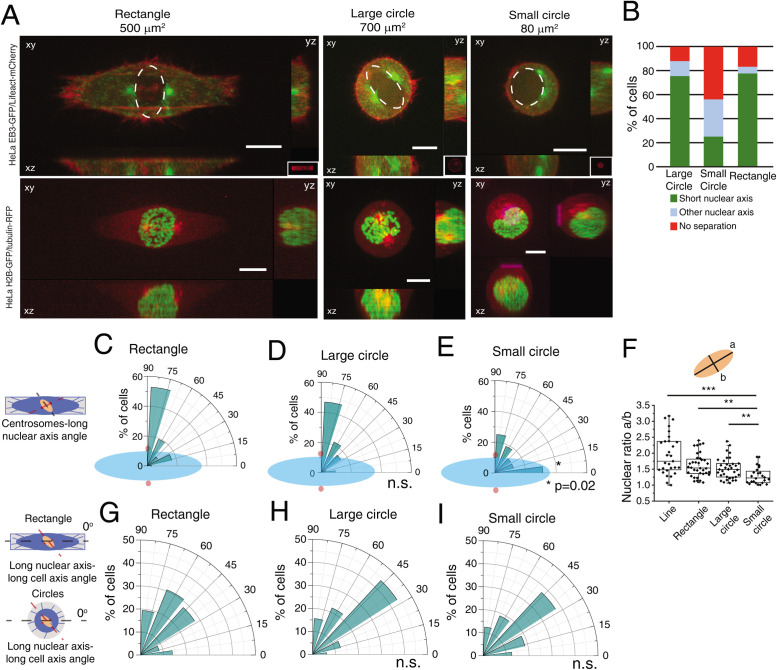
Our initial observations were obtained with cells seeded on line micropatterns that have a highly polarized shape. To determine whether centrosome positioning on the shortest nuclear axis was a result of shape polarization or a more general feature, we compared cells on circles and rectangles during mitotic entry (Figure 3A). Strikingly, changing from a polarized shape such as a rectangle to an unpolarized large circle did not block the capacity of centrosomes to position on the shortest nuclear axis (78% of cells on rectangles and 75% of cells on large circles; Figure 3, A–D). However, correct positioning depended on the initial spreading area, as seeding cells in small circles led to erratic centrosome movement and only 25% of cells placed centrosomes on the short nuclear axis (Figure 3, A, B, and E, *p = 0.02). In addition, although nuclear shape changed when cells were seeded on small circles (Figure 3F; ***p < 0.001), with nuclei becoming more rounded, cell shape did not interfere with nucleus orientation relative to the micropattern (Figure 3, G–I), As a result, cells on small circles lost the coordination of movement between centrosomes and nucleus, when compared with the other micropatterns. Taken together, these data indicated that centrosomes position on the shortest nuclear axis at NEB in a cell area-dependent manner but independently of cell shape.
FIGURE 3:
Centrosome positioning on the shortest nuclear axis depends on cell adhesion area. (A) Representative images from cells at NEB expressing EB3-GFP/Lifeact-mCherry (top panels) or H2B-GFP/alpha-tubulin-RFP (bottom panels) seeded on rectangles (500 μm2; n = 36), large circles (700 μm2; n = 32), or small circles (80 μm2; n = 16) showing lateral projections (xz and yz). Scale bars, 10 μm. Ellipses highlight nuclear shape at NEB. (B) Quantification of centrosome separation behavior at NEB for cells seeded on the different micropatterns. Polar plot quantifying centrosome positioning (red circles) relative to the longest nuclear axis (blue ellipse) at NEB for cells seeded on rectangles (C), large circles (D), or small circles (E). (F) Quantification of nuclear shape asymmetry, as defined by the ratio long nuclear axis/short nuclear axis, for cells seeded on different micropatterns (***p < 0.001; **p < 0.01). Polar plot quantifying alignment of the long nuclear axis with the long cell axis at NEB for cells on rectangles (G), large circles (H), and small circles (I). For cells seeded on circles, 0° was defined horizontally.
Cell rounding allows the centrosomes–nucleus axis to reorient in prophase
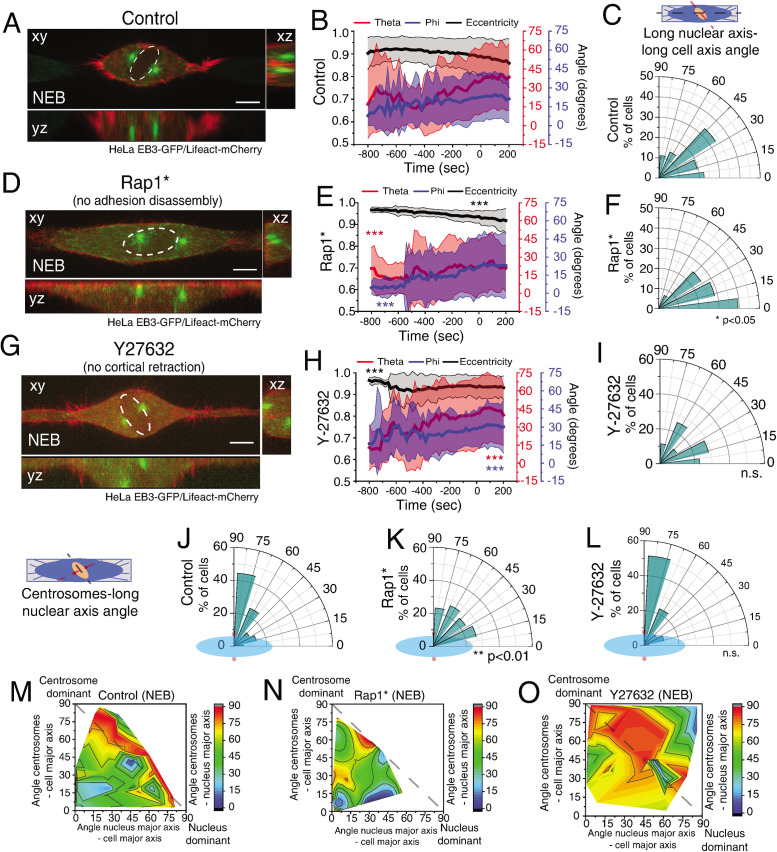
Metaphase spindle orientation is determined by the distribution of actin-based retraction fibers and this depends on extracellular matrix organization (Thery et al., 2005; Fink et al., 2011). Therefore, it would be reasonable to assume that centrosomes should orient according to the same cues during prophase. However, our results showed that prior to NEB (and simultaneously with cell rounding), the centrosomes reoriented away from the underlying retraction fiber distribution imposed by the micropattern, but according to the shortest nuclear axis. This suggests that the rounding process changes the manner in which cells interact with the extracellular matrix. To confirm this, we performed Traction Force Microscopy (TFM) analysis on cells seeded on rectangles (Supplemental Figure S3A). Under these conditions, cells showed a well-defined traction axis that correlated with the initial centrosome separation axis (theta; Supplemental Figure S3, A–C). On mitotic rounding, both cell area and the contractile energy exerted on the substrate decreased (Supplemental Figure S3D), leading us to conclude that mitotic rounding decreases the force exerted by the cell on the substrate. These observations, together with our previous results, suggest that interfering with cellular adhesion during mitotic rounding could affect the centrosome–nucleus axis orientation. Mitotic rounding is comprised of two parallel pathways that involve adhesion disassembly and cortical retraction (Maddox and Burridge, 2003). To test which of these processes is required for centrosome positioning during prophase, we decided to express a mutant form of Rap1 (Rap1Q63E; Rap1*) that interferes with focal adhesion disassembly, effectively blocking mitotic rounding (Dao et al., 2009). As expected, Rap1* expression affected cell rounding (Figure 4, A, B, D, and E) and delayed focal adhesion disassembly (Supplemental Figure S4 and A, B, **p < 0.002). To confirm these results, we decided to deplete the RhoGEF Ect2 by RNAi, as this protein was previously shown to interfere with mitotic cell rounding (Matthews et al., 2012). Accordingly, Ect2-depleted cells showed delayed cell rounding (Supplemental Figure S4C, ***p < 0.001). Importantly, affecting adhesion disassembly was sufficient to impair centrosome (Figure 4B, E; ***p < 0.001) and nuclear reorientation (Figure 4, C and F, *p < 0.05; Supplemental Figure S4D, *p < 0.03), when compared with controls. This eventually resulted in a failure to position centrosomes on the shortest nuclear axis at NEB (Figure 4, J, K, M, and N, **p < 0.01; Supplemental Figure S4E, **p < 0.002). Inversely, interfering with cortical retraction by inhibiting ROCK with Y27632 did not impair centrosome movement or nuclear reorientation (Figure 4, G–I, ***p < 0.001), which allowed correct positioning of centrosomes (Figure 4, L and O). We concluded that adhesion disassembly in prophase is required for positioning of centrosomes on the shortest nuclear axis by allowing both nuclear reorientation and efficient centrosome movement.
FIGURE 4:
Adhesion complex disassembly is required to establish the centrosomes–nucleus axis at NEB. Representative time frame from a movie of HeLa cells expressing EB3-GFP/Lifeact-mCherry treated with DMSO (A; n = 38), Rap1* (D; n = 28), or Y-27632 (G; n = 35) at NEB. Correlation of cell membrane eccentricity (black), theta (red), and phi (blue) for controls (B), Rap1* (E) and Y-27632 (H). Lines correspond to average values and shaded areas correspond to SD. Inhibiting adhesion disassembly limits centrosome movement (***p < 0.001). Polar plot quantifying alignment of the long nuclear axis with the long cell axis at NEB for DMSO (C), Rap1* (F; *p < 0.05), and Y-27632 (I). Polar plot quantifying centrosome positioning (red circles) relative to the longest nuclear axis (blue ellipse) at NEB for controls (J), Rap1* (K; **p < 0.01) or Y-27632 (L). Quantification of the contribution of centrosome displacement (angle between centrosomes-long cell axis) and nucleus displacement (angle nucleus long axis-long cell axis) for centrosome positioning on the shortest nuclear axis (angle centrosomes-long nuclear axis) for controls (M), Rap1* (N), or Y-27632 (O) at NEB. All experiments were replicated at least three times.
Dynein on the NE is required for nuclear rotation during prophase
Next, we wanted to systematically dissect which factors influence the positioning of centrosomes on the shortest nuclear axis by either regulating nuclear rotation or centrosome movement. It is known that kinesin-5 is essential for centrosome separation (Whitehead et al., 1996; Kapoor et al., 2000). To assess whether it is also required to position centrosomes on the shortest nuclear axis, we treated cells with an Eg5 inhibitor (STLC) when centrosomes were already on opposite sides of the nucleus (Late stage) or when centrosomes were still not fully separated (Early stage). Early Eg5 inhibition significantly decreased pole-to-pole distance, preventing centrosome positioning on the shortest nuclear axis (Supplemental Figure S5, A–C). When Eg5 was inhibited in the Late stage, centrosomes moved toward the shortest nuclear axis, simultaneously with mitotic cell rounding (Supplemental Figure S5D, dashed line). We concluded that kinesin-5 is required for initial centrosome separation but not directionality.
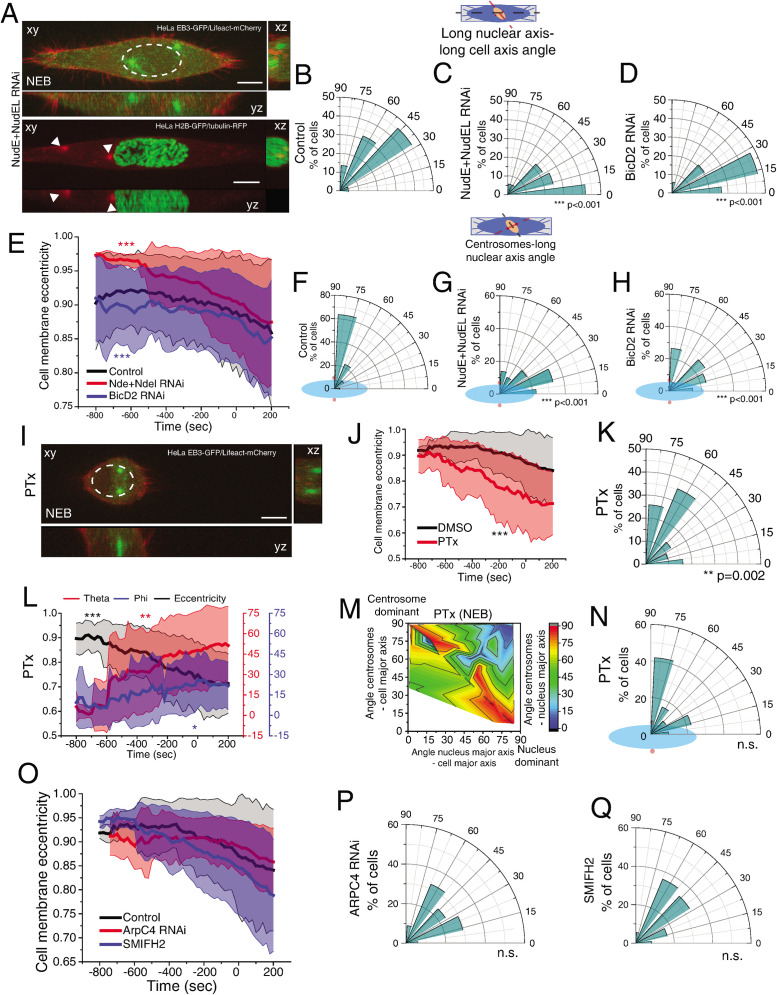
In addition to kinesin-5, additional factors contribute to early centrosome separation such as Dynein (Raaijmakers et al., 2012; van Heesbeen et al., 2013), actin (Whitehead et al., 1996; Wang et al., 2008; Cao et al., 2010; Winkler et al., 2015), and microtubules (Whitehead et al., 1996; Tanenbaum and Medema, 2010), making these likely candidates to mediate centrosomes–nucleus orientation. We started by depleting total Dynein Heavy Chain (DHC), which induced defects in centrosome positioning, probably due to a delayed cell rounding (Supplemental Figure S6, A–C, ***p < 0.001). However, in mitosis Dynein can be found in different subcellular localizations. During prophase, Dynein is loaded on the NE through two pathways involving RanBP2-BicD2 and Nup133-CENP-F-NudE/NudEL (Splinter et al., 2010; Bolhy et al., 2011; Hu et al., 2013; Baffet et al., 2015). Later on, it localizes to the cell cortex through the LGN-Gαi-NuMA complex, which can be prevented by inhibiting Gαi activity with pertussis toxin (PTx) (Woodard et al., 2010; Kotak et al., 2012). To determine if prophase centrosome positioning required any of these specific pools of Dynein, we interfered with NE Dynein by depleting either NudE/NudEL or BicD2 by RNAi (Supplemental Figure S6, D and E). Depletion of NudE+NudEL, but not BicD2, led to a small but significant detachment of centrosomes from the NE (Figure 5A and Supplemental Figure S6F, ***p < 0.001; Supplemental Movie S2). However, both treatments impaired reorientation of the nucleus relative to the long cell axis (Figure 5, B–D, ***p < 0.001). This effect was independent of cell rounding, as depleting NudE+NudEL or BicD2 had opposing effects on cell rounding (Figure 5E, ***p < 0.001) but showed a similar defect in nuclear reorientation. Consequently, centrosomes no longer positioned on the shortest nuclear axis (Figure 5, F–H, ***p < 0.001). Notably, this effect was independent of centrosomes distance to the nucleus (Supplemental Figure S6, F and G), indicating that nuclear reorientation requires Dynein-mediated forces. Next, to determine whether cortical Dynein was also required for nuclear rotation, we treated cells with PTx during mitotic entry (Figure 5I). Under these conditions, Dynein was selectively removed from the cortex but not the NE (Supplemental Figure S6, H and I). When Dynein was prevented from accumulating in the cortex, cells rounded up prematurely (Figure 5J, ***p < 0.001), leading to increased nuclear rotation (Figure 5K, **p = 0.002) and centrosome movement (Figure 5L; theta, **p < 0.01; phi, *p = 0.04). Overall, this did not affect centrosome positioning on the shortest nuclear axis at NEB (Figure 5, M and N), which is in agreement with the presence of Dynein on the NE following PTx treatment. Moreover, Dynein accumulation on the NE was independent on cell rounding, as treating cells with PTx or expressing Rap1* had opposite effects on rounding, but did not interfere with NE Dynein (Supplemental Figure S6, H–K).
FIGURE 5:
Dynein on the NE is required for nuclear rotation. (A) Images of HeLa cells expressing EB3-GFP/Lifeact-mCherry (top panel; n = 36) or H2B-GFP/tubulin-RFP (bottom panel; n = 25) treated with NudE+NudEL RNAi. White arrowheads indicate centrosome position. Time lapse is 20 s. Scale bar, 10 μm. Polar plot quantifying alignment of the long nuclear axis with the long cell axis at NEB for controls (B), NudE+NudEL RNAi (C; ***p < 0.001), or BicD2 RNAi (D; ***p < 0.001; n = 34). (E) Cell membrane eccentricity for controls, NudE+NudEL RNAi, and BicD2 RNAi. Lines correspond to average values and shaded areas correspond to SD (***p < 0.001). Polar plot quantifying centrosome positioning (red circles) relative to the longest nuclear axis (blue ellipse) at NEB for controls (F), cells treated with RNAi for NudE+NudEL (G), or BicD2 (H). (I) Time frame from cells expressing EB3-GFP/Lifeact-mCherry treated with PTx (n = 31). Scale bar, 10 μm. (J) Cell membrane eccentricity for controls and PTx-treated cells. Lines correspond to average values and shaded areas correspond to SD (***p < 0.001). (K) Polar plot quantifying alignment of the long nuclear axis with the long cell axis at NEB for cells treated with PTx. (L) Correlation of cell membrane eccentricity (black), theta (red), and phi (blue) for PTx cells. (M) Quantification of the contribution of centrosome displacement (angle between centrosomes-long cell axis) and nucleus displacement (angle nucleus long axis-long cell axis) for centrosome positioning on the shortest nuclear axis (angle centrosomes-long nuclear axis) at NEB for cells treated with PTx. (N) Polar plot quantifying centrosome positioning (red circles) relative to the longest nuclear (blue ellipse) at NEB for cells treated with PTx. (O) Cell membrane eccentricity for controls (n = 38), ArpC4 RNAi (n = 34), and SMIFH2 (n = 36). Lines correspond to average values and shaded areas correspond to SD (***p < 0.001). Polar plot quantifying alignment of the long nuclear axis with the long cell axis at NEB for ArpC4 RNAi (P) and SMIFH2 (Q). All experiments were replicated at least three times.
Movie S2.
HeLa cell expressing H2B‐GFP/tubulin‐RFP treated with NudE+NudEL RNAi, seeded on a line micropattern, related to Figure 5
Next, we wanted to determine whether the actin cytoskeleton was required for nuclear reorientation during prophase, since it is involved in nuclear rotation during interphase (Kumar et al., 2014). For that reason, we interfered with the activity of actin nucleators Arp2/3 or formins by depleting the Arp2/3 subunit, ArpC4, with RNAi or with the small-molecule inhibitor of formins, SMIFH2. Interfering with either actin nucleator did not affect cell rounding (Figure 5O) or nuclear orientation (Figure 5, P and Q). Similar results regarding nuclear orientation were obtained when we used a small-molecule inhibitor of Arp2/3 (CK666; our unpublished observations). Overall, we concluded that NE Dynein, coupled with loss of cortical Dynein, ensured the correct centrosome–nucleus axis at NEB by facilitating nuclear reorientation.
Arp2/3 activity is required for centrosome orientation during prophase
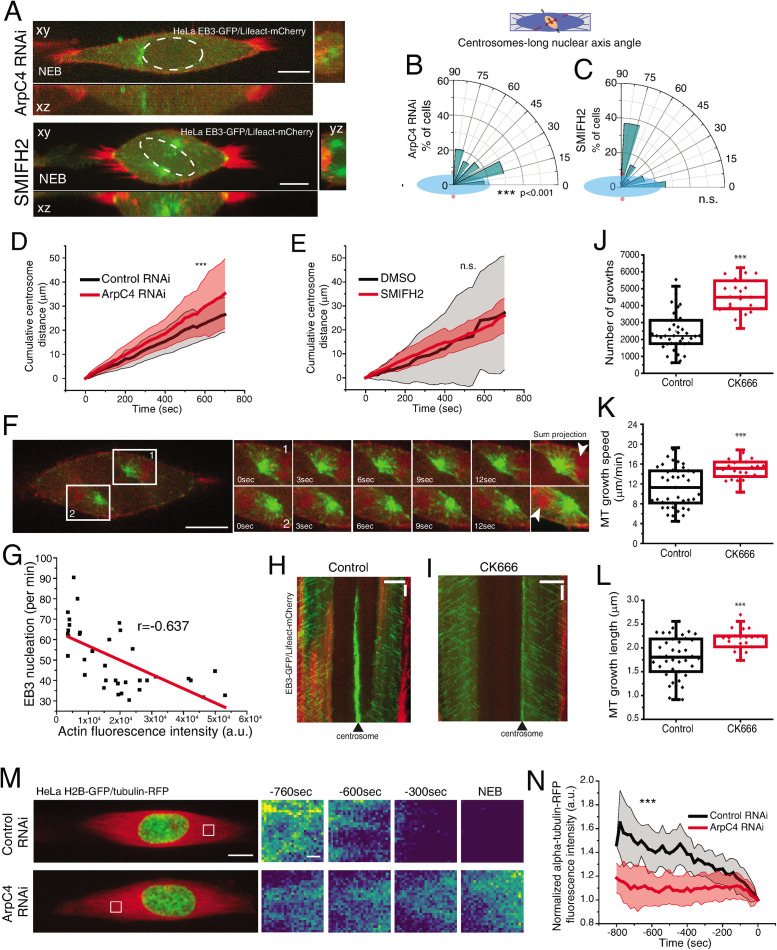
The experiments above support a model where Dynein-mediated forces act on the NE to induce nuclear reorientation. However, they do not take into account the contribution of centrosome movement for positioning on the shortest nuclear axis. This is particularly relevant since we showed that Arp2/3 activity did not affect nuclear orientation (Figure 5, O–Q) but was required for centrosome positioning on the shortest nuclear axis (Figure 6, A–C, ***p < 0.001, Supplemental Movie S3). It is possible the Arp2/3-mediated branched actin network directly affects centrosome movement and positioning, in line with previously published in vitro work (Colin et al., 2018). Accordingly, we observed that depleting ArpC4 by RNAi leads to increased centrosome movement (Figure 6D, ***p < 0.001). Inversely, interfering with formins activity (Figure 6E) or with any of the Dynein pools (Supplemental Figure S7, A–C) did not affect centrosome movement, suggesting this process required the presence of a branched actin network, but was independent of NE Dynein or centrosome distance to the NE. Interestingly, expression of Rap1* did not affect overall centrosome movement, indicating that adhesion disassembly is primarily required for nuclear reorientation (Supplemental Figure S7D and Figure 4F).
FIGURE 6:
Arp2/3 activity regulates centrosome movement. (A) Representative images of HeLa cells expressing EB3-GFP/Lifeact-mCherry after treatment with ArpC4 RNAi or SMIFH2 at NEB. Scale bar, 10 μm. (B) Polar plots quantifying centrosome positioning (red circles) relative to the longest nuclear axis (blue ellipse) at NEB for cells treated with ArpC4 RNAi (B) or SMIFH2 (C). Cumulative centrosome distance measured for cells treated with ArpC4 RNAi (D) or SMIFH2 (E). (F) HeLa cell expressing EB3-GFP/Lifeact-mCherry filmed with a 500-ms time lapse to show how the presence of actin limits microtubule growth. Scale bar, 10 μm. Time is in seconds. (G) Correlation between actin fluorescence intensity (a.u.) and EB3 nucleation (per minute). Kymograph of cells expressing EB3-GFP/Lifeact-mCherry for control (H) or CK666-treated (I) cells. Horizontal scale bar, 10 μm; vertical scale bar, 30 s. Microtubule dynamics parameters for control (79,899 tracks; 41 cells) and CK666-treated cells (79,957 tracks; 23 cells; ***p < 0.001) indicating number of growths (J), growth speed (K; μm/min), and growth length (L; μm).(M) Representative images of HeLa cells expressing H2B-GFP/tubulin-RFP filmed during mitotic entry. Scale bar, 10 μm. White box corresponds to area used for insets on the right. Insets represent tubulin fluorescence intensity visualized with a Viridis LUT. Scale bar, 1 μm. (N) Quantification of normalized tubulin-RFP fluorescence intensity for controls and ArpC4 RNAi-treated cells (***p < 0.001).
Movie S3.
HeLa cell expressing H2B‐GFP/tubulin‐RFP treated with ArpC4 RNAi, related to Figure 6
It was recently shown, using reconstituted in vitro approaches, that the Arp2/3-mediated branched actin network controls centrosome movement by regulating microtubule dynamics (Colin et al., 2018). To assess whether centrosome positioning depended on Arp2/3 regulation of microtubule dynamics, we started by performing high-resolution imaging of cells expressing EB3-GFP/Lifeact-mCherry during prophase (Figure 6F). We consistently observed the presence of actin near the centrosome that correlated with a decrease in microtubule nucleation (Figure 6, F, white arrowheads, and G). Next, to interfere with the Arp2/3-mediated actin network, we treated cells with the Arp2/3 inhibitor, CK666, and analyzed astral microtubule dynamics using the plusTipTracker software (Applegate et al., 2011). Treatment with CK666 during prophase increased the number of microtubule growths, as well as microtubule speed and length, when compared with controls (Figure 6, H–L, ***p < 0.001), in line with the microtubule-destabilizing effect of branched actin networks (Colin et al., 2018; Farina et al., 2019; Inoue et al., 2019). Microtubule destabilization during early mitosis is crucial to allow proper spindle assembly (Zhai et al., 1996; McHedlishvili et al., 2018). To determine whether Arp2/3 activity was required for microtubule cytoskeleton reorganization and early spindle assembly, we filmed HeLa cells expressing H2B-GFP/tubulin-RFP during mitotic entry and quantified the levels of tubulin-RFP fluorescence intensity in controls and ArpC4 RNAi cells. In controls, we observed a decrease in tubulin fluorescence intensity over time (Figure 6, M and N, ***p < 0.001), reflecting the disassembly of the interphase microtubule cytoskeleton (McHedlishvili et al., 2018). By comparison, tubulin fluorescence intensity in ArpC4-depleted cells did not vary significantly (Figure 6, M and N), supporting a role for Arp2/3 activity in prophase microtubule destabilization. To further confirm that microtubule destabilization was important for centrosome movement and positioning during prophase, we treated cells with low doses of the microtubule stabilizer, Taxol. As expected, stabilizing microtubules with Taxol treatment led to increased centrosome movement (Supplemental Figure S7E, ***p < 0.001) and incorrect centrosome positioning at NEB (Supplemental Figure S7F, **p = 0.008), independently of cell rounding (Supplemental Figure S7H) or nuclear reorientation (Supplemental Figure S7G). Thus, we concluded that Arp2/3 regulates microtubule dynamics to facilitate cytoskeleton reorganization, centrosome movement, and correct positioning during prophase.
Centrosome positioning on the shortest nuclear axis facilitates spindle assembly
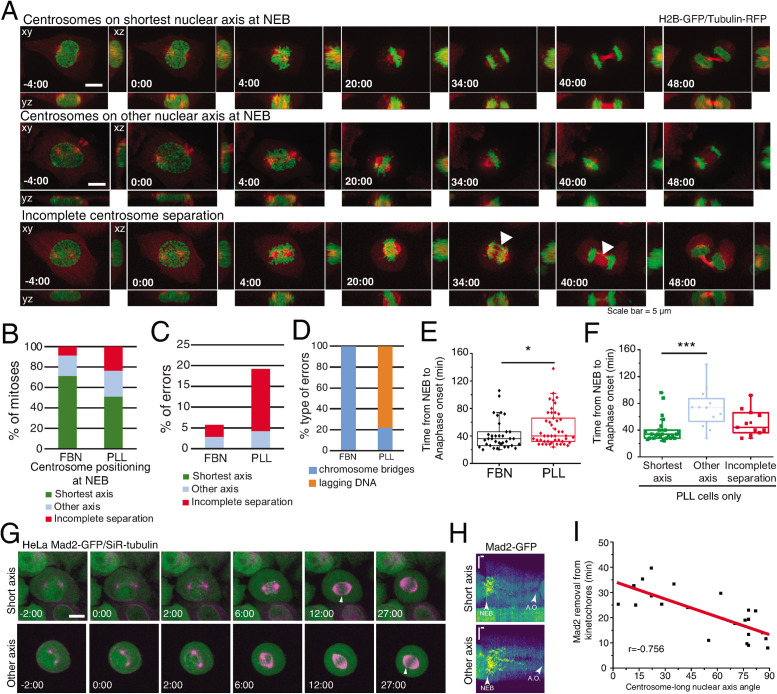
Centrosome positioning at NEB is known to affect mitotic fidelity (Kaseda et al., 2012; Silkworth et al., 2012). Here, we tested specifically whether positioning on the shortest nuclear axis affected spindle assembly efficiency. We imaged cells on FBN or PLL and correlated centrosome positioning, mitotic timing, and missegregation events (Figure 7A; Supplemental Movies S4 and S5). Seeding cells on PLL affected centrosome separation (“incomplete separation;” 23% for PLL and 8% for FBN) and positioning on the shortest nuclear axis (51% for PLL and 72% for FBN), when compared with FBN (Figure 7B). Consequently, PLL-seeded cells had increased missegregation events (19.2%) when compared with FBN (5.7%) (Figure 7C). These were mainly lagging chromosomes in cells with incomplete centrosome separation (Figure 7D), as was described previously (Silkworth et al., 2012). We concluded that the extent of centrosome separation increases chromosome segregation fidelity. Next, we determined whether centrosome positioning affects mitotic timing. Cells on PLL had a significant delay in anaphase onset when compared with cells on FBN (Figure 7E, *p < 0.05). This delay was due to cells that separate, but do not position centrosomes on the shortest nuclear axis (Figure 7F, “other axis;” 72 ± 29 min, ***p < 0.001), as opposed to cells with centrosomes on the “shortest axis” (40 ± 18 min). To confirm if the increased mitotic timing observed was due to a delay in Spindle Assembly Checkpoint (SAC) satisfaction, we filmed HeLa cells expressing Mad2-GFP/SiR-tubulin. By monitoring Mad2 removal from kinetochores, we can determine how efficiently microtubules attach to kinetochores (Waters et al., 1998), leading to SAC satisfaction. We seeded cells in PLL and followed them as they progressed through mitosis (Figure 7G). We then correlated Mad2 removal from kinetochores with the centrosome–nucleus axis at NEB (Figure 7, H and I). Interestingly, we observed that cells which place their centrosomes on the shortest nuclear axis remove Mad2 faster from the kinetochores (Figure 7, G–I, r = –0.756). We concluded that centrosome positioning on the shortest nuclear axis ensures timely progression through mitosis by ensuring faster SAC satisfaction.
FIGURE 7:
Centrosome positioning on the shortest nuclear axis facilitates spindle assembly. (A) HeLa cells expressing H2B-GFP/alpha-tubulin-RFP during mitosis. The top panel represents a cell with centrosomes on the shortest nuclear axis at NEB, the middle panel represents a cell with centrosomes on a random nuclear axis, and the bottom panel represents a cell with incomplete centrosomes separation. Time lapse is 2 min. Scale bar, 5 μm. (B) Proportion of cells that place centrosomes on shortest axis (green), other axis (blue), or have incomplete separation (red) at NEB, depending on the coating (FBN or PLL). Quantification of the proportion (C) and type of missegregation events (D) for cells seeded on FBN (n = 35) or PLL (n = 47). (E) Quantification of the time between NEB and anaphase onset for cells seeded on FBN or PLL (***p < 0.001). (F) Timings from NEB to anaphase onset, for cells seeded on PLL, according to their centrosome separation status (***p < 0.001; n.s., not significant). (G) HeLa cells expressing Mad2-GFP/SiR-tubulin during mitosis. The top panel represents a cell with centrosomes on shortest nuclear axis at NEB and the bottom panel represents a cell with centrosomes on a random nuclear axis. White arrowheads indicate Mad2-positive kinetochores. Time lapse is 20 s. Time is in minutes:seconds. Scale bar, 10 μm. (H) Representative kymographs of Mad2-GFP (Viridis LUT) for cells with centrosomes on the shortest nuclear axis (top panel) and cells with centrosomes on a random axis (bottom panel). Horizontal scale bar, 100 s. Vertical scale bar, 5 μm. A.O., anaphase onset. (I) Correlation between the time for Mad2 removal from kinetochores and centrosome-long nuclear axis and NEB, with a Pearson´s correlation coefficient = –0.756.
Movie S4.
HeLa cell expressing H2B‐GFP/tubulin‐RFP showing centrosomes on shortest nuclear axis, related to Figure 7
Movie S5.
HeLa cell expressing H2B‐GFP/tubulin‐RFP, showing a missegregation event, related to Figure 7
DISCUSSION
At the transition from G2 to mitosis, interphase adhesion complexes disassemble (Dao et al., 2009; Marchesi et al., 2014; Dix et al., 2018), cell margins retract (Cramer and Mitchison, 1997; Maddox and Burridge, 2003), and microtubule organization and dynamics change (Zhai et al., 1996; McHedlishvili et al., 2018). This leads to a global reorganization of cellular geometry that is required to form a stiff mitotic cortex (Kunda et al., 2008) and help bipolar spindle assembly (Lancaster et al., 2013). During this stage, Dynein is loaded on the NE through multiple pathways mediated by RanBP2 (Splinter et al., 2010) and Nup133 (Bolhy et al., 2011). This allows centrosome tethering to the NE which assists in centrosome separation (Raaijmakers et al., 2012), NEB (Beaudouin et al., 2002; Salina et al., 2002), and early spindle assembly (Bolhy et al., 2011). How these processes are spatiotemporally coordinated to ensure efficient spindle assembly remained unclear.
Here, we propose a model in which efficient spindle assembly requires a combination of centrosome movement and nuclear reorientation to position centrosomes on the shortest nuclear axis at NEB. Our observations suggest that microtubule remodeling during early mitosis is essential for centrosome movement, partly through the action of Arp2/3-mediated microtubule regulation, but also likely through the inactivation of interphase microtubule stabilizers (McHedlishvili et al., 2018). Although the Arp2/3 complex does not seem to be relevant for mitotic progression (Lancaster et al., 2013) or mitotic cortex structure (Chugh et al., 2017), its activity was recently proposed to assist in microtubule remodeling at mitotic exit (Farina et al., 2019). In particular, recruitment of Arp2/3 to the centrosome during this stage leads to increased actin filament nucleation, accompanied by decrease in centrosomal microtubule density. Our results (Figure 6) suggest that Arp2/3 may function in a similar manner during mitotic entry to decrease microtubule density and assist in cytoskeletal reorganization. This is in line with previous works that propose a role for Arp2/3 in fine-tuning changes in microtubule dynamics (Colin et al., 2018; Inoue et al., 2019) and could contribute to the microtubule reorganization observed during the G2/M transition (Zhai et al., 1996). Globally, this would trigger centrosome repositioning during prophase.
In combination with centrosome movement, we propose that Dynein-mediated forces on the NE also contribute to correct centrosome positioning by regulating nuclear reorientation, as was already described in other systems (Pomerat, 1953; Levy and Holzbaur, 2008; Wu et al., 2011). During prophase, centrosomes separate to opposite sides of the nucleus (Tanenbaum and Medema, 2010). This event creates an asymmetry in the centrosomal microtubule arrays due to the presence of an intact nucleus between them, biasing microtubule nucleation along the surface of the NE. At the same time, Dynein is loaded on the NE (Splinter et al., 2010; Bolhy et al., 2011). This allows Dynein to capture centrosomal microtubules, likely stabilizing them (Hendricks et al., 2012) and generating pulling forces that are sufficient to assist in NEB (Beaudouin et al., 2002; Salina et al., 2002). Due to the asymmetry in the centrosomal microtubule arrays and the dynamic nature of microtubules, these pulling forces exert a rotational torque on the prophase nucleus (Levy and Holzbaur, 2008; Wu et al., 2011). This torque is then translated into nuclear reorientation, once cell rounding occurs. This hypothesis is supported by our data showing that adhesion disassembly or PTx-triggered cell rounding is sufficient to allow nuclear rotation (Figures 4 and 5), whereas blocking deadhesion constrains nuclear reorientation (Figure 4, Supplemental Figure S4, and Versaevel et al., 2012).
Importantly, the positioning of centrosomes on the shortest nuclear axis during prophase is clearly distinct from the mechanism driving metaphase spindle orientation. During metaphase, external cues (Thery et al., 2005; Toyoshima and Nishida, 2007; Fink et al., 2011) activate cortical force generators (Thery et al., 2007; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012) and transmit pulling forces to astral microtubules (Cytrynbaum et al., 2003; Grill et al., 2003; Fink et al., 2011; Bosveld et al., 2016) to ensure robust centrosome positioning. However, before NEB, the mitotic cortex is not fully assembled (Ramanathan et al., 2015; Rosa et al., 2015) and cortical force generators are not yet in place (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012). Consequently, at this stage, centrosomes do not align according to external cues but follow internal cues provided by the cytoskeleton and nucleus.
Chromosome capture during early mitosis was proposed to occur through a “search-and-capture” mechanism (Kirschner and Mitchison, 1986). Subsequent work demonstrated that timely spindle assembly could not rely solely on the search-and-capture mechanism (Wollman et al., 2005), but depended on the contribution of kinetochore-driven microtubule nucleation (Maiato et al., 2004), kinetochore compaction (Magidson et al., 2015), and chromosome motion (Paul et al., 2009) and distribution (Magidson et al., 2011). However, these facilitating mechanisms often require that centrosomes deviate from the external cues (Toso et al., 2009; Magidson et al., 2011) to ensure efficient chromosome capture. These reports are in line with our observations that the centrosome–nuclear axis orients away from the underlying micropattern during early spindle assembly. We propose that centrosome positioning on the shortest nuclear axis favors the assembly of a bipolar spindle scaffold to ensure maximum exposure of kinetochores to microtubules (Lancaster et al., 2013). In combination with the spatial distribution of chromosomes in a ring configuration (Magidson et al., 2011), this would accelerate spindle assembly, minimizing the probability of generating erroneous attachments. In agreement with this hypothesis, we observed that centrosome mispositioning significantly delayed mitosis and Mad2 removal from kinetochores (Figure 7), whereas failure to separate centrosomes altogether generated chromosome missegregation events, as previously described (Silkworth et al., 2012). In summary, we propose a model where centrosomes–nucleus axis orientation during prophase facilitates the formation of an initial bipolar spindle scaffold to ensure mitotic fidelity.
MATERIALS AND METHODS
Cell lines and transfections
Cell lines were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (FBS; Life Technologies) and grown in a 37°C humidified incubator with 5% CO2. HeLa cell line expressing histone H2B-GFP/mRFP-α-tubulin was generated in our lab using lentiviral vectors. For this purpose, HEK293T cells at 50–70% confluence were cotransfected with lentiviral packaging vectors (16.6 µg of Pax2 and 5.6 µg of pMD2) and 22.3 µg of LV-H2B-GFP (a gift from Elaine Fuchs, Addgene plasmid 25999) or pRRL-mRFP-α-tubulin plasmids, using 30 µg of Lipofectamine2000 (Life Technologies). After transfection, the virus-containing supernatant was collected, centrifuged, filtered, and stored at –80°C. HeLa parental cells were then transduced with each lentivirus in the presence of polybrene (1:1000) in standard culture media for 24 h. The lentiviruses were used individually, giving time for cells to recover between transductions. After the second transduction, H2B-GFP/mRFP-α-tubulin double-positive cells were isolated by fluorescence-activated cell sorting (FACS; FACS Aria II). For transient overexpression of pRK5-Rap1[Q63E] plasmids (a gift from Jean de Gunzburg), cells were transfected with the corresponding plasmid using Lipofectamine2000 (Life Technologies). Briefly, cells at 50–70% confluence were incubated for 6 h with 5 µl of Lipofectamine 2000 and 0.6 µg/ml DNA. DNA–lipid complexes were previously diluted in Opti-Minimal Essential Medium (Opti-MEM; Alfagene) and incubated for 30 min before adding to the cells. Prior to and during transfection, cell medium was changed to a reduced serum medium (DMEM supplemented with 5% FBS). Cells were analyzed 48 h after transfection. HeLa Mad2-GFP cells were previously described (Schweizer et al., 2013).
Micropatterning
Micropatterns to control individual cell shape and adhesion pattern were produced as previously described (Azioune et al., 2009). Briefly, glass coverslips (22 × 22 mm No. 1.5, VWR) were activated with plasma (Zepto Plasma System, Diener Electronic) for 1 min and incubated with 0.1 mg/ml PLL(20)-g[3,5]-PEG(2) (SuSoS) in 10 mM HEPES at pH 7.4, for 1 h, at RT. After rinsing and air-drying, the coverslips were placed on a synthetic quartz photomask (Delta Mask), previously activated with deep-UV light (PSD-UV, Novascan Technologies) for 5 min; 3 µl of MiliQ water were used to seal each coverslip to the mask. The coverslips were then irradiated through the photomask with the UV lamp for 5 min. Afterward, coverslips were incubated with 25 μg/ml FBN (Sigma-Aldrich) and 5 μg/ml Alexa 546– or 647–conjugated fibrinogen (Thermo Fisher Scientific) in 100 mM NaHCO3 at pH 8.6, for 1 h, at RT. Cells were seeded at a density of 50,000 cells/coverslip and allowed to spread for ∼10–15 h before imaging. Nonattached cells were removed by changing the medium ∼2–5 h after seeding.
Drug treatments
PTx was used at 40 nM (Merck). To inhibit the activity of the Arp2/3 complex, we used 100 µM of CK666 (Tocris Bioscience). ROCK inhibitor Y-27632 was used at a concentration of 20 µM (Sigma-Aldrich). The small molecule inhibitor of formin-mediated actin assembly, SMIFH2, was used at 30 μM. To interfere with the microtubule cytoskeleton, we used Taxol (20 nM) (Sigma-Aldrich). To inhibit Eg5, STLC was added at 5 μM. All the drugs used were added to the culture medium 30 min–1 h before live-cell imaging or fixation. Control cells were treated with DMSO (Sigma-Aldrich) only.
RNAi experiments
Cells were transfected with small interfering RNAs (siRNAs) using Lipofectamine RNAi Max (Life Technologies). Specifically, 5 μl of Lipofectamine and 20 nM of each siRNA were diluted and incubated in Opti-MEM (Alfagene) for 30 min. The siRNA–lipid complexes were then added to 50–70% confluence cells cultured, during transfection (6 h), in reduced serum medium (DMEM supplemented with 5% FBS). Commercial ON-TARGETplus SMARTpool siRNAs (Dharmacon) were used for BICD2, ARPC4, and Ect2 depletions. For DHC depletion, the following oligos were ordered: 5′-GAACUAGACUUGGUUAAUU-3′ and 5′-AAUUAACCAAGUCUAGUUC-3′. For combined NudE+NudEL, depletion the following oligos were ordered: 5’-GCUUGAAUCAGGCCAUCGA-3’ and 5’-UCGAUGGCCUGAUUCAAGC-3’ for NudE and 5’-GGAUGAAGCAAGAGAUUUA-3’and 5’-UAAAUCUCUUGCUUCAUCC-3’for NudEL. Commercial ON-TARGETplus SMARTpool nontargeting siRNAs and mock transfections were used as controls. For all siRNAs used, cells were analyzed 72 h after transfection, except for Ect2 RNAi where cells were analyzed 24–48 h posttransfection. Protein depletion efficiency was monitored by immunoblotting and phenotypic analysis.
Time-lapse microscopy
For time-lapse microscopy, 12–24 h before the experiments 1.5 × 105 cells were seeded on coverslips coated with FBN (25 μg/ml; F1141, Sigma) or PLL (25 μg/ml; F1141, Sigma). When micropatterns were used, 5 × 104 cells were seeded on coverslips coated with FBN (25 μg/ml; F1141, Sigma). Prior to each experiment, cell culture medium was changed from DMEM with 10% FBS to Leibovitz’s-L15 medium (Life Technologies) supplemented with 10% FBS and Antibiotic-Antimycotic 100X (AAS; Life Technologies). When SiR-dyes were used, they were added to the culture medium 30 min–1 h before acquisition (20 nM Sir-tubulin or 10 nM Sir-DNA; Spirochrome). Live-cell imaging was performed using temperature-controlled Nikon TE2000 microscopes equipped with a modified Yokogawa CSU-X1 spinning-disk head (Yokogawa Electric), an electron multiplying iXon+ DU-897 EM-CCD camera (Andor), and a filter wheel. Three laser lines were used for excitation at 488, 561, and 647 nm. For nuclear pore fluctuation analysis, an oil-immersion 100× 1.4 NA Plan-Apo DIC (Nikon) was used. All the remaining experiments were done with an oil-immersion 60× 1.4 NA Plan-Apo DIC (Nikon). Image acquisition was controlled by NIS Elements AR software. For centrosome tracking, 17–21 z-stacks with a 0.5-µm separation were collected every 20 s. For mitotic timing quantifications, 13 z-stacks with a 0.7-µm separation were collected every 2 min. For microtubule dynamics measurements, a single z-stack was collected every 500 ms.
Quantitative analysis of centrosomes, cell membrane, and nucleus membrane
Detailed quantitative analysis of centrosomes location and membranes topology (cell and nucleus) was performed using custom-made MATLAB scripts (MathWorks, USA; R2018a). The image analysis took advantage of the different labeling for centrosomes, cell membrane, and nuclear membrane. The scripts were separated into three modules with specific workflows: 1) centrosomes tracking and 2) nuclear and cellular membrane reconstruction. Tracking of centrosomes position/trajectories was performed in 3D space using image stacks with a pixel size of 0.190 μm and z-step of 0.7 μm. Images were preprocessed using a Laplacian of Gaussian filter with a user-defined kernel size, associated with the centrosome radius in pixels. Image segmentation was performed using Otsu’s method, and morphological operators were used to improve the mask and obtain the centrosomes 3D coordinates. Error correction methods, such as automatic thresholding adjustment or in the limit frame elimination, were implemented to take care of frames where the standard method was unable to uniquely identify two centrosomes. For the visualization of the centrosomes trajectories (space and time), the centrosomes coordinates were interpolated using cubic splines. Different metrics, such as the distance between centrosomes (pole-to-pole), were calculated to analyze and characterize the trajectories. Cellular and nuclear membranes were reconstructed in 3D space taking advantage of specific labeling. For each membrane, a mask was produced using Otsu’s method and improved with a sequence of morphological operators (namely image close, dilation and erosion, small objects removal). The orientation axis for the membranes were calculated using principal components analysis (PCA) of a large sample of membrane surface points. This method using PCA was found to be more robust than ellipsoid fitting to the membrane surface (followed by extraction of the axis vectors). From the centrosomes locations and nuclear membrane reconstruction, it was possible to calculate the angle between the centrosomes axis and the nucleus major axis.
Preparation of micropatterned hydrogels with nanobeads
First, 32-mm coverslips were plasma cleaned for 30 s and then incubated with a drop of PLL-PEG 0.1 mg/ml in HEPES 10 mM, pH 7.4, for 30 min at RT as described previously (Vignaud et al., 2014). Coverslips are then put upright to let the excess PLL-PEG run off and placed on a rectangle shape quartz photomask (Toppan) on a 3-μl drop of MilliQ water. The coverslips on the photomask are then exposed to deep UV for 5 min. Then, coverslips are detached from the photomask and incubated with 20 μg/ml FBN (Sigma) and 20 μg/ml Alexa 546–conjugated fibrinogen (Invitrogen) in phosphate-buffered saline (PBS) for 30 min at RT. To prepare the gels, a 42-μl drop of a 40-KPa mix of polyacrylamide and bisacrylamide (Sigma) containing 0.1 μl carboxylate-modified polystyrene fluorescent beads (Invitrogen) is placed onto the FBN-coated coverslips and then covered with a second coverslip and pretreated with Bind-silane solution (100% ethanol solution containing 18.5 μl Bind Silane; GE Healthcare Life Science) and 161 μl 10% acetic acid (Sigma) for 5 min. Gels are polymerized for 30 min and finally the gel is retrieved with the silanized coverslip. FBN proteins are trapped within the acrylamide mesh. Gels are stored in PBS at 4°C.
TFM imaging and analyses
For TFM live-cell imaging, rectangle micropatterned coverslips are mounted in dedicated chambers and supplemented with L-15/10% FBS medium. A Leica SP8 confocal microscope was used to acquire the images using a 40× objective (oil immersion, numerical aperture 1.3) with a temperature control chamber set at 37°C. Cells were imaged every 3 min; 488- and 533-nm lasers were used in sequential scanning mode. All the laser parameters and imaging setups are controlled through the LAS X system. Cellular traction forces were calculated using a method previously described (Mandal et al., 2014). Briefly, at each time point, the image of the fluorescent beads embedded in the substrate was compared with a reference image corresponding to a relaxed substrate and taken after washing away the cells. After correcting for experimental drift, the displacement field was obtained by a two-step process consisting of cross-correlation on 9.6-μm subimages followed by particle tracking to improve the spatial resolution. The final displacement field was interpolated to a regular grid with 1.2-μm spacing. Traction stress reconstruction was performed with the assumption that the substrate is a linear elastic half-space using Fourier transform traction cytometry and zeroth order regularization. The stress map was defined on the same 1.2-μm-period grid. From this stress map and the cell mask, we checked that the out of equilibrium force is less than 10% of the sum of forces magnitude, as a quality criterion for all cells and time points. The contractile energy, which is the mechanical energy transferred from the cell to the substrate, was computed from the traction map by integrating the scalar product of the displacement and stress vectors over the cell surface. To determine the principal direction of contraction of each cell, we calculated and diagonalized the first moment tensor of the stress. The eigenvector corresponding to the larger eigenvalue gives the direction of the main force dipole. The degree of force polarization is obtained by comparing both eigenvalues. All the calculations are performed in MATLAB (MathWorks, USA; R2018a).
Immunofluorescence
Cells were fixed with 4% PFA in cytoskeleton buffer (274 mM NaCl, 2.2 mM Na2HPO4, 10 mM KCL, 0.8 mM KH2PO4, 4 mM EDTA, 4 mM MgCl2, 10 mM Pipes, 10 nM glucose, pH 6.1) and subsequently permeabilized with 0.5% Triton X-100 (Sigma- Aldrich) in 1× PBS for 5 min. After washing in 0.1% Triton X-100, cells were blocked with 10% FBS in 0.1% Triton X-100 in PBS for 30 min. All the primary antibodies were diluted in blocking solution and incubated for 1 h at room temperature. After this incubation, the cells were washed with 0.1% Triton X-100 in 1× PBS and incubated with the respectively secondary antibody for 1 h at room temperature. The secondary antibodies were diluted in blocking solution. DNA was stained with DAPI, which was added to the secondary antibodies solution (1 µg/ml, Sigma-Aldrich). After incubation with the secondary antibodies and DAPI the coverslips were washed with 0.1% Triton X-100 in 1× PBS and sealed on glass slides mounted with 20 mM Tris, pH 8, 0.5 N-propyl gallate, and 90% glycerol. The following primary antibodies were used: rat anti-alpha Tubulin (1:500 Bio-Rad) and pFAK(Y397) (1:100, Invitrogen). Alexa Fluor 488, 568, and 647 (1:2000, Invitrogen) were used as secondary antibodies. Where indicated, SiR-actin was used at a concentration of 20 nM (Spirochrome). Images were acquired using an AxioImager Z1 (63×, Plan oil differential interference contract objective lens, 1.4 NA; all from Carl Zeiss) which is coupled with a CCD camera (ORCA-R2; Hamamatsu Photonics) using the Zen software (Carl Zeiss).
Western blotting
HeLa cell extracts were collected after trypsinization and centrifuged at 1200 rpm for 5 min, washed, and resuspended in 30–50 μl of lysis buffer (NP-40, 20 nM HEPES/KOH, pH 7.9; 1 mM EDTA, pH 8; 1 mM EGTA; 150 mM NaCl; 0.5% NP40; 10% glycerol, 1:50 protease inhibitor; 1:100 Phenylmethylsulfonyl fluoride). The samples were then flash frozen in liquid nitrogen and kept on ice for 30 min. After centrifugation at 14,000 rpm for 8 min at 4°C, the supernatant was collected and protein concentration determined by the Bradford protein assay (Bio-Rad). The proteins were run on 7–15% SDS–PAGE gels (50 μg/lane) and transferred using the iBlot Gel Transfer Device (Thermo Scientific) to a nitrocellulose Hybond-C membrane. With the exception of DHC RNAi samples, all others were transferred using a wet blot apparatus for 3 h at 70 V, with constant amperage. Afterward, the membranes were blocked with 5% milk in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBS-T) for 1 h at room temperature. The primary antibodies used were: mouse anti-Nde1 (1:500, Abnova), rabbit anti-ARPC4 (1:500, Bethyl Laboratories), rabbit anti-BICD2 (1:250, Atlas Antibodies), mouse anti-Dynein intermediate chain 74.1 (1:1000, Merck), and rat anti-alpha tubulin (1:1000, Bio-Rad). All primary antibodies were incubated overnight at 4°C with shaking. After three washes in TBS-T, the membranes were incubated with the secondary antibody for 1 h at room temperature. The secondary antibodies used were anti-mouse-HRP and anti-rabbit-HRP at 1:5000. After several washes with TBS-T, the detection was performed with Clarity Western ECL Substrate (Bio-Rad).
Statistical analysis and data presentation
Each experiment was repeated independently at least three times and sample sizes are defined in each figure legend. We used three to six independent experiments or biologically independent samples for statistical analysis. For knockdown experiments, the knockdown efficiency of each experiment was measured by quantifying immunoblots. When data are represented as box-whisker plots, the box size represents 75% of the population and the line inside the box represents the median of the sample. The size of the bars (whiskers) represents the maximum (in the upper quartile) and the minimum (in the lower quartile) values. Statistical analysis for multiple group comparison was performed using a parametric one-way analysis of variance (ANOVA) when the samples had a normal distribution. When the sample did not have a normal distribution, multiple group comparison was done using a nonparametric ANOVA (Kruskal–Wallis). All pairwise multiple comparisons were subsequently analyzed using either post-hoc Student–Newman–Keuls (parametric) or Dunn’s (nonparametric) tests. When comparing only two experimental groups, a parametric t test was used when the sample had a normal distribution, or a nonparametric Mann–Whitney test was used for samples without normal distribution. Distribution normalities were assessed using the Kolmogorov–Smirnov test. No power calculations were used. All statistical analyses were performed using SigmaStat 3.5 (Systat Software).
Supplementary Material
Acknowledgments
The authors thank Iain Cheeseman for the DHC-GFP cell line, Jean de Gunzburg for the pRK5-Rap1[Q63E] construct, Elaine Fuchs for the LV-H2B-GFP vector, and Richard Vallee for the Nde antibody. We thank all members of the CID lab for discussions and suggestions. The authors thank Edgar Gomes, Tiago Dantas, Reto Gassman, Bernardo Orr, and António Pereira for critical reading of the manuscript. This work was funded by grants from FEDER—Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020—Operacional Programme for Competitiveness and Internationalization (POCI), Portugal 2020, and by Portuguese funds through FCT—Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior in the framework of the project PTDC/BEX-BCM/1758/2014 (POCI-01–0145-FEDER-016589). This work was also partially supported by a grant PHC-Pessoa Campus France/Fundação para a Ciência e Tecnologia. V.N. is supported by grant PD/BD/135545/2018 from the BiotechHealth FCT-funded PhD program. M.D. is supported by grant PD/BD/135548/2018 from the BiotechHealth FCT-funded PhD program. Work in the laboratory of H.M. is funded by the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 681443).
Abbreviations used:
- ANOVA
one-way analysis of variance
- DHC
Dynein Heavy Chain
- FBN
fibronectin
- FBS
fetal bovine serum
- NE
nuclear envelope
- NEB
nuclear envelope breakdown
- PBS
phosphate-buffered saline
- PCA
principal components analysis
- PLL
poly- l-lysine
- PTx
pertussis toxin
- SAC
Spindle Assembly Checkpoint
- siRNA
small interfering RNA
- TBS
Tris-buffered saline
- TFM
Traction Force Microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0047) on April 15, 2020.
REFERENCES
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. (2011). plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol , 168–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azioune A, Storch M, Bornens M, Thery M, Piel M. (2009). Simple and rapid process for single cell micro-patterning. Lab Chip , 1640–1642. [DOI] [PubMed] [Google Scholar]
- Baffet AD, Hu DJ, Vallee RB. (2015). Cdk1 activates pre-mitotic nuclear envelope dynein recruitment and apical nuclear migration in neural stem cells. Dev Cell , 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell , 83–96. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. (2011). A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol , 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth AJR, Yue Z, Eykelenboom JK, Stiff T, Luxton GWG, Hochegger H, Tanaka TU. (2019). Contractile acto-myosin network on nuclear envelope remnants positions human chromosomes for mitosis. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosveld F, Markova O, Guirao B, Martin C, Wang Z, Pierre A, Balakireva M, Gaugue I, Ainslie A, Christophorou N, et al. (2016). Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature , 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau V, Chen R, Edwards A, Sulaimain M, Maddox PS. (2019). PP2A-B55/SUR-6 collaborates with the nuclear lamina for centrosome separation during mitotic entry. Mol Biol Cell mbcE18100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Crest J, Fasulo B, Sullivan W. (2010). Cortical actin dynamics facilitate early-stage centrosome separation. Curr Biol , 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F. (2008). Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J Cell Biol , 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PC, Hsu RY, Liu CW, Lai CC, Chen HC. (2014). Adducin-1 is essential for mitotic spindle assembly through its interaction with myosin-X. J Cell Biol , 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, Roux PP, Charras G, Salbreux G, Paluch EK. (2017). Actin cortex architecture regulates cell surface tension. Nat Cell Biol , 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin A, Singaravelu P, Thery M, Blanchoin L, Gueroui Z. (2018). Actin-network architecture regulates microtubule dynamics. Curr Biol , 2647–2656.e2644. [DOI] [PubMed] [Google Scholar]
- Cramer LP, Mitchison TJ. (1997). Investigation of the mechanism of retraction of the cell margin and rearward flow of nodules during mitotic cell rounding. Mol Biol Cell , 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytrynbaum EN, Scholey JM, Mogilner A. (2003). A force balance model of early spindle pole separation in Drosophila embryos. Biophys J , 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao VT, Dupuy AG, Gavet O, Caron E, de Gunzburg J. (2009). Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J Cell Sci , 2996–3004. [DOI] [PubMed] [Google Scholar]
- De Simone A, Nedelec F, Gonczy P. (2016). Dynein transmits polarized actomyosin cortical flows to promote centrosome separation. Cell Rep , 2250–2262. [DOI] [PubMed] [Google Scholar]
- Dix CL, Matthews HK, Uroz M, McLaren S, Wolf L, Heatley N, Win Z, Almada P, Henriques R, Boutros M, et al. (2018). The role of mitotic cell-substrate adhesion re-modeling in animal cell division. Dev Cell , 132–145.e133. [DOI] [PubMed] [Google Scholar]
- Farina F, Gaillard J, Guerin C, Coute Y, Sillibourne J, Blanchoin L, Thery M. (2016). The centrosome is an actin-organizing centre. Nat Cell Biol , 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina F, Ramkumar N, Brown L, Samander-Eweis D, Anstatt J, Waring T, Bithell J, Scita G, Thery M, Blanchoin L, et al. (2019). Local actin nucleation tunes centrosomal microtubule nucleation during passage through mitosis. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J, Carpi N, Betz T, Betard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D, Piel M. (2011). External forces control mitotic spindle positioning. Nat Cell Biol , 771–778. [DOI] [PubMed] [Google Scholar]
- Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. (2003). The distribution of active force generators controls mitotic spindle position. Science , 518–521. [DOI] [PubMed] [Google Scholar]
- Grill SW, Hyman AA. (2005). Spindle positioning by cortical pulling forces. Dev Cell , 461–465. [DOI] [PubMed] [Google Scholar]
- Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur EL. (2012). Dynein tethers and stabilizes dynamic microtubule plus ends. Curr Biol , 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig O. (1884). Das Problem der Befruchtung und der Isotropie des Eies. Eine Theorie der Vererbung. Jenaische Zeitschrift fur Naturwissenschaft . [Google Scholar]
- Hu DJ, Baffet AD, Nayak T, Akhmanova A, Doye V, Vallee RB. (2013). Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell , 1300–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Obino D, Pineau J, Farina F, Gaillard J, Guerin C, Blanchoin L, Lennon-Dumenil AM, Thery M. (2019). Actin filaments regulate microtubule growth at the centrosome. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol , 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda K, McAinsh AD, Cross RA. (2012). Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol Open , 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell , 329–342. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. (2012). Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol , 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. (2012). Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol , 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Maitra A, Sumit M, Ramaswamy S, Shivashankar GV. (2014). Actomyosin contractility rotates the cell nucleus. Sci Rep , 3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Pelling AE, Liu T, Baum B. (2008). Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol , 91–101. [DOI] [PubMed] [Google Scholar]
- Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B. (2013). Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell , 270–283. [DOI] [PubMed] [Google Scholar]
- Levy JR, Holzbaur EL. (2008). Dynein drives nuclear rotation during forward progression of motile fibroblasts. J Cell Sci , 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock JG, Jones MC, Askari JA, Gong X, Oddone A, Olofsson H, Goransson S, Lakadamyali M, Humphries MJ, Stromblad S. (2018). Reticular adhesions are a distinct class of cell-matrix adhesions that mediate attachment during mitosis. Nat Cell Biol , 1290–1302. [DOI] [PubMed] [Google Scholar]
- Maddox AS, Burridge K. (2003). RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J Cell Biol , 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. (2011). The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell , 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, McEwen BF, Mogilner A, Khodjakov A. (2015). Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol , 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. (2004). Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol , 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal K, Wang I, Vitiello E, Orellana LA, Balland M. (2014). Cell dipole behaviour revealed by ECM sub-cellular geometry. Nat Commun , 5749. [DOI] [PubMed] [Google Scholar]
- Marchesi S, Montani F, Deflorian G, D’Antuono R, Cuomo A, Bologna S, Mazzoccoli C, Bonaldi T, Di Fiore PP, Nicassio F. (2014). DEPDC1B coordinates de-adhesion events and cell-cycle progression at mitosis. Dev Cell , 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews HK, Delabre U, Rohn JL, Guck J, Kunda P, Baum B. (2012). Changes in Ect2 localization couple actomyosin-dependent cell shape changes to mitotic progression. Dev Cell , 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BD, Thodeti CK, Tytell JD, Mammoto A, Overby DR, Ingber DE. (2010). Ultra-rapid activation of TRPV4 ion channels by mechanical forces applied to cell surface beta1 integrins. Integr Biol (Camb) , 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHedlishvili N, Matthews HK, Corrigan A, Baum B. (2018). Two-step interphase microtubule disassembly aids spindle morphogenesis. BMC Biol , 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner A. (2009). Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc Natl Acad Sci USA , 15708–15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessner M, Knerr J, Grosse R. (2019). Centrosomal actin assembly is required for proper mitotic spindle formation and chromosome congression. iScience , 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerat CM. (1953). Rotating nuclei in tissue cultures of adult human nasal mucosa. Exp Cell Res , 191–196. [DOI] [PubMed] [Google Scholar]
- Prosser SL, Pelletier L. (2017). Mitotic spindle assembly in animal cells: a fine balancing act. Nature reviews. Mol Cell Biol , 187–201. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RG, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. (2012). Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J , 4179–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan SP, Helenius J, Stewart MP, Cattin CJ, Hyman AA, Muller DJ. (2015). Cdk1-dependent mitotic enrichment of cortical myosin II promotes cell rounding against confinement. Nat Cell Biol , 148–159. [DOI] [PubMed] [Google Scholar]
- Rosa A, Vlassaks E, Pichaud F, Baum B. (2015). Ect2/Pbl acts via Rho and polarity proteins to direct the assembly of an isotropic actomyosin cortex upon mitotic entry. Dev Cell , 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J, Cramer LP, Baum B, McGee KM. (2004). Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell , 361–372. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Tulu US, Fagerstrom C, Wadsworth P. (2002). Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J Cell Biol , 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell , 97–107. [DOI] [PubMed] [Google Scholar]
- Schweizer N, Ferras C, Kern DM, Logarinho E, Cheeseman IM, Maiato H. (2013). Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J Cell Biol , 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. (2012). Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell , 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, et al. (2010). Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol , e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. (2010). Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell , 797–806. [DOI] [PubMed] [Google Scholar]
- Thery M, Jimenez-Dalmaroni A, Racine V, Bornens M, Julicher F. (2007). Experimental and theoretical study of mitotic spindle orientation. Nature , 493–496. [DOI] [PubMed] [Google Scholar]
- Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. (2005). The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol , 947–953. [DOI] [PubMed] [Google Scholar]
- Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. (2009). Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol , 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. (2007). Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J , 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RG, Raaijmakers JA, Tanenbaum ME, Medema RH. (2013). Nuclear envelope-associated dynein cooperates with Eg5 to drive prophase centrosome separation. Commun Integr Biol , e23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaevel M, Grevesse T, Gabriele S. (2012). Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat Commun , 671. [DOI] [PubMed] [Google Scholar]
- Vignaud T, Ennomani H, Thery M. (2014). Polyacrylamide hydrogel micropatterning. Methods Cell Biol , 93–116. [DOI] [PubMed] [Google Scholar]
- Wang W, Chen L, Ding Y, Jin J, Liao K. (2008). Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J Cell Sci , 1334–1343. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol , 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead CM, Winkfein RJ, Rattner JB. (1996). The relationship of HsEg5 and the actin cytoskeleton to centrosome separation. Cell Motil Cytoskeleton , 298–308. [DOI] [PubMed] [Google Scholar]
- Winkler F, Gummalla M, Kunneke L, Lv Z, Zippelius A, Aspelmeier T, Grosshans J. (2015). Fluctuation analysis of centrosomes reveals a cortical function of Kinesin-1. Biophys J , 856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. (2005). Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr Biol , 828–832. [DOI] [PubMed] [Google Scholar]
- Woodard GE, Huang NN, Cho H, Miki T, Tall GG, Kehrl JH. (2010). Ric-8A and Gi alpha recruit LGN, NuMA, and dynein to the cell cortex to help orient the mitotic spindle. Mol Cell Biol , 3519–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lee KC, Dickinson RB, Lele TP. (2011). How dynein and microtubules rotate the nucleus. J Cell Physiol , 2666–2674. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. (1996). Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol , 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.