Abstract
This single-center, retrospective study aimed to explore the immune characteristics of COVID-19 and biomarkers to predict the severity of this disease. Patients infected with SARS-CoV-2 (n = 215) treated at the First Affiliated Hospital of Nanchang University from January 24 to March 12, 2020, were included in the study and classified into severe and non-severe groups. Peripheral immunocyte count and cytokine statuses were compared. The correlation between immune status, cytokine levels, and disease severity was analyzed. Leukocyte numbers were normal in both groups; however, they were relatively high (7.19 × 109/L) in patients of the severe group. Leukocyte distributions differed between the two groups; the severe group had a higher percentage of neutrophils and lower percentage of lymphocytes compared with the non-severe group, and absolute lymphocyte numbers were below normal in both groups, and particularly deficient in patients in the severe group. Lymphocyte counts have negative correlation with duration of hospital period whereas neutrophil count has no significant correlation with it. Of tested cytokines, IL-6 levels were significantly higher in the severe group (P = 0.0418). Low level of lymphocyte predicts severity of COVID-19. IL-6 levels were significantly higher in the severe group, especially in some extremely severe patients. But we did not detect the significant correlation between severity of COVID-19 with IL-6 level which may be due to limited case numbers. Our observations encourage future research to understand the underlying molecular mechanisms and to improve treatment outcome of COVID-19.
Electronic supplementary material
The online version of this article (10.1007/s12026-020-09156-2) contains supplementary material, which is available to authorized users.
Keywords: SARS-CoV-2, Lymphocyte subsets, Cytokine
Introduction
In December 2019, the emergence of a novel coronavirus, SARS-CoV-2, led to the ongoing outbreak of pneumonia called COVID-19 in Wuhan City, Hubei Province, China [1]. SARS-CoV-2 is an enveloped RNA Betacoronavirus, belonging to the subgenus Sarbecovirus, subfamily Orthocoronavirinae, and is the seventh member of the coronavirus family identified that can infect humans [2]. The original host is likely to be bats; however, whether or not another intermediate host facilitated viral infection of humans remains unclear [3]. Accumulated evidence indicates that person-to-person transmission probably occurs via respiratory droplets produced by infected individuals [4–8]. Angiotensin converting enzyme 2 (ACE2) is the receptor for SARS-CoV-2 entry into host cells [3, 9], and infection can cause acute respiratory disease, with a median incubation period of 3 days (range, 0 to 24 days).
The clinical manifestations of patients infected with SARS-CoV-2 are variable, ranging from asymptomatic, through mild cold symptoms (such as fever and cough), to severe pneumonia and death. The incubation period and severity of symptoms may be associated with age and physical condition; however, there are currently shortages of biomarkers to predict severity of COVID-19. Patients in severe conditions often develop multiple organ complications, besides pneumonia, and have reduced numbers of peripheral blood lymphocytes [1, 10, 11]. The mechanisms by which the immune system responds against the SARS-CoV-2 infection and whether these are associated with disease severity are unclear. Here, we analyzed peripheral blood immune test data of patients infected with SARS-CoV-2, by comparing the numbers of different types of lymphocyte and cytokines between patients with non-severe and severe symptoms. This study improves our understanding on the immune response against this new coronavirus infection, which may help reveal the pathogenic mechanisms underlying COVID-19.
Material and methods
Patients and samples
This retrospective study was approved by the ethics board of the First Affiliated Hospital of Nanchang University (Ethics No. 2020013). Cases of laboratory-confirmed SARS-CoV-2 infection (n = 215) admitted to the hospital from January 24 to March 12, 2020, were enrolled. Oral consent was obtained from patients.
The First Affiliated Hospital of Nanchang University is located in Nanchang, Jiangxi Province, and is the main hospital responsible for treatment of COVID-19, as assigned by the Jiangxi Province Government. In addition to patients with non-severe symptoms, the majority of severe cases of COVID-19 in Jiangxi province were admitted to our hospital. All patients with COVID-19 enrolled in this study were diagnosed according to the WHO (World Health Organization) interim guidance [12]. A confirmed case of COVID-19 was defined as a positive result of real-time reverse transcriptase polymerase chain reaction (RT-PCR) assay of pharyngeal swab specimens. Laboratory-confirmed patients were classified into non-severe and severe COVID-19 disease groups on admission, according to the WHO interim guidance [12] and Novel coronavirus pneumonia diagnosis and treatment protocol in China (7th trial version) [13]. We included patients with mild illness and common pneumonia in the non-severe group and severe pneumonia and acute respiratory distress syndrome patients in the severe group. The classification criteria are as follows:
Mild illness—patients have mild symptoms and no pneumonia signs on chest image.
Pneumonia—patients have signs of pneumonia (fever and/or respiratory syndrome) but no signs of severe pneumonia. There are signs of pneumonia on chest image.
Severe pneumonia—adult (meet one of the following): (1) fast breathing (in breaths/min), ≥ 30 breath/min; (2) at rest state, SpO2 ≤ 93%; and (3) PaO2/FiO2 ≤ 300 mmHg. Chest imaging: pneumonia signs progress > 50% within 24–48 h.
Acute respiratory distress syndrome—meet one of the following: (1) respiratory failure, mechanical ventilation is required; (2) shock; and (3) combine other organ failure, ICU treatments are required.
Basic and epidemiological information, including age, Wuhan contact history, incubation period, and durations of hospital stay period, as well as laboratory test results, was collected from electronic medical records. Peripheral blood samples were obtained on the day of admission and immune tests conducted in the clinical laboratory. Immune laboratory tests consisted of leukocyte count; lymphocyte, monocyte, neutrophil, eosinophil, and basophil count; and lymphocyte subset count. Lymphocyte subsets were analyzed by Cytomics FC 500 flow cytometer (Beckman Coulter, USA). Cytokine (IL-1β, IL-2R, IL-6, IL-8, and IL-10) levels were measured by NovoCyte (ACEA Bioscience) flow cytometer using the Cytokine 12 kit (Raisecare, China) according to the manufacturer’s instructions. For cytokine assay, the plasma was used without diluting, and the value was acquired by comparing with standard curve and analyzed by NovoExpress software (ACEA). The minimum detectable concentrations of cytokines were 5 pg/mL for IL-1β, 2 pg/mL for IL-6, 5 pg/mL for IL-8, and 5 pg/mL for IL-10, respectively. We presented data as median (range), and values below the cutoff defined as 0 pg/mL.
Statistical analysis
All statistical analyses were performed using SPSS 23.0 and GraphPad Prism 5. Continuous variables are expressed as mean ± standard deviation (SD), median and interquartile range (IQR), and range values. Means were analyzed using the Mann-Whitney test. Categorical variables are described as frequency rates and percentages. Chi-square tests were used to evaluate differences between frequencies. Correlation analysis results are expressed as the Spearman correlation coefficient. P < 0.05 was considered statistically significant.
Results
Demographic and general clinical characteristics
The 215 hospitalized patients included in this study were classified into the non-severe (n = 159) and severe (n = 56) groups. Their demographic and general clinical characteristics are presented in Table 1. Patients in the severe group were significantly older than the patients in the non-severe group (median age, 56.5 vs. 43 years, P < 0.0001) and had a significantly longer median hospital stay duration (20 vs. 4 days, P < 0.0001). Further, the incubation period was significantly longer in the severe group (median, 5 vs. 4 days, P = 0.0079). By March 12, 2020, all the patients were discharged from hospital, and no patient died.
Table 1.
Baseline characteristics of COVID-19 patients
| No. (%) | P value | ||
|---|---|---|---|
| Non-severe (n = 159) | Severe (n = 56) | ||
| Age, median (IQR), years | 44 (32–52) | 56.5 (20–72) | < 0.0001a* |
| Sex | |||
| Female | 68 (42.8%) | 20 (35.7%) | 0.356b |
| Male | 91 (57.2%) | 36 (64.3%) | |
| Wuhan contact | 110 (69.2%) | 35 (62.5%) | 0.359b |
| Incubation period, median (IQR), days | 4 (2–6) | 5 (3–8) | 0.0079a* |
| Duration of hospital stay, median (IQR), days | 4 (2–6) | 20 (16.5–25) | < 0.0001a* |
P values indicate the differences between non-severe and severe patients. P < 0.05 was considered statistically significant
a*Mann-Whitney test
bPearson’s chi-square test
Peripheral leukocyte characteristics of COVID-19 patients
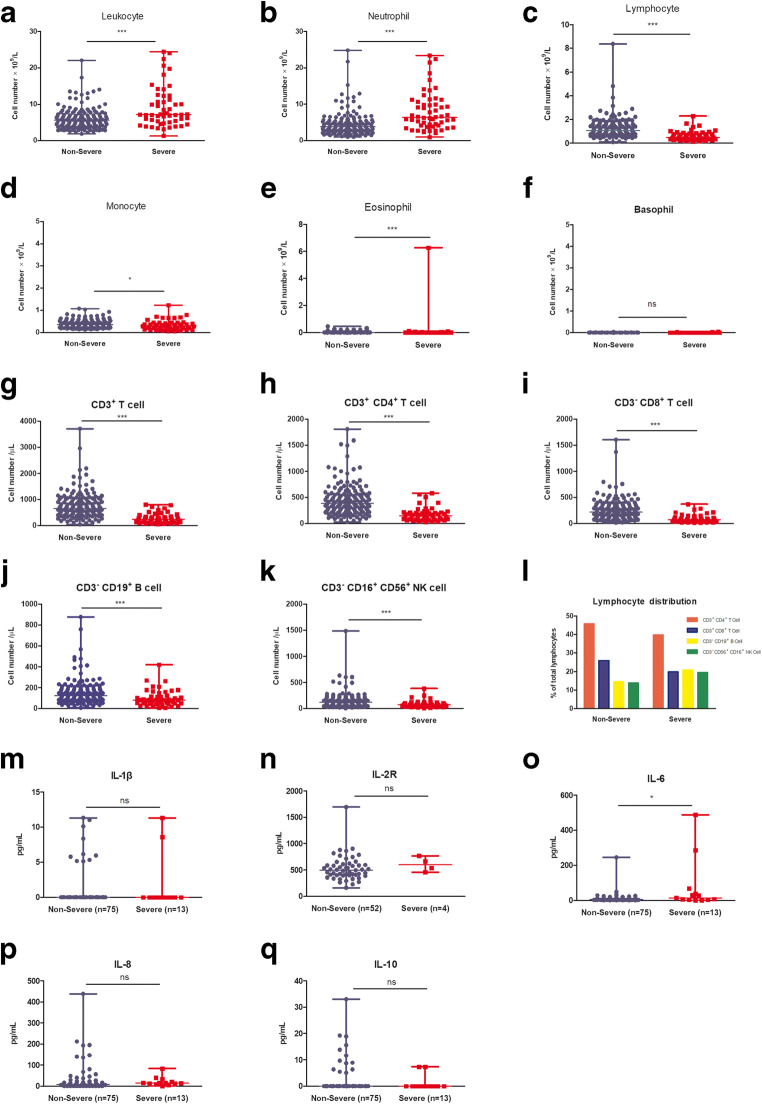
Patients in the severe group had significantly higher leukocyte (7.19 vs. 5.66 × 109/L) and neutrophil (6.4 vs. 3.8 × 109/L) counts (Fig. 1a, b; Table 2). Although both leukocyte counts were within the normal range (3.5–9.5 × 109/L), neutrophil counts of the severe group were a little bit above the high end of the normal range (1.8–6.3 × 109/L). The median of eosinophil and monocyte counts of patients in the severe group was significantly lower than those in the non-severe group (Fig. 1d, e). Notably, lymphocyte counts were dramatically decreased in the severe group (median 0.475 × 109/L, Fig. 1c), which was lower than half of the low end of the normal range (1.1–3.2 × 109/L). Lymphocyte counts in the non-severe group were slightly lower than the normal range (median 1.06 × 109/L). The median of basophil counts was similar between two groups (Fig. 1f).
Fig. 1.
Peripheral immunocyte counts and cytokine comparison between the non-severe and severe COVID-19 groups. a–f Leukocyte and five immunocyte subset counts. g–k Lymphocyte subset counts. l NK cell, CD4+ T cell, CD8+ T cell, and B cell distribution. m–q IL-1β, IL-2R, IL-6, IL-8, and IL-10 concentration. The cell count and cytokine concentration were presented as median with range, respectively; each point indicates one sample cell count or cytokine concentration. P < 0.05 was considered statistically significant difference between the non-severe and severe groups. *P < 0.05, ***P < 0.0001
Table 2.
Immunocyte comparison between non-severe and severe COVID-19 patients
| Normal range | Median (IQR) | P value | ||
|---|---|---|---|---|
| Non-severe (n = 159) | Severe (n = 56) | |||
| Leukocytes (× 109/L) | ||||
| Leukocyte | 3.5–9.5 | 5.66 (4–7.36) | 7.19 (5.15–12.1) | 0.0001* |
| Lymphocyte | 1.1–3.2 | 1.06 (0.83–1.49) | 0.475 (0.33–0.75) | < 0.0001* |
| Monocyte | 0.1–0.6 | 0.36 (0.25–0.46) | 0.30 (0.20–0.43) | 0.015* |
| Neutrophil | 1.8–6.3 | 3.8 (2.63–5.29) | 6.4 (4.018–10.49) | < 0.0001* |
| Eosinophil | 0.02–0.52 | 0.01 (0.00–0.04) | 0.005 (0.00–0.02) | 0.0003* |
| Basophil | 0–0.06 | 0.00 (0.00–0.01) | 0.00 (0.00–0.01) | 0.0543 |
| Lamphocyte subsets (/μL) | ||||
| CD3+ T lymphocyte | 1185–1901 | 644 (418–1014) | 242 (128–339) | < 0.0001* |
| CD3+ CD4+ T lymphocyte | 561–1137 | 383 (252–566) | 148 (83–210) | < 0.0001* |
| CD3+ CD8+ T lymphocyte | 404–754 | 218 (123–352) | 74 (43–120) | < 0.0001* |
| CD3− CD19+ B lymphocyte | 180–324 | 122 (79–183) | 78 (49–114) | < 0.0001* |
| CD19− CD56+ CD16+ NK cell | 175–567 | 116 (73–180) | 73 (42–101) | < 0.0001* |
P values indicate the differences between non-severe and severe patients. P < 0.05 was considered statistically significant
*Mann-Whitney test
Lymphocyte subset cytopenias are characteristics of COVID-19 patients, especially patients with severe symptoms
The median of CD3+ T, CD3− CD19+ B, and CD56+ CD16+ NK cell counts of COVID-19 patients was below the normal range (Table 2). The patients in the severe group were more affected as the CD3+ T cell count was decreased to 20% of lower end of the normal range, and CD3− CD19+ B cell and CD56+ CD16+ NK were nearly half the lower end of the normal range, respectively (Fig. 1g, j, k; Table 2). Lymphocyte subset analysis revealed that the median CD4+ T cell count of the non-severe group was slightly lower than the normal range, whereas that in the severe group was more reduced, representing a significant difference (Fig. 1h); CD8+ T cell counts were shown to be more affected, and the median is at 218/μL in the non-severe group and 74/μL (IQR 43–120/μL) in the severe group patients, which was dramatically decreased (Fig. 1i).
We also analyzed the distribution of lymphocyte subsets among total lymphocyte populations and found T cell percentage was decreased, and B cell and NK cell percentage were increased in the severe group, compared with the non-severe group (Fig. 1l).
Cytokine detection
The median of IL-1β and IL-10 was the same between the non-severe and severe groups. The median of IL-2R, IL-8, and IL-6 was higher in the severe group than in the non-severe group. The difference of IL-2R and IL-8 was not significant. The difference of IL-6 was significant; P value was 0.0418 (Fig. 1m–q, Table.S2). There was a small population of strong signals in each cytokine. To check if these high signals came from the same patients, we checked each strong signal and found that they were not always overlapped; most of them were from different patients (Table.S1). In the severe group, there were two very strong IL-6 signals (Fig. 1o); one of them has the highest IL-8 signal, but IL-1β and IL-10 were normal (Table.S1). This patient was extremely severe and stayed in the ICU for 36 days. Other strong signal patients also have longer duration of hospital stay compared with the median value of their own group (data not shown).
Correlations between duration of hospital stay period and immunological indexes
To explore the correlation between immunology indexes and severity of disease, we analyzed the correlations among patient duration of hospital stay period and immunological indexes by two-tailed Spearman’s correlation analysis. We found no significant correlation among duration of hospital stay period with age (r = 0.092, P = 0.201), absolute leukocyte count (r = − 0.023, P = 0.752), absolute neutrophil count (r = 0.007, P = 0.917), IL-6 (r = 0.089, P = 0.414), IL-8 (r = − 0.181, P = 0.094), and IL-2R (r = 0.203, P = 0.134), respectively. In contrast, analysis of duration of hospital stay and absolute lymphocyte count identified a significant negative correlation (r = − 0.187, P = 0.008). Further, absolute CD3+ T cell count (r = − 0.228, P = 0.002) and absolute CD3−CD19+ B cell count (r = − 0.151, P = 0.039) also have significant negative correlation with patients’ duration of hospital stay, respectively.
Discussion
This retrospective study described the immune characteristics of COVID-19 patients and explored the biomarkers predicting the severity of this disease. SARS-CoV-2 is the most recently discovered coronavirus, which causes COVID-19. The severity of disease may depend on both the age and immune status of an infected individual. The patients in the severe group were significantly older than those in the non-severe group, but there was no correlation between age and duration of hospital stay. Age may not be the important factor to determine the disease severity; the severity of COVID-19 depends more on the individual immune status, as lymphocyte level, including absolute T cell and B cell numbers. We identified significant negative correlation between lymphocyte count and duration of hospital stay period. Further, absolute CD3+ T cell count and absolute CD3−CD19+ B cell count also have negative correlations with duration of hospital stay.
SARS-CoV-2 enters the epithelial cells of the respiratory tract mucosal surface via ACE2. Infected epithelial cells release types I and III interferon and IL-1, which in turn activate three pathways that counter virus infection: autophagy, innate immunity, and adaptive immunity [14]. Myeloid lineage and NK cells are major effector cells in the innate immune system, while T and B lymphocytes function in adaptive immunity. Our analysis of peripheral leukocytes revealed that leukocyte numbers remained in the normal range, although the value was relatively high in the severe group (7.19 × 109/L, IQR 5.15–12.1 × 109/L) compared with the non-severe group (5.66 × 109/L, IQR 4–7.36 × 109/L), consistent with recently published data [1]. Innate immunity is the primary barrier when virus invades the human body, with neutrophils as the first responders to cytokines and chemokines released by the infected epithelial cells [15]. In the early stage of SARS-CoV-2 infection, neutrophils are involved in the immune response against the virus and the presence of higher levels of invading virus requires the production of more of this type of cell. Therefore, we suspected the increased level of neutrophils in COVID-19 patients, especially in the patients of the severe group. The increased neutrophils may be attributable to high virus loads and associated with the severity of disease. We analyzed the correlation between neutrophils and duration of hospital stay, but found no significant correlation between them. We speculate that lymphocytes played more important role than neutrophils in determining the severity of COVID-19.
Lymphopenia was observed in COVID-19 patients, especially in the patients of the severe group (Fig. 1c, Table 2). The mechanism of lymphocyte reduction in severe patients remains unclear. We conclude possible explanations for lymphocyte reduction as follows: (1) Lymphocyte migration—T cells are activated by virus infection and migrate to the infected location. For SARS-CoV-2, the respiratory tract, particularly the lung, is the main viral infection site. Activated CD8+ T cells migrate to the lung, where they recognize and kill the infected cells directly [16]. In our experiments, lymphocytes were detected in the peripheral blood, rather than the alveolar fluid. Analysis of lymphocytes from both sites should provide more informative data. (2) Lymphocyte depletion—If the first explanation is found not to be the case, it is possible that SARS-CoV-2-infected patients may have lymphocyte depletion. More severe disease would likely be associated with more serious depletion. Such depletion could be caused by excessive lymphocyte activity, due to virus infection or immune dysregulation. (3) Bone marrow suppression may also be the case.
In this study, we found IL-2R, IL-6, and IL-8 levels were increased in the severe group (Fig. 1m–q). High levels of IL-2R, lL-6, and IL-8 are common in patients with severe symptoms, which may be associated with “cytokine storm syndrome” (CSS). CSS is characterized by systemic inflammation, ferritinemia, hemodynamic instability, and multiple organ failure, and is caused by an uncontrolled immune response. Overactivation of immune responses causes sustained activation of lymphocytes and macrophages, which secrete numerous cytokines, leading to cytokine storm. Patients with severe symptoms have a high risk of CSS, due to high levels of cytokines, particularly IL-6, which are significantly different between the non-severe group and severe group. But we did not detect the significant correlation between IL-6 with length of hospital stay which may be due to limited case numbers. Still, IL-6 levels may be potential biomarkers that predict the progress of infection and inform the treatment decisions of physicians.
In summary, patients with COVID-19 severe symptoms have longer duration of hospital stay period and lymphopenia. Severity of COVID-19 is negatively correlated with lymphocyte number. Lymphocyte number may serve as prediction markers for disease prognosis. Further detailed test and patient follow-up are needed to clarify the immune characteristics of SARS-CoV-2 infection.
Electronic supplementary material
(DOCX 139 kb)
(DOCX 69 kb)
Authors’ contributions
Xiaolei Li: conceived study, analyzed data, and wrote the paper
Yang Liu: performed research, contributed methods
Junming Li and Lagen Wan: performed research
Longhua Sun, Jibin Yang, Fei Xu and Jing Zhou: performed research
Xinping Xu, Aiping Le: writing review and editing
Wei Zhang: project administration, writing review
Funding
Emergency Science and Technology Project for COVID-19 of Jiangxi Province (No. 202011-2); Nanchang Science and Technology Bureau (No. 20203306)
Data availability
Not applicable
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All research methods were approved by the medical ethics committee of the First Affiliated Hospital of Nanchang University and were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent to participate
All subjects were explained about the purpose, method, and potential risks and signed an informed consent form.
Consent for publication
All authors were informed of the submission.
Code availability
Not applicable
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaolei Li and Yang Liu contributed equally to this work.
Contributor Information
Xiaolei Li, Email: xiaoleili1225@163.com.
Aiping Le, Email: leaiping@126.com.
Wei Zhang, Email: zhangweiliuxin@163.com.
References
- 1.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. Lancet: China; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020. [DOI] [PMC free article] [PubMed]
- 4.Phan LT, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020. [DOI] [PMC free article] [PubMed]
- 5.Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wölfel R, Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, D., et al., Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA, 2020. [DOI] [PMC free article] [PubMed]
- 11.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Organization, W.H . Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected. 2020. [Google Scholar]
- 13.China, N.H.C.o., Novel coronavirus pneumonia diagnosis and treatment plan in China (Version 7). 2020.
- 14.Braciale TJ, Hahn YS. Immunity to viruses. Immunol Rev. 2013;255(1):5–12. doi: 10.1111/imr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol. 2017;8:550. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt ME, Varga SM. The CD8 T cell response to respiratory virus infections. Front Immunol. 2018;9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 139 kb)
(DOCX 69 kb)
Data Availability Statement
Not applicable