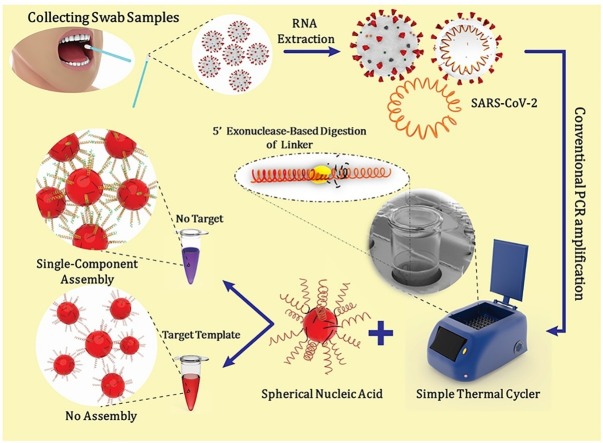
Graphical abstract
Keywords: Coronavirus disease 2019 (COVID-19), Post-PCR detection, 5’-exonuclease activity, Spherical nucleic acids (SNAs), Colorimetric detection, DNA-programmable assembly
Highlights
-
•
The amplification power of PCR is coupled with programmability of SNA assembly for detection of COVID-19.
-
•
A linker-based single-component assembly system based on E gene of SARS-CoV-2 RNA provided sensitive colorimetric response.
-
•
The palindromic linker is cleaved based on 5′-exonuclease activity in the course of PCR amplification.
-
•
The positive and negative viral COVID-19 samples are simply distinguished via different colors by naked eyes.
Abstract
Continuous identification of suspected infectious cases is crucial to control the recent pandemic caused by the novel human coronavirus SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). Real-time polymerase chain reaction (real-time PCR) technology cannot be implemented easily and in large scale in some communities due to lack of resources and infrastructures. Here, we report a simple colorimetric strategy derived from linker-based single-component assembly of gold nanoparticle-core spherical nucleic acids (AuNP-core SNAs) for visual detection of PCR products of SARS-CoV-2 ribonucleic acid (RNA) template. A palindromic linker is designed based on SARS-CoV-2 specific E gene to program the identical colloidal SNAs into large assemblies along with a distinct red-to-purple color change. The linker acts as a probe of SARS-CoV-2 RNA in conventional PCR reaction. In the presence of the correct template the palindromic linker, which is complementary to a short region within the target amplicon, is cleaved by 5′-exonuclease activity of deoxyribonucleic acid (DNA) polymerase. Cleavage of the palindromic linker during the amplification process inhibits the single-component assembly formation of SNAs. So, positive and negative viral samples produce simply red and purple colors in the post PCR colorimetric test, respectively. Evaluation of the samples obtained from cases with laboratory-confirmed SARS-CoV-2 infection revealed that our assay can rival with real-time PCR method in sensitivity.
1. Introduction
In the late December 2019, an ongoing spread of a viral pneumonia due to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported in Wuhan, China which quickly erupted into a worldwide pandemic of COVID-19 disease [[1], [2], [3], [4]]. The continuous rise in the number of infected cases and deaths as a result of COVID-19 caused this disease to rapidly become a global public health concern. Extensive screening for COVID-19 has been considered as a crucial strategy in controlling this infectious disease. Unfortunately, due to presenting non-specific influenza-like clinical symptoms or the likelihood of being asymptomatic at onset of illness [2,5,6], it is difficult to early diagnose SARS-CoV-2 infection through clinical symptoms. So, laboratory diagnosis of COVID-19 is the main current issue and challenge for public health systems to prevent the vast spread of the disease.
Computed tomography (CT) imaging is an important tool to diagnose or exclude a disease. Inflammatory lung diseases especially pneumonia cases [7] present abnormalities in lung CT images. Recently, analysis of chest CT images for finding abnormal features has also widely been used for both screening and diagnosis of COVID-19 [5,[8], [9], [10], [11], [12]]. But CT systems cannot specifically diagnose COVID-19 because of large nonspecific overlap of CT imaging features between different viral pneumonias with similar symptoms to COVID-19.
Molecular techniques based on nucleic acid hybridization strategies, in virtue of targeting specific pathogens, are more suitable for accurate diagnosis of COVID-19. The standard molecular method for COVID-19 diagnosis is real-time reverse transcription polymerase chain reaction (rRT-PCR) in which RNA from SARS-CoV-2 is detected. This technique is based on the amplification of complementary DNA (cDNA), synthesized from a conserved region of the viral genome, in a PCR reaction using primer pairs and a dual-labeled fluorophore-quencher probe targeting the cDNA sequence of interest. During the amplification cycles the probe is cleaved, resulting in the fluorescence signal. For molecular techniques to be applied, the complete genetic composition of pathogen (i.e. viral genome sequences) must be ascertained.
SARS-CoV-2 is a single-stranded, positive-sense RNA virus with a genome of approximately 29.8 kb in size [13] that has been enclosed inside an enveloped virion containing helical nucleocapsid [14]. The coronavirus genomes encode five major open reading frames (ORFs), including a 5′ frameshifted polyprotein (ORF1a/ORF1ab) and four structural genes that encode the four major structural proteins (the spike surface glycoprotein (S), small envelope protein (E), matrix protein (M), and nucleocapsid protein (N)) [13]. It has been proved that in SARS-CoV-2 RNA the same as other RNA viruses, the core residues of viral proteins are highly conserved within structural proteins [[15], [16], [17]].
The early sequencing of the SARS-CoV-2 (Accession number in Genbank: MN908947), allowed for the design of primers and probes needed for the progress of several rRT-PCR assays based on targeting different genomic regions, including the RdRp gene (RNA-dependent RNA polymerase, part of the ORF1ab gene), E gene and N gene regions for SARS-CoV-2 specific testing. Among all provided assays, the E gene and RdRp gene assays demonstrated higher detection sensitivity [10,[18], [19], [20]]. Despite the inherent advantages of real-time PCR, many communities are deficient in required laboratory infrastructures to implement real-time PCR technology. Moreover, the availability of the relevant real-time PCR reagent kits is not balanced with demand for high sample throughput diagnosis to cope with COVID-19 in the time of crisis. Therefore, further investigations for methods to detect SARS-CoV-2 nucleic acid are ongoing.
Nanothechnology relies on the binding between nanoparticles and target molecules to provide a detectable signal for detection of pathogens like SARS-CoV-2 [[21], [22], [23]]. Recently, a combination of plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) in a dual functional plasmonic biosensor has been used for detection of SARS-CoV-2 nucleic acid [24]. A chip with two-dimensional gold nanoislands functionalized by thiol-complementary DNA receptor of RdRp, ORF 1ab, or the E gene sequence can detect the selected sequences of SARS-CoV-2 via nucleic acid hybridization. The chip has the capacity of generating local PPT heat that improves the in situ hybridization kinetics of strands for specific nucleic acid detection.
A colorimetric assay for detection of SARS-CoV-2 has also been developed to detect the presence of the virus visually using the color change of functionalized AuNPs with thiol modified oligonucleotides specific for N-gene (nucleocapsid phosphoprotein) of SARS-CoV-2 [22]. In the presence of the target RNA of SARS-CoV-2, AuNPs modified with thiolated oligoucleotide for nucleocapsid phosphoprotein selectively agglomerate and a change in surface plasmon resonance with a red shift in the absorbance spectra happens. This bioassay was totally selective so that no distinct change in absorbance was found with Middle East respiratory syndrome coronavirus (MERS-CoV) RNA.
Spherical nucleic acids (SNAs) are a new class of nanostructures, primarily introduced by Mirkin and coworkers [25,26]. They were made by a dense shell and highly oriented nucleic acids covalently attached to the surface of gold nanoparticles. The binding of DNA to nanoparticles leads to materials which can act as building blocks in assembly processes. SNAs are good diagnostic tools because, through programmable base-pairing interactions, they can rationally guide the formation of macroscopic assemblies.
Palindromes, a famous concept in DNA, consist of nucleic acid sequences which read exactly the same from the 5′ end to the 3′ end on one strand and from 3′ to 5′ end on the complementary strand. These sequences have the fascinating property of inter-molecular hybridization with each other i.e. self-complementarity within the inverted repeated region [[27], [28], [29]]. Previous studies have shown that using the appropriate linker sequences, the assembly of both single- and binary-component SNAs can properly be programmed [30,31]. In linker-mediated assembly systems a free palindromic tail in the linker provides the indispensable need for programming single component self-assembly.
Recently, we developed a simple colorimetric method to detect human immunodeficiency virus-1 (HIV-1) viral RNA based on a single-component assembly system of AuNP-core SNAs [32]. Therein, a 6-nucleotide (6-nt) palindromic linker designed as an oligo-probe of HIV-1 genome. Thereinafter, we found that the kinetics of assembly formation in the linker-based single-component systems depended on the length and the guanine-cytosine content (GC content) of the palindromic tail (unpublished results). We came to the result that a linker with 4-nt palindromic sequence at the 5′ end led to the faster assembly kinetics and the more intense color change of SNAs than one with 6-nt palindromic sequence (100 % GC content). In the present work, it has been shown that nanotechnology can be applied to develop a new nanodiagnosis method for COVID-19. A simple PCR-based colorimetric strategy is proposed by taking the advantages of both amplification power of PCR and DNA-based programmability of SNA assembly to propose a reliable and affordable diagnostic assay for SARS-CoV-2 RNA.
A single-component assembly system based on a 4-nt palindromic linker was designed to provide a fast and intense colorimetric response after PCR run. The palindromic linker was designed such that it could hybridize exactly a sequence of the target (SARS-CoV-2 RNA) between the two PCR primers for E gen amplicon. During polymerization and synthesis of the complementary strand, the palindromic linker is disintegrated by 5′ to 3′ exonuclease activity of the polymerase. So the post-analysis of PCR product for visual colorimetric detection of SARS-CoV-2 RNA via our designed single component SNA system shows no assembly and consequent color change of dispersed SNAs if SARS-CoV-2 RNA target exists in the original sample. Otherwise, the red to purple color change is simply an indication of the absence of RNA target. To the best of our knowledge, this work is the first report of utilization of 5′-exonuclease activity in conventional PCR coupled with the programmability of SNA assembly for colorimetric detection of nucleic acids. The method presents a powerful diagnostic tool for COVID-19 in view of the higher availability of conventional PCR devices in the laboratories and can reduce the pressure on real-time RT-PCR tests.
2. Experimental section
2.1. Instrumentation and apparatus
Ultraviolet–visible (UV–vis) absorption spectra were recorded on a SPECORD 210 spectrophotometer (Analytik Jena) equipped with a Peltier temperature-controlled cell holder. All spectrophotometric measurements were implemented in a quartz ultra-micro cell cuvette with polytetrafluoroethylene (PTFE) stopper and 1.0 cm path length. Transmission electron microscopy (TEM) images were taken on a Philips EM 208S microscope operating at electron energy of 100 kV. Amplification processes were performed using LightCycler® 96 System from Roche Life Science for real-time RT-PCR and Applied Biosystems™ Veriti™ Dx 96-well from Thermo Fisher Scientific for conventional RT-PCR.
2.2. Chemicals and oligonucleotide sequences
All designed oligonucleotides, PCR primers and probes were synthesized by Bioneer, Inc. (Korea) with Bio-reversed phase (Bio-RP) purification (except for dual-labeled fluorophore-quencher probe for real-time RT-PCR that was purified by high-performance liquid chromatography (HPLC)). All oligonucleotide sequences are provided in Table 1 . Oligonucleotide with phosphorothioate modification of the backbone was used for functionalization of AuNPs to make SNAs. Hydrogen tetrachloroaurate (III) trihydrate, HAuCl4.3H2O, 99.99 % and sodium citrate were purchased from Sigma-Aldrich. Sodium chloride, 4-(2-hydroxyethyl) piperazine-1-ethanesulfonate (HEPES), Tris-Borate-EDTA (TBE) buffer (Molecular Biology Grade) and agarose powder for gel electrophoresis were purchased from Merck. Dried oligonucleotides were resuspended in RNase-, and DNase-free water, and ultrapure water was used to synthesize AuNPs. All buffers and other solutions were prepared in ultrapure water.
Table 1.
Oligonucleotide sequences used in this study.
| DNA name | Sequences and modifications (from 5′ to 3′) |
|---|---|
| E gene SNA-DNAa | A*A*A*A*A*A*A*A*AAAA GTTACACTAGCCATCCTTAC |
| E gene linker probeb | GCGCAGTAAGGATGGCTAGTGTAAC |
| Forward primer | ACAGGTACGTTAATAGTTAATAGCGT |
| Reverse primer | ATATTGCAGCAGTACGCACACA |
| E gene real-time probec | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ |
Phosphorothioate modification is denoted by “*”.
The underlined bases represent the palindromic sequence.
FAM: 6-carboxyfluorescein; BBQ: blackberry quencher.
2.3. Synthesis and characterization of SNA
AuNP cores were synthesized through the standard citrate reduction of HAuCl4 described elsewhere [33] with slight modifications. Briefly, aqueous solution of 1.0 mM HAuCl4 was first heated to boil for 20−30 min in a 100 mL round-bottom flask equipped with a condenser. Then, a total of 5.0 mL of 38.8 mM sodium citrate was rapidly added to the boiled HAuCl4 solution with vigorous stirring. The color of solution changed from pale yellow to deep red in about 1 min. The solution was allowed to reflux for an additional 12 min under vigorous stirring. Then the heating was turned off and the solution was allowed to cool to room temperature under stirring. This colloidal AuNP solution was stored at 4 °C for further use. The functionalization of the synthesized AuNP cores with the desired DNA (SNA-DNA in Table 1) was performed according to the low pH assisted method developed by Liu et al. [34,35] with some modifications. In brief, 500 μL of the already prepared AuNPs was mixed with 15 μL of the stock solution of SNA-DNA (100 μM, resuspended in nuclease-free water) in an autoclaved microcentrifuge tube by a brief vortexing. After 5 min, 10 μL of 500 mM trisodium citrate−HCl buffer (pH = 3) was added to the AuNP/DNA mixture so that the final concentration of sodium citrate was 10 mM. After 3 min incubation, the concentration of NaCl was brought to 50 mM by adding specific amount of NaCl solution. After another 3 min incubation, 40 μL of HEPES buffer (500 mM, pH 7.6) was added to the tube by a brief vortexing in order to bring the pH value of the AuNP solution back to neutral. To remove the excess amount of non-conjugated DNA, the mixture was centrifuged at 10,000 rpm (revolutions per minute) for 4 min, and the supernatant was removed, The pellet was resuspended in 10 mM HEPES solution. After 3 times repeating this post-conjugation washing, the pellet was resuspended in 300 μL of 10 mM HEPES (in order to adjust the concentration of SNAs around 15 nM for further use). The functionalization of AuNPs with phosphorothioate DNA is facile and rapid so that SNAs can be reproducibly prepared daily or weekly. Characterization of AuNP-core SNAs is available in Supplementary Information (SI) in details.
2.4. Sample collection and RNA extraction
All upper respiratory specimens used in this study were taken from suspected infectious patients who had referred to Farzan Molecular and Pathobiology Laboratory, Hamedan, Iran. According to the declaration of Farzan lab a record of the patients signed their informed consent form for research use is available. Dacron flocked swabs were used to collect samples from upper respiratory tract and the collected samples were kept in the viral transport medium until RNA extraction. Extraction of viral RNA from upper respiratory specimens was performed using the MagNA Pure 96 system (Roche) according to the instructions of manufacturer, and all extracted RNA samples were stored frozen at -80℃ until use.
2.5. Amplification process
Reverse transcription and PCR amplification were performed in one step for both the real-time and conventional RT-PCR assays using Add-Probe RT-PCR Kit (Product Code: 74311) and QIAGEN One-Step RT-PCR Kit (Cat. Number: 210210), respectively, according to the manufacturer’s instructions. The same specific primer pair was used for amplifying the E gene in both RT-PCR methods. In a 25-μL total volume of the amplification reaction, the primers were used at an optimized concentration of 0.4 μM, while the linker DNA probe for conventional RT-PCR and the fluorophore-quencher labeled probe for real-time RT-PCR were used at final concentrations of 0.1 and 0.2 μM, respectively. The real-time PCR program with a 20-min reverse transcription step at 50 °C and a 10-min denaturation step at 95 °C to activate the Taq DNA polymerase followed by 45 amplification cycles with a denaturation temperature of 95 °C for 10 s, annealing and extension temperatures of 55 °C for 45 s has been applied. Also, thermal cycling for conventional RT-PCR was performed at 50 °C for 30 min followed by 95 °C for 15 min for reverse transcription and DNA polymerase activation, respectively. Then 45 amplification cycles were run at 94 °C for 15 s, 55 °C for 45 s. Reaction components and optimized concentrations are shown in Table S2.
2.6. Gel electrophoresis
A part of PCR products (7.0 μL) were mixed with 1.0 μL DNA gel loading dye (6X) (Thermo Scientific) and the mixtures were then loaded to different lanes of an agarose minigel (5%) and run at 80 V for 90 min. The 5% agarose minigels were freshly prepared each time before application using 1X TBE buffer, and stained by SYBR Safe dye. TBE buffer (1X) was also used as running buffer. After separation of DNA fragments of the PCR products, the gel images were recorded using a Quantum VilberLourmat gel documentation system. A 50 bp DNA ladder (Invitrogen, Cat. number: 10416014) was used as DNA size marker to determine the band sizes.
2.7. Post-PCR colorimetric assay
The used clinically positive and negative samples had formerly been confirmed by SARS-CoV-2 RNA detection kit (PCR-Fluorescence Probing, EUL 0493-141-00. Da An Gene Co.) according to the manufacturer’s instructions in that ORF1ab and N genes were selected as amplification target regions. RNA extraction and conventional RT-PCR amplification (45 cycles) were carried out according to the procedures described in sections 2.4 and 2.5, respectively. Then, the post-PCR colorimetric assay was achieved by mixing 7.5 μL of the PCR product (which contains 2.5 mM MgCl2 from QIAGEN One Step RT-PCR Kit) with as-prepared SNAs in an eppendorf PCR tube followed by adding NaCl in a total volume of 30 μL (with the final concentration of SNAs, linker probe, MgCl2, and NaCl 7.5 nM, 25 nM, 0.625 mM, and 380 mM, respectively). After 10 min incubation at 25 °C, the color changes of the solutions were recorded using a digital camera Nikon D7200.
3. Results and discussion
3.1. Design principles of single-component system for SARS-CoV-2 nucleic acid detection
The first step in designing a single-component assembly system based on AuNP-core SNAs is the selection of a linker DNA to play both as oligo-probe for the nucleic acid target and self-assembler of SNA particles. The second step is to design a proper di-block oligonucleotide to form the nucleic acid shell on gold nanoparticles to construct SNAs. The structure of the linker consists of three main parts; a non-palindromic region (region 1, complementary to DNA sequences attached to AuNPs (SNA-DNA)), a palindromic region at the 5′ end (region 2, with self-complementary property), and a non-binding single-nucleotide region, called the flexor (generally adenosine, A) which separates the two regions of the linker and gives flexibility to the linker during assembly process [32,36,37].
To design a proper linker probe, the conserved and specific regions of E gene of SARS-CoV-2 (227-nt E gene sequence) were analyzed to look for appropriate palindromic sequences. The palindromic sequences, that could potentially be useful in designing a palindromic linker, are listed in Table S1. We chose the 4-nt 5′-GCGC-3′ sequence (because of faster assembly kinetics) and the 21 nucleotides behind it (towards the 5′ end) in the E gene region for designing a palindromic linker (see Table 1). The selected linker targets the conserved and specific regions of E gene of SARS-CoV-2 RNA and partially overlaps the TaqMan probe in real-time RT-PCR for E gene assay [19,20].
The di-block oligonucleotides comprising SNAs (that we named it SNA-DNA) consist of an attachment moiety of 12-adenines (poly-A) as anchor block and a programmable recognition block that is complementary to the non-palindromic region of the linker. The anchor block and the recognition block are separated by the spacer group, here 4 adenines, which pushes the recognition block away from nanoparticle surface. The anchor block is tandemly modified at the first-eight phosphates by phosphorothioate (PS) where a sulfur atom was substituted for one of non-bridging oxygen atoms on the phosphate backbone. The PS modification has been previously used to immobilize DNA on AuNPs to propose a fast, cost-effective and pre-treatment free method for DNA conjugation [32,35].
3.2. Simulation and characterization of the designed system
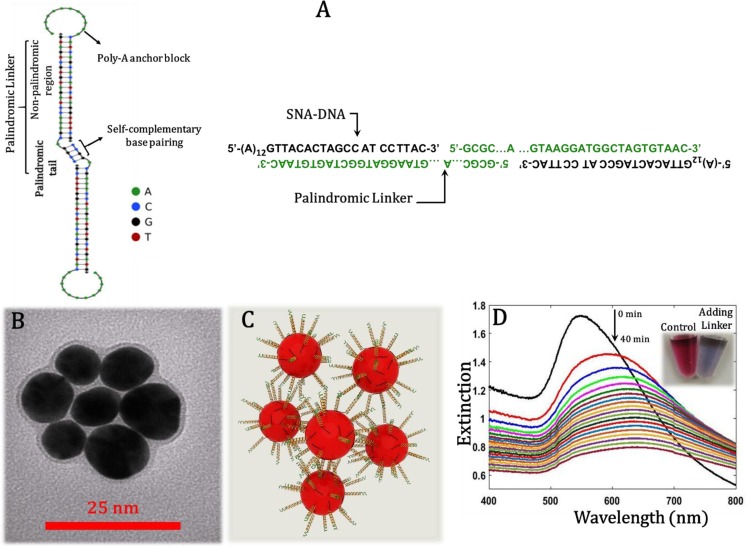
After selecting the linker and SNA-DNA oligonucleotide we used NUPAK Analysis Algorithms [[38], [39], [40]] to simulate hybridization between the selected palindromic linker and the designed SNAs. By using NUPACK we are able to determine whether the expected assembly structure between the interacting nucleic acid strands can be formed as the most dominant structure (with minimum free energy). Structural evaluation of sequences in NUPACK is done by a polymer graph of a secondary structure. Fig. 1 A depicts the polymer graph for the possible secondary structure with minimum free energy (MFE) between the predesigned strands of palindromic linker and SNA-DNAs. The obtained polymer graph demonstrates that the predicted self-complementary assembly structure can be successfully formed via the palindromic base pairing of the linker-hybridized adjacent SNAs.
Fig. 1.
Simulative and experimental characterization of the designed single-component assembly system. (A) The polymer graph of the assembly-guided structure simulated by NUPACK Analysis Algorithms (left) and DNA sequences forming single-component assembly (right). (B) TEM image of flower-like structure of single-component assembly (obtained from13-nm AuNP core SNAs and the used palindromic linker). (C) A 3D schematic illustration of flower-like structure of single-component assembly. (D) Time-dependent changes in the UV–vis spectra of 13-nm AuNP- core SNAs in the presence of palindromic linker over 40 min at 25 °C (60 μL of 10 mM HEPES buffer, pH = 7.6, containing 0.45 pmol SNAs, 5 pmol linker and 400 mM of NaCl). The inset is a photograph of the dispersed colloidal SNAs as control and the aggregated SNAs after 10 min of the addition of palindromic linker. Dispersed SNA particles are red, whereas aggregated SNA particles are purple. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
The self-assembly formation was then investigated by transmission electron microscopy (TEM) and spectrophotometric methods. The mixture of linker/SNAs was deposited on TEM grid, dried and imaged. Remarkably, TEM image demonstrated a flower-like configuration (Fig. 1 B) that revealed each particle (i.e. SNA) can be bound to every other SNA with equal possibility. A scheme of the single-component assembly of SNAs is also shown in Fig. 1 C for better visualization. Kinetics of the single-component self-assembly formation in the presence of the used palindromic linker was monitored by UV–vis spectroscopy since the colorimetric response of a DNA-based nanoparticle assembly system depends critically on the kinetics of the assembly formation. Fig. 1 D shows the absorbance spectra vs. time for self-assembly of 13-nm AuNP-core SNAs in the presence of the designed palindromic linker up to 40 min. The corresponding photographs of the dispersed colloidal SNAs as control and the aggregated SNAs after 10 min of the addition of palindromic linker are shown as inset. As shown in Fig. 1 D, upon addition of the linker the extinction spectra of colloidal SNAs decreases with time, accompanied by a red shift in the maximum of the AuNPs plasmon band to longer wavelengths. This indicates the formation of larger structures due to self-assembly between linker-hybridized adjacent SNAs through the free sticky ends of the palindromic linker. The kinetics of assembly formation is relatively fast and the color change is visible to naked eyes after 10 min. So, this single-component system can provide sensitive colorimetric responses.
3.3. 5′-exonuclease-based post-PCR assay and workflow for SARS-CoV-2 RNA colorimetric detection
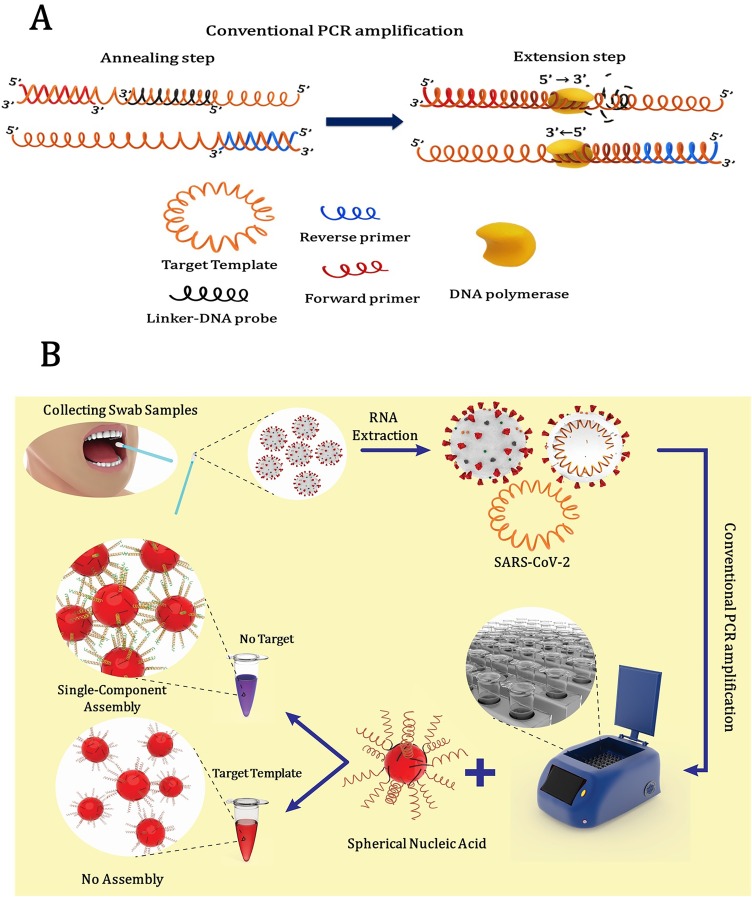
Exonuclease-based conventional PCR utilizes 5′→3′ exonuclease activity of DNA polymerase as the amplification reaction proceeds through the oligonucleotide probe (here the palindromic linker probe) which cleaves the probe during the extension phase of PCR. Our central idea, captured by the concept of DNA-programmable assembly, is that SNA assembly is inhibited when the linker probe is cleaved in the DNA polymerase amplification process. The used palindromic linker probe is a short unmodified single-stranded DNA (ssDNA) that is complementary to the sequence of bases in the targeted nucleic acid strand as PCR template. In the presence of target template, the pre-formed duplex of the linker and the target DNA, in the region between the two specific PCR primers of the E gene amplicon, subjected to 5′- exonuclease activity of DNA polymerase [41,42]. As PCR proceeds through the region where the linker is annealed, the linker DNA is cleaved and removed from its partner during the amplification reaction (Fig. 2A ).
Fig. 2.
A) Scheme of the 5′-exonuclease activity of the DNA polymerase in conventional PCR which cleaves the linker DNA associated with programmable assembly of SNAs. During the annealing step, linker-DNA probe and amplification primers are simultaneously hybridized to the target template and during the extension step, linker-DNA probe is displaced and cleaved by DNA polymerase. (B) Schematic diagram of the PCR-based colorimetric method workflow for SARS-CoV-2 RNA detection.
The total strategy is graphically represented in Fig. 2 B. In general, the upper respiratory tract samples are collected by nasopharyngeal, nasal or throat swabs from the patients. Then, viral RNA is extracted from these specimens. Isolated RNA is reverse transcribed to complementary DNA (cDNA) and subsequently specific regions of cDNA amplified by conventional PCR in the presence of the palindromic linker that is designed as a specific probe for SARS-CoV-2 RNA. In the presence of the target template, cleavage of the linker probe which is formerly annealed to the target template occurrs by the 5′-exonuclease activity of the DNA polymerase during the amplification process. Finally, an aliquot of the PCR products is added to the single-component SNA solution (see experimental section). After 5−10 min incubation at room temperature, the colorimetric response can be readily observed by the naked eye. The color of the SNAs keeps red for the positive samples and turns to purple in the case of negative samples.
3.4. Optimization of assay performance
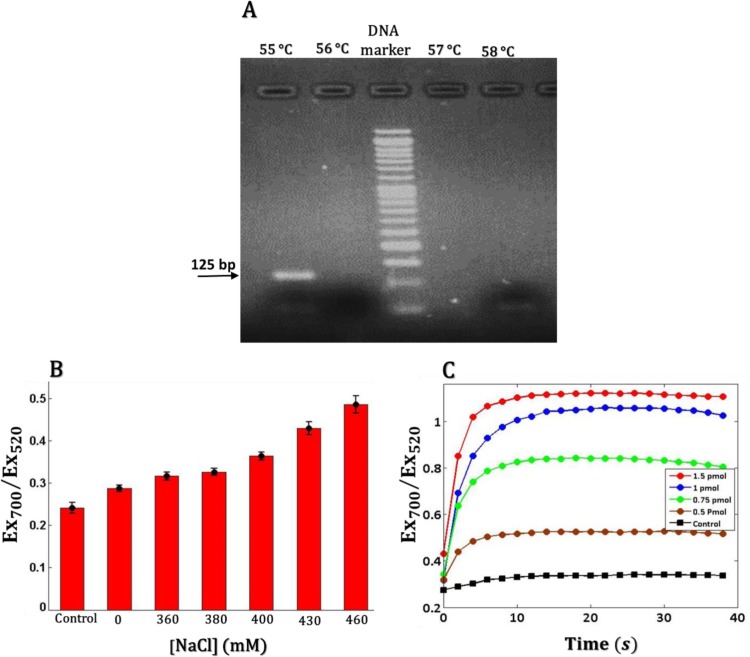
Since our assay provides a post-PCR colorimetric detection, the assay performance is dependent upon both optimum PCR amplification condition and the best colorimetric response associated with the single-component assembly of SNAs. The annealing temperature in PCR amplification process, the ionic strength of the SNA solution and the linker concentration in colorimetric detection are some important factors which must be optimized to get the best performance of the assay. As we targeted the same region in E gene which has been appointed in real-time RT-PCR method for detection of SARS-CoV-2, we used the same target-specific primer pairs [20]. The RNA templates were reverse transcribed using one-step RT-PCR Kit. Then, 45 cycles were performed at different annealing temperatures to amplify the reverse-transcribed DNA templates. Eventually, the PCR products were analyzed using gel electrophoresis (Fig. 3 A). The gel electrophoretic analysis of PCR products at the annealing temperatures of 55, 56, 57, and 58 °C, shows a unique-sharp band only in the lane corresponding to the annealing temperature of 55 °C. The details of PCR reaction components are available in Table S2.
Fig. 3.
Laboratory-based optimization. (A) Setting up the annealing temperature of the PCR reaction. PCR products for different annealing temperatures (written at the top of each lane) were checked by 5 % agarose gel electrophoresis. (B) Effect of NaCl concentration on the colloidal stability of SNAs. Extinction ratio Ex700/ Ex520 was monitored as a function of NaCl concentration in the presence of 0.625 mM Mg2+ (Note that the UV–Vis spectra were recorded 10 min after adding NaCl and control was pure SNAs. Error bars are standard deviations n = 3). (C) Evaluation of assembly growth via time dependency of Ex700/ Ex520 ratio at the various amounts of linker. The Ex700/ Ex520 ratios higher than 0.8 are coincident with the red-to-purple color change which could easily be observed by the naked eye.
In DNA-based nanoparticle assembly, the binding stability of the sticky ends that connects adjacent SNAs together, at a given temperature, mainly depends on the salt concentration. So, the colorimetric response followed by assembly formation due to self-complementarity of the free palindromic tails is also a function of ionic strength of the medium and the amount of the linker. We observed that with increasing ionic strength and linker concentration, the assembly growth rate also increased i.e. hybridization of the linker and SNAs became more favorable. Besides the need for stable interconnection of SNAs in the presence of linker for accurate diagnosis, colloidal stability of SNAs alone (with no linker) is another important aspect that also depends on salt concentration [35]. Addition of salt more than the salt tolerance of SNA in solution causes irreversible aggregation of SNAs before analysis. Therefore, the highest salt concentration that provides both colloidal SNAs stability and a fast colorimetric response sufficient for visual detection, in the presence and absence of the correct DNA template, respectively must be determined. On the other hand, the minimum amount of the linker which can sufficiently induce self-assembly of SNAs and observable color change, in the absence of target should also be obtained. This amount helps to get high sensitivity and minimum probability of false negative results in low viral load specimens. As mentioned in section 3.3, the linker is cleaved and removed by the 5′-exonuclease activity of DNA polymerase in conventional PCR amplification process. If high amounts of linker are used, in cases of low viral load samples most of the linker will stay intact and contributes in aggregation of SNA in post PCR colorimetric detection and leads to false negative results. Therefore, the lowest appropriate amount of linker to get a visual color change in the absence of the correct DNA template and avoid from false negative outcome in low viral load samples must be determined.
To optimize the concentration of NaCl to achieve the above aforementioned condition, the colloidal stability of SNAs at different concentrations of NaCl was first monitored spectrophotometrically. Since the PCR product contains 2.5 mM of MgCl2 (QIAGEN One Step RT-PCR Kit), the colloidal stability of SNAs was investigated at a given concentration of MgCl2 (15 μL of 2.5 mM MgCl2 in 60 μL total volume) while changing the concentration of NaCl up to 460 mM (Fig. 3 B). Higher extinction ratio at 700 nm over 520 nm (Ex700/Ex520 ratio) at greater than 400 mM NaCl, was associated with aggregation of SNAs and irreversible red-to-purple color change. Therefore, in order to ensure the stability of SNAs, the 380 mM was chosen as the optimum concentration of NaCl. Further, in order to optimize the amount of linker, the variations of Ex700/Ex520 ratio as a function of time were monitored for linker/SNA mixture with various amounts of linker (Fig. 3 C). The Ex700/Ex520 ratios higher than 0.8 leads to a visible color change with the naked eye. So we selected the 1.5 pmol linker (25 nM, total volume 60 μL) to get an easily observable color change in about 10 min. Overall, the optimum performance of our PCR-based colorimetric method was obtained under the following conditions: at the annealing temperature of 55 °C in the presence of 25 nM of the linker probe for PCR amplification, and 380 mM of NaCl for post-PCR colorimetric detection step in a total volume of 30 μL.
3.5. Verification and validation of the assay
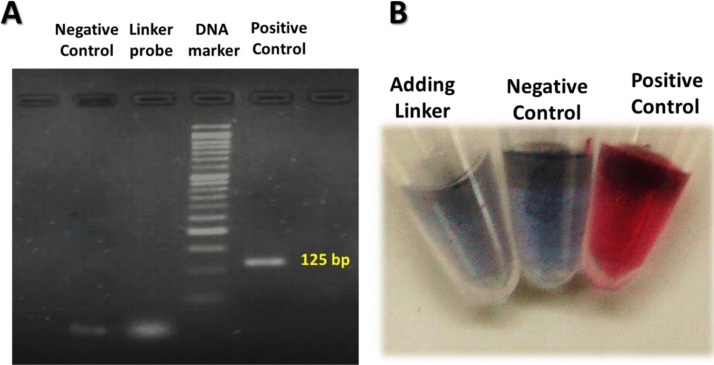
In this work, we used the same specific primers as those commonly used to amplify the E gene sequence of SARS-CoV-2 in real-time RT-PCR. Since the E gene assay has been often recommended for screening SARS-CoV-2, the specificity of the assay has already been established so that no cross-reactivity has been reported with other human coronaviruses [19]. To evaluate the ability of DNA polymerase to degrade the palindromic linker probe (which pre-hybridized with the target template) during amplification of the E gene amplicon, Post-PCR gel electrophoresis was run on 5 % agarose gel (Fig. 4 A).Whereas, only an intense band of the expected size for the E gene amplicon is observed in the positive control lane, a weak band is observable in the negative control lane with roughly the same size as the band of the linker probe lane. As a proof of the original idea, the colloidal SNAs were added to three PCR tubes containing 25 nM of the palindromic linker, the PCR products of negative and positive control, respectively. As shown in Fig. 4 B, a distinct red- to-purple color change is observed in the negative control and linker probe tubes, whereas for the positive control tube, the color of SNAs remains red. These data verify that, in the case of the positive control sample, during the amplification process the annealed linker probe is digested by the 5′-exonuclease activity of DNA polymerase in conventional PCR. However, the linker probe is intact in the negative control sample and the red to purple color changes in the negative control tube indicating the presence of the intact palindromic linkers during the amplification process.
Fig. 4.
Experimental verification of the basic idea behind the strategy. (A) The gel electrophoresis image of PCR products obtained for positive and negative control samples in the presence of 0.25 μM of the linker probe after 45 cycles of PCR. (B) Photograph of the color changes corresponding to the same PCR product samples after adding SNAs (picture was taken using a digital camera Nikon D7200). In gel electrophoresis experiment, 1 μM of the linker probe was implemented alongside the positive and negative control samples. Note, the positive control had been confirmed by real-time RT-PCR with the Ct value of 29.58, and the negative control was contained nuclease-free water instead of the target template.
The accuracy of the assay was evaluated in a local clinical lab (Farzan Molecular and Pathobiology Laboratory, Hamedan, Iran) using 9 samples of extracted RNA from patients including 3 positive, 3 low positive, and 3 negative samples that had already been analyzed by the real-time RT-PCR assay. Positive samples are those with positive real-time RT-PCR results targeting amplification of E gene with cycle threshold (Ct) values between 20–25. The low positive samples had been diluted to obtain Ct values between 34–38. All the samples were correctly diagnosed with the proposed assay. To test the assay reproducibility, the PCR product of a positive sample was aliquotted to three individual PCR tubes and frozen at -20 C. The three replicates were analyzed by freshly prepared SNAs on three different days. All three repetitions gave the same result.
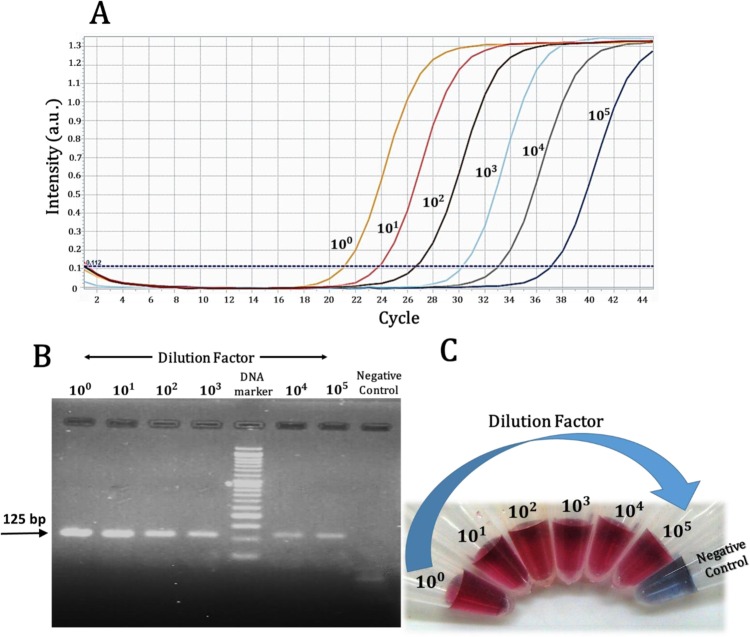
The sensitivity of the method was compared with real-time RT-PCR for the E gene region using serial dilutions (10-fold dilution series with five dilution steps) of the same extracted RNA samples (Table 2 ). The real-time RT-PCR and post-PCR colorimetric results have been reported in the form of Ct value and a Boolean answer + or −, respectively. The Ct value is the number of amplification cycles required to detect a real signal from the samples. At this point the target gene exceeds a threshold level and the fluorescence signal emitted during amplification exponentially intersects the threshold line (or background level). The Ct values are inversely proportional to the viral loads so that lower Ct values represents the higher viral RNA copy number. Generally, a Ct value of more than 40 is not clinically defined as a positive result. In contrast, A Ct value less than 40 is interpreted as positive for SARS-CoV-2 RNA. Except for sample 1, the detection performance of real-time RT-PCR in the fifth pipetting step (dilution factor of 105) was poor. Whereas, the post-PCR colorimetric method was applicable to all three samples in all five pipetting steps. For further comparison, the amplification curves of real-time RT-PCR, the agarose gel electrophoresis of conventional PCR products, and a photograph of the post-PCR colorimetric detection for five steps of 10-fold dilution series of sample 1 are represented in Fig. 5 A–C, respectively. Again the band related to the intact linker is obvious in the negative control lane. As the results imply, it can be claimed that the sensitivity of the post PCR colorimetric method has been increased at least 10 times compared to real-time RT-PCR method.
Table 2.
Sensitivity comparison of the real-time RT-PCR and post-PCR colorimetric assays.
| Dilution Factor | Resultsa |
|||||
|---|---|---|---|---|---|---|
| Real time RT-PCR (Ct value) |
Past-PCR colorimetric method |
|||||
| No. 1 | No. 2 | No. 3 | No. 1 | No. 2 | No. 3 | |
| 1 | 21.24 | 23.76 | 23.98 | + | + | + |
| 10 | 23.96 | 26.44 | 27.33 | + | + | + |
| 100 | 26.85 | 30.06 | 31.17 | + | + | + |
| 1000 | 28.31 | 33.07 | 33.97 | + | + | + |
| 10,000 | 33.12 | 36.86 | 37.60 | + | + | + |
| 100,000 | 37.21 | NAb | NA | + | + | + |
Positive and negative results are indicated by "+" and "−".
NA, Not Applicable.
Fig. 5.
Real-time and post PCR analysis of amplification products corresponding to the five-step 10-fold serial dilutions of the positive sample 1. (A) Real-time amplification curves (the dilution factors are shown at the top of each curve). (B) The gel electrophoresis images and, (C) Photographs showing colorimetric response (pure linker probe was implemented alongside the serial dilutions).
4. Conclusions
Overall, the proposed method is conceptually simple and technically applicable. The simultaneous 5′-exonuclease activity and polymerase activity of the DNA polymerase persuaded us to develop a simple PCR-based platform, which enables practical implementation of the DNA-programmable assembly of SNAs as a colorimetric molecular diagnostic method. This 5′-exonuclease activity allows PCR cycles to digest the assembler linker DNA while amplifying target template DNA. Hence, linker DNA digestion followed by assembly inhibition can be considered as a specific consequence of the presence of the target template DNA. The far-reaching distribution of conventional PCR equipment in most of clinical laboratories makes the conventional PCR an available and affordable platform for molecular diagnosis of viral pathogens. Therefore, our PCR-based colorimetric method can partly contribute to meet the demand for wide-scale screening for suspected infectious patients. Herein, our attention was focused on the detection of SARS-CoV-2 RNA since the COVID-19 outbreak has become a global challenge with apprehension. We designed a single-component assembly system based on 4-nt palindromic linker for probing the E gene region of SARS-CoV-2 RNA that provides a fast and intense colorimetric response as post-PCR detection. The results convincingly showed that our method could rival at sensitivity with real-time RT-PCR method. Considering the high distribution of palindromic sequences in all genomes, we hope that this proposed strategy could be extended to detect other nucleic acid targets.
CRediT author statement
Abbas Karami designed and performed the experiments, drafted the manuscript and designed the figures.
Masoumeh Hasani supervised the project and wrote the current version of the manuscript.
Farid Azizi Jalilian designed and directed the molecular experiments that were conducted.
Razieh Ezati performed molecular experiments such as RNA extraction and PCR reactions.
Declaration of Competing Interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are grateful to Bu-Ali Sina University Research Council for financial support.
Biographies
Masoumeh Hasani received her PhD in Analytical Chemistry at Shiraz University, Shiraz, Iran. She is currently an academic member of Bu-Ali Sina University, Hamedan Iran. Her research interests include kinetic and thermodynamic studies of complexation equilibria, nanoscience including DNA-based sensors, chemical and bio nanosensors.
Abbas Karami is currently a Ph.D. student at Bu-Ali Sina University, Hamedan, Iran. His research interests are engineering of DNA-based biosensors for molecular diagnosis and chemometrics.
Farid Azizi Jalilian is currently an academic member of Hamedan University of Medical science. His research interests are molecular virology, diagnostic, Vaccine, cancer.
Razieh Ezati received her M.S from National Institute of Genetic Engineering and Biotechnology, Tehran, Iran. She currently works in Department of Molecular Diagnosis, Farzan Molecular and Pathobiology Laboratory, Hamedan, Iran.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2020.128971.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Zumla A., Hui D.S., Azhar E.I., Memish Z.A., Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet. 2020;395:e35–e36. doi: 10.1016/S0140-6736(20)30305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020;12:1–6. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. China Novel Coronavirus, I., Research, T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Vol. 395. Elsevier; 2020. pp. 1054–1062. (Clinical Course and Risk Factors for Mortality of Adult Inpatients With COVID-19 in Wuhan, China: a Retrospective Cohort Study). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiting P., Singatullina N., Rosser J. Computed tomography of the chest: I. Basic principles. Bja Educ. 2015;15:299–304. [Google Scholar]
- 8.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x. Clinical characteristics of 2019 novel coronavirus infection in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 11.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acheson N.H. John Wiley & Sons, Inc; 2011. Fundamentals of Molecular Virology. [Google Scholar]
- 15.Patel H., Kukol A. Evolutionary conservation of influenza A PB2 sequences reveals potential target sites for small molecule inhibitors. Virology. 2017;509:112–120. doi: 10.1016/j.virol.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Hulswit R.J., Lang Y., Bakkers M.J., Li W., Li Z., Schouten A. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Mo S. Structural genomics and interactomics of 2019 Wuhan novel coronavirus, 2019-nCoV, indicate evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoecklin S.B., Rolland P., Silue Y., Mailles A., Campese C., Simondon A. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Euro Surveill. 2020;25(6) doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman V., Bleicker T., Brünink S., Drosten C., Zambon M. World Health Organization; 2020. Diagnostic Detection of 2019-nCoV by Real-time RT-PCR. Jan, 17. [Google Scholar]
- 21.Rabiee N., Bagherzadeh M., Ghasemi A., Zare H., Ahmadi S., Fatahi Y. Point-of-use rapid detection of sars-cov-2: nanotechnology-enabled solutions for the covid-19 pandemic. Int. J. Mol. Sci. 2020;21:5126. doi: 10.3390/ijms21145126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moitra P., Alafeef M., Dighe K., Frieman M., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talebian S., Wallace G.G., Schroeder A., Stellacci F., Conde J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020;15:618–621. doi: 10.1038/s41565-020-0751-0. [DOI] [PubMed] [Google Scholar]
- 24.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 25.Mirkin C.A., Letsinger R.L., Mucic R.C., Storhoff J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 26.Cutler J.I., Auyeung E., Mirkin C.A. Spherical nucleic acids. J. Am. Chem. Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 27.Lu L., Jia H., Dröge P., Li J. The human genome-wide distribution of DNA palindromes. Funct. Integr. Genomics. 2007;7:221–227. doi: 10.1007/s10142-007-0047-6. [DOI] [PubMed] [Google Scholar]
- 28.Lewis S.M., Chen S., Strathern J.N., Rattray A.J. New approaches to the analysis of palindromic sequences from the human genome: evolution and polymorphism of an intronic site at the NF1 locus. Nucleic Acids Res. 2005;33(22):e186. doi: 10.1093/nar/gni189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruss A.D., Ross H.F., Novick R.P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park S.Y., Lytton-Jean A.K., Lee B., Weigand S., Schatz G.C., Mirkin C.A. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 31.Laramy C.R., O’Brien M.N., Mirkin C.A. Crystal engineering with DNA. Nat. Rev. Mater. 2019;4:201–224. [Google Scholar]
- 32.Karami A., Hasani M. A palindromic-based strategy for colorimetric detection of HIV-1 nucleic acid: single-component assembly of gold nanoparticle-core spherical nucleic acids. Anal. Chim. Acta. 2020;1102:119–129. doi: 10.1016/j.aca.2019.12.050. [DOI] [PubMed] [Google Scholar]
- 33.Liu J., Lu Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Servos M.R., Liu J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 2012;134:7266–7269. doi: 10.1021/ja3014055. [DOI] [PubMed] [Google Scholar]
- 35.Zhou W., Wang F., Ding J., Liu J. Tandem phosphorothioate modifications for DNA adsorption strength and polarity control on gold nanoparticles. ACS Appl. Mater. Interfaces. 2014;6:14795–14800. doi: 10.1021/am504791b. [DOI] [PubMed] [Google Scholar]
- 36.Nykypanchuk D., Maye M.M., Van Der Lelie D., Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlane R.J., Lee B., Hill H.D., Senesi A.J., Seifert S., Mirkin C.A. Assembly and organization processes in DNA-directed colloidal crystallization. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10493–10498. doi: 10.1073/pnas.0900630106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dirks R.M., Pierce N.A. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 2003;24:1664–1677. doi: 10.1002/jcc.10296. [DOI] [PubMed] [Google Scholar]
- 39.Dirks R.M., Pierce N.A. An algorithm for computing nucleic acid base‐pairing probabilities including pseudoknots. J. Comput. Chem. 2004;25:1295–1304. doi: 10.1002/jcc.20057. [DOI] [PubMed] [Google Scholar]
- 40.Dirks R.M., Bois J.S., Schaeffer J.M., Winfree E., Pierce N.A. Thermodynamic analysis of interacting nucleic acid strands. Siam Rev. 2007;49:65–88. [Google Scholar]
- 41.Roh K., Kim D.-M., Lee E.H., Kim H., Park H.S., Jang J.-H. A simple PCR-based fluorometric system for detection of mutant fusion DNAs using a quencher-free fluorescent DNA probe and graphene oxide. Chem. Commun. 2015;51:6960–6963. doi: 10.1039/c5cc00263j. [DOI] [PubMed] [Google Scholar]
- 42.Navarro E., Serrano-Heras G., Castaño M., Solera J. Real-time PCR detection chemistry. Clin. Chim. Acta. 2015;439:231–250. doi: 10.1016/j.cca.2014.10.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.