Graphical abstract
A highly sensitive SERS assay is proposed for detecting DENV gene fragment via a cascade signal amplification strategy of localized catalytic hairpin assembly and hybridization chain reaction.
Keywords: SERS assay, DENV gene, Cascade signal amplification, Localized catalytic hairpin assembly, Hybridization chain reaction
Highlights
-
•
A highly sensitive SERS assay of DENV gene is proposed via a cascade signal amplification strategy of LCHA and HCR.
-
•
The SERS assay possesses high sensitivity with a linear calibration curve from 1 fM to 10 nM and a LOD low to 0.49 fM.
-
•
The SERS assay can provide a reliable tool for early diagnosis of virus infections.
Abstract
Pathogenic viruses with worldwide distribution, high incidence and great harm are significantly and increasingly threatening human health. However, there is still lack of sufficient, highly sensitive and specific detection methods for on-time and early diagnosis of virus infection. In this work, taking dengue virus (DENV) as an example, a highly sensitive SERS assay of DENV gene was proposed via a cascade signal amplification strategy of localized catalytic hairpin assembly (LCHA) and hybridization chain reaction (HCR). The SERS assay was performed by two steps, i.e., the operation of cascade signal amplification strategy and the following SERS measurements by transferring the products on SERS-active AgNRs arrays. The sensitivity of the cascade signal amplification strategy is significantly amplified, which is 4.5 times that of individual CHA, and the signal-to-noise ratio is also improved to 5.4 relative to 1.8 of the CHA. The SERS sensing possesses a linear calibration curve from 1 fM to 10 nM with the limit of detection low to 0.49 fM, and has good specificity, uniformity and recovery, which indicates that the highly sensitive SERS assay provides an attractive tool for reliable, early diagnosis of DENV gene and is worth to be popularized in a wide detection of other viruses.
1. Introduction
Viruses as a class of pathogens cause diseases in animals, plants and humans, which sometimes lead to severe morbidity and mortality [[1], [2], [3], [4]]. In recent decades, several viruses have spread from animals to humans and triggered international pandemics, such as dengue virus (DENV) [5], avian influenza virus (AIV) [6], severe acute respiratory syndrome-associated coronavirus (SARS-CoV) [7,8], Middle East respiratory syndrome coronavirus (MERS-CoV) [9], and the 2019 severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) [10], causing a massive loss of life. A particularly important action to be taken in the fight against virus is the accurate and early detection of virus infection, so that to prevent and control the virus pandemics and avoid the loss of life and property [[11], [12], [13]]. Taking DENV as an example, it can cause severe illness such as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) with more than 100 million infections annually [14]. Since there is less information about pathognomonic features of DENV infection from other febrile illnesses, the diagnosis of this virus in early stages of disease is very crucial [[15], [16], [17]]. The traditional pathogen detection methods are based on reverse transcriptase polymerase chain reaction (RT-PCR), which offers sensitive assay of virus genes [18,19]. However, the traditional PCR-based assays are usually limited by complex operations, time consuming, requirement of professionals, and the introduction of contaminating nucleic acids which can also be amplified during the test and leads to false positive results [[20], [21], [22]]. In response to the urgent need to develop new and more reliable detection methods, currently some recent advances in nanosensing have opened up new opportunities for developing novel optical and electrical methods for DENV detection, such as fluorescence [[23], [24], [25]], surface plasmon resonance (SPR) [26], surface-enhanced Raman spectroscopy (SERS) [27,28], and electrochemistry [29,30]. Among them, SERS as a molecular fingerprint spectrum which can achieve ultrasensitive detection even to single molecular level is considered to be a potential and powerful analysis tool [[31], [32], [33]]. For instance, Huh et al. reported an on-chip SERS by integrating capture probe immobilized AuNPs and dye labeled target DNA for the detection of DENV-related nucleic acid sequences with a detection limit of 30 pM [34]. Vo-Dinh’ s group proposed a SERS bioassay-on-chip to achieve a detection limit low to pM level, which was operated basing on the change in the distance between the Raman molecules on the molecular beacons and the metal surface before and after DENV gene assay [27,35]. Paul et al. developed a bioconjugated SERS probe for the detection of DENV gene based on the plasmon coupling in nanoassembly [36]. More recently, Singh’ s group reported a SERS-based diagnosis and classification of DENV positive, negative, and healthy clinical blood samples by testing DENV gene on silver nanorods array substrates [28]. Though several SERS-based DENV gene assays were proposed and shown good sensitivity, the more sensitive and convenient SERS sensing strategies are still required urgently in order to achieve early detection of DENV infection.
Recently, various signal amplification strategies have been introduced into the detection of virus-associated nucleic acids [[37], [38], [39]], including nuclease-assisted rolling circle amplification (RCA), duplex-specific nuclease (DSN), enzyme-free catalyzed hairpin assembly (CHA) and hybridization chain reaction (HCR), etc. Among them, CHA and HCR are isothermal enzyme-free nucleic acid circuits, which require only the facile design of reacting hairpins. As two efficient and homogeneous amplification strategies, CHA assembles into a series of double-stranded DNA (dsDNA) by the target-catalyzed hairpins, while HCR involves an autonomous cross-hybridization of hairpins to generate a long nicked dsDNA [40,41]. Meanwhile, various cascaded enzyme-free strategies have been constructed with significantly amplified sensing performance, including HCR-HCR [42], CHA-CHA [43], and CHA-HCR [44,45]. Furthermore, a localized system, named localized catalytic hairpin assembly (LCHA), has been deemed to possess much more possibilities to be triggered by adjacent molecules, thereby better controlling over directing reaction pathways and enhancing the sensitivity and accuracy of assay [[46], [47], [48]]. Thus, the SERS assays integrating signal amplification strategies are expected to achieve ultrasensitive SERS sensing of DENV genes. However, so far few SERS strategies with signal amplifications have been proposed, and to the best of our knowledge, there is no SERS assay of DENV gene by combining two cascaded amplification strategies.
In order to develop novel SERS strategy for ultrasensitive detection of DENV gene, herein, a cascade enzyme-free signal amplification strategy integrating the localized catalytic hairpin assembly (LCHA) and hybridization chain reaction (HCR) was proposed for SERS assay of DENV gene fragment corresponding to nucleotides 10699-10723 (GenBank No. KT187556.1) of the DENV serotype 2 which is one of the four Dengue virus serotypes [24]. The SERS assay was performed by two steps. Firstly, the cascade enzyme-free signal amplification strategy of LCHA and HCR was executed in a PCR tube. Secondly, the SERS assay was performed by transferring the HCR product on AgNRs arrays with the surface blocked by 6-mercapto-1-hexanol (MCH). The feasibility of the specially designed cascade signal amplification strategy was characterized by electrophoresis, and the improved sensing performance of the cascade signal amplification strategy was investigated by comparing the SERS assays based on the individual CHA, LCHA, and the cascade amplification strategy of LCHA and HCR. Additionally, the MCH blocking was also optimized to obtain the optimal sensing conditions. Finally, the performance of the proposed SERS strategy on detecting DENV gene was investigated, such as calibration curve, limit of detection, specificity and recovery rate.
2. Experimental section
2.1. Reagents
All the single-stranded DNAs (ssDNAs) were synthesized and purified by Sangon Biotech (Shanghai, China), and their names, name abbreviations, nucleotide sequences and roles are listed in Table S1. The special designs of the base sequences of the ssDNAs were described in the Supplementary materials. The 6-mercapto-1-hexanol (MCH, ≥97 %) was purchased from Sigma-Aldrich (Shanghai, China). The magnesium chloride (MgCl2·6H2O, ≥98 %) and tris (hydroxymethyl) aminomethane (C4H11NO3, ≥98 %) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China) to prepare TM buffer solution (10 mM Tris-HCl and 1 mM MgCl2, pH 8.0) for DNA hybridization. All solutions were prepared with ultrapure Millipore water (18.2 MΩ cm).
2.2. Apparatus
A confocal Raman microscope (InVia, Renishaw, England) was utilized to record SERS spectra under conditions of 633 nm incident light (1% laser power), 20× objective and 1 s integration time. Unless specified description, the SERS assay of each sample was achieved by recording the SERS signals at ten spots on the substrate to obtain an averaged SERS spectrum with the error bar representing the standard error. Before statistical analysis, the spectral baselines were subtracted by the software Wire 4.0. A Bio-Rad electrophoretic apparatus (Bio-Rad Laboratories, USA) was used to operate the polyacrylamide gel electrophoresis (PAGE) in TM buffer for 110 min with the voltage of 80 V, and the electrophoretic patterns were imaged by a GeneSys system (Syngene, UK).
2.3. SERS sensing strategy via cascade LCHA and HCR
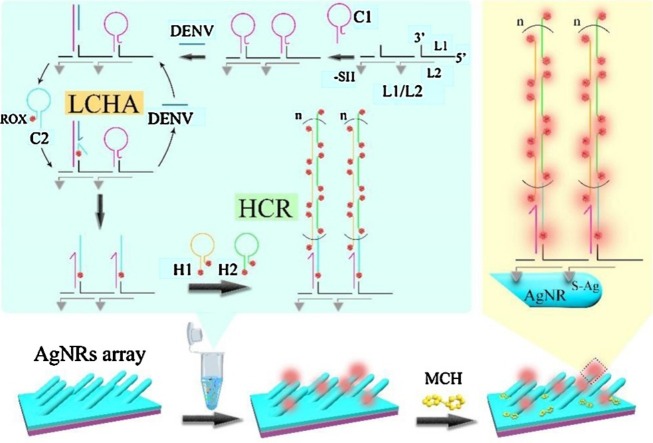
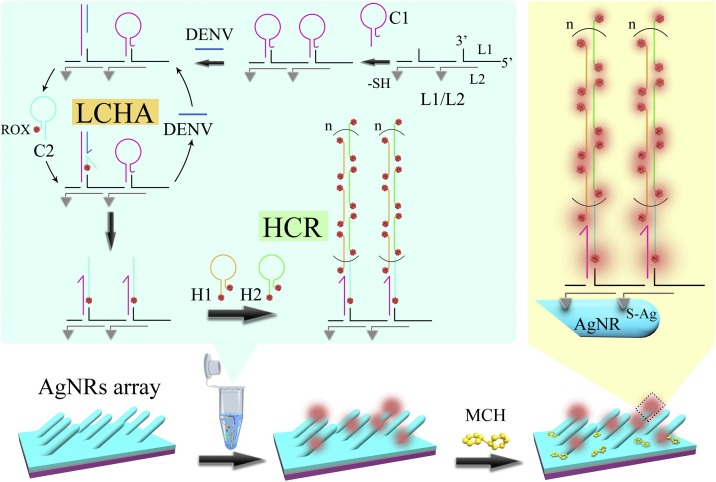
The SERS assay of DENV gene fragment was performed by two steps as shown in Scheme 1 . In the first step, the cascade enzyme-free signal amplification strategy of localized catalytic hairpin assembly (LCHA) and hybridization chain reaction (HCR) was carried out in a PCR tube as the green shadow-marked sketch map shown in Scheme 1. In the second step, the HCR products were dropped onto SERS-active AgNRs arrays and the AgNRs were then blocked by MCH, followed by SERS measurements. The AgNRs array was prepared by physical vapor deposition (PVD) to coat 20 nm Ti film, 200 nm Ag film, and the following oblique angle deposition (OAD) to deposit 2000 nm AgNRs layer from the bottom up on a glass slide, followed by molding a premopolydimethylsiloxane (PDMS) layer with 4 × 10 arrayed wells (4 mm in diameter, 1 mm in height) on the AgNRs array substrate. The arrayed circular sensing interface in each well can not only facilitate the detection with uniform sampling areas but also improve the detection reproducibility. Fig. S1 shows a SEM image of AgNRs array and a photo of PDMS wells-patterned AgNRs array substrate, and the preparation process in details can be found in our previous papers [[49], [50], [51]].
Scheme 1.
Schematic illustration of SERS assay of DENV gene via a cascade enzyme-free signal amplification strategy of localized catalytic hairpin assembly (LCHA) and hybridization chain reaction (HCR).
In the first step (i.e., the standard protocol of cascade signal amplification strategy of LCHA and HCR), equivalent amounts (10 μM, 2 μL) of L1 and L2 were incubated in a PCR tube for 60 min to assemble L1/L2 double-stranded DNAs (i.e., DNA skeleton) with the 20 nucleotides (20-nts) of toeholds (20 adenines (Ploy A) marked in underline in Table S1) placing on the double strands at equal distance. Unless otherwise specified, the incubation temperature of the DNA hybridization was 25 °C. Then, the hairpin DNA C1 (10 μM, 2 μL) was added into the tube and incubated with the L1/L2 for 60 min to load C1 on L1/L2 by binding the toeholds via the hybridization between Ploy A and the Ploy T (20 thymines of C1 marked in red in Table S1). Subsequently, the ROX-labeled C2 (10 μM, 2 μL) and 2 μL DENV gene fragment (DENV) with a certain concentration were added into the tube together and incubated for 90 min. During this period, the C1 hybridized with DENV and released the sticky-end of C1 (bases of C1 marked in italics in Table S1) to trigger the catalytic hairpin assembly between C1 and C2. As a result the DENV was released into the solution for cyclic utilization. Since the C1 hairpins were localized on the L1/L2, the DENV-triggered enzyme-free localized catalytic hairpin assembly (LCHA) was achieved. Then, equivalent amounts (10 μM, 2 μL) of hairpin DNAs H1 and H2 with ROX labeling at both 3’- and 5’-ends were added into the tube and incubated for 90 min. During this period the hybridization chain reaction (HCR) was triggered by the sticky-ends of the C1-C2 complex (bases of C2 marked in bold in Table S1) on the L1/L2 and then the H1-H2 DNA concatamers containing numerous ROX molecules were formed. At this point, the cascade enzyme-free signal amplification strategy of LCHA and HCR was achieved.
In the second step, the final solution in first step was dropped in the wells (20 μL per well) on the AgNRs array substrate and incubated at 25 °C for 3 h in order to immobilize the DNA concatamers-decorated L1/L2 onto the AgNRs by the strong Ag-S covalent bonds. After TM buffer washing, the nonspecifically adsorbed DNAs were removed by adding 20 μL of 1 mM MCH into the well and incubating for 10 min. The DNA concatamers-immobilized AgNRs array substrate (i.e., the magnified sketch map shown in Scheme 1) was obtain after water washing and the SERS measurements were then performed after air-drying of the substrate.
To illustrate the advantages of the cascade signal amplification strategy of LCHA and HCR, the SERS assays of 10 nM DENV by three types of signal amplification strategies were investigated, i.e., individual CHA (without the local effect), individual LCHA, and the cascade signal amplification strategy of LCHA and HCR. Both the individual CHA and LCHA were performed in the absence of H1 and H2 (i.e., HCR). For LCHA-based SERS sensing, the protocol was the same as the proposed cascade signal amplification strategy but without the HCR. For CHA-based SERS sensing, the performance was different from the LCHA introduced in the experimental section, i.e., without the usage of L1/L2. Briefly, 10 nM DENV were incubated with the mixture including equivalent amounts (10 μM, 2 μL) of thiol-modified C1 (C1-SH) and ROX-labeled C2 at 25 °C for 150 min to achieve the CHA, and then the C1-C2 complexes in the final solution (20 μL) were transferred in a well on the AgNRs array substrate. After the same MCH blocking, the SERS measurements were performed.
3. Results and discussion
3.1. Characterization of the cascade signal amplification strategy
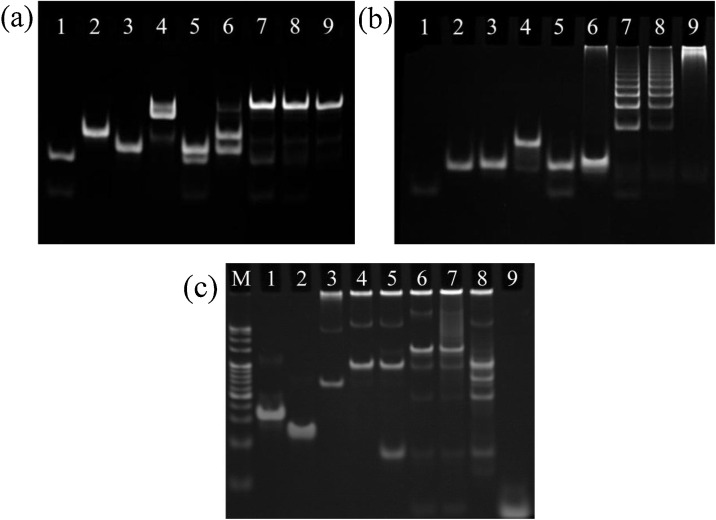
The feasibility of the cascade signal amplification strategy was characterized by the PAGE. The catalytic hairpin assembly (CHA) of C1 and C2 in the presence of DENV was characterized first and the electrophoretic pattern is shown in Fig. 1 a. The electrophoretic stripes of individual DNEV, C1 and C2 are shown in the lanes 1–3, respectively. When C1 was incubated with DENV (C1 + DENV in lane 4), a bright strip can be observed with a much lower migration rate than the single strands of C1 and DENV in lanes 1 and 2 respectively, indicating the formation of the DENV-C1 complex. In contrast, the mixture of C2 and DENV in lane 5 (C2 + DENV) shows two distinct strips similarly as the two strips in lanes 1 and 3, which means C2 cannot hybridize with DENV directly. The two obvious strips belonging to C1 and C2 respectively shown in lane 6 (C1 + C2) indicate that the hairpin C1 almost did not hybridize with the hairpin C2. Compared to lane 6, lane 7 (C1 + C2 + DENV) including the mixture of equivalent amounts of DENV, C1 and C2 illustrates that the DENV triggered the hybridization between C1 and C2 and simultaneously released DENV by referencing the strip shown in lane 1. The distinct strip of C1-C2 complex is also observed from lane 8 or lane 9 by adding 0.5 times or 0.1 times DENV relative to the amount of DENV in lane 7. The results show in lanes 7–9 indicate the effective CHA of C1 and C2 triggered by DENV. Fig. 1b shows the electrophoretic pattern of HCR characterization. In order to imitate the trigger of HCR by the sticky-end of the C1-C2 complex, a specially designed T-C2 including the same sticky-end was used to characterize the feasibility of HCR. The lanes 1–3 present the electrophoretic strips of T-C2, H1 and H2, respectively. The lanes 4 and 5 indicate that the T-C2 can hybridize with H1 rather than H2. The lane 6 demonstrates that the hairpin H1 almost did not hybridize with the hairpin H2. The HCR strips shown in lanes 7–9 indicate the efficient performance of HCR, which is similar as the lanes 7–9 of CHA shown in Fig. 1a. Therefore, the PAGE characterizations confirm the feasibility and high-efficiency of the CHA and HCR. Fig. 1c shows the electrophoretic pattern of the cascade signal amplification. The lanes 1 and 2 present the single electrophoretic stripes of L1 and L2, respectively, and the mixture of L1 and L2 (L1/L2) in lane 3 exhibits a clear stripe with a lower migration rate relative to individual L1 or L2, which indicates the formation of the L1-L2 double-stranded DNAs (i.e., DNA skeleton), and the obvious stipe in the sample injection port means some DNA skeletons with large structures were also formed. The slowly moved stipe in lane 4 (L1 + L2+C1) of the mixture of C1 and L1/L2 illustrates the load of C1 on the L1/L2. The mixture of L1/L2, C1 and C2 in lane 5 (L1 + L2+C1+C2) shows two obvious strips belonging to L1 + L2+C1 and C2 respectively, indicating that C2 almost did not hybridize with the C1 loaded on L1/L2. Once the mixture of L1 + L2+C1 and C2 was incubated with DENV, the bright strip in lane 6 (L1 + L2+C1+C2+DENV) demonstrates the occurrence of LCHA reaction and the weak strip with the same position of DENV in lane 9 indicates the release of DENV during the LCHA reaction. The following cascaded HCR in lane 7 (L1 + L2+C1+C2+H1+H2+DENV) shows the successful operation of HCR by comparing to the mixture (L1 + L2+C1+C2+H1+H2) in lane 8 without DENV.
Fig. 1.
PAGE characterizations of the CHA, HCR and cascade signal amplification strategy of LCHA and HCR. (a) Electrophoretic pattern of the CHA characterization (10 % PAGE). Lane 1: DNEV; lane 2: C1; lane 3: C2; lane 4: C1 + DENV; lane 5: C2 + DENV; lane 6: C1 + C2; lane 7: C1 + C2 + DENV; lane 8: C1 + C2 + DENV (0.5 times); lane 9: C1 + C2 + DENV (0.1 times). (b) Electrophoretic pattern of the HCR characterization (10 % PAGE). Lane 1: T-C2; lane 2: H1; lane 3: H2; lane 4: H1 + T-C2; lane 5: H2 + T-C2; lane 6: H1 + H2; lane 7: H1 + H2 + T-C2; lane 8: H1 + H2 + T-C2 (0.5 times); lane 9: H1 + H2 + T-C2 (0.1 times). (c) Electrophoretic pattern of the cascade signal amplification strategy of LCHA and HCR (8 % PAGE). Marker (20-500 bp); lane 1: L1; lane 2: L2; lane 3: L1 + L2; lane 4: L1 + L2+C1; lane 5: L1 + L2+C1+C2; lane 6: L1 + L2+C1+C2+DENV; lane 7: L1 + L2+C1+C2+H1+H2+DENV; lane 8: L1 + L2+C1+C2+H1+H2; lane 9: DENV.
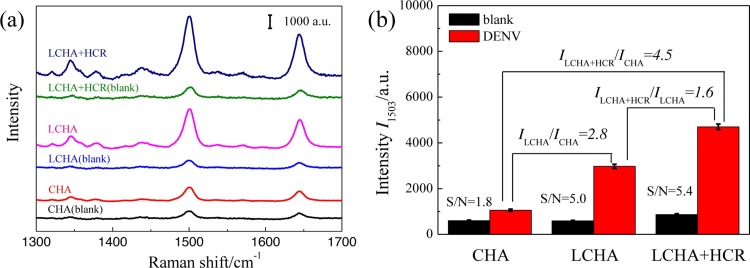
The feasibility of the sensing strategy was also characterized by SERS detections and the advantage of the proposed cascade signal amplification strategy was investigated. Fig. 2 shows the SERS assays of 10 nM DENV by three types of signal amplification strategies, i.e., CHA, LCHA, and the cascade signal amplification strategy of LCHA and HCR (LCHA + HCR), respectively. Both the CHA and LCHA were performed in the absence of H1 and H2 (i.e., HCR). Fig. 2a shows the SERS spectra collected from the assays of DENV and blank control (without any DENV, i.e., TM buffer) by the three amplification strategies, respectively. The corresponding peak intensities at 1503 cm−1 were plotted in Fig. 2b. The SERS intensity of LCHA assay is about 2.8 times (I LCHA/I CHA) that of CHA, and the intensity of the cascade signal amplification strategy of LCHA and HCR is about 1.6 times (I LCHA+HCR/I LCHA) that of LCHA strategy. Totally, the SERS intensity of the cascade signal amplification strategy of LCHA and HCR (i.e., LCHA + HCR) is significantly amplified, which is 4.5 times that of CHA. Meanwhile, the signal-to-noise ratio (S/N, I DENV/I blank) of the cascade signal amplification strategy (S/N = 5.4) is also improved relative to the ones of the individual CHA (S/N = 1.8) or LCHA (S/N = 5.0) assay. These results clearly demonstrate that the cascade amplification strategy of LCHA and HCR is beneficial for highly sensitive detection.
Fig. 2.
SERS assays of 10 nM DENV by three types of signal amplification strategies, i.e., CHA, LCHA, and the cascade amplification strategy of LCHA and HCR. (a) SERS spectra. (b) SERS intensities at 1503 cm−1 corresponding to the SERS spectra shown in (a).
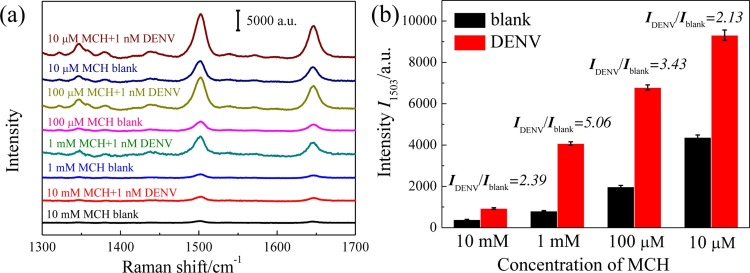
3.2. Optimization of MCH blocking
To avoid the unspecific absorption and minimize the background, MCH was used to block the AgNRs array substrate and remove the physically adsorbed DNAs. The MCH blocking was optimized by using 20 μL of 10 mM, 1 mM, 100 μM and 10 μM MCH for 10 min, respectively, and the corresponding SERS spectra collected from the assays of 1 nM DENV and blank were shown in Fig. 3 a. The SERS intensities gradually decreased with the increase of MCH concentration, since the high concentration of MCH not only removed the physically adsorbed DNAs but also replaced the Ag-S covalently bound DNAs. The S/N of the assays using the different MCH concentrations were analyzed and plotted in Fig. 3b. The S/N value by using 1 mM MCH is 5.06, which was the highest value with respect to the other three MCH blockings. Therefore, the 1 mM MCH was the optimal blocking concentration to obtain the optimal sensitivity.
Fig. 3.
Optimization of MCH blocking. (a) SERS spectra of the assays of 1 nM DENV or TM buffer (blank) by using 10 mM, 1 mM, 100 μM and10 μM MCH blocking, respectively. (b) SERS intensities at 1503 cm−1 corresponding to the spectra shown in (a).
3.3. Calibration curve and limit of detection
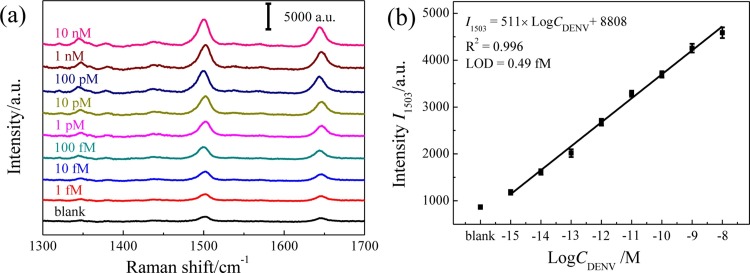
The calibration curve of the SERS assay was built by testing different concentrations of DENV, i.e., 0 (TM buffer as blank control), 1 fM, 10 fM, 100 fM, 1 pM, 10 pM, 100 pM, 1 nM, and 10 nM, respectively. Fig. 4 a shows the concentration-dependent SERS spectra of the assays, and the SERS signal at 1503 cm−1 increases gradually with the increasing concentration of DENV as plotted in Fig. 4b. The corresponding linear relationship between SERS intensity (I 1503) and the logarithm of DENV concentration (C DENV) was also obtained from 1 fM to 10 nM, i.e., I 1503 = 511× Log C DENV + 8808 (R2 = 0.996), and the limit of detection (LOD) was calculated to be 0.49 fM (S/N = 3). The proposed SERS sensing strategy possesses a wider detection range and lower LOD for DENV gene relative to the previous reported assays of virus genes (Table S2), which exhibits significant advantage for DENV gene detection.
Fig. 4.
Calibration curve of the SERS assay of DENV. (a) Concentration-dependent SERS spectra. (b) Plot of the SERS intensities at 1503 cm−1 corresponding to the spectra shown in (a) (n = 10), and the fitted linear calibration curve.
3.4. Specificity, uniformity and repeatability of SERS assay
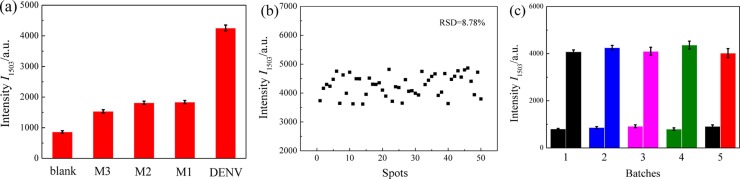
The specificity of the SERS assay via the cascade signal amplification strategy of LCHA and HCR was investigated by testing 1 nM DENV, TM buffer (blank) and 10 nM mismatched DNAs including single-base mismatched (M1), double-base mismatched (M2) and three-base mismatched (M3) DNAs, respectively. As shown in Figs. 5 a and S2, the SERS sensing can obviously distinguish the DENV from the blank and the three types of mismatched DNAs, which confirms the good specificity of the proposed sensing strategy. The uniformity of the SERS sensing was investigated by detecting 1 nM DENV and collecting the SERS signals from 50 random spots. Fig. 5b plots the corresponding SERS intensities at 1503 cm−1 which shows a small variation with a relative standard deviation (RSD) of 8.78 %, indicating that the proposed SERS sensing has good uniformity. The repeatability of the SERS sensing was characterized by testing 1 nM DENV and blank control using five batches of SERS substrates. The SERS intensities at 1503 cm−1 collected from the five SERS substrates are plotted in Fig. 5c, and the small RSDs of the assays of DENV (3.44 %) and blank control (6.70 %) confirm the good repeatability of the SERS assay.
Fig. 5.
Specificity and uniformity of the SERS assay of DENV. (a) Plot of the SERS intensities at 1503 cm−1 by testing 1 nM DENV, TM buffer (blank) and 10 nM mismatched DNAs including single-base mismatched (M1), double-base mismatched (M2) and three-base mismatched (M3) DNAs, respectively. (b) Plot of SERS intensities at 1503 cm−1 collected from 50 random spots on the AgNRs array substrate for assay of 1 nM DENV. (c) SERS intensities at 1503 cm−1 collected from five batches of SERS substrates for sensing 1 nM DENV and blank control.
3.5. Recovery of the SERS assay of DENV
In order to characterize the reliability and reproducibility of the proposed SERS assay, the recovery was investigated by testing the artificially prepared DENV gene solutions with four different concentrations (Table 1 ). By carrying out three parallel detections of each sample, the found values and the corresponding recoveries are shown in Table 1. The recovery ranges from 93.10–114.0% with the RSDs less than 4.62 %, which indicates that the proposed SERS strategy of the cascade signal amplification of LCHA and HCR shows good reliability and reproducibility for DENV gene detection.
Table 1.
Recovery of the SERS assay of DENV gene.
| Sample | Added/M | Found/M | Recovery/% | RSD/% |
|---|---|---|---|---|
| 1 | 3.3 × 10−9 | 3.56 × 10−9 | 107.8 | 2.16 |
| 2 | 8.0 × 10−11 | 9.12 × 10−11 | 114.0 | 4.62 |
| 3 | 6.7 × 10−13 | 6.82 × 10−13 | 102.2 | 3.74 |
| 4 | 2.0 × 10−14 | 1.86 × 10−14 | 93.10 | 3.07 |
4. Conclusions
A highly sensitive SERS assay via a cascade enzyme-free signal amplification strategy of LCHA and HCR was proposed for the detection of DENV gene fragment. The SERS assay was performed by two steps, i.e., the operation of a specially designed cascade enzyme-free signal amplification strategy of LCHA and HCR, and the following SERS measurements by transferring the HCR products on SERS-active AgNRs array substrates with optimal MCH blocking. The well operation of signal amplification strategies is confirmed by electrophoresis, and the optimal MCH blocking is 1 mM. The SERS assays indicate that the sensitivity of LCHA assay is about 2.8 times that of CHA, and the sensitivity of the cascade signal amplification strategy of LCHA and HCR is significantly amplified, which is 4.5 times that of CHA. Besides, the S/N is also improved to 5.4 relative to 1.8 of the individual CHA. The proposed SERS sensing strategy for DENV gene detection possesses a linear calibration curve from 1 fM to 10 nM with the LOD low to 0.49 fM. The SERS assay has good specificity, uniformity, repeatability and reliability, which can provide an attractive tool for reliable, early diagnosis of DENV gene and is worth to be popularized in a wide detection of other viruses.
CRediT authorship contribution statement
Chunyuan Song: Conceptualization, Methodology, Formal analysis, Writing - review & editing. Jingjing Zhang: Formal analysis, Writing - original draft. Yang Liu: Methodology, Investigation. Xiangyin Guo: Methodology. Yan Guo: Formal analysis, Data curation. Xinyu Jiang: Writing - original draft. Lianhui Wang: Project administration, Supervision.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2017YFA0205300), National Natural Science Foundation of China (61871236, 61971207), Jiangsu Provincial Key Research and Development Program (BE2018732), Natural Science Foundation of Jiangsu Province of China (BK20181395), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX19_0937).
Biographies
Chunyuan Song received his PhD degree in Optical Engineering at Southeast University in 2012. Then he joined the Institute of Advanced Materials (IAM), Nanjing University of Posts and Telecommunications (NJUPT). He is currently an associate professor with his research interest focusing on the fabrication of plasmonic nanomaterials and SERS probes/sensors for chemical and biological analysis.
Jingjing Zhang is a PhD candidate at the Institute of Advanced Materials (IAM), Nanjing University of Posts and Telecommunications (NJUPT) under the supervision of Prof. Lianhui Wang and Dr. Chunyuan Song. Her current research interest focuses on the development of SERS sensors for cancer-related biomarker detection.
Yang Liu worked on the SERS-based biosensors and obtained her Master degree in 2019 under the supervision of Dr. Chunyuan Song at the Institute of Advanced Materials (IAM), Nanjing University of Posts and Telecommunications (NJUPT).
Xiangyin Guo is a graduate student at the Institute of Advanced Materials (IAM), Nanjing University of Posts and Telecommunications (NJUPT) under the supervision of Dr. Chunyuan Song. His current research interest focuses on the preparation of SERS-active nanomaterials.
Yan Guo is a graduate student at the Institute of Advanced Materials (IAM), Nanjing University of Posts and Telecommunications (NJUPT) under the supervision of Dr. Chunyuan Song. Her current research interest focuses on the flexible SERS substrate.
Xinyu Jiang is a graduate student at the Institute of Advanced Materials (IAM), Nanjing University of Posts and Telecommunications (NJUPT) under the supervision of Dr. Chunyuan Song. He received his Bachelor’s degree at NJUPT in 2017. His research interest focuses on the development of SERS/SPR dual-mode biosensors for nucleic acid detection.
Lianhui Wang obtained his PhD degree in polymeric chemistry and physics at Zhejiang University in 1998. Then he joined Prof. E. T. Kang’s group at the National University of Singapore (NUS) as a postdoctoral researcher from 1998 to 2000, followed by being a researcher and assistant professor at Institute of Molecular and Cell Biology, NUS. Since June 2005, he joined the faculty of Fudan University as a professor and then moved to Nanjing University of Posts and Telecommunications (NJUPT) in January 2011. Currently, he is a professor at the Institute of Advanced Materials (IAM), NJUPT. His research group works on bioelectronics and nanobiology including synthesis of optoelectronic nanomaterials and their applications for biochemical sensing, multimodal imaging, drug delivery and cancer therapy.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2020.128970.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Person B., Sy F., Holton K., Govert B., Liang A. NCID SARS Community Outreach Team, Fear and stigma: The epidemic within the SARA outbreak. Emerg. Infect. Dis. 2004;10:358–363. doi: 10.3201/eid1002.030750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peiris J.S.M., de Jong M.D., Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 2007;20:243. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X., Chen G., Rodriguez W.R. Micro- and nanotechnology for viral detection. Anal. Bioanal. Chem. 2008;393:487–501. doi: 10.1007/s00216-008-2514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen L.R., Jamieson D.J., Powers A.M., Honein M.A. Zika virus. N. Engl. J. Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 5.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., Myers M.F., George D.B., Jaenisch T., Wint G.R.W., Simmons C.P., Scott T.W., Farrar J.J., Hay S.I. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K., Xu X., Lu H., Zhu W., Gao Z., Xiang N., Shen Y., He Z., Gu Y., Zhang Z., Yang Y., Zhao X., Zhou L., Li X., Zou S., Zhang Y., Li X., Yang L., Guo J., Dong J., Li Q., Dong L., Zhu Y., Bai T., Wang S., Hao P., Yang W., Zhang Y., Han J., Yu H., Li D., Gao G.F., Wu G., Wang Y., Yuan Z., Shu Y. Human infection with a novel avian-origin influenza a (H7N9) virus. N. Engl. J. Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 7.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuiken T., Fouchier R.A.M., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K.S., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S.M., Osterhaus A. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assiri A., McGeer A., Perl T.M., Price C.S., Al-Rabeeah A.A., Cummings D.A.T., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A., Team K.M.C.I. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H.Z., He D.G., Wan K.J., Sheng X.W., Cheng H., Huang J., Zhou X., He X.X., Wang K.M. In situ multiplex detection of serum exosomal microRNAs using an all-in-one biosensor for breast cancer diagnosis. Analyst. 2020;145:3289–3296. doi: 10.1039/d0an00393j. [DOI] [PubMed] [Google Scholar]
- 11.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claudia K., Annika B., Piotr W.D., Aleksandar R., Andreas N., Andreas K. Protocol for metagenomic virus detection in clinical specimens. Emerg. Infect. Dis. 2015;21:48–57. doi: 10.3201/eid2101.140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monto A.S., Comanor L., Shay D.K., Thompson W.W. Epidemiology of pandemic influenza: use of surveillance and modeling for pandemic preparedness. J. Infect. Dis. 2006;194:S92–S97. doi: 10.1086/507559. [DOI] [PubMed] [Google Scholar]
- 14.Lai S.C., Huang Y.Y., Shu P.Y., Chang S.F., Hsieh P.S., Wey J.J., Tsai M.H., Ben R.J., Hsu Y.M., Fang Y.C., Hsiao M.L., Lin C.C. Development of an enzyme-linked immunosorbent assay for rapid detection of dengue virus (DENV) NS1 and differentiation of DENV serotypes during early infection. J. Clin. Microbiol. 2019;57:e00221–00219. doi: 10.1128/JCM.00221-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathakrishnan A., Sekaran S.D. New development in the diagnosis of dengue infections. Expert Opin. Med. Diagn. 2013;7:99–112. doi: 10.1517/17530059.2012.718759. [DOI] [PubMed] [Google Scholar]
- 16.Guzman M.G., Halstead S.B., Artsob H., Buchy P., Farrar J., Gubler D.J., Hunsperger E., Kroeger A., Margolis H.S., Martínez E., Nathan M.B., Pelegrino J.L., Simmons C., Yoksan S., Peeling R.W. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darwish N.T., Sekaran S.D., Khor S.M. Point-of-care tests: a review of advances in the emerging diagnostic tools for dengue virus infection. Sens. Actuators B Chem. 2018;255:3316–3331. [Google Scholar]
- 18.McClellan M.S., Domier L.L., Bailey R.C. Label-free virus detection using silicon photonic microring resonators. Biosens. Bioelectron. 2012;31:388–392. doi: 10.1016/j.bios.2011.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walper S.A., Lasarte Aragonés G., Sapsford K.E., Brown C.W., Rowland C.E., Breger J.C., Medintz I.L. Detecting biothreat agents: from current diagnostics to developing sensor technologies. ACS Sens. 2018;3:1894–2024. doi: 10.1021/acssensors.8b00420. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G.J., Zhang L., Huang M.J., Luo Z.H.H., Tay G.K.I., Lim E.J.A., Kang T.G., Chen Y. Silicon nanowire biosensor for highly sensitive and rapid detection of dengue virus. Sens. Actuators B Chem. 2010;146:138–144. [Google Scholar]
- 21.Singh R.K., Dhama K., Karthik K., Tiwari R., Khandia R., Munjal A., Iqbal H.M.N., Malik Y.S., Bueno-Mari R. Advances in diagnosis, surveillance, and monitoring of zika virus: an update. Front. Microbiol. 2018;8:2677. doi: 10.3389/fmicb.2017.02677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin C.Y., Wu Y.Y., Li X.L., Niu J.J., Lei J., Ding X.D., Xiao D., Zhou C.S. Highly selective, naked-eye, and trace discrimination between perfect-match and mismatch sequences using a plasmonic nanoplatform. Anal. Chem. 2018;90:7371–7376. doi: 10.1021/acs.analchem.8b00756. [DOI] [PubMed] [Google Scholar]
- 23.Gao M., Waggoner J.J., Hecht S.M., Chen S. Selective detection of dengue virus serotypes using tandem toehold-mediated displacement reactions. ACS Infect. Dis. 2019;5:1907–1914. doi: 10.1021/acsinfecdis.9b00241. [DOI] [PubMed] [Google Scholar]
- 24.Xie B.P., Qiu G.H., Hu P.P., Liang Z., Liang Y.M., Sun B., Bai L.P., Jiang Z.H., Chen J.X. Simultaneous detection of dengue and zika virus RNA sequences with a three-dimensional Cu-based zwitterionic metal–organic framework, comparison of single and synchronous fluorescence analysis. Sens. Actuators B Chem. 2018;254:1133–1140. [Google Scholar]
- 25.He Z.Q., Wu J., Qiao B., Pei H., Xia Q.F., Wu Q., Ju H.X. Target-catalyzed assembly of pyrene-labeled hairpins for exponentially amplified biosensing. ACS Appl. Biol. Mater. 2020;3:5342–5349. doi: 10.1021/acsabm.0c00658. [DOI] [PubMed] [Google Scholar]
- 26.Omar N.A.S., Fen Y.W., Abdullah J., Mustapha Kamil Y., Daniyal W., Sadrolhosseini A.R., Mahdi M.A. Sensitive detection of dengue virus type 2 E-proteins signals using self-assembled monolayers/reduced graphene oxide-PAMAM dendrimer thin film-SPR optical sensor. Sci. Rep. 2020;10:2374. doi: 10.1038/s41598-020-59388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngo H.T., Wang H.N., Fales A.M., Nicholson B.P., Woods C.W., Vo-Dinh T. DNA bioassay-on-chip using SERS detection for dengue diagnosis. Analyst. 2014;139:5655–5659. doi: 10.1039/c4an01077a. [DOI] [PubMed] [Google Scholar]
- 28.Gahlaut S.K., Savargaonkar D., Sharan C., Yadav S., Mishra P., Singh J.P. SERS platform for dengue diagnosis from clinical samples employing a hand held Raman spectrometer. Anal. Chem. 2020;92:2527–2534. doi: 10.1021/acs.analchem.9b04129. [DOI] [PubMed] [Google Scholar]
- 29.Tripathy S., Krishna Vanjari S.R., Singh V., Swaminathan S., Singh S.G. Electrospun manganese (III) oxide nanofiber based electrochemical DNA-nanobiosensor for zeptomolar detection of dengue consensus primer. Biosens. Bioelectron. 2017;90:378–387. doi: 10.1016/j.bios.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Cui J.J., Gao L., Chen S.W., Huang Z., Wang X.P. Electrochemical voltammetric behaviors of synthetic dengue virus RNAs at ITO sensing electrode. J. Electroanal. Chem. 2019;851 [Google Scholar]
- 31.Kneipp K., Wang Y., Kneipp H., Perelman L.T., Itzkan I., Dasari R., Feld M.S. Single molecule detection using surface-enhanced Raman scattering (SERS) Phys. Rev. Lett. 1997;78:1667–1670. [Google Scholar]
- 32.Nie S.M., Emery S.R. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 33.Fang W.N., Jia S.S., Chao J., Wang L.Q., Duan X.Y., Liu H.J., Li Q., Zuo X.L., Wang L.H., Wang L.H., Liu N., Fan C.H. Quantizing single-molecule surface-enhanced Raman scattering with DNA origami metamolecules. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aau4506. eaau4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh Y.S., Chung A.J., Cordovez B., Erickson D. Enhanced on-chip SERS based biomolecular detection using electrokinetically active microwells. Lab Chip. 2009;9:433–439. doi: 10.1039/b809702j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngo H.T., Wang H.N., Fales A.M., Vo-Dinh T. Plasmonic SERS biosensing nanochips for DNA detection. Anal. Bioanal. Chem. 2016;408:1773–1781. doi: 10.1007/s00216-015-9121-4. [DOI] [PubMed] [Google Scholar]
- 36.Amber M.P., Zhen F., Sudarson S.S., Yongliang S., Linda L., Fengwei B., Paresh C.R. Bioconjugated gold nanoparticle based SERS probe for ultrasensitive identification of mosquito-borne viruses using Raman fingerprinting. J. Phys. Chem. C. 2015;119:23669–23675. doi: 10.1021/acs.jpcc.5b07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H.Y., Zhang M.J., Zou L., Li R.M., Ling L.S. Sequence specific recognition of HIV-1 dsDNA in the large amount of normal dsDNA based upon nicking enzyme signal amplification and triplex DNA. Talanta. 2017;173:9–13. doi: 10.1016/j.talanta.2017.05.043. [DOI] [PubMed] [Google Scholar]
- 38.Du M.Y., Mao G.B., Tian S.B., Liu Y.C., Zheng J., Ke X.L., Zheng Z.H., Wang H.Z., Ji X.H., He Z.K. Target-induced cascade amplification for homogeneous virus detection. Anal. Chem. 2019;91:15099–15106. doi: 10.1021/acs.analchem.9b03805. [DOI] [PubMed] [Google Scholar]
- 39.Xiao G.Y., Chen B.B., He M., Hu B. Dual-mode detection of avian influenza virions (H9N2) by ICP-MS and fluorescence after quantum dot labeling with immuno-rolling circle amplification. Anal. Chim. Acta. 2019;1096:18–25. doi: 10.1016/j.aca.2019.10.063. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Y.X., Chen F., Li Q., Wang L.H., Fan C.H. Isothermal amplification of nucleic acids. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 41.Zhou H., Liu J., Xu J.J., Zhang S.S., Chen H.Y. Optical nano-biosensing interface via nucleic acid amplification strategy: construction and application. Chem. Soc. Rev. 2018;47:1996–2019. doi: 10.1039/c7cs00573c. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y.J., Yang L., Wei J., Ma K., Gong X., Shang J.H., Yu S.S., Wang F.A. An autonomous nonenzymatic concatenated DNA circuit for amplified imaging of intracellular ATP. Anal. Chem. 2019;91:15229–15234. doi: 10.1021/acs.analchem.9b04126. [DOI] [PubMed] [Google Scholar]
- 43.Yue S.Z., Zhao T.T., Qi H.J., Yan Y.C., Bi S. Cross-catalytic hairpin assembly-based exponential signal amplification for CRET assay with low background noise. Biosens. Bioelectron. 2017;94:671–676. doi: 10.1016/j.bios.2017.03.071. [DOI] [PubMed] [Google Scholar]
- 44.Yang W.T., Zhou X.X., Zhao J.M., Xu W.J. A cascade amplification strategy of catalytic hairpin assembly and hybridization chain reaction for the sensitive fluorescent assay of the model protein carcinoembryonic antigen. Microchim. Acta. 2018;185:100. doi: 10.1007/s00604-017-2620-6. [DOI] [PubMed] [Google Scholar]
- 45.Li S.Y., Zhang F., Chen X.M., Cai C.Q. Efficient dual-amplification system for G-quadruplex-based non-enzymatic fluorescence detection of microRNA. Sens. Actuators B Chem. 2018;263:87–93. [Google Scholar]
- 46.Ren K.W., Xu Y.F., Liu Y., Yang M., Ju H.X. A responsive “nano string light” for highly efficient mRNA imaging in living cells via accelerated DNA cascade reaction. ACS Nano. 2018;12:263–271. doi: 10.1021/acsnano.7b06200. [DOI] [PubMed] [Google Scholar]
- 47.Wei Q.M., Huang J., Li J., Wang J.L., Yang X.H., Liu J.B., Wang K.M. A DNA nanowire based localized catalytic hairpin assembly reaction for microRNA imaging in live cells. Chem. Sci. 2018;9:7802–7808. doi: 10.1039/c8sc02943a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang F., Cheng Y.R., Cao Y., Zhang Y.Y., Dong H.F., Lu H.T., Zhang X.J. MicroRNA triggered DNA “nano wheel” for visualizing intracellular microRNA via localized DNA cascade reaction. Anal. Chem. 2019;91:9828–9835. doi: 10.1021/acs.analchem.9b01487. [DOI] [PubMed] [Google Scholar]
- 49.Song C.Y., Yang Y.J., Yang B.Y., Sun Y.Z., Zhao Y.P., Wang L.H. An ultrasensitive SERS sensor for simultaneous detection of multiple cancer-related miRNAs. Nanoscale. 2016;8:17365–17373. doi: 10.1039/c6nr05504d. [DOI] [PubMed] [Google Scholar]
- 50.Song C.Y., Yang B.Y., Zhu Y., Yang Y.J., Wang L.H. Ultrasensitive sliver nanorods array SERS sensor for mercury ions. Biosens. Bioelectron. 2017;87:59–65. doi: 10.1016/j.bios.2016.07.097. [DOI] [PubMed] [Google Scholar]
- 51.Zhang J.J., Yang Y.J., Jiang X.Y., Dong C., Song C.Y., Han C.Q., Wang L.H. Ultrasensitive SERS detection of nucleic acids via simultaneous amplification of target-triggered enzyme-free recycling and multiple-reporter. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.