Abstract
Atherosclerosis, which is the most common chronic disease of the coronary artery, constitutes a vascular pathology induced by inflammation and plaque accumulation within arterial vessel walls. Both DNA methylation and histone modifications are epigenetic changes relevant for atherosclerosis. Recent studies have shown that the DNA methylation and histone modification systems are closely interrelated and mechanically dependent on each other. Herein, we explore the functional linkage between these systems, with a particular emphasis on several recent findings suggesting that histone acetylation can help in targeting DNA methylation and that DNA methylation may control gene expression during atherosclerosis.
Keywords: DNA methylation, Histones, Acetylation, Atherosclerosis
Introduction
Coronary artery disease remains a leading cause of death globally. Atherosclerosis is the most common chronic disease of the coronary artery. It is characterized by the adherence of cells circulating in the blood to the endothelium, migration to the sub-endothelial layer, and differentiation into macrophages. It is a process by which inflammation, fibrous proliferation, and plaque build-up occur in the arterial wall. Inflammation is the main process underlying atherosclerosis, which is associated with cell-to-cell interactions involving multiple cell types, including endothelial cells, leukocytes, and smooth muscle cells (SMCs).1 Known risk factors for atherosclerosis include elevated circulating low-density lipoprotein (LDL) and triglyceride levels, smoking, obesity, and aging. In response to these stimuli, vascular endothelial cells—the cells composing the inner lining of blood vessels—become activated and recruit monocytes from the circulation. Monocytes differentiate into macrophages upon entering the vascular wall, and following the uptake of lipids, including oxidized low-density lipoprotein (oxLDL), can further differentiate into foam cells, which are retained underneath the endothelial layer.2 Leukocytes, primarily macrophages, which have an important role in LDL uptake, are crucial players in the pathophysiological processes leading to cardiovascular diseases, including myocardial infarction, as they lead to the formation of foam cells in the intima.3
Epigenetics plays a pivotal role in regulating gene expression in development, in response to cellular stress or in disease states, in virtually all cell types. Epigenetic modifications are processes whereby a cell retains a memory of past cellular states and perturbations without altering the DNA sequence itself. By remodeling the chromatin structure or gene expression, epigenetic modifications cooperate with transcription factors and the translational machinery in fine-tuning gene expression.4 Recently, many clinicians and basic researchers have proposed that the pathogenesis of atherosclerosis involves dynamic epigenetic modifications. These epigenetic changes regulate gene expression in a cell type- and stage-specific manner.5,6 The definition of epigenetics is the study of heritable changes in gene expression that do not involve changes to the underlying DNA sequence (i.e., a change in phenotype without a change in genotype). Epigenetic mechanisms include DNA methylation and histone modifications, which consist of acetylation, methylation, phosphorylation, deimination, ubiquitylation, sumoylation, and ADP ribosylation, and these modifications affect the chromatin structure and create affinities for chromatin-associated proteins, thereby modulating gene expression. MicroRNAs (miRNAs) are short, non-coding RNA molecules that mediate RNA silencing and regulate gene expression. Therefore, miRNAs have also been identified as components of the epigenetic machinery.
Much remains to be discovered regarding the epigenetic mechanisms that are operative in atherosclerosis, which is partially due to the difficulty in analyzing specific cell types in the complex milieu of the plaque. However, emerging technological advances, such as single cell assays for transposase-accessible chromatin using sequencing (scATAC-seq) and multiple complementary experimental systems for studying atherosclerosis, including genome-wide association studies (GWAS), are rapidly transforming the approaches that can be utilized to understand the molecular basis of this disease.7 In this review, we primarily focus on the modifications of DNA methylation and histone acetylation in cardiovascular atherosclerotic disease.
DNA METHYLATION
DNA methylation is a key biochemical process for regulating the activity of DNA. It involves adding a methyl group to the fifth position of the pyrimidine ring of cytosine, generating 5-methylcytosine (5meC), or to the number-6 nitrogen of the purine ring of adenine. Methylation usually occurs at the gene promoter, and typically inhibits gene transcription. This change can be inherited through cell differentiation and it is related to the pathogenesis of various diseases. DNA methylation constitutes 3 processes: de novo methylation, 5meC recognition, and active and passive demethylation. It was initially established that de novo DNA methylation occurs at symmetric CpG sites, starts with DNA methyltransferase (DNMT) 3A/3B, and is maintained by DNMT1 (Fig. 1). However, further research revealed that both DNMT3A/3B and DNMT1 are needed for DNA methylation initiation and maintenance. DNA methylation can inhibit gene transcription directly or indirectly. Methylation can directly interfere with the binding of DNA and transcription factors, or it can attract proteins that bind specifically to modify DNA, thereby blocking other transcription factors from binding the site.8 The transcription factors AP-2, c-Myc, cyclic AMP response element binding protein (CREB), EBP-80, and E2F lose their affinity to genes when their binding sites are methylated.9,10,11,12,13,14,15 Some indirect mechanisms exist that repress transcription. When proteins (repressors) bind preferentially to methylated DNA, they reduce gene expression. For example, methyl-CpG-binding protein 1 and 2 (MeCP1 and MeCP2), despite their differences in binding stability to methyl-CpG, both act as repressors that inhibit gene transcription.16,17 The methyl-CpG-binding domain (MBD) and nucleosome remodeling deacetylase (NuRD) protein constitute the chromatin remodeling complex and bind to methylated CpG, which later turns off the transcription of the gene. MeCP2 represses gene expression by introducing histone deacetylase (HDAC) 1 and 2 and Sin3A to the gene, removing acetyl groups from acetylated histone tails. These 2 chromatin remodeling complexes bind to methylated CpGs and repress transcription.18 DNA methylation itself has been thought to be a stable and long-term epigenetic modification, but the discovery of DNA demethylation and its mechanism revealed that this process is more complex and dynamic than was initially envisioned. DNA demethylation can occur either passively or actively.19 Passive DNA demethylation occurs through DNA replication. When methylated DNA is replicated with DNMT1 inhibited or absent, the new strand is synthesized without the DNA methylation, and when this DNA double-strand is replicated once more, the new DNA, synthesized using the strand without methyl group as the template, loses the remaining methyl groups. While the passive DNA demethylation process is straightforward, much remains to be discovered regarding active DNA demethylation, which is still controversial. Active DNA demethylation can be distinguished by whether it is a genome-wide or a gene-specific event. Genome-wide active DNA demethylation occurs in germ cells or gametes associated with reproduction and development. Locus-specific active demethylation, in contrast, occurs in somatic cells as a response to certain signals. Several possible mechanisms of DNA active demethylation have been proposed, including, enzymatic removal of methyl groups,20,21,22 direct excision of 5meC during base excision repair (BER),23,24,25 conversion of 5meC to thymine, deamination with BER of the T/G mismatch by thymine-DNA glycosylase (TDG),26,27,28,29 and oxidative demethylation (Fig. 1).30,31
Fig. 1. The mechanism of DNA methylation and demethylation.
C, cytosine; 5meC, 5-methylcytosine; 5caC, 5-carboxylcytosine; 5fC, 5-formylcytosine; 5hmC, 5-hydroxymethylcytosine; DNMT, DNA methyltransferase; TET, ten-eleven translocation enzyme; TDG, thymine-DNA-glycosylase; BER, base excision repair.
Oxidative demethylation (by an enzyme or ten-eleven translocation [TET]) or deamination (by activation-induced cytidine deaminase or apolipoprotein B messenger RNA [mRNA] editing enzyme, catalytic polypeptide-like) precedes TDG-related demethylation. 5meC is deaminated by the enzymes above to become thymidine, which is further demethylated by TDG through BER.32 As oxidative demethylation proceeds, 5meC is first oxidized by TET 1-3 to become 5-hydroxymethylcytosine (5hmC), which is further oxidized into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by the same TET family.33 During the process and at the end of the process, TDG recognizes 5fC and 5caC, creates an apurinic/apyrimidinic site and restores unmodified cytosine.34,35
CROSSTALK BETWEEN DNA METHYLATION AND HISTONE ACETYLATION
It is well known that DNA methylation and histone deacetylation both repress gene transcription. When histones are acetylated, their electrostatic interactions with DNA become weaker, resulting in relaxed chromatin, which upregulates transcription; the opposite happens when histones are deacetylated by HDAC.36,37 Research has explored the crosstalk between these 2 epigenetic activities, to reveal how one process affects the other, and herein, we present examples of how they work to regulate gene transcription.
As described above, MeCP2 can bind to methylated DNA, and when it binds to methylated cytosines, it also binds to chromatin. This led to the hypothesis that transcriptional repression by DNA methylation might also occur due to chromatin structural changes, with MeCP2 recruiting a chromatin-modifying corepressor. In rat brain nuclear extracts, it was found that precipitates of MeCP2-bound proteins showed deacetylation activity. This finding made it clear that there is an association between DNA methylation and histone deacetylation via a methylated DNA binding protein, MeCP2.38 The induction of histone deacetylation by MeCP2 was further studied and found to have a connection with Sin3A, which interacts with HDAC. Therefore, it has been demonstrated that MeCP2 consists of the MBD and transcriptional repression domain (TRD), and that TRD interacts with Sin3A, which brings HDAC to the histone of methylated DNA.39
In addition to relationship between DNA methylation and acetylation, it was found that histone acetylation can induce DNA demethylation. The model described above, according to which DNA methylation can induce histone deacetylation, can explain gene inactivation, but is insufficient to explain the demethylation of the gene upon its activation. In an attempt to find the link between these 2 processes, a model was proposed according to which active chromatin induces associated DNA demethylation. When human lymphoma cell lines were treated with phorbol myristate acetate and butyrate, which is an HDAC inhibitor, levels of methylated CpG were decreased. This finding means that inhibition of histone deacetylation induced demethylation of DNA.40 Similarly, when HEK 293 cells were treated with trichostatin A (TSA), which inhibits histone deacetylation, genes were demethylated actively in a replication-independent manner.41
DNA MODIFICATIONS IN ATHEROSCLEROSIS
As we described above, atherosclerosis is characterized by the adherence of cells circulating in the blood to the endothelium, migration to the sub-endothelial layer, and differentiation into macrophages. This results in a buildup of plaque inside the artery made up of fat, ions, and proteins, which hardens over time and narrows the vessel. This can result in the blockage of fresh, oxygen-rich blood from reaching the organs, and over time, a plaque can rupture inside the artery, causing the catastrophic clinical manifestation of myocardial infarction or ischemic stroke.42,43 Recent studies of the pathogenesis of the disease have found that chronic inflammation plays a major role in disease initiation and progression.44 Since the pathogenesis of atherosclerosis consists of endothelial dysfunction, intimal lipid deposition, SMC proliferation, cell apoptosis and necrosis, and local and systemic inflammation,45 we describe herein the relationships between these pathogenic processes and epigenetic modifications.
Since atherosclerosis is an inflammation-related disease, like other cardiovascular diseases, research into the role of epigenetics in the inflammation process is relevant for understanding atherosclerosis. A study of patients with chronic kidney disease (CKD), which is relevant since dialysis patients often experience the complication of cardiovascular disease, compared oxidative stress biomarkers, homocysteine, and global DNA methylation in peripheral blood leukocytes according to the level of inflammation. The results showed that patients with inflammation had global DNA hypermethylation, which was significantly associated with cardiovascular mortality even correcting for other factors such as age.46
Leukocytes are not the only cell type to have shown a correlation between DNA hypermethylation and atherosclerosis. The genomic DNA methylation status of peripheral lymphocytes in angiographically confirmed coronary artery disease (CAD) patients and control individuals was compared utilizing a methylation-sensitive restriction enzyme. The results of that study showed that genomic DNA methylation in the patients was significantly higher than in the controls, and also found a positive correlation between DNA hypermethylation and plasma levels of homocysteine in CAD patients.47 It is clear that atherosclerosis is inherently related to epigenetic changes, especially DNA methylation, in light of the result that CAD patients had higher DNA methylation levels, which are also correlated with homocysteine levels, a risk factor for atherosclerosis. Moreover, as mentioned above, CKD patients on dialysis, who are also susceptible to atherosclerosis, have higher DNA methylation levels. Later in this section, we will discuss studies of DNA methylation and specific pathogenic steps of atherosclerosis.
The first step in the pathogenesis of atherosclerosis is endothelial cell activation. Endothelial cells, as the first barrier of the disease, regulate hemostasis and thrombosis, local vascular tone and redox balance, and acute and chronic inflammatory reactions. When they start to malfunction, and their balance is disrupted, it leads to a downstream cascade of the disease, including the permeation of blood monocytes to the endothelial cell layer and infiltration into the intima and subintima. Endothelial cell activation is mediated by metabolic risk factors such as hyperlipidemia,48 and is linked with disturbed blood flow (DBF). It is also known that prevalent abnormal blood flow at arterial branches and curvatures is a predictor of early atherosclerotic lesions.49 Further studies have found that DBF-related atherogenesis is mediated by DNA methylation changes.50,51,52 In an animal model of atherosclerosis induced by DBF, significant upregulation of DNMT1 transcription was observed. To support the hypothesis that DNMT1 plays a major role in the pathogenesis of atherosclerosis, DNMT1 activity was inhibited, which resulted in amelioration of endothelial dysfunction and atherosclerosis. In an attempt to elucidate the mechanism of DBF-induced DNMT1 upregulation, signaling pathways that can take part in the conversion of mechanical stimuli into electrochemical activity of the cells have been studied. Mammalian target of rapamycin (mTOR) was demonstrated to be a major factor. When endothelial cells are pretreated with rapamycin, inhibiting the mTOR pathway before DBF, phosphorylation of the mTOR effector p70S6K was blocked. Additionally, DNMT1 expression was blocked in comparison with the control group. These findings revealed that DBF-induced DNMT1 upregulation is mediated by mTOR/p70S6K signaling. Further experiments screened for flow-sensitive genes highly expressed in atherosclerotic plaques. In that study, cyclin A and connective tissue growth factor induced the expression of DNMT1 and it was found that it was upregulated upon BDF.52 These results suggest that endothelial cell activation and BDF can induce DNMT expression and DNA methylation, which may change the gene expression pattern and aggravate atherosclerosis.
Endothelial dysfunction then results in the permeation of blood monocytes to the endothelial cell layer and infiltration into the intima and subintima. The infiltrated monocytes then differentiate into macrophages. In the process of macrophage differentiation, macrophages are polarized from alternatively activated (M2) to classically activated (M1). M2 expresses anti-inflammatory cytokines (e.g., interleukin [IL]-10 and arginase 1), while M1 expresses proinflammatory cytokines (e.g., tumor necrosis factor-alpha [TNF-α], IL-6, and IL-1β). Peroxisome proliferator-activated receptor-gamma (PPAR-γ) plays an important role in macrophage polarization, and the epigenetic mechanisms behind this were studied. When DNMT1 was macrophage-specifically overexpressed in transgenic mice of atherosclerosis model, proinflammatory cytokines in the macrophage and plasma were increased compared to the control group and the progression of the disease was accelerated. In that study, it was found that the proximal promoter of PPAR-γ was hypermethylated by DNMT1. Pharmacological activation of PPAR-γ decreased proinflammatory cytokine production and successfully prevented atherosclerosis development. The monocytes isolated from the peripheral blood of atherosclerosis showed increased DNMT1 and decreased PPAR-γ expression. These results confirmed the detailed mechanism of how DNA methylation increases inflammation in atherosclerosis (Table 1).53
Table 1. Epigenetic modifications in atherosclerosis.
| Enzymes | Function | Links to atherosclerosis |
|---|---|---|
| DNMT1 | DNA methylation | Induces endothelial cell dysfunction under disrupted blood flow |
| Macrophage differentiation–PPAR-γ | ||
| Accumulation of oxLDL–BRCA1 | ||
| p300 acetyltransferase | H4 acetylation | Increases IL-8 and MCP1 expression |
| H3K14 acetylation | ||
| H3K9 acetylation | Increases MMP-1, -3, -10, -12, -13 expression | |
| H3K9 acetylation | Increases levels of acetylation in proportion to atherosclerosis lesion | |
| H3K27 acetylation | ||
| H3K9 acetylation | Increases IL-6 expression | |
| H3K14 acetylation | ||
| HAT1 | H3K27 acetylation | Increases NOX5 expression |
| HDAC1 | H3K9 deacetylation | Accumulation of cholesterol |
| HDAC | H3K9 and H3K27 deacetylation | Decreases IL-6 and CCL5 expression |
| Reduces the SMC proliferation rate | ||
| H4 deacetylation | Decreases CD36, SRA, TNF-α, and VCAM-1 expression | |
| Increases IL-1β, IL-6 levels |
DNMT, DNA methyltransferase; PPAR-γ, peroxisome proliferator-activated receptor-gamma; oxLDL, oxidized low density lipoprotein; IL, interleukin; MCP, monocyte chemoattractant protein; MMP, matrix metalloproteinase; HAT, histone acetyltransferase; NOX5, NADPH5 oxidase; HDAC, histone deacetylase; CCL5, C-C motif chemokine 5; SMC, smooth muscle cell; CD, cluster of differentiation; SRA, scavenger receptor A; TNF-α, tumor necrosis factor-alpha; VCAM-1, vascular cell adhesion molecule-1.
These differentiated and polarized macrophages then take up lipids and form foam cells. The accumulated foam cells, along with the activated immune cells, release cytokines and form atherosclerotic plaque. In the process of macrophage lipid uptake, oxLDL plays a crucial role and the breast cancer type 1 (BRCA1) gene acts as a gatekeeper of atherosclerosis basally expressed in endothelial cells. The role of BRCA1 in an atherosclerotic model was studied using an endothelial cell-specific knockout of BRCA1 (endothelial cell-BRCA1−/−) in atherosclerosis-induced mice. When BRCA1 was overexpressed in the transgenic mice model, BRCA1 expression could rescue the cells from the effects of oxLDL.54 A study identifying differentially methylated regions of atherosclerosis patients revealed that the BRCA1 gene was significantly more methylated than in healthy controls.55 These 2 results imply that the hypermethylated BRCA1 gene in atherosclerosis patients has an impact on the pathogenesis of atherosclerosis by changing the function of the gene, making it susceptible to the effects of oxLDL.
In addition to DNA methylation, DNA demethylation is also an important mechanism explaining the pathogenesis of atherosclerosis. As shown in an atherosclerotic mice model, TET2 overexpression significantly reduced atherosclerotic lesion formation, potentially by oxidatively demethylating 5meC to 5hmC in the endothelial vessel wall. This TET2-induced rescue occurs via the upregulation of autophagy, as TET2 overexpression decreased the methylation level of the promoters of autophagic flux-related genes.56
HISTONE MODIFICATIONS
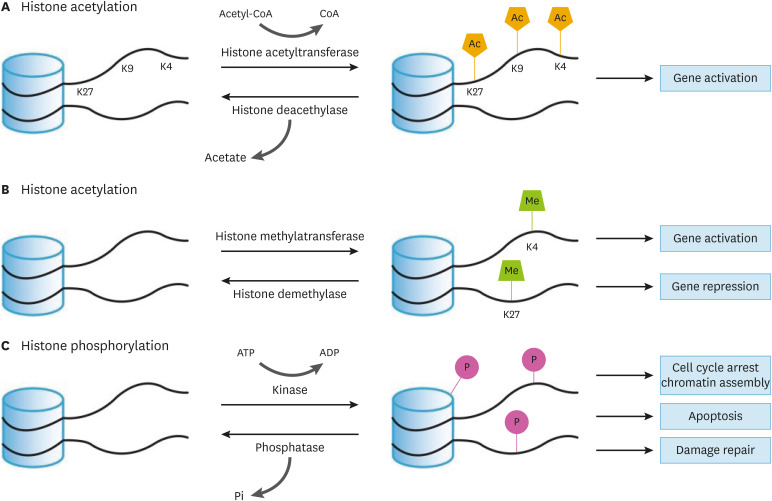
Histones are fundamental components of the transcriptional regulatory machinery. H2A, H2B, H3, and H4 histones assemble as an octamer and wrap DNA. Each histone is composed of a C-terminal globular domain and an N-terminal tail domain. This tail domain is unstructured and flexible, so this feature allows the histone tail to be altered by various epigenetic modifiers, which function as a major component of gene expression regulation, thereby controlling a number of biological processes such as proliferation, DNA replication, and cellular death. The mechanisms through which histone modifications control gene expression have been characterized. First, epigenetic modifications alter the binding capacity between DNA and histones by changing their electrostatic charges. Second, they serve as docking sites for transcriptional factors. Third, they reconstitute the structure of chromatin. There are several epigenetic modifications, such as acetylation, methylation, and phosphorylation, which all have distinct biological functions (Fig. 2). Despite their significant function in controlling gene expression, the precise mechanism through which they contribute to atherosclerosis remains elusive. Here, we focus on the interaction between histone acetylation and atherosclerosis.
Fig. 2. A schematic diagram of histone modification.
(A) Histone acetylation: Acetylation and deacetylation are catalyzed by histone acetyltransferase and histone deacetylase, respectively. Acetyl-CoA is a source and co-factor of acetylation. Histone acetylation occurs at lysine residues and it increases gene expression in general. (B) Histone methylation: Methylation is catalyzed by histone methyltransferase. Histone demethylase reverses methylation. Methylation activates or represses gene expression depending on which residue is methylated. K4 methylation activates gene expression. K27 methylation represses gene expression. (C) Histone phosphorylation; Phosphorylation is catalyzed by kinases. Phosphorylation takes place on the C-terminal tail and N-terminal structure. ATP is a source of phosphorylation. Histone phosphorylation is not only associated with gene expression control, but also chromatin condensation following cell cycle and DNA damage. Phosphatases remove phosphate groups.
Ac, acetylation; Me, methylation; P, phosphorylation.
Histone acetylation
Histones are positively charged proteins due to their high content of lysine and arginine residues. Acetylation usually occurs on lysine residues, neutralizing their positive charge and thereby causing histones to drift away from DNA, which has a negative charge. The released structure facilitates access to transcriptional machinery such as transcription factors and RNA polymerase II.57 Thus, acetylation induces and enhances gene expression in general. Histone acetylation and deacetylation are catalyzed by histone acetyltransferases (HATs) and HDACs, respectively. HATs consist of 2 types: type A and type B. Type A HATs are mainly localized at the nucleus and include the Gcn5-related N-acetyltransferases, MYST (MOZ, Ybf2, Sas2 and Tip60), and p300/CBP families. They acetylate nucleosomal histones by transferring acetyl group from acetyl-CoA. Meanwhile, type B HATs are located in the cytosol and acetylate free histones or non-histone proteins. HDACs are classified into 4 classes: class 1 (HDAC1,2,3,8), class 2 (2a: HDAC4,5,7,9; 2b: HDAC6,10), class 3 (SIRT), and class4 (HDAC11). They remove acetyl groups from acetylated proteins, consequently repressing gene expression by condensing nucleosomes. HATs and HDACs are recruited to their distinct target regions by specific transcriptional factors. In promoter regions, HATs acetylate histones and recruit HAT-containing complexes to activate the transcriptional process. For instance, H3K9ac and H3K27ac levels are associated with promoter and enhancer activities. Furthermore, H3K27ac enhances not only the kinetics of transcriptional activation, but also accelerates the transition of RNA polymerase II from the initiation state to the elongation state.58 However, some researchers have revealed that histone acetylation can also repress gene expression and is associated with heterochromatin assembly. Indeed, high levels of H4K20ac were found at the transcriptional start site of minimally expressed genes and silenced genes. This marker prevents access by transcriptional activators such as c-Myc, STAT3, and p300 and recruits transcriptional repressors such as neuron-restrictive silencer factor and repressor element 1—silencing transcription.59 Furthermore, H3K4ac is localized at pericentromeric heterochromatin. In this situation, H3K4ac is located just after H3K9me2 during the cell cycle. When H3K4 is not acetylated, the RNA-induced transcriptional silencing complex is tightly bonded with H3K9me2 using Chp1, a chromodomain-containing protein with high-affinity binding for H3K9me2. However, the acetylation of H3K4 destabilizes this interaction and sequentially recruits heterochromatin protein 1 homolog Swi6/Chp2, causing heterochromatin to form. In other words, H3K4ac contributes to heterochromatin formation by switching Chp/Clr4 to Swi6/Chp2 at adjacent H3K9me2.60
HISTONE ACETYLATION IN ATHEROSCLEROSIS
Numerous studies have attempted to elucidate the significance of epigenetic regulation as a cause and result in the process of atherosclerosis. Therefore, studies have been conducted on when and how histone modifications control the expression of genes, thereby exacerbating, accelerating, or delaying the progress of atherosclerotic lesions. Because it has been assumed that epigenetic changes are closely related with atherosclerosis, we will discuss the effects of histone acetylation on atherosclerosis.
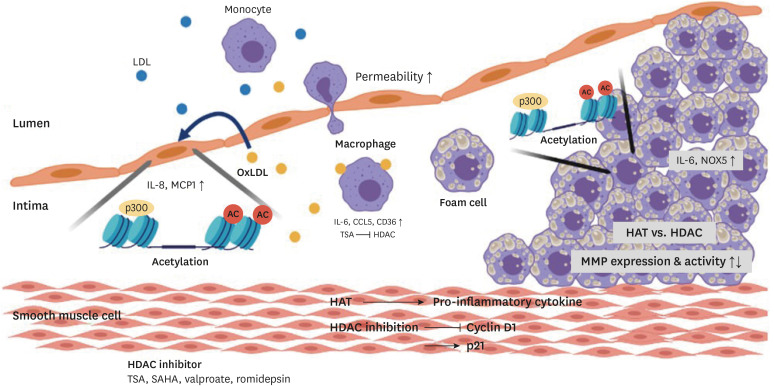
Dysfunction of vascular epithelial cells has been considered the initiation of atherosclerosis. Among several stimuli, oxLDL is a key risk factor for activation of the endothelium. Increased oxLDL levels induce the expression of adhesion molecules and the release of cytokines and chemokines, enhancing the permeability of the endothelium. Monocytes are then attracted and migrate. The infiltrated monocytes differentiate into macrophages and convert into foam cells by taking up oxLDL. Emerging evidence shows that histone post-transcriptional modifications, especially histone acetylation, trigger these processes (Fig. 3). oxLDL induces the acetylation of IL-8, a pro-inflammatory cytokine, and the chemokine ligand monocyte-chemoattractant protein-1 (MCP-1) promoter by recruiting p300 in endothelial cells. In this process, the lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1) accepts oxLDL and transmits the signal through extracellular signal-regulated kinases 1/2 to activate transcription. Furthermore, oxLDL reduces HDAC1 and HDAC2 expression and their binding affinities. Collectively, these mechanisms result in inflammatory activation. As clinically applicable HMG-CoA reductases, statins partially reverse these effects by reducing nuclear factor (NF)-κB p65/RelA, CREB, and RNA polymerase II recruitment at the IL8 and MCP-1 gene promoter.61
Fig. 3. Histone acetylation changes during the process of atherosclerosis.
Histone acetylation occurs at the promoter of pro-inflammatory cytokines. oxLDL increases IL-8 and MCP1 expression by histone acetylation in the endothelium. TSA inhibits HDAC activity and it upregulates CD36, IL-6, and CCL5 in macrophages. Accumulated foam cells release IL-6 and increase NOX5 expression. Histone acetyltransferase and histone deacetylase alter MMP expression and activity.
LDL, low density lipoprotein; oxLDL, oxidized low density lipoprotein; IL, interleukin; MCP1, monocyte chemoattractant protein 1; CCL5, C-C motif chemokine 5; CD, cluster of differentiation; NOX5, NADPH5 oxidase; MMP, matrix metalloproteinase; AC, acetylation; HAT, histone acetyltransferase; HDAC, histone deacetylase; TSA, trichostatin A; SAHA, suberoylanilide hydroxamic acid.
Inflammatory processes are accompanied by macrophage foam cell formation in the artery wall, but the relationship between macrophage lipid loading and their response to inflammatory stimuli is controversial. Recently it has been reported that oxLDL downregulates TLR-induced pro-inflammatory responses in macrophages at the late stages of TLR-induced proinflammatory gene expression through the reduction of NF erythroid 2 related factor 2 binding at IL-6 and the C-C motif chemokine 5 (CCL5) promoter. Although oxLDL does not modulate lipopolysaccharide (LPS)-induced NF-κB or MAPK signaling, it reduces p65/RelA binding affinity to IL-6 and the CCL5 promoter in LPS-stimulated macrophages. The HDAC inhibitor, TSA, rescues this inhibitory effect.62 Furthermore, TSA also increases the H4 acetylation level of the cluster of differentiation (CD) 36 promoter and oxLDL uptake. Thus, histone acetylation is closely associated with the inflammation process in atherosclerosis. In addition, scavenger receptor A, TNF-α, and vascular cell adhesion molecule-1 RNA levels were elevated, but IL-1β and IL-6 levels were reduced, exacerbating atherosclerosis. 63 Therefore, oxLDL loading of macrophages negatively regulates the transcription of proinflammatory gene expression and implicates epigenetic mechanisms such as HDAC activity. As a result, histone acetylation levels are altered by oxLDL, and the effect of inflammation depends on the stage of atherosclerosis.
Accumulated foam cells and activated immune cells release cytokines, chemokines such as IL-6, interferon gamma, CCL5, and CCL11, inducing the activation of SMCs. Activated SMCs proliferate, migrate and form a fibrous cap to stabilize the plaque. During these processes, significant upregulation of H3K9 and H3K27 acetylation in SMCs has been reported,64 and histone acetyltransferases and p300 enhance pro-inflammatory IL-6 expression.65 Under inflammatory conditions, levels of p300 and HAT1 increase, and they catalyze acetylation of the Nox5 gene promoter. Nox5 is a novel NADPH oxidase that generates superoxide. Conversely, the HAT inhibitor CPTH2 and C646 downregulate LPS-activated Nox5. This finding indicates that histone acetylation occurs during atherosclerosis development and can stimulate environmental factors that accelerate atherosclerosis.66 Furthermore, TSA inhibits proliferation of vascular SMCs via induction of p21.67 The other 2 HDAC inhibitors, sodium valproate and MS-275, prevent the expression of CD1a, a hallmark of mature dendritic cells that disturbs proper dendritic cell differentiation by reducing the release of cytokines that are needed for differentiation.68 Mitogenic stimuli induce HDAC1, HDAC2, and HDAC3 expression in SMCs, increasing the proliferation of SMCs. HDAC inhibition prevents mitogen-induced SMC proliferation by modifying the expression of cyclin dependent kinase inhibitors.69 In hyperhomocysteinemia-related atherosclerosis, HDAC is increased, thereby decreasing H3K9ac levels and promoting the accumulation of total cholesterol, free cholesterol, and triglycerides in foam cells.70
In the late stage of atherosclerosis, apoptosis of SMCs and activation of metalloproteases cause plaque rupture, resulting in significant morbidity and mortality. Matrix metalloproteinases (MMPs) disassemble extracellular components, and therefore are considered to be important enzymes in the disease process. The presence of MMP-1 has been reported in atherosclerotic lesions.71 And the increased MMP-3 levels have been found to cause plaque rupture.72 Several epigenetic studies have revealed that histone acetylation is connected with MMP expression. Therefore, at this stage, histone acetylation makes atherosclerosis aggressive via MMP expression. In human dermal fibroblasts, p300 acetyltransferase recruits the MMP-1 promoter, leading to induction of MMP-1 transcription.73 MMP cluster genes are coordinately increased in proportion to H3K9 acetylation.74 In addition, sodium butyrate, an HDAC inhibitor, increases the secretion of pro-MMP9 and pro-MMP2 via H4 hyperacetylation.75 The increased expression and activity of MMPs are obviously found in atherosclerosis plaques and promote plaque rupture.72,76,77 However, in other studies, 2 HDAC inhibitors (TSA and sodium butyrate) inhibited the induction of MMP-1 and MMP-13.78 Conversely, when HDAC levels were decreased in macrophages, stabilization of atherosclerotic lesions was reported.79 In other words, the regulation of MMP genes according to histone acetylation shows different patterns depending on the cell type and the timing of atherosclerosis, and the precise mechanisms should be further studied.
LDL receptor knockout mice showed a reduction of atherosclerosis lesions, an effect that was accompanied by systemic deletion of HDAC9. This resulted in the downregulation of inflammatory genes, such as those coding for IL-1β and Mcp1, and the upregulation of lipid homeostatic genes such as Abca1, Abcg1, and Pparγ.80 In HDAC3 conditional knockout mouse, myeloid Hdac3 deficiency enhanced transforming growth factor-beta secretion to promote collagen deposition, thereby inducing stable plaque formation.79
FUTURE CONSIDERATIONS
Much remains to be discovered regarding the epigenetic mechanisms that are operative in atherosclerosis, and this is due in part to the difficulty in analyzing specific cell types in the complex milieu of the plaque. However, emerging technological advances such as scATAC-seq and multiple complementary experimental systems for studying atherosclerosis, including GWAS, can be utilized to understand the molecular basis of this disease. The pathogenesis of atherosclerosis is very complex and this disease occurs through multiple processes. Therefore, a genome-wide analysis should be done in all samples prepared from stages in the multi-step process of atherosclerosis.
CONCLUSION
Based on a synthesis of recent studies, DNA methylation and histone acetylation in all stages of atherosclerosis play a key role in regulating the expression of inflammatory cytokines and chemokines. They also control epigenetic modifications through a reciprocal mechanism. In addition, anti-inflammatory treatment using drugs that control histone acetyltransferases or HDAC has been tried in various studies. Much remains to be discovered regarding exact role of DNA methylation and histone acetylation in atherosclerosis. However, it is indisputably clear that epigenetic modifications play an important role in atherosclerosis, and further studies will provide valuable information for atherosclerosis treatment.
Footnotes
Funding: This study was supported by grant No. 03-2017-0390 from the SNUH Research Fund.
Conflict of Interest: The authors have no conflicts of interest to declare.
- Conceptualization: Kwon YW.
- Funding acquisition: Kwon YW.
- Project administration: Kwon YW.
- Supervision: Kwon YW.
- Validation: Kwon YW.
- Writing - original draft: Lee HT, Oh S, Ro DH, Yoo H, Kwon YW.
- Writing - review & editing: Kwon YW.
References
- 1.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 2.Tabas I, García-Cardeña G, Owens GK. Recent insights into the cellular biology of atherosclerosis. J Cell Biol. 2015;209:13–22. doi: 10.1083/jcb.201412052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chistiakov DA, Melnichenko AA, Myasoedova VA, Grechko AV, Orekhov AN. Mechanisms of foam cell formation in atherosclerosis. J Mol Med (Berl) 2017;95:1153–1165. doi: 10.1007/s00109-017-1575-8. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi M, Renzini A, Adamo S, Moresi V. Coordinated actions of microRNAs with other epigenetic factors regulate skeletal muscle development and adaptation. Int J Mol Sci. 2017;18:840. doi: 10.3390/ijms18040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaina S, Heyn H, Carmona FJ, Varol N, Sayols S, Condom E, et al. DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet. 2014;7:692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 7.Khyzha N, Alizada A, Wilson MD, Fish JE. Epigenetics of atherosclerosis: emerging mechanisms and methods. Trends Mol Med. 2017;23:332–347. doi: 10.1016/j.molmed.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22:3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. Molecular biology of the gene. 7th ed. New York (NY): Pearson Higher Ed USA; 2004. [Google Scholar]
- 10.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 12.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 13.Falzon M, Kuff EL. Binding of the transcription factor EBP-80 mediates the methylation response of an intracisternal A-particle long terminal repeat promoter. Mol Cell Biol. 1991;11:117–125. doi: 10.1128/mcb.11.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovesdi I, Reichel R, Nevins JR. Role of an adenovirus E2 promoter binding factor in E1A-mediated coordinate gene control. Proc Natl Acad Sci U S A. 1987;84:2180–2184. doi: 10.1073/pnas.84.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inamdar NM, Ehrlich KC, Ehrlich M. CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol Biol. 1991;17:111–123. doi: 10.1007/BF00036811. [DOI] [PubMed] [Google Scholar]
- 16.Ehrlich KC, Cary JW, Ehrlich M. A broad bean cDNA clone encoding a DNA-binding protein resembling mammalian CREB in its sequence specificity and DNA methylation sensitivity. Gene. 1992;117:169–178. doi: 10.1016/0378-1119(92)90726-6. [DOI] [PubMed] [Google Scholar]
- 17.Nan X, Cross S, Bird A. Gene silencing by methyl-CpG-binding proteins. Novartis Found Symp. 1998;214:6–16. discussion 16–21, 46–50. doi: 10.1002/9780470515501.ch2. [DOI] [PubMed] [Google Scholar]
- 18.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 19.Meehan RR, Stancheva I. DNA methylation and control of gene expression in vertebrate development. Essays Biochem. 2001;37:59–70. doi: 10.1042/bse0370059. [DOI] [PubMed] [Google Scholar]
- 20.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 22.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 23.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 25.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 27.Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–20712. [PubMed] [Google Scholar]
- 28.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 29.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 30.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 31.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 35.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 36.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr Opin Genet Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 38.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 40.Szyf M, Eliasson L, Mann V, Klein G, Razin A. Cellular and viral DNA hypomethylation associated with induction of Epstein-Barr virus lytic cycle. Proc Natl Acad Sci U S A. 1985;82:8090–8094. doi: 10.1073/pnas.82.23.8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- 42.Chen YC, Huang AL, Kyaw TS, Bobik A, Peter K. Atherosclerotic plaque rupture: identifying the straw that breaks the camel's back. Arterioscler Thromb Vasc Biol. 2016;36:e63–e72. doi: 10.1161/ATVBAHA.116.307993. [DOI] [PubMed] [Google Scholar]
- 43.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 44.Hansson GK, Robertson AK, Söderberg-Nauclér C. Inflammation and atherosclerosis. Annu Rev Pathol. 2006;1:297–329. doi: 10.1146/annurev.pathol.1.110304.100100. [DOI] [PubMed] [Google Scholar]
- 45.Herrero-Fernandez B, Gomez-Bris R, Somovilla-Crespo B, Gonzalez-Granado JM. Immunobiology of atherosclerosis: a complex net of interactions. Int J Mol Sci. 2019;20:5293. doi: 10.3390/ijms20215293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, et al. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007;261:488–499. doi: 10.1111/j.1365-2796.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- 47.Sharma P, Kumar J, Garg G, Kumar A, Patowary A, Karthikeyan G, et al. Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol. 2008;27:357–365. doi: 10.1089/dna.2007.0694. [DOI] [PubMed] [Google Scholar]
- 48.Gimbrone MA, Jr, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiu JJ, Chien S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol Rev. 2011;91:327–387. doi: 10.1152/physrev.00047.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Li YS, Chien S. Shear stress-initiated signaling and its regulation of endothelial function. Arterioscler Thromb Vasc Biol. 2014;34:2191–2198. doi: 10.1161/ATVBAHA.114.303422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest. 2014;124:3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang YP, Huang YT, Huang TS, Pang W, Zhu JJ, Liu YF, et al. The mammalian target of rapamycin and DNA methyltransferase 1 axis mediates vascular endothelial dysfunction in response to disturbed flow. Sci Rep. 2017;7:14996. doi: 10.1038/s41598-017-15387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J, Qiu Y, Yang J, Bian S, Chen G, Deng M, et al. DNMT1-PPARγ pathway in macrophages regulates chronic inflammation and atherosclerosis development in mice. Sci Rep. 2016;6:30053. doi: 10.1038/srep30053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh KK, Shukla PC, Quan A, Al-Omran M, Lovren F, Pan Y, et al. BRCA1 is a novel target to improve endothelial dysfunction and retard atherosclerosis. Thorac Cardiovasc Surg. 2013;146:949–960.e4. doi: 10.1016/j.jtcvs.2012.12.064. [DOI] [PubMed] [Google Scholar]
- 55.Istas G, Declerck K, Pudenz M, Szic KS, Lendinez-Tortajada V, Leon-Latre M, et al. Identification of differentially methylated BRCA1 and CRISP2 DNA regions as blood surrogate markers for cardiovascular disease. Sci Rep. 2017;7:5120. doi: 10.1038/s41598-017-03434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng J, Yang Q, Li AF, Li RQ, Wang Z, Liu LS, et al. Tet methylcytosine dioxygenase 2 inhibits atherosclerosis via upregulation of autophagy in ApoE−/− mice. Oncotarget. 2016;7:76423–76436. doi: 10.18632/oncotarget.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 58.Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 59.Kaimori JY, Maehara K, Hayashi-Takanaka Y, Harada A, Fukuda M, Yamamoto S, et al. Histone H4 lysine 20 acetylation is associated with gene repression in human cells. Sci Rep. 2016;6:24318. doi: 10.1038/srep24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dje N'Guessan P, Riediger F, Vardarova K, Scharf S, Eitel J, Opitz B, et al. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:380–386. doi: 10.1161/ATVBAHA.108.178319. [DOI] [PubMed] [Google Scholar]
- 62.Jongstra-Bilen J, Zhang CX, Wisnicki T, Li MK, White-Alfred S, Ilaalagan R, et al. Oxidized low-density lipoprotein loading of macrophages downregulates TLR-induced proinflammatory responses in a gene-specific and temporal manner through transcriptional control. J Immunol. 2017;199:2149–2157. doi: 10.4049/jimmunol.1601363. [DOI] [PubMed] [Google Scholar]
- 63.Choi JH, Nam KH, Kim J, Baek MW, Park JE, Park HY, et al. Trichostatin A exacerbates atherosclerosis in low density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2005;25:2404–2409. doi: 10.1161/01.ATV.0000184758.07257.88. [DOI] [PubMed] [Google Scholar]
- 64.Greißel A, Culmes M, Burgkart R, Zimmermann A, Eckstein HH, Zernecke A, et al. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc Pathol. 2016;25:79–86. doi: 10.1016/j.carpath.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Sahar S, Reddy MA, Wong C, Meng L, Wang M, Natarajan R. Cooperation of SRC-1 and p300 with NF-kappaB and CREB in angiotensin II-induced IL-6 expression in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2007;27:1528–1534. doi: 10.1161/ATVBAHA.107.145862. [DOI] [PubMed] [Google Scholar]
- 66.Vlad ML, Manea SA, Lazar AG, Raicu M, Muresian H, Simionescu M, et al. Histone acetyltransferase-dependent pathways mediate upregulation of NADPH oxidase 5 in human macrophages under inflammatory conditions: a potential mechanism of reactive oxygen species overproduction in atherosclerosis. Oxid Med Cell Longev. 2019;2019:3201062. doi: 10.1155/2019/3201062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okamoto H, Fujioka Y, Takahashi A, Takahashi T, Taniguchi T, Ishikawa Y, et al. Trichostatin A, an inhibitor of histone deacetylase, inhibits smooth muscle cell proliferation via induction of p21(WAF1) J Atheroscler Thromb. 2006;13:183–191. doi: 10.5551/jat.13.183. [DOI] [PubMed] [Google Scholar]
- 68.Nencioni A, Beck J, Werth D, Grünebach F, Patrone F, Ballestrero A, et al. Histone deacetylase inhibitors affect dendritic cell differentiation and immunogenicity. Clin Cancer Res. 2007;13:3933–3941. doi: 10.1158/1078-0432.CCR-06-2903. [DOI] [PubMed] [Google Scholar]
- 69.Findeisen HM, Gizard F, Zhao Y, Qing H, Heywood EB, Jones KL, et al. Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arterioscler Thromb Vasc Biol. 2011;31:851–860. doi: 10.1161/ATVBAHA.110.221952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao Q, Li S, Li N, Yang X, Ma S, Yang A, et al. miR-34a targets HDAC1-regulated H3K9 acetylation on lipid accumulation induced by homocysteine in foam cells. J Cell Biochem. 2017;118:4617–4627. doi: 10.1002/jcb.26126. [DOI] [PubMed] [Google Scholar]
- 71.Ye S, Gale CR, Martyn CN. Variation in the matrix metalloproteinase-1 gene and risk of coronary heart disease. Eur Heart J. 2003;24:1668–1671. doi: 10.1016/s0195-668x(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 72.Galis ZS, Sukhova GK, Lark MW, Libby P. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1994;94:2493–2503. doi: 10.1172/JCI117619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS One. 2009;4:e4864. doi: 10.1371/journal.pone.0004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ujfaludi Z, Tuzesi A, Majoros H, Rothler B, Pankotai T, Boros IM. Coordinated activation of a cluster of MMP genes in response to UVB radiation. Sci Rep. 2018;8:2660. doi: 10.1038/s41598-018-20999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodríguez-Salvador J, Armas-Pineda C, Perezpeña-Diazconti M, Chico-Ponce de León F, Sosa-Sáinz G, Lezama P, et al. Effect of sodium butyrate on pro-matrix metalloproteinase-9 and -2 differential secretion in pediatric tumors and cell lines. J Exp Clin Cancer Res. 2005;24:463–473. [PubMed] [Google Scholar]
- 76.Lenglet S, Mach F, Montecucco F. Role of matrix metalloproteinase-8 in atherosclerosis. Mediators Inflamm. 2013;2013:659282. doi: 10.1155/2013/659282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newby AC. Metalloproteinases promote plaque rupture and myocardial infarction: a persuasive concept waiting for clinical translation. Matrix Biol. 2015;44-46:157–166. doi: 10.1016/j.matbio.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 78.Young DA, Lakey RL, Pennington CJ, Jones D, Kevorkian L, Edwards DR, et al. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res Ther. 2005;7:R503–R512. doi: 10.1186/ar1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoeksema MA, Gijbels MJ, Van den Bossche J, van der Velden S, Sijm A, Neele AE, et al. Targeting macrophage Histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol Med. 2014;6:1124–1132. doi: 10.15252/emmm.201404170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao Q, Rong S, Repa JJ, St Clair R, Parks JS, Mishra N. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arterioscler Thromb Vasc Biol. 2014;34:1871–1879. doi: 10.1161/ATVBAHA.114.303393. [DOI] [PMC free article] [PubMed] [Google Scholar]