Abstract
Purpose
Ocular anterior segment disorders (ASDs) are clinically and genetically heterogeneous, and genetic diagnosis often remains elusive. In this study, we demonstrate the value of a combined analysis protocol using phenotypic, genomic, and pedigree structure data to achieve a genetic conclusion.
Methods
We utilized a combination of chromosome microarray, exome sequencing, and genome sequencing with structural variant and trio analysis to investigate a cohort of 41 predominantly sporadic cases.
Results
We identified likely causative variants in 54% (22/41) of cases, including 51% (19/37) of sporadic cases and 75% (3/4) of cases initially referred as familial ASD. Two-thirds of sporadic cases were found to have heterozygous variants, which in most cases were de novo. Approximately one-third (7/22) of genetic diagnoses were found in rarely reported or recently identified ASD genes including PXDN, GJA8, COL4A1, ITPR1, CPAMD8, as well as the new phenotypic association of Axenfeld–Rieger anomaly with a homozygous ADAMTS17 variant. The remainder of the variants were in key ASD genes including FOXC1, PITX2, CYP1B1, FOXE3, and PAX6.
Conclusions
We demonstrate the benefit of detailed phenotypic, genomic, variant, and segregation analysis to uncover some of the previously “hidden” heritable answers in several rarely reported and newly identified ocular ASD-related disease genes.
Keywords: ocular anterior segment dysgenesis, exome and genome sequencing, genomic medicine, ophthalmology, eye
INTRODUCTION
Ocular anterior segment disorders (ASDs) encompass conditions with broad clinical and genetic heterogeneity that affect the structures anterior to the vitreous surface of the eye. There are multiple conditions grouped clinically under the term ASD including aniridia, iris hypoplasia (IH), Axenfeld–Rieger anomaly (ARA) and syndrome (ARS), primary congenital glaucoma (PCG), Peters anomaly (PA), and sclerocornea, and there are many syndromal associations. Phenotypic features may overlap, and there are complex embryonic, genetic, and environmental factors involved in the pathogenesis of this group of disorders. In addition, several genes contribute to multiple phenotypes, adding to the complexity of the phenotype–genotype correlations and genetic diagnostic accuracy.
Despite the successful adoption of next-generation sequencing (NGS) in many genetic conditions, there is a lack of systematic investigation of the diagnostic utility of NGS in the full group of ocular ASD patients that may present for genetic diagnosis. Most studies focus on a particular phenotypic subset or gene set, and such studies suggest there may be a detection rate of <10–40% in the broader cohort.1–3 Our earlier work has highlighted novel genotype–phenotype correlations4 in the ocular ASDs, and the benefit of analysis of a broader group of genes using genomic approaches to find “missing” genetic diagnoses. In this study, we applied a combination of genomic, phenotypic, and pedigree structure and segregation analyses, aimed at maximizing the genetic diagnostic detection rate in this complex patient group.
MATERIALS AND METHODS
Forty-one probands with a variety of ASD phenotypes were investigated for genetic diagnosis at a major pediatric referral hospital in Sydney, Australia, over a 12-year period. In these patients the predominant presenting phenotype was ocular ASD, and included ARA, ARS, IH, PA, and sclerocornea, with overlapping features such as microphthalmia, cataract or coloboma in some cases, and occasional presence of nonocular features such as intellectual disability, ataxia, or autism (Table 1 and Supplementary Table 1). Cases with the distinct phenotypes of aniridia, known to be predominantly caused by variants in PAX6, or primary congenital glaucoma were not included in this study. The majority (35/41) were from a Caucasian background, with a small minority with Asian (3) or Middle Eastern (3) heritage. The majority (37/41) initially presented as sporadic cases, while 4 had a family history suggesting an autosomal dominant mode of inheritance (Supplementary Table 1). Ophthalmological details and samples for genomic DNA extraction were collected from family members when available.
Table 1.
Patients with likely causative variants.
| Patient number | Inheritance before/after testing | Phenotype | Platform | Gene (NM) | Nucleotide change (heterozygous, except where otherwise specified) | Amino acid change | gnomAD MAF | In silico: SIFT, MutTaster, PolyPhen, PhyloP (respectively) | ACMG criteria | Segregation | Novel |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Spor/new AD | IH & PA | Array | FOXC1 NM_001453.2 | chr6 del :1595464–1716115 | n/a | n/a | n/a | n/a | De novo | No |
| 2 | Spor/new AD | ARS | Array | PITX2 NM_153427.1 | chr4 del: 111445336–112392782 | n/a | n/a | n/a | n/a | De novo | No |
| 5 | Spor/new AD | Sclero | ES | GJA8 NM_005267.4 | c.281G>A | p.(Gly94Glu) | Nil | D, D, P, highly conserved | LP (PM2, PM6, PP2, PP3) | De novo | No |
| 9 | Spor/AR | ARA | ES | ADAMTS17 NM_139057.2 | hom c.526C>T | p.(Arg176*) | Nil | n/a | P (PVS1, PM2, PM4) | Segregates | Yes |
| 10 | Spor/Fam (AD) | ARA | ES | FOXC1 NM_001453.2 | c.516_518dupGCG | p.(Arg173dup) | Nil | n/a | LP (PM1, PM2, PP1, PP3) | Segregates | Yes |
| 11 | Fam (AD)/AD | PA | GS CNV | PAX6 NM_000280.4 | del chr11:31822357–31823717 | n/a | n/a | n/a | n/a | Segregates | Yes |
| 12 | Fam (AD)/AD | ARA | ES | FOXC1 NM_001453.2 | c.518G>A | p.(Arg173His) | Nil | D, D, P, highly conserved | LP (PM1, PM2, PP2, PP3) | Segregates | Yes |
| 14 | Spor/likely new AD | ARA | ES | PITX2 NM_153427.1 | c.341dup | p.(Asn115Glnfs*84) | Nil | n/a | P (PVS1, PM2, PM4) | n/k | Yes |
| 15 | Spor/new AD | Sclero | ES | GJA8 NM_005267.4 | c.280G>C | p.(Gly94Arg) | Nil | D, D, P, highly conserved, | LP (PM2, PM6, PP2, PP3) | De novo | No |
| 19 | Spor/likely new AD | ARS | ES | PITX2 NM_153427.1 | c.250C>T | p.(Arg84Trp) | Nil | D, D, P, highly conserved | P (PS1, PM1, PM2, PP2, PP3) | n/k | No |
| 21 | Spor/AR | Sclero | ES | PXDN NM_012293.2 | Hom c.4085_4086delAG | p.(Gln1362Argfs*22) | 1/249,252 (het only) | n/a | P (PVS1, PM2, PM4) | n/k | No |
| 23 | Spor/likely new AD | PA | ES | FOXC1 NM_001453.2 | c.1399C>T | p.(Gln467*) | Nil | n/a | P (PVS1, PM2, PM4) | n/k | Yes |
| 25 | Spor/new AD | IH | GS trio | ITPR1 NM_001168272.1 | c.7615G>A | p.(Gly2539Arg) | 1/249,244 (het only) | D, D, P, highly conserved | LP (PS1, PM2, PP2, PP3) | De novo | No |
| 26 | Spor/Fam (AD) | PA | ES | COL4A1 NM_001845.5 | c.634G>A | p.(Gly212Ser) | Nil | D, D, P, highly conserved | LP (PM1, PM2, PP2, PP3) | Segregates | No |
| 28 | Spor/AR | ARA | GS trio | CPAMD8 NM_015692.2 | Comp Het c.4549–1G>A | Splice p.(=) | 1/249,494 (het only) | n/a | P (PVS1, PS3, PM2, PP5) | Segregates (mat) | No |
| c.3149G>T | p.(Gly1050Val) | 1/249,372 (het only) | D, D, P, highly conserved | LP (PM2, PM3, PP2, PP3) | Segregates (pat) | Yes | |||||
| 29 | Spor/AR | PA | ES | CYP1B1 NM_000104.3 | Hom c.171G>A | p.(Trp57*) | 42/233,224 (het only) | n/a | P (PVS1, PM2, PM4) | n/k | No |
| 30 | Spor/AR | PA | ES | CYP1B1 NM_000104.3 | Comp Het c.171G>A; | p.(Trp57*) | 42/233,224 (het only) | n/a | P (PVS1, PM2, PM4) | n/k | No |
| c.1331G>A | p.(Arg444Gln) | Nil | D, D, P, highly conserved | P (PS3, PM2, PM3, PP1, PP5) | n/k | No | |||||
| 32 | Fam(AD)/fam (AR) | Multiple ASD | GS | FOXE3 NM_012186.2 | Hom c.720C>A | p.(Cys240*) | 7/43,132 (het only) | n/a | P (PVS1, PS3, PM2, PPS5) | Segregates | No |
| 36 | Spor/likely new AD | Sclero | GS | PITX2 NM_153427.1 | c.185G>A | p.(Arg62His) | Nil | D, D, P, highly conserved | P (PS1, PM1, PM2, PP2, PP3) | n/k | No |
| 38 | Spor/AR | PA | ES | CYP1B1 NM_000104.3 | Comp Het c.171G>A | p.(Trp57*) | 21/50,846 (het only) | n/a | P (PVS1, PM2, PM4) | Segregates | No |
| c.1200_1209dup | p.(Thr404Serfs*30) | Nil | n/a | P (PVS1, PM2, PM4) | Segregates | No | |||||
| 39 | Spor/new AD | PA | ES | PAX6 NM_000280.4 | c.152G>T | p.(Gly51Val) | Nil | D, D, P, highly conserved | LP (PM2, PM6, PP2, PP3) | De novo | No |
| 41 | Spor/new AD | IH | ES | FOXC1 NM_001453.2 | c.478_482dup | p.(Met161Ilefs*22) | Nil | n/a | P (PVS1, PM2) | De novo | Yes |
Human genome reference GRCh37/HG19 used and NCBI gene reference sequences (NM) provided. gnomAD database v2.1.1 was used (https://gnomad.broadinstitute.org/). ACMG criteria according to ref. 9 References for previously published variants also included in table. In Silico: D, damaging; P, pathogenic.
ACMG American College of Medical Genetics and Genomics, AD autosomal dominant, AR autosomal recessive, ARA Axenfeld–Rieger anomaly, ARS Axenfeld–Rieger syndrome, ASD anterior segment disorder, ES exome sequencing, Fam familial, GS genome sequencing, Het heterozygous, Hom homozygous, IH iris hypoplasia, LP likely pathogenic, MAF minor allele frequency, P pathogenic, PA Peters anomaly, Sclero sclerocornea, Spor sporadic.
Ethics statement
Informed consent was obtained, including the publishing of photographs where applicable, and the study was approved by the Human Research Ethics Committee of Sydney Children’s Hospitals Network, Sydney, Australia.
Structural variation analysis
All probands underwent copy-number variant (CNV) analysis with chromosomal microarray (CMA), on a 400K comparative genomic hybridization (CGH) array platform (Agilent SurePrint G3 Human Microarray, Santa Clara, CA, USA). Samples that underwent genome sequencing (GS) were analyzed for structural variants (SVs) with ClinSV (Minoche et al., in prep; https://github.com/KCCG/ClinSV), utilizing evidence from split-reads, discordant pairs, and depth of coverage to obtain rare, high confidence structural and CNV calls, in accordance with best practice guidelines. Multiplex ligation dependant probe amplification (MLPA) was also performed to validate any deletions (MRC-Holland, Amsterdam, Netherlands).
Next-generation sequencing
NGS was performed using exome sequencing (ES) with Illumina TruSight One Clinical Exome (Illumina, USA) or Agilent SureSelect Exome (Agilent SureSelect V4, Macrogen Inc, Seoul, South Korea). GS was performed on a number of ES-negative samples, new probands, and family samples for segregation analysis, on the Illumina TruSeq Nano HT kit with the Illumina HiSeq X (Illumina Inc, and Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, Australia).
The library preparation, genomic alignment, variant calling, and annotation were performed as previously described5–8 with variant filtering undertaken for specific anterior segment, cataract, and microphthalmia/anophthalmia disease genes, as in our previous studies and review of ASD genes4,5,8 (Supplementary Table 2). Average coverage of the key ASD genes was 93% and 92% above 20× in ES and GS platforms respectively. For negative cases, as well as trio and family samples, rare variants of interest based on in silico analysis, conservation, population databases, and phenotypic data (including pedigree structure) were also examined and manually reviewed for pathogenicity, according to American College of Medical Genetics and Genomics (ACMG) guidelines.9 All variants reported in this paper have been submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/).
RESULTS
Likely causative variants in 54% of probands, including in 51% of sporadic cases
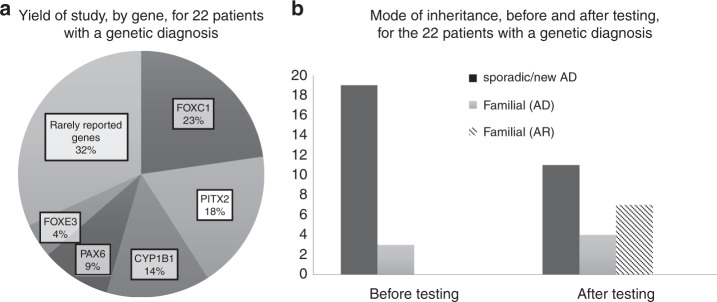
Overall, pathogenic and likely pathogenic variants in known genes9 were found in the majority of cases (22/41; 54%), with a detection rate in sporadic cases of 51% (19/37) as well as genetic diagnosis in a very high proportion (75%; 3/4) of familial cases from initial referral (Table 1). Pathogenic or likely pathogenic variants were found in 11 genes (Fig. 1a). Interestingly, 32% (7/22) of the likely causative variants were in the rarely reported genes PXDN, COL4A1, GJA8 (2), CPAMD8, and ITPR1, and for the first time reported in ADAMTS17. The rest of the likely causative variants were identified in the well-known ocular ASD genes (number of cases) FOXC1 (5) and PITX2 (4), as well as CYP1B1 (3), PAX6 (2), and FOXE3 (1) (Fig. 1a).
Fig. 1. Yield of study and mode of inheritance.
a Yield of study, by gene, for 22 patients with a genetic diagnosis. The proportion of genetic diagnoses found in the relevant genes is shown in this chart. The group of rarely reported genes includes six genes: COL4A1, PXDN, CPAMD8, ADAMTS17, ITPR1, and GJA8 (two variants). b Mode of inheritance, before and after testing, for the 22 patients with a genetic diagnosis. This figure demonstrates the breakdown of inheritance among the 22 solved cases. On referral, 19/22 were thought to be sporadic, and 3 familial with autosomal dominant (AD) inheritance. After testing, of the sporadic cases, 11 were found to be due to de novo autosomal dominant variants, 6 were due to autosomal recessive (AR) inheritance, and 2 were familial autosomal dominant cases with subtle clinical features in parents. Of the 3 familial cases, 2 were confirmed as familial autosomal dominant and one was found to be due to autosomal recessive inheritance in an inbred population group leading to pseudodominance. Hence overall after testing, there were 11 de novo autosomal dominant cases, 4 familial autosomal dominant cases, and 7 autosomal recessive cases.
Of the 19 initially referred sporadic cases where causative variants were found, most were due to de novo autosomal dominant inheritance (Fig. 1b) while the rest were inherited. Two were found to have autosomal dominant parental inherited variants in COL4A1 and FOXC1 respectively, and on re-examination, the parents were found to have subtle features of ocular ASD (Fig. 1b). In six, autosomal recessive inheritance was identified due to variants in CYP1B1 (3), CPAMD8, PXDN, and ADAMTS17 (Fig. 1b). Of the three cases initially referred as familial where causative variants were identified, two were confirmed as autosomal dominant, and one was found to have the same homozygous variant in FOXE3 in a son and his father, in a case of pseudodominance from a highly inbred population group (Fig. 1b).
Overall, of the 22 cases where causative variants were identified, 3 were due to SVs, and 19 were due to single-nucleotides (SNVs) (Table 1). Two of the SVs were found on CMA, and one SV was found on GS, and this was further validated on MLPA.
Notable variants with a role in collagen and extracellular matrix integrity including COL4A1, PXDN, CYP1B1, and the newly identified ARA phenotype finding due to ADAMTS17 variation
We identified a number of variants in collagen-related proteins, emphasizing the importance of these in the formation of the ocular anterior segment. In an individual (patient 26) with reportedly sporadic bilateral PA, a heterozygous COL4A1 p.(Gly212Ser) missense variant was found (Fig. 2a, 3a). This variant was found to be maternally inherited, and the mother was found to have mild iris hypoplasia with iris strands, consistent with a diagnosis of Rieger anomaly (Fig. 2b). This recently reported10 p.Gly212Ser variant affects a key glycine (G-X-Y) residue of the COL4A1 protein and is expected to disrupt collagen IV heterotrimer formation with COL4A2, as noted for other missense pathogenic COL4A1 variants associated with ASD (Fig. 3a).11
Fig. 2. Representative clinical images of this cohort demonstrate broad range of severity across multiple genotypes.
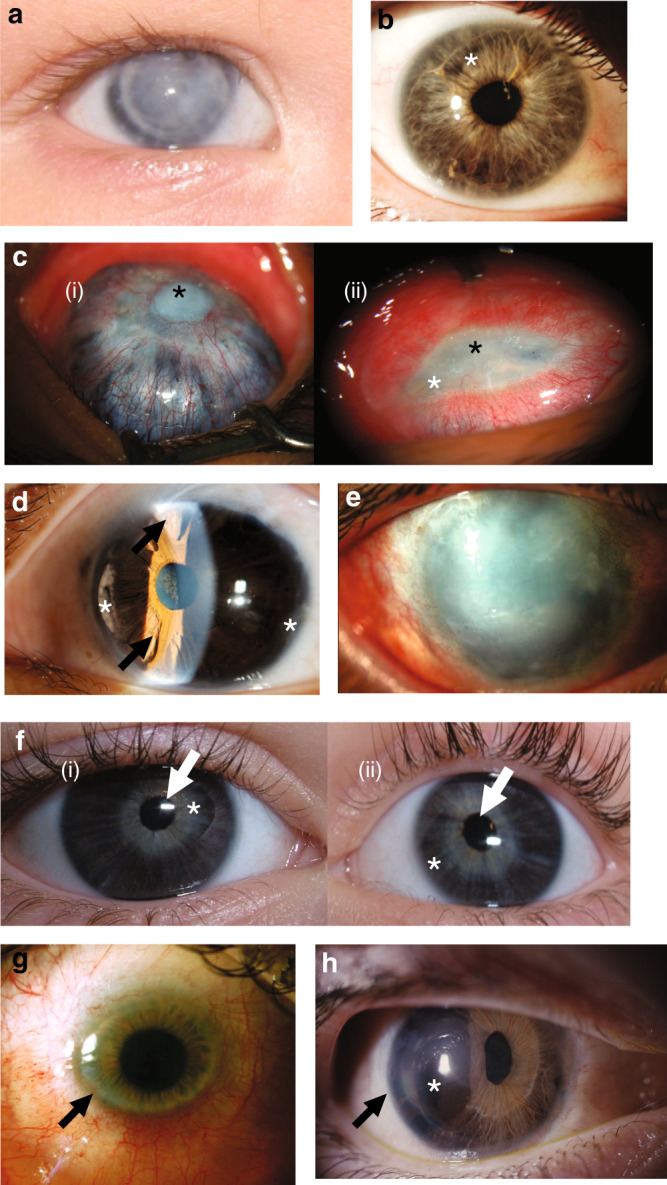
a Patient 26 with COL4A1 heterozygous variant. Photograph of right eye with Peters anomaly and failed corneal graft. b Previously undiagnosed affected mother of patient 26, with right eye showing mild features of anterior segment disorder (ASD) including Rieger anomaly, with strands of iris adhesions to the overlying cornea and mild iris hypoplasia (white asterisk). c Patient 21 with PXDN homozygous variant. Right eye (i) has previously undergone penetrating keratoplasty at age 4 years. Now failed corneal graft with central corneal opacity (black asterisk). Scleromalacia surrounding this with choroidal tissue visible through the residual sclera. Left eye (ii) shows sclerocornea with a residual small oval opaque central corneal tissue (black asterisk) with injected and dilated superficial corneoscleral vessels. No clear view of iris structures through cornea. d Patient 9 with homozygous variant in ADAMTS17. Left eye slit lamp view of anterior segment demonstrating features of Axenfeld–Rieger anomaly: corectopia, polycoria (arrows), peripheral iridocorneal adhesions, anterior iris stroma hypoplasia (white asterisk). ( e) Patient 30 with CYP1B1 variants: left eye shows generalized corneal opacification. In addition, surgical scarring is visible superiorly from previous glaucoma filtration surgery. f Patient 28 with CPAMD8 variants: right (i) and left (ii) eyes of proband showing corectopia of pupils (white arrows) and iris hypoplasia with the iris sphincter muscle visible (white asterisk). g Patient 36 with PITX2 heterozygous variant: image shows the clinical features of primary congenital corneal opacification, commonly termed sclerocornea. This case has central area of clear cornea, which on corneal topography has low (flat) keratometry in the range meeting definition of cornea plana. The peripheral cornea is scleralized (arrow) making identification of the peripheral iris difficult as well. h Patient 14 with PITX2 heterozygous variant: image shows features of Axenfeld–Rieger anomaly with iris hypoplasia (asterisk), corectopia, polycoria, and posterior embryotoxon (black arrow).
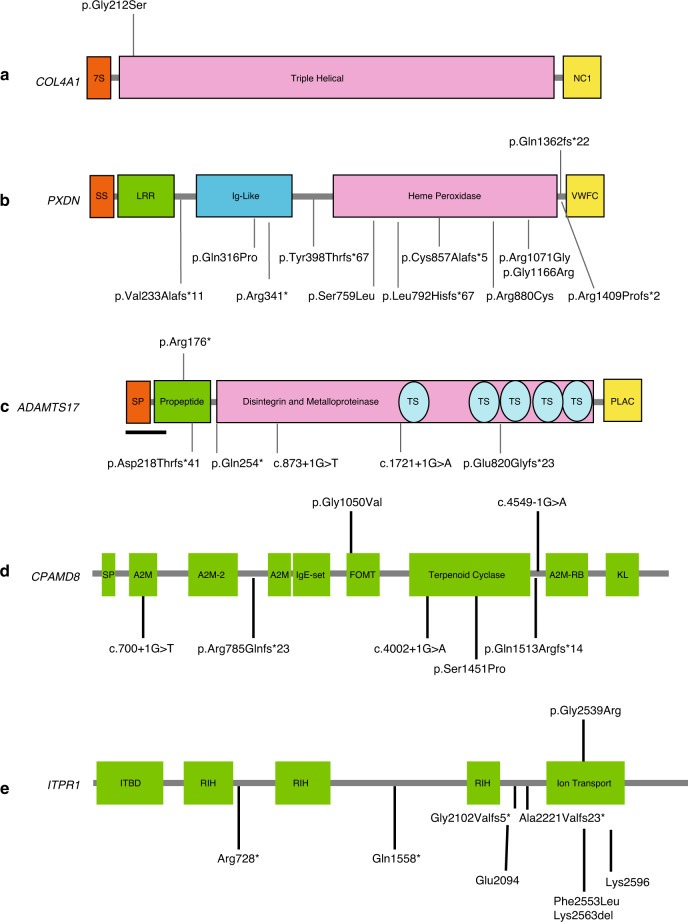
Fig. 3. Variants in extracellular matrix-associated genes COL4A1, PXDN, ADAMTS17, and recently reported anterior segment disorder (ASD) genes CPAMD8 and ITPR1.
Variants reported in the rarely reported ASD genes a COL4A1, b PXDN, c ADAMTS17, d CPAMD8, e ITPR1. Variants above the gene were found in this study, and previously reported variants are listed underneath the gene diagrams. Note: in COL4A1 over 50 missense variants, mostly involving glycine residues in the triple helical domain, have been reported in the literature. Several well reported variants are displayed. Also, the ADAMTS17 variant we report is the first associated with an Axenfeld–Rieger anomaly (ARA) phenotype.
PXDN is a rarely reported gene associated with ocular ASD and sclerocornea, as well as cataract, microcornea, glaucoma, and microphthalmia. It has a key role in collagen IV cross-linking in the basement membrane, another clue in the vital role of this protein in the extracellular matrix (ECM) of the eye. In a proband (patient 21) with severe bilateral sclerocornea (Fig. 2c), glaucoma, and severe developmental delay, a previously reported12,13 pathogenic homozygous frameshift variant was found in PXDN (c.4085_4086delAG, p.[Gln1362Argfs*22]) (Fig. 3b).
A novel likely pathogenic variant in another ECM-related gene, ADAMTS17, demonstrates further the role of collagen-related proteins in anterior segment development. In a patient referred with ARA, a novel homozygous nonsense variant in ADAMTS17 was identified (patient 9, Table 1, Fig. 2d) on manual curation of data for potentially significant pathogenic autosomal recessive variants (Fig. 3c). Homozygous deleterious (frameshift, canonical splice site, nonsense) variants in ADAMTS17 were found in families with a skeletal and eye phenotype overlapping with Weil–Marchesani syndrome (WMS).14 WMS is associated with microspherophakia, ectopia lentis, and myopia, but there are no previous reports of an association with ARA. Our patient also had short stature (adult height 150 cm) and ectopia lentis, and the presence of ARA in him broadens the known phenotype of this gene, adding variants in this gene as a new cause of ARA.
In addition, three sporadic cases with severe PA were found with homozygous or compound heterozygous pathogenic variants affecting CYP1B1 (patients 29, 30, 38, Table 1.8,15 The cyp1b1 deficient mouse model displays marked loss of collagen and degeneration of the trabecular meshwork.16 This gene was originally identified as a causative disease gene in autosomal recessive PCG, and findings from this study confirm its significant additional contribution to causation in PA.8,15 Patient 30 had a very severe Peters phenotype (Fig. 2e), and patient 29 also had severe corneal opacification and iridocorneal adhesions.
Novel variants in rarely reported ocular ASD gene CPAMD8, and a syndromal diagnosis in ITPR1 highlight the importance of deep phenotyping and data reanalysis
Several novel variants were found in rarely reported ocular ASD-associated genes. For these families, availability of pedigree structure information and deep phenotyping greatly aided identification of these causative variants.
At the time of initial ES in patient 28 (Table 1, Fig. 2f) with subluxed lenses, corectopia and iris hypoplasia, no likely pathogenic variants were found. Subsequent trio GS with our updated gene list including CPAMD817 demonstrated compound heterozygous variants, one a maternally inherited previously reported pathogenic splice site (c.4549–1G>A) variant, and another a novel missense variant (p.[Gly1050Val]) (Fig. 3d), inherited from the father. Interestingly, the father also had milder ocular ASD features with posterior embryotoxon, and some iridocorneal strands. No other known ocular ASD-related variants were found in his genomic analysis. While posterior embryotoxon is known to occur in 10–15% of the general population, the presence of the iridocorneal strands may indicate that heterozygous carriers for CPAMD8 variants may carry milder ocular changes, as has been noted for other autosomal recessive developmental ocular conditions.18
Similarly, reinterrogation of clinical and ES/GS data yielded a pathogenic variant in the recently and rarely reported syndromal gene ITPR1. In patient 25, initial analysis of ES data was negative. Subsequent manual review of GS trio data identified a de novo, heterozygous, previously reported19 pathogenic ITPR1 variant (c.7615G>A; p.[Gly2539Arg]) in the proband (Table 1, Fig. 3e). This variant was considered in the light of a concurrent clinical re-review at the time, which indicated presence of severe global developmental delay, iris hypoplasia with scalloped margins, and ataxia consistent with the diagnosis of Gillespie syndrome, known to be caused by variants in ITPR1.
Novel variants in FOXC1, and novel genotype–phenotype correlation in PITX2 with sclerocornea
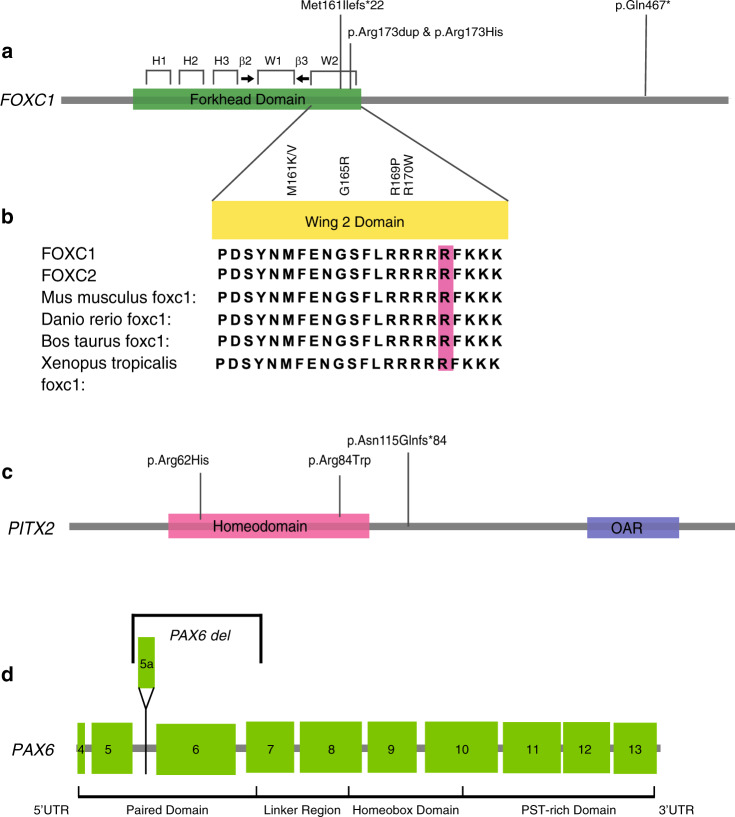
Although FOXC1 is one of the most well-studied genes in ASD, we found four novel variants in this gene. Two novel variants affecting p.Arg173 emphasize the importance of the Wing 2 region of the forkhead domain of FOXC1. Case 12 had a dominantly inherited missense (c.518G>A, p.[Arg173His]) variant, and the second proband (case 10) had an in-frame duplication of p.Arg173 (Table 1, Fig. 4a). The duplication variant was also found in the asymptomatic father of case 10, and subsequent detailed examination revealed that he actually had a mild form of ARA with posterior embryotoxon and evidence of iris adhesions. It is noteworthy that p.Arg173 lies in a group of five conserved arginine residues in the highly conserved Wing 2 region of the forkhead domain of FOXC1, which is conserved across species and related FOX proteins (Fig. 4b). Also, even in the more distantly related FOX proteins such as FOXL1/FOXS1 there is a very high degree of homology in this part of the gene across multiple species.20 Missense variants affecting the nearby p.Arg169 and p.Arg170 have also been identified in patients with ARS,21,22 and crystal structure analysis and biochemical studies of p.Arg169 indicate a role in the FOXC1 wing domain DNA binding and transactivation abilities.22,23 Our work further highlights the likely critical nature of the wing 2 domain in FOXC1 DNA binding and transactivation.
Fig. 4. Novel variants in FOXC1 Wing 2 domain, PITX2, and a PAX6 deletion identified on genome sequencing (GS).
(a) FOXC1 gene, with key domains and regions of the major forkhead domain, with wing 2 (W2) region highlighted. b Alignment demonstrates the highly conserved residues of this domain across the FOXC1 paralog FOXC2, and the phylogenetic tree. Previously reported pathogenic variants in the wing 2 domain are labeled, and the Arg173 highlighted in pink. c PITX2 gene and three novel variants found in this study. d PAX6 deletion is also demonstrated, with exon numbering in the diagram.
We also report two novel deleterious pathogenic variants in FOXC1 in children with ASD and intellectual disability (ID), indicating a possible role for this gene in learning disorders. A girl (patient 23) with PA, intellectual delay, and absent septum pellucidum harbored a novel nonsense pathogenic variant in FOXC1 (c.1399C>T, p.[Gln467*]) (Table 1, Fig. 4a). A novel frameshift variant (p.[Met161Ilefs*22]) was found in a young boy (patient 41) with iris hypoplasia, corectopia and glaucoma, global developmental delay, and bilateral sensorineural hearing loss (Table 1, Fig. 4a). For both patients, other causes of intellectual delay were not found following baseline testing with chromosome microarray, urine metabolic screen, and fragile X testing, or ES reanalysis for causative variants in known ID genes (Genomics England PanelApp, https://panelapp.genomicsengland.co.uk, Intellectual disability [Version 2.1046]). There have been reports of FOXC1 deletions in combination with deletions of surrounding genes, causing brain malformations such as Dandy–Walker malformation, partial agenesis of the corpus callosum, and intellectual disability.24 In a prior study, 1/13 SNV patients, who had a missense p.Met109Val in the forkhead domain of FOXC1, was reported to have learning difficulties.25 This, combined with our cases, highlights a potential new link between variants in this gene and ID.
Case 36 with a clinical diagnosis of peripheral sclerocornea had a missense pathogenic variant in PITX2 (c.185G>A, p.[Arg62His]), which was previously found in a family reported to have “ring dermoid” of the cornea. Notably the phenotypic images in this family also showed peripheral sclerocornea,26 very similar to our patient (Fig. 2g, 4c, Table 1). This highlights peripheral sclerocornea as a new phenotypic link to variation in PITX2. In PITX2, a novel pathogenic frameshift variant (c.341dup; p.[Asn115Glnfs*84]) was found causing premature truncation of the protein in a patient with ARA (patient 14, Fig. 2h, 4c, Table 1).
CNV detection in PAX6 facilitated by GS
A novel 1360-bp PAX6 deletion, was found in a proband (patient 11, Table 1) with PA, and his affected mother who had bilateral partial iris hypoplasia and right ectopia lentis. Initial investigation with ES in the proband was negative. GS was undertaken in the proband, his affected mother, and his two unaffected siblings and no causative SNVs were found. Subsequent SV analysis of GS data found a small 1.4-kb deletion in PAX6 in the proband and his mother, with breakpoints indicating a deletion of exons 5a, 6, and part of 7 in the paired domain (Fig. 4d) and the deletion was confirmed using MLPA (MRC-Holland P219-B1 PAX6 probemix). Review of CMA data could not find this deletion, as it was beyond the resolution of the Agilent 400K CGH microarray used. Similar deletions are reported in the literature, including one study with an exon 6–7 deletion and sporadic aniridia.27 Missense variants of the paired domain have also been reported in atypical/milder aniridia and also in PA, as well as congenital cataracts, foveal hypoplasia, keratitis, and optic nerve abnormalities.28
DISCUSSION
Deep phenotyping and variant curation for maximization of genetic diagnoses in the ocular ASDs
This study highlights the importance of detailed phenotypic information, pedigree structure, and careful variant curation to find “hidden” variants in the ocular ASD patient cohort that may not be apparent on first analysis. In over 1/3 of the cases (8/22) where causative variants were identified, the variants were novel and required careful clinical review and manual review of sequence data for their identification (Table 1). In addition, approximately one-third of the likely causative variants found in this study were in rarely reported ocular ASD genes with one, in ADAMTS17, where this association was identified for the first time (Figs. 1a, 2d, 3c). Identified variants revealed new genotype–phenotype correlations in rarely reported and syndromal ocular ASD genes (Fig. 1a and Fig. 2c, d, f), or were novel variants in key ocular ASD genes revealing new functional domain elements or phenotypic features (Figs. 4a–c, 2g), or in one case required GS analysis for CNV detection (Fig. 4d). This study demonstrates that accurate phenotypic information on the proband and family members, in conjunction with careful analysis of ES/GS variants, and trio and segregation analysis of variants, greatly benefits genetic diagnosis in the ocular ASDs. This approach had a vital role in discovering new genotype–phenotype correlations, broadening the list of candidate disease-causing genes and identifying additional likely causative variants.
In two families, pathogenic variants were found in recently reported ocular ASD-related genes CPAMD8 (patient 28) and ITPR1 (patient 25). In CPAMD8, the similar clinical features reported in the literature, with absence of posterior embryotoxon, corneal opacity, or any extraocular features, highlight a unique combination of features specific to this gene and a newly emerging genotype–phenotype correlation. Recently an additional family with a homozygous frameshift variant in CPAMD8 was found in a PCG cohort from Saudi Arabia,29 with PCG and lens subluxation. With our family, this increases the number of reported families with variants in this gene to five. Similarly, patient 25 initially referred with iris hypoplasia was found to have a syndromal form of ASD with clear genotype–phenotype correlation due to an ITPR1 variant.
Variants in collagen and extracellular matrix–associated genes highlight novel genotype–phenotype correlations
For three families in this study, variants were found in ECM-related genes COL4A1 (case 26, Fig. 2a, b, 3a), ADAMTS17 (case 9, Figs. 2d, 3c) and PXDN (case 21, Figs. 2c, 3b). Variants were also found in three cases in CYP1B1 (cases 29, 30, 38, Fig. 2e), and this gene has emerging evidence for its role in the ECM.30 In addition, variants in ECM genes of the TGFβ pathway have been found in other anterior segment abnormalities, including FBN1 (ectopia lentis), LTBP2 (primary congenital glaucoma), and ADAMTS10 (ectopia lentis).
Our findings highlight the importance of ECM, collagen, and related genes, in the formation of the anterior segment and scaffolding of tissues to the basement membrane. The ECM has a crucial role in regulating cell adhesion, cellular migration, and tissue morphogenesis. There is increasing evidence pointing to the importance of the periocular ECM in the morphogenesis of the optic cup,31 with cross-linking of laminin, collagen IV, and other ECM components noted as critical factors.32 In conditions associated with COL4A1 variants, eye abnormalities, including ASD, are the second commonest presenting feature.33 COL4A1 forms a heterotrimer with COL4A2, and is secreted into the ECM to form a scaffolding, with key bonds to other heterotrimers.11,34 Crucial to the stable formation of these bonds are sulfilamine bonds, and these are catalyzed in a peroxidase reaction by peroxidasin, or PXDN.35 This may explain why the phenotype associated with PXDN may be so severe, as demonstrated in case 21 (Fig. 2c) of this study, and as shown in the mouse pxdn nonsense variant model.36 Also, the association with PXDN and intellectual delay has been reported in siblings37 highlighting the broadening of the reported phenotype with this gene. In addition, ADAMTS17 belongs to a family of secreted metalloproteases with an important role in ECM remodeling.38
Implications for genetic information for patients and families, and genomic workflow in ASD
Few studies have been performed utilizing NGS in the area of ocular ASD with large cohorts consisting of patients with the variety of ASD clinical diagnoses that present to the clinic. A study using ES in PA3 found a genetic diagnosis in only a small proportion, and previous studies utilizing conventional sequencing of cohorts with a high proportion of phenotypically defined ARS cases1 led to a diagnosis rate of around 40% by examining PITX2/FOXC1. In ASD patients with a variety of ASD phenotypic subtypes that present for clinical genetic information, whether familial or sporadic, our work highlights the important role of ES/GS and SV analysis, as this leads to a high overall rate of diagnosis (22/41; 54%). Using this approach the highest yields of testing were in ARS (2/3; 67%), ARA (5/9; 56%), and PA (7/11; 64%) (Table 1).
Of clinical utility and importance for families, this approach led to a revision of the mode of inheritance in 8/19 (42%) of referred sporadic cases where a responsible variant was found, and in 1/4 (25%) of referred autosomal dominant cases (Fig. 1b). Finding a de novo autosomal dominant pathogenic variant may provide reproductive confidence for families. However, finding an autosomal recessive variant in a supposedly sporadic case means 25% recurrence risk for parents, which happened in six families (cases 9, 21, 28, 29, 30, 38; Fig. 1b). In one of these families (patient 28) with autosomal recessive CPAMD8 variants, subsequent detailed clinical review revealed subtle ASD features in the father, and without our genetic diagnostic findings, this could have been interpreted as an autosomal dominant family with variable expression. Also, two cases presenting as sporadic, and therefore low recurrence risk, were found to be familial autosomal dominant cases with 50% recurrence risk. This was after genotype and careful phenotype review revealed an affected parent (families 10 and 26). The parental features were milder, raising the possibility of somatic mosaicism. However, we could find no evidence of this on careful review of the genomic coverage, allele frequency data, and the Sanger sequencing traces.
This is the first study to evaluate the role of ES, GS, and CMA in ASD. Considering the findings of a large meta-analysis in children with suspected genetic diseases,39 it would have been expected there may be a diagnostic yield of CMA of around 10%, and ES of 36%, with an additional 5% detected by GS. In our study, CMA had a yield of 2/41 (5%). Upon initial analysis of ES data, the vast majority (15/19) of SNV variants were found and with GS, there were an additional 4 SNVs and a single CNV identified. However, the 4 SNVs found on GS would all have been found from ES, if it had been performed first-line rather than GS (cases 32 and 36), and clinical correlation and an updated gene list had been prioritized (cases 25 and 28). Hence, from a platform perspective of our cohort, using ES first, followed by the other approaches, would lead to a 46.3% (19/41) diagnostic yield with ES, with CMA finding pathogenic deletions in an additional two cases (extra 4.9%) and GS an additional one case (extra 2.4%), to give an overall genetic diagnosis rate of 22/41 (54%). If GS had been used as the first-line and only platform, then it would be expected to pick up all cases giving the same overall detection rate. At this stage, it is difficult to assess the yield and cost effectiveness of ES versus GS in a cohort examined over time with many confounding factors, as has been raised elsewhere in the literature.40 Apart from discovering small SVs, the enhanced yield of GS may lie in the discovery of additional potential regulatory variants, which would require further studies for variant interpretation. We would advise that clinical genetics and genetic eye services should consider performing either ES or GS as a first-line test in ASD, with SV analysis and trio/segregation analysis for additional yield. In addition, patients should first have detailed ophthalmic and clinical review for syndromal features, as well as parental review for undiagnosed autosomal dominant disease.
Conclusion
We have identified a high diagnostic rate in a cohort of families with ASD, using ES and GS with CNV analysis. These findings add to our understanding of the genetics of ASD, and also highlight the complex heterogeneous nature of this condition, with many syndromal associations and novel genotype–phenotype correlations. This study also has significant implications in recurrence risk counseling for families, and highlights the importance of relying on an accurate genomic diagnosis, rather than pedigree information alone, for genetic counseling. Some of our cohort remain unsolved, and this may be due to additional undiscovered genes, or further variants within known genes such as 5’UTR promoter, cis-regulatory, deep intronic, and other difficult to interpret regions of the genome. These conditions warrant further study to uncover their full genetic basis.
Supplementary information
Acknowledgements
R.V.J. and J.R.G. acknowledge support from National Health and Medical Research Council (NHMRC) grant APP1116360, the NSW Office of Health and Medical Research, Costco, the Sydney Research Excellence Initiative, and the Ophthalmic Research Institute of Australia. We thank the Kinghorn Centre for Clinical Genomics for assistance with production and processing of genome sequencing data. M.J.C. was supported by a NSW Health Early-Mid Career Fellowship. We thank the families involved in this study and supporting clinicians and laboratory staff for their time and efforts.
Disclosure
The authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-020-0854-x) contains supplementary material, which is available to authorized users.
References
- 1.Reis LM, Semina EV. Genetics of anterior segment dysgenesis disorders. Curr Opin Ophthalmol. 2011;22:314–324. doi: 10.1097/ICU.0b013e328349412b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reis LM, Tyler RC, Volkmann Kloss BA, et al. PITX2 and FOXC1 spectrum of mutations in ocular syndromes. Eur J Hum Genet. 2012;20:1224–1233. doi: 10.1038/ejhg.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weh E, Reis LM, Happ HC, et al. Whole exome sequence analysis of Peters anomaly. Hum Genet. 2014;133:1497–1511. doi: 10.1007/s00439-014-1481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma AS, Grigg JR, Prokudin I, Flaherty M, Bennetts B, Jamieson RV. New mutations in GJA8 expand the phenotype to include total sclerocornea. Clin Genet. 2018;93:155–159. doi: 10.1111/cge.13045. [DOI] [PubMed] [Google Scholar]
- 5.Ma AS, Grigg JR, Ho G, et al. Sporadic and familial congenital cataracts: mutational spectrum and new diagnoses using next-generation sequencing. Hum Mutat. 2016;37:371–384. doi: 10.1002/humu.22948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma AS, Grigg JR, Jamieson RV. Phenotype-genotype correlations and emerging pathways in ocular anterior segment dysgenesis. Hum Genet. 2018;138:899–915. doi: 10.1007/s00439-018-1935-7. [DOI] [PubMed] [Google Scholar]
- 7.Nash BM, Symes R, Goel H, et al. NMNAT1 variants cause cone and cone-rod dystrophy. Eur J Hum Genet. 2018;26:428–433. doi: 10.1038/s41431-017-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prokudin I, Simons C, Grigg JR, et al. Exome sequencing in developmental eye disease leads to identification of causal variants in GJA8, CRYGC, PAX6 and CYP1B1. Eur J Hum Genet. 2014;22:907–915. doi: 10.1038/ejhg.2013.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matias-Perez D, Garcia-Montano LA, Cruz-Aguilar M, et al. Identification of novel pathogenic variants and novel gene-phenotype correlations in Mexican subjects with microphthalmia and/or anophthalmia by next-generation sequencing. J Hum Genet. 2018;63:1169–1180. doi: 10.1038/s10038-018-0504-1. [DOI] [PubMed] [Google Scholar]
- 11.Meuwissen ME, Halley DJ, Smit LS, et al. The expanding phenotype of COL4A1 and COL4A2 mutations: clinical data on 13 newly identified families and a review of the literature. Genet Med. 2015;17:843–853. doi: 10.1038/gim.2014.210. [DOI] [PubMed] [Google Scholar]
- 12.Patel N, Khan AO, Alsahli S, et al. Genetic investigation of 93 families with microphthalmia or posterior microphthalmos. Clin Genet. 2018;93:1210–1222. doi: 10.1111/cge.13239. [DOI] [PubMed] [Google Scholar]
- 13.Zazo-Seco C, Plaisancie J, Bitoun P, et al. Novel PXDN biallelic variants in patients with microphthalmia and anterior segment dysgenesis. J Hum Genet. 2020;65:487–491. doi: 10.1038/s10038-020-0726-x. [DOI] [PubMed] [Google Scholar]
- 14.Morales J, Al-Sharif L, Khalil DS, et al. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85:558–568. doi: 10.1016/j.ajhg.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent A, Billingsley G, Priston M, et al. Phenotypic heterogeneity of CYP1B1: mutations in a patient with Peters’ anomaly. J Med Genet. 2001;38:324–326. doi: 10.1136/jmg.38.5.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teixeira LB, Zhao Y, Dubielzig RR, Sorenson CM, Sheibani N. Ultrastructural abnormalities of the trabecular meshwork extracellular matrix in Cyp1b1-deficient mice. Vet Pathol. 2015;52:397–403. doi: 10.1177/0300985814535613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheong SS, Hentschel L, Davidson AE, et al. Mutations in CPAMD8 cause a unique form of autosomal-recessive anterior segment dysgenesis. Am J Hum Genet. 2016;99:1338–1352. doi: 10.1016/j.ajhg.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng WY, Pasutto F, Bardakjian TM, et al. A puzzle over several decades: eye anomalies with FRAS1 and STRA6 mutations in the same family. Clin Genet. 2013;83:162–168. doi: 10.1111/j.1399-0004.2012.01851.x. [DOI] [PubMed] [Google Scholar]
- 19.McEntagart M, Williamson KA, Rainger JK, et al. A restricted repertoire of de novo mutations in ITPR1 cause Gillespie syndrome with evidence for dominant-negative effect. Am J Hum Genet. 2016;98:981–992. doi: 10.1016/j.ajhg.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4:345–352. doi: 10.1186/1479-7364-4-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gripp KW, Hopkins E, Jenny K, Thacker D, Salvin J. Cardiac anomalies in Axenfeld–Rieger syndrome due to a novel FOXC1 mutation. Am J Med Genet A. 2013;161A:114–119. doi: 10.1002/ajmg.a.35697. [DOI] [PubMed] [Google Scholar]
- 22.Murphy TC, Saleem RA, Footz T, Ritch R, McGillivray B, Walter MA. The wing 2 region of the FOXC1 forkhead domain is necessary for normal DNA-binding and transactivation functions. Invest Ophthalmol Vis Sci. 2004;45:2531–2538. doi: 10.1167/iovs.04-0167. [DOI] [PubMed] [Google Scholar]
- 23.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 24.Aldinger KA, Lehmann OJ, Hudgins L, et al. FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat Genet. 2009;41:1037–1042. doi: 10.1038/ng.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Haene B, Meire F, Claerhout I, et al. Expanding the spectrum of FOXC1 and PITX2 mutations and copy number changes in patients with anterior segment malformations. Invest Ophthalmol Vis Sci. 2011;52:324–333. doi: 10.1167/iovs.10-5309. [DOI] [PubMed] [Google Scholar]
- 26.Xia K, Wu L, Liu X, et al. Mutation in PITX2 is associated with ring dermoid of the cornea. J Med Genet. 2004;41:e129. doi: 10.1136/jmg.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bobilev AM, McDougal ME, Taylor WL, Geisert EE, Netland PA, Lauderdale JD. Assessment of PAX6 alleles in 66 families with aniridia. Clin Genet. 2016;89:669–677. doi: 10.1111/cge.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzoulaki I, White IM, Hanson IM. PAX6 mutations: genotype–phenotype correlations. BMC Genet. 2005;6:27. doi: 10.1186/1471-2156-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsaif HS, Khan AO, Patel N, et al. Congenital glaucoma and CYP1B1: an old story revisited. Hum Genet. 2018;138:1043–1049. doi: 10.1007/s00439-018-1878-z. [DOI] [PubMed] [Google Scholar]
- 30.Safari I, Suri F, Haji-Seyed-Javadi R, Yazdani S, Elahi E. The p.Gly61Glu mutation in CYP1B1 affects the extracellular matrix in glaucoma patients. Ophthalmic Res. 2016;56:98–103. doi: 10.1159/000443508. [DOI] [PubMed] [Google Scholar]
- 31.Kwan KM. Coming into focus: the role of extracellular matrix in vertebrate optic cup morphogenesis. Dev Dyn. 2014;243:1242–1248. doi: 10.1002/dvdy.24162. [DOI] [PubMed] [Google Scholar]
- 32.Bryan CD, Casey MA, Pfeiffer RL, Jones BW, Kwan KM. Optic cup morphogenesis requires neural crest-mediated basement membrane assembly. Development. 2020;147:dev181420. doi: 10.1242/dev.181420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zagaglia S, Selch C, Nisevic JR, et al. Neurologic phenotypes associated with COL4A1/2 mutations: expanding the spectrum of disease. Neurology. 2018;91:e2078–e2088. doi: 10.1212/WNL.0000000000006567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao M, Kiss M, Ou Y, Gould DB. Genetic dissection of anterior segment dysgenesis caused by a Col4a1 mutation in mouse. Dis Model Mech. 2017;10:475–485. doi: 10.1242/dmm.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirokmany G, Kovacs HA, Lazar E, et al. Peroxidasin-mediated crosslinking of collagen IV is independent of NADPH oxidases. Redox Biol. 2018;16:314–321. doi: 10.1016/j.redox.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan X, Sabrautzki S, Horsch M, et al. Peroxidasin is essential for eye development in the mouse. Hum Mol Genet. 2014;23:5597–5614. doi: 10.1093/hmg/ddu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi A, Lao R, Ling-Fung Tang P, et al. Novel mutations in PXDN cause microphthalmia and anterior segment dysgenesis. Eur J Hum Genet. 2015;23:337–341. doi: 10.1038/ejhg.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hubmacher D, Schneider M, Berardinelli SJ, et al. Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Sci Rep. 2017;7:41871. doi: 10.1038/srep41871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clark MM, Stark Z, Farnaes L, et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20:1122–1130. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.