Abstract
Purpose
Carrier status associates strongly with genetic ancestry, yet current carrier screening guidelines recommend testing for a limited set of conditions based on a patient’s self-reported ethnicity. Ethnicity, which can reflect both genetic ancestry and cultural factors (e.g., religion), may be imperfectly known or communicated by patients. We sought to quantitatively assess the efficacy and equity with which ethnicity-based carrier screening captures recessive disease risk.
Methods
For 93,419 individuals undergoing a 96-gene expanded carrier screen (ECS), correspondence was assessed among carrier status, self-reported ethnicity, and a dual-component genetic ancestry (e.g., 75% African/25% European) calculated from sequencing data.
Results
Self-reported ethnicity was an imperfect indicator of genetic ancestry, with 9% of individuals having >50% genetic ancestry from a lineage inconsistent with self-reported ethnicity. Limitations of self-reported ethnicity led to missed carriers in at-risk populations: for 10 ECS conditions, patients with intermediate genetic ancestry backgrounds—who did not self-report the associated ethnicity—had significantly elevated carrier risk. Finally, for 7 of the 16 conditions included in current screening guidelines, most carriers were not from the population the guideline aimed to serve.
Conclusion
Substantial and disproportionate risk for recessive disease is not detected when carrier screening is based on ethnicity, leading to inequitable reproductive care.
Keywords: carrier screening, genetic ancestry, medical guidelines, ethnicity, recessive disease
INTRODUCTION
Carrier screening identifies couples at increased risk for conceiving fetuses affected by serious conditions that appreciably reduce lifespan, result in intellectual disability, and/or benefit from prenatal or perinatal intervention. Carrier screening is typically performed by first identifying female partners who are carriers for autosomal recessive and/or X-linked conditions, and subsequently testing their reproductive partners for condition(s) for which the female was a carrier.1 A couple is considered at risk if the female carries a pathogenic variant that would cause an X-linked disease in a male child or if both partners are carriers of pathogenic variants in the same gene associated with an autosomal recessive disease.
Current professional guidelines by the American College of Medical Genetics and Genomics (ACMG) and the American College of Obstetricians and Gynecologists (ACOG) recommend pan-ethnic carrier screening for cystic fibrosis and spinal muscular atrophy.2–4 In addition, these two professional societies have long recommended carrier screening for a partially overlapping and expanded set of conditions based on a patient’s self-reported ethnicity (SRE).
In the past decade, the evolution of carrier screening methodology (e.g., next-generation sequencing, [NGS]) has enabled scalable screening of many serious Mendelian diseases simultaneously, termed expanded carrier screening (ECS). Current commercially available ECS panels range in size from tens to hundreds of diseases.5 Pan-ethnic ECS has established analytical validity,6 clinical validity,7 clinical utility,8 and cost-effectiveness.9 Though ACOG considers ECS an “acceptable strategy” and both ACOG and ACMG generally acknowledge the complexities of race, ethnicity, ancestry, and coupling patterns in their carrier screening recommendations,4,10–12 both groups currently stop short of recommending that ECS be preferentially offered and instead continue to endorse SRE-based screening.
A patient’s SRE combines genetic factors with cultural factors like traditions, lifestyle, diet, religious beliefs, and values, making SRE an indirect proxy for elevated carrier risk. A more direct relationship exists between carrier risk and genetic ancestry because both are based exclusively upon genetic variation. The analysis of genetic ancestry has been enabled by the genotyping of populations across the world (e.g., the Yoruba and Mandinka populations)13,14 from which allele frequencies of inferred ancestral populations (e.g., sub-Saharan African) are derived. An individual patient’s genetic ancestry can therefore be calculated by applying methods15,16 that ascertain the quantitative genetic ancestry proportion from each inferred ancestral population. In contrast, SRE is often qualitative, subjective, and ambiguous. For instance, 39.6% of individuals cannot identify the ancestry of all four grandparents,17 limiting the ability of SRE to reflect genetic ancestry. Further, the design of test requisition forms (e.g., binary choice vs. multiple categories) on which SRE is collected, and the stage at which the requisition is completed (i.e., pre- vs post-test), have been shown to influence SRE choice.18,19 The inability of SRE to accurately reflect genetic ancestry implies that ethnicity-based carrier screening recommendations may not achieve their intended effect, i.e., to identify those at highest risk of carrying serious conditions.
Genetic ancestry in the context of ECS was explored in a 2017 study of approximately 9000 individuals.19 Here we employ a ten times larger cohort to specifically evaluate the validity and impact of ethnicity-based carrier screening clinical guidelines. We analyzed the genetic ancestry of 93,419 individuals undergoing routine ECS for over 90 serious Mendelian disorders via NGS. We investigated the relationship between SRE and genetic ancestry, as well as the relationship between genetic ancestry and carrier status for the studied genetic disorders.
MATERIALS AND METHODS
Study cohort
The study included individuals undergoing ECS (Foresight, Myriad Women’s Health) via NGS between 1 January 2014 and 8 September 2016 who had consented to anonymized research. The protocol for this study was reviewed and designated as exempt by Western Institutional Review Board and complied with the Health Insurance Portability and Accountability Act (HIPAA). Patients could select among the following SRE options on the test requisition form: Northern European, Southern European, French Canadian or Cajun, Ashkenazi Jewish, Mixed or Other Caucasian, East Asian, South Asian, Southeast Asian, African or African American, Hispanic, Middle Eastern, Native American, Pacific Islander, or Unknown. Patients choosing multiple self-reported ethnicities were counted as Mixed or Other Caucasian, and their multiple individual SRE selections (e.g., Hispanic and Northern European) were not accessible during the analysis. Patients with SRE of Unknown were excluded, as were those with SRE of Finnish, Pacific Islander, Native American, or French Canadian/Cajun due to insufficient sample size (<500 individuals in each subpopulation). An additional 621 individuals were excluded due to complex genetic ancestry (see below), resulting in a final analyzed cohort of 93,419 individuals.
Carrier screening
All patients were analyzed with short-read NGS of exons and certain intronic regions, as well as a specialized assay for fragile X syndrome.6 DNA was extracted from blood or saliva. The 96 diseases and corresponding genes included for analysis are listed in SI Table 1. Conditions for which there are multiple manifestations of disease severity (e.g., classical (severe) and nonclassical (moderate) forms of 21-hydroxylase-deficient congenital adrenal hyperplasia) were analyzed as a single, combined condition. A carrier was defined as a patient harboring a pathogenic or likely pathogenic variant in a gene of interest. Certain high-frequency variants or variant combinations involving complex interpretation or mild phenotypes—e.g., the NM_000500.7(CYP21A2):c.955C>T(Q319*) variant in combination with a duplication for congenital adrenal hyperplasia (CYP21A2-related), or the NM_000060.2(BTD):c.1330G>C(D444H) variant for biotinidase deficiency—were considered benign for population-level carrier rate analyses and are enumerated in SI Table 2.
When analyzing guidelines, we took an inclusive approach: if either ACMG or ACOG recommended that patients should be screened or that screening should be considered for a disease, we considered the disease to be part of current guidelines (SI Table 3).
Genetic ancestry analysis
On- and off-target reads obtained during ECS were used for genetic ancestry analysis after filtering was applied for read mapping quality and base-quality scores, as described by Wang et al.20
Genetic ancestry was inferred using a proprietary analysis method (patent application WO2018144135A1). In brief, the method is based on sparse non-negative matrix factorization (with seven components) and was used to estimate ancestry shared with hypothetical ancestral populations using an imputed reference panel of 51 populations and an additional imputed reference panel of the Ashkenazi Jewish population (GEO accession number GSE23636).21 Ancestry was estimated using distances along pair-wise ancestry clines between reference populations (e.g., a European-African cline): each patient was assigned a score from 0 (0%) to 1 (100%) for each genetic ancestry component, with at most two nonzero components. The genetic ancestry components determined were (sub-Saharan) African, Ashkenazi Jewish, European, Middle Eastern, South Asian, East Asian, Native American, and Pacific Islander. For finer resolution of genetic ancestry among patients with >87.5% South Asian, Middle Eastern, Ashkenazi Jewish, or European genetic ancestry from the analysis using the first reference panel, we used a second reference panel containing only those populations (four components).
Six hundred twenty-one individuals (0.66%) could not be confidently categorized as belonging to a single genetic ancestry cline between two populations, mostly due to ancestry from three or more sources (e.g., African, European, and Native American genetic ancestry). Individuals who were not included in subsequent analyses mostly self-reported as Hispanic (506, 81%), Mixed or Other Caucasian (71, 11%), and African or African American (32, 5%), with the remaining 12 individuals characterized by other self-reported ethnicities.
Matching populations among guidelines, self-reported ethnicity, and genetic ancestry
The ethnicity categories used in guidelines and in the ECS test requisition forms do not match perfectly (e.g., a patient can self-report as Southern European, but this is not an ethnicity category for ACOG or ACMG). Furthermore, due to constraints of the ancestry analysis, not all self-reported ethnicities for which there are guidelines were associated with a single genetic ancestry component. The correspondences among self-reported ethnicities and genetic ancestries included in ACOG/ACMG guidelines, the test requisition forms, and our genetic ancestry analysis are listed in SI Table 3.
Categorizing genetic ancestry as low, medium, or high
For all ancestries, we defined a “low” amount of genetic ancestry as being less than 1/32. We then considered genetic ancestry in increments of 1/32 and defined a threshold for a “high” amount of genetic ancestry (g) based on the lowest range of genetic ancestry—g…g + 1/32—in which more than half of individuals self-reported the associated ethnicity. For instance, African genetic ancestry between 15/32 and 16/32 was the lowest genetic ancestry range in which over 50% of patients had a SRE of African or African American; thus, g = 15/32 was the threshold between “high” and “medium” African genetic ancestry. We defined a “medium” amount of genetic ancestry as being between the two threshold values (i.e., more than “low” threshold of 1/32 but less than the “high” threshold at g). The threshold for European genetic ancestry was determined based on both Northern and Southern Europeans.
RESULTS
Self-reported ethnicity is an imperfect indicator of genetic ancestry
We quantified the agreement between SRE and genetic ancestry by categorizing individuals according to the source of the genetic ancestry component responsible for >50% of their ancestry (GAmajority). Overall, 9% of patients (comprising 2475 carriers) had a GAmajority that was unexpected based on the SRE (excluding those self-reporting as Mixed or Other Caucasian). Concordance between SRE and GAmajority was highest in those who self-reported as Northern European (96.9%), Hispanic (96.4%), South Asian (96.3%), or Southeast Asian (95.8%) (Fig. 1a). Concordance was lowest (<90%) in those self-reporting as Middle Eastern (59.2%), Ashkenazi Jewish (80.2%), or Southern European (84.0%) (Fig. 1a). We also observed wide distributions of the genetic ancestry proportions within SRE groups both above and below the 50% threshold (Fig. 1b), suggesting that genetic ancestry level can vary among individuals in the same SRE and that substantial genetic ancestry may still be present even if GAmajority and SRE do not match. Nearly one-third of patients (31%) had a SRE of Mixed or Other Caucasian (either by directly selecting this option on the requisition form or by selecting more than one ethnicity; see “Materials and Methods”), with most having GAmajority of European (Fig. 1a, Other row). Supplemental Text 1 describes these and further observations in more detail.
Fig. 1. Imperfect agreement between self-reported ethnicity and genetic ancestry.
(a) Individuals were categorized according to the source of the genetic ancestry component responsible for >50% of their ancestry (GAmajority). Each row corresponds to a self-reported ethnicity group and sums to 100% (Pacific Islander genetic ancestry not displayed). “Expected” genetic ancestry roughly corresponds to the diagonal and is marked with asterisks (*). (b) Distribution of genetic ancestry (subpanel x-axis) among self-reported ethnicity groups (subpanel titles). Vertical lines within violin plots indicate the 25th, 50th, and 75th percentiles of genetic ancestry. The highlighted region to the right of the vertical 50% line indicates agreement between self-reported ethnicity and GAmajority. EA East Asian, SA South Asian, ME Middle Eastern, AJ Ashkenazi Jewish, EUR European, AF African, NA Native American genetic ancestry.
Carrier risk enrichment in those with substantial but unreported ancestry
We next explored whether limitations of SRE affect carrier identification. Specifically, we hypothesized that individuals with medium genetic ancestry who did not self-report the corresponding ethnicity may have an increased carrier risk, yet their elevated risk would not be captured by ethnicity-specific guidelines. For each SRE, we first partitioned individuals into +SRE (i.e., selected the ethnicity on the requisition form) and -SRE (i.e., did not select the ethnicity) groups, and then further subdivided the groups based on the corresponding genetic ancestry, resulting in high, medium, and low genetic ancestry subgroups (Fig. 2; see “Materials and Methods”).
Fig. 2. For several conditions, patients who have medium genetic ancestry and do not self-report the associated ethnicity have significantly elevated carrier risk.
For progressively higher levels of genetic ancestry (in 32 bins from 0% to 100% on the x-axis), the left panels of (a–d) indicate the relative proportion of patients who self-report the associated ethnicity (see key at top for color scheme). The cutoff between medium genetic ancestry and high genetic ancestry is described in “Materials and Methods”. For the indicated conditions, panels on the right show the relative carrier rate of patients in each subcohort (colored as in the left panels). Percentages shown in yellow in the right panel indicate the proportion of non–self-reporting individuals with medium genetic ancestry screened for the given disease, with an additional annotation indicating the relative carrier risk value. A condition is plotted if the carrier rate was significantly greater in (1) the +SRE population, (2) the -SRE medium genetic ancestry population, and (3) the -SRE high genetic ancestry population, compared to the -SRE population based on the 95% confidence interval of the risk ratio (estimated using a Z-test of the log-transformed risk ratio). While cystic fibrosis met these criteria in the Northern European population, it is recommended for pan-ethnic screening by both the American College of Medical Genetics and Genomics (ACMG) and American College of Obstetricians and Gynecologists (ACOG) and was thus omitted from the figure. GA genetic ancestry, SRE self-reported ethnicity.
We observed that a substantial proportion of individuals fell into the “medium genetic ancestry” subgroup. For example, while 7100 (7.6%) patients in the entire cohort self-reported as African or African American, 2800 (3.0%) other patients had medium African genetic ancestry and did not self-report as being African or African American. The latter group would not have been screened under ethnicity-specific guidelines, yet they had a 3.6-fold higher risk of being a carrier of Hb β chain–related hemoglobinopathy and a 3.0-fold higher risk for being a carrier of ɑ-thalassemia relative to -SRE individuals (Fig. 2a). Among all patients with medium African genetic ancestry, just 16% self-reported as African or African American; the others tended to self-report as Hispanic (47%) or Mixed or Other Caucasian (25%) (SI Fig. 2), meaning that SRE-based guidelines would not have captured their elevated risk for being carriers of hemoglobinopathies.
Importantly, the insufficiency of SRE as a means to identify elevated carrier risk for particular conditions was true for other ancestries as well, including the Ashkenazi Jewish (Fig. 2b), East Asian (Fig. 2c), and South Asian (Fig. 2d) populations. Our observation of carrier rate enrichment among -SRE patients with medium genetic ancestry in Fig. 2 would be trivial if it were composed of Mixed or Other Caucasian patients where one of their several reported ethnicities was the ethnicity of interest (see “Materials and Methods”); however, we observed comparable carrier rate enrichment among -SRE patients with medium genetic ancestry even when we altogether excluded Mixed or Other Caucasian patients (SI Fig. 2). These data suggest that many patients had elevated carrier risk due to their genetic ancestry and did not self-report an ethnicity that would have indicated their risk.
Patients with high levels of genetic ancestry did not always self-report the associated ethnicity, and carrier rates were comparable irrespective of self-reporting (blue vs. gray, Fig. 2). When patients did not self-report the ethnicity associated with their high genetic ancestry, they typically self-reported as Mixed or Other Caucasian or as a related population (e.g., those with high East Asian genetic ancestry self-reporting as Southeast Asian) (SI Fig. 3). Although we do not know why the patients in our cohort reported an ethnicity discordant with their high genetic ancestry, they nonetheless may not have received screening appropriate for their risk if ethnicity-based guidelines had been strictly followed.
Both ethnicity- and ancestry-based guidelines miss substantial proportions of carriers
We hypothesized that carriers of conditions with ethnicity-specific guidelines could be missed because of deficiencies in self-reporting (i.e., those with medium or high genetic ancestry do not self-report the related ethnicity) and/or because many carriers exist outside of the presumed high-risk ancestries (i.e., an abundance of carriers in the low genetic ancestry group, which has many patients and a low carrier rate).
We observed that within the +SRE group, most carriers for conditions in current ACOG and ACMG guidelines had medium or high genetic ancestry, while in the -SRE group the proportion of carriers from the low genetic ancestry group was substantial for all diseases (Fig. 3). This suggests that a large proportion of carriers would still be missed if current ethnicity-based guidelines were adapted to be ancestry-based instead (Fig. 3, comparing the hatched and unhatched regions).
Fig. 3. Distributions of carriers for diseases in the American College of Medical Genetics and Genomics (ACMG) or American College of Obstetricians and Gynecologists (ACOG) guidelines in the +SRE vs. -SRE (blue vs. red) and low vs. medium/high genetic ancestry (hatched vs. unhatched) groups show that both ethnicity- and ancestry-specific screening miss a substantial proportion of carriers.
For diseases with multiple guideline ethnicities (ɑ-thalassemia and Hb β chain–related hemoglobinopathies), a person was considered in the “low genetic ancestry” group if all the related ancestries (shown in parentheses on each row) were low (<1/32). For the Southeast Asian guidelines, determination of low vs. high/medium genetic ancestry was based on South Asian genetic ancestry instead of East Asian genetic ancestry (giving a smaller proportion of carriers in the -SRE, low genetic ancestry group). For 7 of the 16 diseases, most carriers were in the -SRE population; for 7 diseases, at least 25% of carriers were in the low genetic ancestry population. Cystic fibrosis and spinal muscular atrophy were not included, as pan-ethnic carrier screening is recommended for these diseases. EA East Asian, SA South Asian, ME Middle Eastern, AJ Ashkenazi Jewish, EUR European, AF African, NA Native American genetic ancestry.
The distributions of frequencies for variants observed in the low versus medium/high genetic ancestry groups differed, with each group typically having some distinct variants and the medium/high genetic ancestry group having relatively less variant diversity (as expected in the case of founder or ancestry-related variants) (SI Fig. 4; SI Table 5). These data suggest that targeted genotyping carrier screening that interrogates only a subset of variants (as opposed to novel variant detection via NGS) would miss carriers outside of the target population.
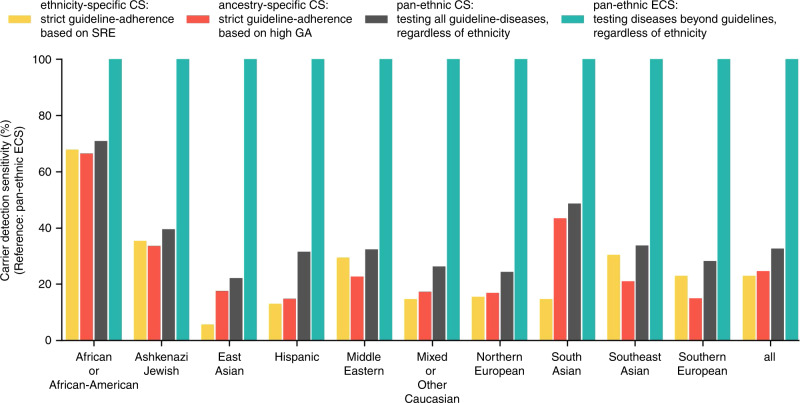
Overall, relative to pan-ethnic ECS, ethnicity-based guidelines would have identified only 23% of carriers in the study cohort, with large differences per ethnicity (Fig. 4, SI Table 4). Just over two-thirds (68%) of carriers among African Americans would have been identified based on screening guidelines from ACMG and ACOG; that number decreases to only 13% of carriers identified among Hispanics when guidelines are followed. Missed carriers were associated with 69 to 94 distinct diseases, depending on the ethnicity (SI Table 4). Furthermore, depending on the ethnicity, only 7–49% of guideline-missed carriers were accounted for by a single disease (SI Table 4), suggesting that the incremental addition of single diseases to current guidelines is not sufficient to close the gap in carrier identification. An ancestry-specific screening approach would have a marginal improvement over ethnicity-specific carrier screening in some ethnicities, yet it would perform worse in other ethnicities (red vs. yellow bars, Fig. 4). A pan-ethnic carrier screening approach would identify the carriers of diseases currently in guidelines but would miss carriers for all other diseases (gray vs. teal bars, Fig. 4). These data suggest that current guidelines do not identify most carriers for serious recessive diseases, and that either pan-ethnic ECS (teal bars, Fig. 4) or the addition of many disease–ethnicity pairs to current guidelines would be needed to adequately identify carriers in the population.
Fig. 4. Comparison of carrier detection sensitivity for different carrier screening scenarios across self-reported ethnicities (SREs).
Ethnicity-specific (yellow) and ancestry-specific (red) carrier screening also include pan-ethnic screening of cystic fibrosis and spinal muscular atrophy. Ancestry-specific carrier screening considers a person eligible for testing if they have “high” levels of any relevant genetic ancestry for a given guideline disease. CS carrier screening, ECS expanded carrier screening.
DISCUSSION
Here we evaluated current carrier screening guidelines by exploring the relationship between SRE, genetic ancestry, and carrier status for a large cohort tested with a 96-gene ECS panel. For the following reasons, we conclude that the current practice of ethnicity-based carrier screening has consequential deficiencies that impact patient care. First, our data demonstrate that SRE is an imperfect indicator of genetic ancestry, which is problematic if the former guides who is offered screening but the latter guides who is a carrier. The gradation of risk by genetic ancestry and the wide distribution of genetic ancestry within an SRE group suggests that the binary nature of SRE may be too coarse to capture clinically meaningful risk enrichment. Second, even if ethnicity-specific guidelines were revised to recommend more diseases in particular ethnicities, carriers would still be missed because many individuals with elevated levels of a genetic ancestry do not self-report the associated ethnicity. Third, for 7 of the 16 diseases in guidelines, the majority of the carriers identified in our cohort were outside the population with a guideline. In several cases, the risk was not due to self-reporting limitations but rather to the nonzero carrier rate in nonguideline populations. Pan-ethnic screening—currently deemed acceptable but not yet recommended by medical societies—overcomes these limitations of ethnicity-based screening.
In past decades, when carrier screening panels were costly to offer and challenging to expand with content, ethnicity-specific screening could provide efficient allocation of health-care resources by screening for a limited set of diseases in high-risk populations. This approach had important successes: ethnicity-based screening for Tay–Sachs disease in the Ashkenazi Jewish population has reduced the number of affected pregnancies in that population so precipitously that there are now relatively more Tay–Sachs disease–affected pregnancies in non–Ashkenazi Jewish ethnicities. But, for several reasons, the original motivations for limiting the recipients and content of carrier screening panels are no longer compelling. First, risk is typically not concentrated in specific ethnicities and is being further spread among populations due to changing demographics and increased intergroup marriage. This increased admixture is reflected in our results: 31% were Mixed or Other Caucasian, and genetic ancestry varied widely in certain SRE. Second, it is not clear that ethnicity-based guidelines are efficient, as a previous study found that an average of 2–5 minutes of office visits were consumed with selection of the ethnic origins of patients undergoing prenatal sickle cell screening.18 Third, Condit et al.17 showed that a large proportion of patients cannot accurately identify the ethnicities of their ancestors. And fourth, the cost of screening for additional conditions is now marginal due to advancements in DNA sequencing methodologies. These factors suggest that modern pan-ethnic ECS panels are an efficient and equitable means to assess reproductive risk for recessive disease.
Our data show that challenges would remain even if a genetic ancestry–based approach, rather than the current SRE-based approach, were used to administer carrier screening. Such genetic ancestry–based guidelines (e.g., screen for condition X if the patient has ancestry for population Y above threshold Z) may still perpetuate the inequities and stigmatization associated with ethnicity-based screening because they restrict screening to only certain populations.22,23 It is also not clear how a genetic ancestry–based carrier screening approach could be efficiently executed on a population scale. For example, if genetic ancestry assessment preceded carrier screening, detailed specifications would be needed to describe how to transmit a patient’s genetic ancestry information to carrier screening laboratories and how the resulting panels should be selected. If genetic ancestry assessment were performed by the carrier screening laboratory, the panel selection challenges would be compounded by ethical issues, such as potentially needing to notify patients of unexpected genetic ancestry in their lineage. For these reasons, as well as results from this study showing that a genetic ancestry approach would have suboptimal carrier detection in some ethnicities, we submit that pan-ethnic ECS is the most effective strategy to simplify administration of population-wide carrier screening and maximize identification of at-risk carrier couples. Results from Ben-Shachar et al. support this notion, showing that ethnicity-based screening found only 18% of the at-risk couples that ECS identified.24 However, using genetic ancestry as a means to calculate patient-tailored residual risk (i.e., by using a proportional combination of detection rates and incidences based on the patient’s ancestral components rather than based on SRE) upon screening negative may have promising potential.
Our results generally provide support for pan-ethnic ECS, but we have not directly explored herein the potentially negative consequences of pan-ethnic ECS, such as increased provider burden for partner testing, an increased rate of variants of uncertain significance (VUS), reduced perceived clinical utility for screening conditions not historically included in guidelines, and reduced cost-effectiveness. However, these possible consequences have been explored and refuted elsewhere. First, two recent studies24,25 highlighted how addition of more carrier screening conditions to a panel does not linearly increase the frequency with which a carrier’s partner would need to be tested: on a 176-condition panel, pan-ethnic screening of just the 18 (10.2%) most prevalent conditions would disproportionately account for 61% of carriers and 84% of at-risk couples detected by the entire panel. Second, though VUS classifications are more likely for rare conditions without substantial evidence of cases in the literature, such variants are not reported with ECS, and consensus of variant classifications was shown to remain high even for rare ECS conditions.7 Third, in a clinical utility study of ECS, patients’ pursuit of alternative reproductive options was driven largely by the severity of the condition for which their children were at risk, not by whether the condition was part of guideline-based screening; indeed, 72% of the at-risk couples who planned or pursued an alternative reproductive option were carriers for conditions outside of guidelines.8 Finally, cost-effectiveness of NGS-based CS has been explored in two publications, one on a guideline-based panel of 14 conditions26 and another on a 176-gene ECS panel.9 Both panels were shown to be cost-effective, and due to life-years gained from screening for a broader array of conditions, a 176-gene panel showed higher cost-effectiveness at multiple price points relative to screening for only cystic fibrosis (by genotyping) and spinal muscular atrophy.9
Despite the ability of ECS to most effectively identify risk across all ethnicities, most carrier screening in the United States does not use an expanded panel. A recent clinical experience study of more than 300,000 screened patients in the United States reported that 52% of carrier screens tested for four genes or fewer, and 80% of screens used a panel with fewer than 30 genes.27 Though many factors contribute to the observation that only 1 in 5 individuals received an ECS panel with >30 genes, we submit that two are most impactful: medical professional societies do not currently recommend such panels, and, accordingly, large commercial payers do not provide coverage for them. Indeed, in July 2019, the largest US commercial payer stopped covering ECS from commercial laboratories, specifically citing the lack of supportive guidelines and insufficient evidence of efficacy.28 Our current study adds to the evidence of ECS efficacy and quantifies the clinical shortcomings of the ethnicity-based approach to carrier screening.
The results of this study could be further refined in the future. For instance, the set of conditions and populations analyzed could be expanded, empowering genetic ancestry analysis that interrogates subancestries with greater precision. In particular, the delineation of individuals with joint European–South Asian ancestry versus those with joint European–Middle Eastern ancestry could be improved upon (miscategorizing these ancestry clines could partially underlie the 28% of self-reported Middle Easterners with South Asian GAmajority; the reversed combination—i.e., high proportion of self-reported South Asians with Middle Eastern GAmajority—was not observed). Further investigation is also warranted for individuals reporting multiple ethnicities on the requisition form, who were considered as belonging to the Mixed or Other Caucasian SRE group. While our findings indicate that over 90% of these patients have European GAmajority, factors influencing the choice of multiple ethnicities are of interest but could not be investigated here.
Taken together, our observations—coupled with studies demonstrating the analytical validity, clinical validity, clinical utility, and cost-effectiveness of ECS—elucidate the merit of pan-ethnic ECS as a means to provide equitable reproductive care and maximize the impact of carrier screening.
Supplementary information
Acknowledgements
The authors thank the patients who made this study possible. We also thank and acknowledge Katie Johansen Taber, Aishwarya Arjunan, and Anna Gardiner for their thorough comments on and assistance with the manuscript.
Disclosure
K.E.K., I.S.H., J.D.G., and D.M. are current or former employees of Myriad Women’s Health. I.S.H. is a former employee of Freenome, Inc. and is currently employed by Recursion Pharmaceuticals. L.P.S declares no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41436-020-0869-3) contains supplementary material, which is available to authorized users.
References
- 1.Pletcher BA, Bocian M. Preconception and prenatal testing of biologic fathers for carrier status. Genet Med. 2006;8:134–135. doi: 10.1097/01.gim.0000200948.58427.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior TW, Professional P, Guidelines C. Carrier screening for spinal muscular atrophy. Genet Med. 2008;10:840–842. doi: 10.1097/GIM.0b013e318188d069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grody WW, Thompson BH, Gregg AR, et al. ACMG position statement on prenatal/preconception expanded carrier screening. Genet Med. 2013;15:482–483. doi: 10.1038/gim.2013.47. [DOI] [PubMed] [Google Scholar]
- 4.Committee on Genetics. Committee opinion no. 691: carrier screening for genetic conditions. Obstet Gynecol. 2017;129:e41–e55. doi: 10.1097/AOG.0000000000001952. [DOI] [PubMed] [Google Scholar]
- 5.Chokoshvili D, Vears D, Borry P. Expanded carrier screening for monogenic disorders: where are we now? Prenat Diagn. 2018;38:59–66. doi: 10.1002/pd.5109. [DOI] [PubMed] [Google Scholar]
- 6.Hogan GJ, Vysotskaia VS, Beauchamp KA, et al. Validation of an expanded carrier screen that optimizes sensitivity via full-exon sequencing and panel-wide copy number variant identification. Clin Chem. 2018;64:1063–1073. doi: 10.1373/clinchem.2018.286823. [DOI] [PubMed] [Google Scholar]
- 7.Kaseniit KE, Collins E, Lo C, et al. Inter-lab concordance of variant classifications establishes clinical validity of expanded carrier screening. Clin Genet. 2019;96:236–245. doi: 10.1111/cge.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansen Taber KA, Beauchamp KA, Lazarin GA, Muzzey D, Arjunan A, Goldberg JD. Clinical utility of expanded carrier screening: results-guided actionability and outcomes. Genet Med. 2019;21:1041–1048. doi: 10.1038/s41436-018-0321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beauchamp KA, Johansen Taber KA, Muzzey D. Clinical impact and cost-effectiveness of a 176-condition expanded carrier screen. Genet Med. 2019;21:1948–1957. doi: 10.1038/s41436-019-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Committee on Genetics American College of Obstetricians and Gynecologists. ACOG committee opinion. Number 325, December 2005. Update on carrier screening for cystic fibrosis. Obstet Gynecol. 2005;106:1465–1468. doi: 10.1097/00006250-200512000-00055. [DOI] [PubMed] [Google Scholar]
- 11.Grody WW, Cutting GR, Klinger KW, et al. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–154. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Edwards JG, Feldman G, Goldberg J, et al. Expanded carrier screening in reproductive medicine-points to consider: a joint statement of the American College of Medical Genetics and Genomics, American College of Obstetricians and Gynecologists, National Society of Genetic Counselors, Perinatal Quality Foundation, and Society for Maternal-Fetal Medicine. Obstet Gynecol. 2015;125:653–662. doi: 10.1097/AOG.0000000000000666. [DOI] [PubMed] [Google Scholar]
- 13.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 14.Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Nyunoya T, Leng S, Belinsky SA, Tesfaigzi Y, Bruse S. Softwares and methods for estimating genetic ancestry in human populations. Hum Genomics. 2013;7:1. doi: 10.1186/1479-7364-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Condit C, Templeton A, Bates BR, Bevan JL, Harris TM. Attitudinal barriers to delivery of race-targeted pharmacogenomics among informed lay persons. Genet Med. 2003;5:385–392. doi: 10.1097/01.GIM.0000087990.30961.72. [DOI] [PubMed] [Google Scholar]
- 18.Dyson SM, Culley L, Gill C, et al. Ethnicity questions and antenatal screening for sickle cell/thalassaemia [EQUANS] in England: a randomised controlled trial of two questionnaires. Ethn Health. 2006;11:169–189. doi: 10.1080/13557850500460348. [DOI] [PubMed] [Google Scholar]
- 19.Shraga R, Yarnall S, Elango S, et al. Evaluating genetic ancestry and self-reported ethnicity in the context of carrier screening. BMC Genet. 2017;18:99. doi: 10.1186/s12863-017-0570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet. 2015;96:926–937. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci USA. 2010;107:16222–16227. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henneman L, Borry P, Chokoshvili D, et al. Responsible implementation of expanded carrier screening. Eur J Hum Genet. 2017;25:1291. doi: 10.1038/ejhg.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Hout S, Holtkamp KC, Henneman L, de Wert G, Dondorp WJ. Advantages of expanded universal carrier screening: what is at stake? Eur J Hum Genet. 2016;25:17–21. doi: 10.1038/ejhg.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Shachar R, Svenson A, Goldberg JD, Muzzey D. A data-driven evaluation of the size and content of expanded carrier screening panels. Genet Med. 2019;21:1931–1939. doi: 10.1038/s41436-019-0466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo MH, Gregg AR. Estimating yields of prenatal carrier screening and implications for design of expanded carrier screening panels. Genet Med. 2019;21:1940–1947. doi: 10.1038/s41436-019-0472-7. [DOI] [PubMed] [Google Scholar]
- 26.Azimi M, Schmaus K, Greger V, Neitzel D, Rochelle R, Dinh T. Carrier screening by next-generation sequencing: health benefits and cost effectiveness. Mol Genet Genomic Med. 2016;4:292–302. doi: 10.1002/mgg3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westemeyer M, Saucier J, Wallace J, et al. Clinical experience with carrier screening in a general population: support for a comprehensive pan-ethnic approach. Genet Med. 2020. 10.1038/s41436-020-0807-4. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 28.UnitedHealthcare. Carrier testing for genetic diseases: commercial medical policy. 2019. https://www.uhcprovider.com/content/dam/provider/docs/public/policies/comm-medical-drug/carrier-testing-for-genetic-diseases.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.