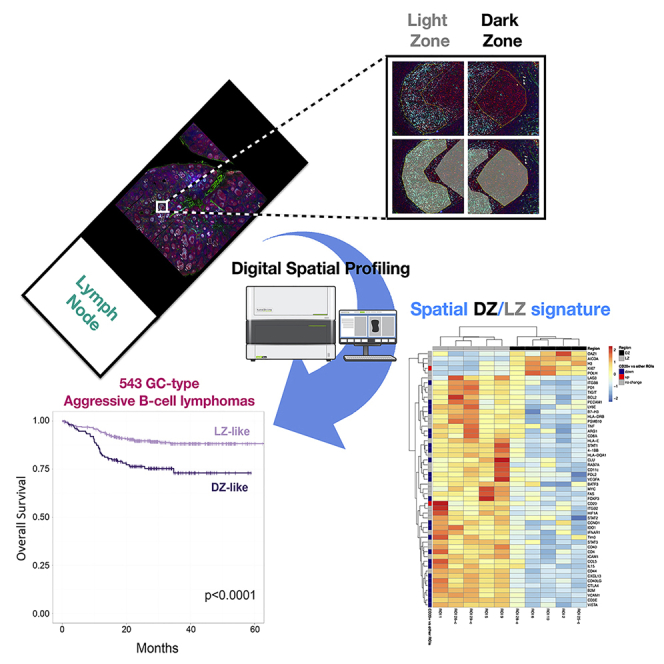
Summary
We applied digital spatial profiling for 87 immune and stromal genes to lymph node germinal center (GC) dark- and light-zone (DZ/LZ) regions of interest to obtain a differential signature of these two distinct microenvironments. The spatially resolved 53-genes signature, comprising key genes of the DZ mutational machinery and LZ immune and mesenchymal milieu, was applied to the transcriptomes of 543 GC-related diffuse large B cell lymphomas and double-hit (DH) lymphomas. According to the DZ/LZ signature, the GC-related lymphomas were sub-classified into two clusters. The subgroups differed in the distribution of DH cases and survival, with most DH displaying a distinct DZ-like profile. The clustering analysis was also performed using a 25-genes signature composed of genes positively enriched in the non-B, stromal sub-compartments, for the first time achieving DZ/LZ discrimination based on stromal/immune features. The report offers new insight into the GC microenvironment, hinting at a DZ microenvironment of origin in DH lymphomas.
Subject Areas: Cancer Systems Biology, Cancer, Transcriptomics
Graphical Abstract
Highlights
-
•
Digital Spatial Profiling identifies an immune/stromal GC dark-zone/light-zone signature
-
•
The DZ/LZ signature highlights prognostic clusters in aggressive GC B cell lymphomas
-
•
Digital Spatial Profiling reveals GC DZ microenvironment programs in double-hit lymphomas
Cancer Systems Biology; Cancer; Transcriptomics
Introduction
Within the category of aggressive B cell lymphomas, diffuse large B cell lymphomas (DLBCL) and high-grade lymphomas (HGBL) with or without MYC, BCL2, and/or BCL6 (indicated as double- or triple-hit, DH/TH lymphomas) gene rearrangements are comprised. DLBCL represent a highly heterogeneous disease entity that encompasses both lymphomas expressing germinal center (GC) B cell markers and others lacking signs of GC transit (the distinction underlying the cell of origin—COO—classification of DLBCL) (Alizadeh et al., 2000). The distinction between the GC and non-GC DLBCL refers to genetic, epigenetic and transcriptional, and phenotypic differences, which, all together, impact on the clinical course, prognosis, and response to treatment (Chapuy et al., 2018; Schmitz et al., 2018). Although generally GC-DLBCL have a more favorable prognosis, a considerable proportion of them display a more aggressive course (Pasqualucci and Dalla-Favera, 2018). Recently, Wright and co-workers, using the LymphGen algorithmic tool to classify DLBCL, highlighted that GC-DLBCL genetic subtypes (defined by mutational patterns) are strikingly different in the response to standard immuno-chemotherapy and possibly to targeted therapies (Wright et al., 2020). The heterogeneous clinical behavior of GC-related aggressive B cell lymphomas has been partly explained by the inclusion in this group of DH cases (Ennishi et al., 2019a). DH HGBL have significantly unfavorable outcomes and show poor response to conventional immuno-chemotherapy regimens; the different course of these lymphomas has been mostly ascribed to the peculiar biology of the B cell clones undergoing lymphomagenesis, but no clues have so far emerged regarding the stromal/immune imprint of DH (Scott et al., 2015). Here, we aimed at probing in situ distinct immune and stromal gene expression signatures in two functionally compartmentalized regions of the non-neoplastic GC, namely, the dark zone (DZ) and the light zone (LZ), in which B cell proliferation, immunoglobulin genes' somatic hypermutation, and antigen-driven B cell selection events occur. In situ gene expression was investigated to achieve a differential signature of the two microenvironments, which included genes involved in B cell proliferation and mutational activity, myeloid cell activation, antigen presentation and suppressive/regulatory functions, T cell identity and immune checkpoint, follicular dendritic cell (FDC) and other mesenchymal cell markers, and cytokine and chemokine signaling. Through a spatially resolved region of interest (ROI) selection-based approach, we investigated transcriptional features reflective of biological differences in the regulation of B cell/stroma interfaces within the DZ and LZ functional microenvironments of the non-neoplastic GC. The resulting signature was applied to GC-related DLBCL and HGBL transcriptomes to establish a possible relationship with the GC microenvironment of origin. Our hypothesis-driven experiment sheds light on the underlying heterogeneity of GC-related aggressive lymphomas, revealing an immunologically cold DZ-like microenvironment characteristic of DH lymphomas.
Results
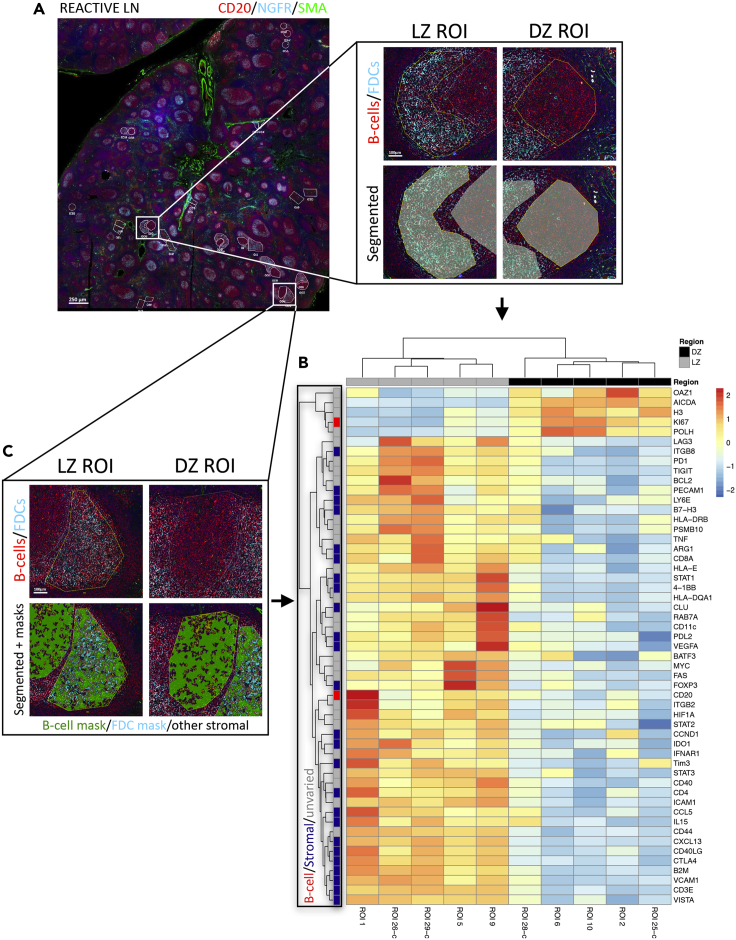
ROIs were identified and selected on reactive lymph nodes characterized by follicular hyperplasia and clear-cut DZ/LZ polarization, based on multiplexed immunofluorescence on a Nanostring GeoMx Digital Spatial Profiler (Nanostring Technologies Inc., USA). GCs and extra-follicular regions were identified according to the expression of the CD20 B cell marker, the FDC meshwork highlighted by CD271 (NGFR), and the reticular fibroblastic cell meshwork highlighted by smooth muscle actin. Within polarized GC foci, DZ and LZ ROIs were selected and segmented for ROI-targeted gene expression (Figures 1A–1C). A customized version of the Human Immuno-Oncology RNA Panel including 87 immune and stromal genes was developed and applied using a Nanostring GeoMx (Merritt et al., 2020). A list of the 87 genes and of their main biological pathways according to Gene Ontology Annotation (https://www.ebi.ac.uk/GOA/) is reported in Table S1.
Figure 1.
Identification of a DZ/LZ Microenvironment Differential Signature by Digital Spatial Profiling on Reactive Lymph Node ROIs
(A) Multiplexed immunofluorescence for CD20 (red signal), nerve growth factor receptor (NGFR)/CD271 (cyan signal), smooth muscle actin (SMA, green signal), and Syto83 (blue signal) has been performed to drive selection of ROIs corresponding to LZ and DZ, for segmentation and gene expression analysis. Scale bar, 250 micrometers.
(B) Unsupervised clustering of the gene expression profiles generated from DZ and LZ segmented ROIs according to a custom mRNA panel, showing the separation of DZ and LZ ROIs based on the expression of the 53 deregulated genes.
(C) DZ and LZ ROIs segmentation with the application of masks for B cells (CD20+NGFR−), FDCs (CD20−NGFR+), and other stromal and immune components (CD20-NGFR−) allows identification of genes significantly upmodulated in B cells (red), not significantly different between the B cell and stromal compartment (gray), and upmodulated in stromal cells (blue).
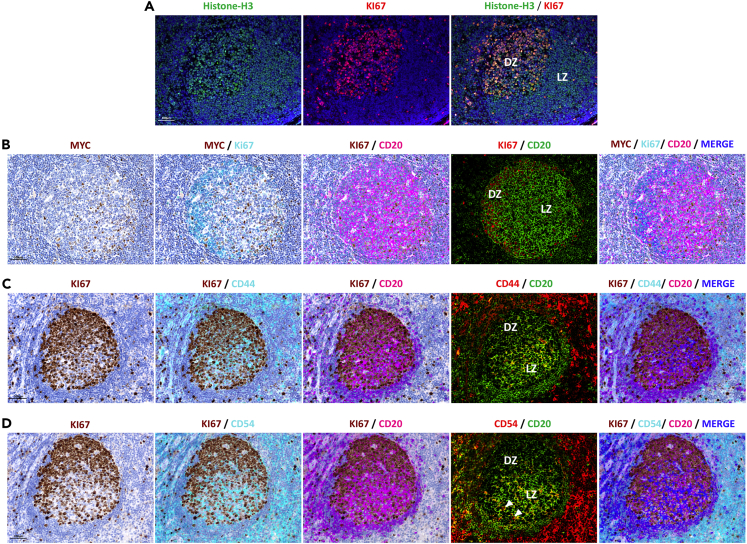
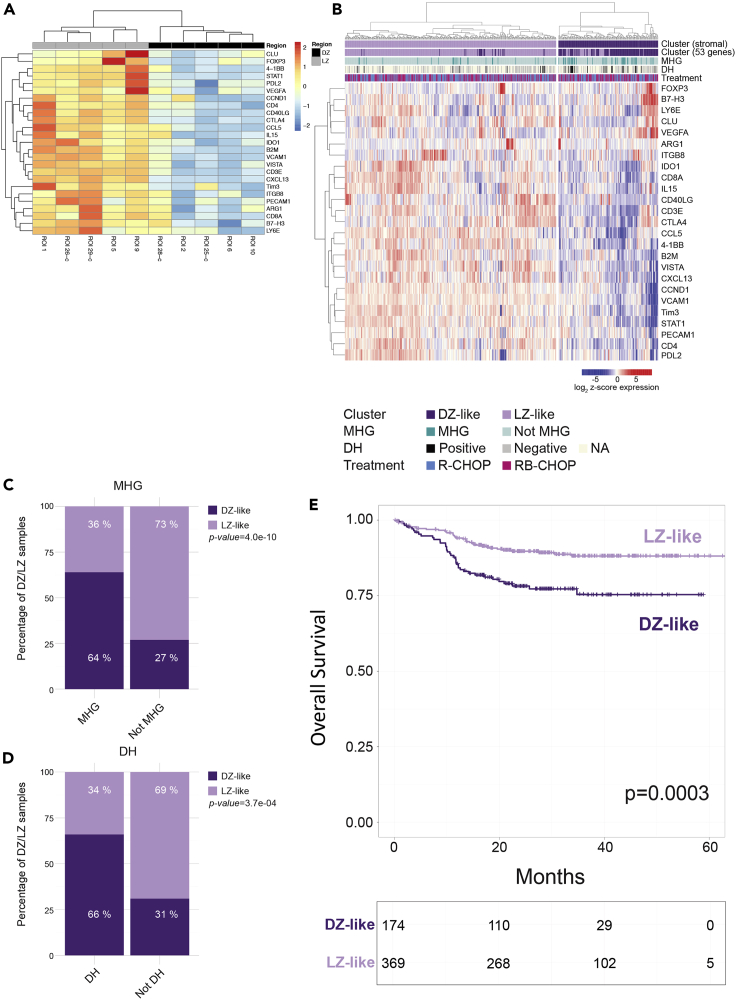
Based on the differential gene expression, 53 genes significantly discriminated between DZ and LZ ROIs. Unsupervised clustering analysis of normalized gene expression profiles within unsegmented ROIs (n = 10) was performed, which highlighted sharp DZ and LZ separation (Figure 1B and Table S2). Among these genes, those involved in cell proliferation, somatic hypermutation, and chromatin remodeling, such as AICDA, MKI67, H3F3B, and OAZ1, were upregulated in the DZ ROIs, consistently with the DZ being a tissue microenvironment hosting GC B cell proliferation and immunoglobulin gene hypermutation events induced by activation-induced cytidine deaminase (AID) activity. The expression of the histone H3 in the GC DZ and LZ was probed at the protein level using a double-marker immunofluorescence approach, which highlighted the bright fluorescence signal of H3 in Ki-67+ DZ cells (Figure 2A), indicating the association of H3 expression with cell replication in the DZ. Genes involved in antigen presentation by major histocompatibility complex (MHC) classes I and II (B2M, CD74, HLA-E, HLA-DRB1, and HLA-DQA1), immune checkpoints (TIGIT, CTLA4, LAG-3, PDCD1, and PDCD1LG2), and FDC biology (CLU, CXCL13, and VCAM1) were upregulated in the LZ ROIs, in view of the ongoing immune recognition and selection dynamics characterizing the LZ. The transcriptional regulator MYC also resulted significantly upregulated in the LZ, and its spatial enrichment was validated at the protein level by triple-marker immunostaining including CD20 and Ki-67, which revealed that, within the GC, the scattered MYC+ elements were predominantly localized within the Ki-67-low areas corresponding to the LZ (Figure 2B). Among the genes predominantly expressed in the LZ ROIs, we found the transcript relative to the adhesion receptor CD44, originally retained, to be differentially expressed in LZ and DZ B cells (Kremmidiotis and Zola, 1995). Interestingly, CD44 protein expression was not observed as a differential feature of the LZ B cell subset purified by cell sorting in the seminal study by Victora and colleagues, which provided reference DZ and LZ B cell profiles for classification of B cell malignancies (Victora et al., 2012). The positive enrichment of CD44 in the LZ was validated in situ, by a triple-marker immunolocalization analysis, which showed that CD44 expression was stronger in the CD20+Ki-67- LZ than in the CD20+Ki-67+ DZ B cells within the GC (Figure 2C). This result highlights the relevance of spatial profiling in the exploration of transcriptional network elements possibly controlled in their expression by the cross talk between different cell types sharing the same microenvironment, as in the case of B cell and accessory cells (T, myeloid, and mesenchymal) populating the LZ. To better investigate the contribution of B cell-intrinsic and B cell-extrinsic (i.e., stromal and immune) transcripts to the DZ/LZ discrimination, we analyzed the expression of the 53 DZ/LZ differentially expressed genes in 15 additional ROIs relative to DZ and LZ regions to which B-cell (CD20+NGFR-) and stromal (CD20-NGFR+ and CD20-NGFR-) sub-compartment segmentation masks were applied (Figure 1C). Among the genes differentially expressed between DZ and LZ ROIs, we identified a list of 25 genes that showed significant upmodulation in the stromal sub-compartment masks when compared with the B cell masks (Figures 1C and S1, Table S3). These 25 genes, hereinafter referred to as stromal signature, included key genes regulating T cell identity and activation, such as CD3E, CD4, CD8A, FOXP3, CD40L, 4-1BB, B7-H3, VISTA, CTLA4, and TIM3; genes related with myeloid/macrophage regulatory function such as IL-15, IDO1, and ARG1; and FDC/mesenchymal cell-related genes such as CLU, PECAM-1, CXCL13, VCAM-1, and VEGFA. Interestingly, two of the myeloid cytokines included in the stromal signature have a consolidated immunoregulatory/suppressive function (IDO1 and ARG1), whereas IL-15 has been involved in the induction of a T follicular helper phenotype (Cooley et al., 2015). Besides the 25-genes stromal signature, 26 genes were not significantly modulated between the B cell and stromal sub-compartment masks (e.g., CD44, ICAM1, CD40, STAT3), and only two genes (CD20 and MKI67) were significantly upregulated in the B cell masks (Figures 1B, 1C, and S1, Table S3). This result indicated that several genes included in the 53-genes DZ/LZ signature and contributed by B cells were also expressed by other stromal elements (e.g., ICAM1). Accordingly, ICAM-1 in situ protein validation by triple-marker immunolocalization analysis in combination with Ki-67 and CD20 showed modulation between the DZ and LZ with a higher expression in the latter, where it showed co-localization with CD20 in B cells and also expression in CD20- stromal elements (Figure 2D, arrowheads).
Figure 2.
In Situ Protein Validation of Selected Markers from DZ/LZ Differential Gene Signature in Reactive Lymph Node GCs
(A) Double-marker immunofluorescence analysis of Histone H3 expression in the GC showing bright nuclear expression of H3 (green signal) in Ki-67+ (red signal) proliferating cells within the GC DZ.
(B) Triple-marker combined immunohistochemistry/immunofluorescence analysis of MYC (brown signal) expression in the GC showing that scattered MYC+ cells are detected in CD20+ (cyan and red signals) cells within the Ki-67-low (purple and green signals) area corresponding to the LZ.
(C) Triple-marker combined immunohistochemistry/immunofluorescence analysis of CD44 (cyan and red signals) expression in the GC showing that CD44 expression is more intense in Ki-67-low (brown signal) B cells characterized by a brighter CD20 expression (purple and green signals), corresponding with the LZ.
(D) Triple-marker combined immunohistochemistry/immunofluorescence analysis of CD54/ICAM-1 (cyan and red signals) expression in the GC showing that CD54 signal is more intense in Ki-67-low (brown signal) area corresponding to the LZ, where it marks both CD20+ (purple and green signals) and negative (arrowheads) cells. All the representative microphotographs have a 200× original magnification. Scale bars, 100 micrometers.
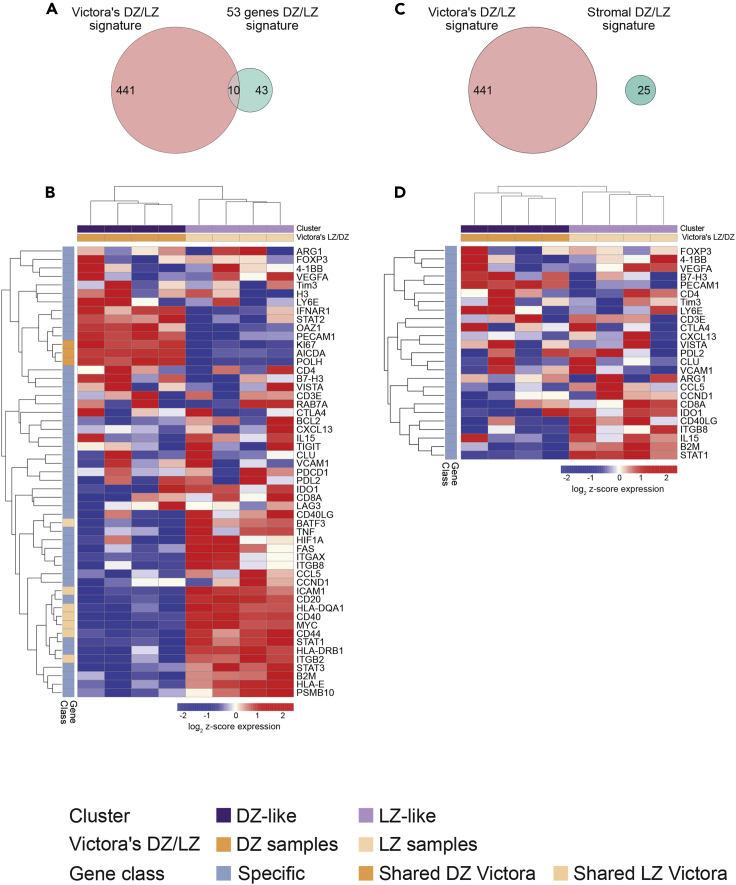
We then investigated whether genes modulated in our spatial DZ/LZ profiling could be found consistently modulated between the transcriptional profiles of DZ and LZ from purified human B cells identified by Victora et al. (2012). To this aim, we performed an unsupervised clustering analysis of their dataset, using our 53-genes DZ/LZ spatial signature. Interestingly, our signature assigned samples in total agreement with Victora's groups. We could verify the shared and concordant modulation of KI-67, AICDA, and POLH that were higher in DZ B cells and of BART3, ICAM-1, HLA-DQA1, CD40, CD44, ITGB2, and MYC that showed higher expression in LZ B cells (Figures 3A and 3B). We also assessed whether the 25-genes stromal signature that we derived from transcripts significantly enriched in non-B spatial sub-compartments (CD20−NGFR+ and CD20−NGFR-) when compared with B cell sub-compartments (CD20+NGFR−) had overlap with the differential DZ/LZ B cell genes reported by Victora et al. Indeed, none of the 25 genes were found among the DZ/LZ B cell differential genes. Nonetheless, the clustering of the Victora et al. samples according to our 25-genes stromal signature was consistent (Figures 3C and 3D). Prompted by the successful discrimination of DZ/LZ ROIs according to the spatial 53-genes signature, we aimed at probing the transcriptome of GC-related aggressive B cell lymphomas in the attempt to trace transcriptional clusters reminiscent of the two distinct GC microenvironments. We therefore applied the DZ/LZ signature to a large dataset of 543 GC-DLBCL including DH cases treated with RCHOP or RCHOP + bortezomib (RB-CHOP), in the context of the REMoDL-B clinical trial (Sha et al., 2019). The trial had shown no significant benefit with the addition of bortezomib to R-CHOP, but represents a valuable cohort for biomarkers of standard front-line treatment, as in the identification of features associated with molecular high-grade (MHG) lymphomas (Sha et al., 2019). We found that according to the expression of the DZ/LZ differential genes, two subgroups of GC-related lymphomas could be identified on unsupervised clustering analysis: one characterized by a transcriptional profile resembling that of the spatial LZ signature (LZ-like) and the other more related to the spatial DZ profile (DZ-like) (Figure 4A). This finding indicated that the heterogeneity intrinsic to the gene expression profiles of GC-related aggressive lymphomas is at least in part organized according to DZ/LZ microenvironment-linked transcriptional features.
Figure 3.
Performance of the DZ/LZ 53-Genes Signature and of the 25-Genes Stromal Signature in Victora's Dataset (GSE38697)
(A and C) Venn diagrams representing the overlap of the two spatial signatures (the 53-genes differential DZ/LZ signature and the 25-genes signature) with Victora's DZ/LZ B cell signature. The 53-genes DZ/LZ signature (A) displays an overlap of 10 genes (3 upregulated in the DZ and 7 upregulated in the LZ) with the 441 genes of Victora's DZ/LZ B cell signature, whereas the 25-genes stromal signature (C) shows no overlapping genes.
(B and D) Unsupervised hierarchical clustering analysis based on the 53-genes DZ/LZ signature (B) and on the 25-genes signature (D) in Victora's dataset (genes are listed in rows, and samples are shown as columns).
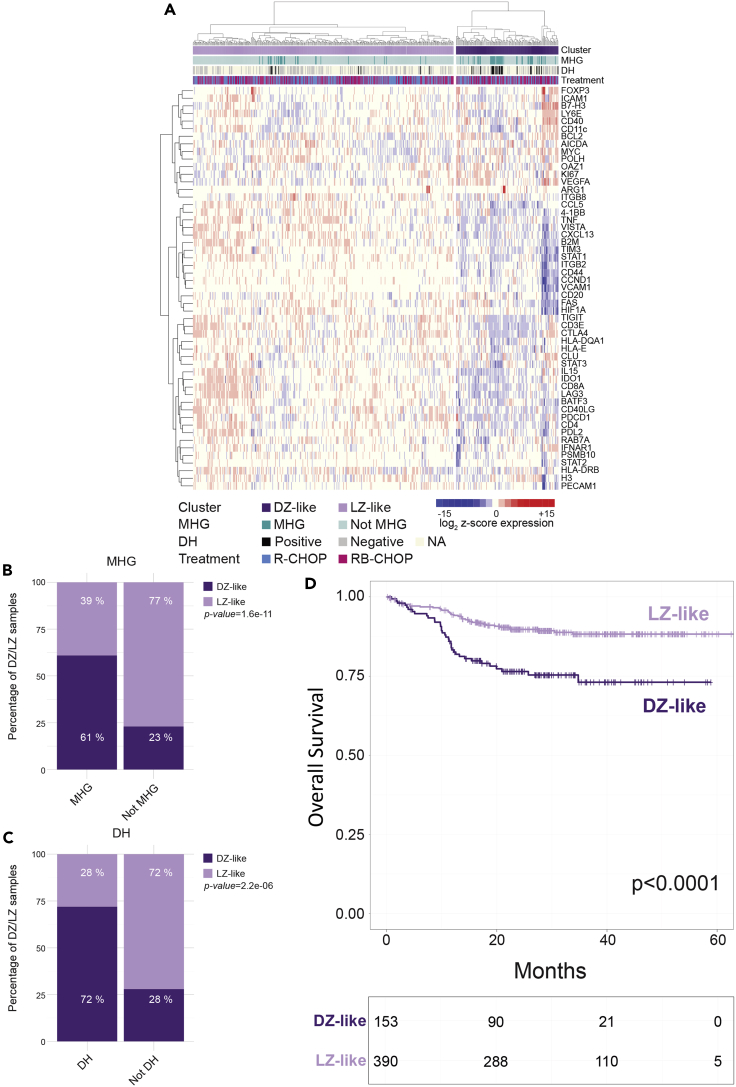
Figure 4.
Application of a Spatially Resolved 53-Genes Signature Discriminating DZ and LZ to 543 GC-Related DLBCL and DH Cases (GSE117556)
(A) Unsupervised clustering analysis of the 543 GC-DLBCL and DH cases based on the DZ/LZ signature identifies two distinct clusters (DZ-like and LZ-like).
(B and C) Bar plots showing the differential enrichment in molecular high-grade (MHG, B) and double-hit (DH, C) cases between the DZ-like and LZ-like clusters.
(D) Survival analysis showing significantly different overall survival in the LZ-like and DZ-like clusters.
Notably, the LZ-like and DZ-like subgroups identified in the cohort of 543 lymphomas of GC origin showed a significantly different enrichment for DH cases characterized by genetic rearrangement of MYC and BCL2 or BCL6 and for MHG cases classified according to a gene expression classifier (Figures 4B and 4C) (Sha et al., 2019). Indeed, the DZ-like cluster was positively enriched in DH and MHG cases when compared with the LZ-like cluster. Consistently, the cases falling in the DZ-like cluster showed a significantly worse overall survival (OS) when compared with the predominant LZ-like cluster (Figure 4D). Actually, the prognostic difference of the two clusters was dependent on the presence of DH cases prominently contributing to the DZ-like cluster, as the exclusion of DH from the prognostic analysis resulted in a striking downsizing of the cluster (from n = 153 to n = 24).
To validate that our spatial DZ/LZ signature could identify DZ-like lymphomas, we extended our analysis by applying the 53-genes DZ/LZ signature to the Burkitt lymphoma (BL) setting. We performed an integrated analysis including DLBCL and BL samples from Sha's dataset by applying unsupervised hierarchical clustering. The analysis revealed that the majority of BLs clustered with MHG GCB-like DLBCL samples based on shared gene expression profile resembling DZ phenotype (Figure S2). In particular, 56 of 70 BLs (80%) were clustered in the DZ-like cluster according to the 53-genes DZ/LZ signature. This finding was of particular interest if we consider that BL represents a rather homogeneous disease as far as the biological features of the malignant clone are concerned, and that the recently reported variability in BL immune landscape (Granai et al., 2020) may configure different microenvironment scenarios more similar to or distinct from the immunologically cold DZ.
To further investigate the contribution of primarily stromal and immune cell transcripts to the clustering of the 543 GC-related lymphoma cases, we applied the 25-genes stromal signature obtained from the analysis of ROI sub-compartments (Figure 5A). Interestingly, the 25-genes signature also subdivided the 543 GC-related aggressive lymphoma cases into two distinct LZ-like and DZ-like clusters (Figure 5B) with significantly different MHG and DH enrichment (Figures 5C and 5D), and the two clusters also showed significantly different OS (Figure 5E). This finding revealed the possibility to discriminate GC-related lymphomas differently enriched in MHG and DH according to stromal features, indicating that DH lymphomas, despite a common GC origin with other non-DH GC-DLBCL display an immunological and mesenchymal microenvironment reminiscent of the GC DZ.
Figure 5.
Application of a 25-Genes Stromal Signature Discriminating DZ and LZ to 543 GC-Related DLBCL and DH Cases (GSE117556)
(A) Unsupervised clustering of DZ and LZ ROIs according to 25 genes significantly more expressed in stromal (CD20−NGFR+ FDCs and CD20−NGFR− remaining stromal and immune cells) ROI masks when compared with B cell (CD20+NGFR−) masks (see Figure S1).
(B) Unsupervised clustering analysis of the 543 GC-DLBCL cases based on the 25-stromal/immune genes DZ/LZ signature identifies two distinct clusters (DZ-like and LZ-like).
(C and D) Bar plots showing the differential enrichment in molecular high-grade (MHG, C) and double-hit (DH, D) cases between the two clusters identified according to the 25-genes signature.
(E) Survival analysis showing significantly different overall survival in the LZ-like and DZ-like clusters identified according to the 25-genes stromal signature.
We finally addressed whether our 25-genes stromal signature was in part overlapping with two consolidated stromal microenvironment signatures of prognostic significance identified by Lenz and colleagues in DLBCL (Lenz et al., 2008). We therefore investigated the degree of overlap of the 25-genes DZ/LZ stromal signature with Lenz's stromal signatures and found that only one gene, CLU, was shared with the Stromal-1 and one gene, PECAM1, with the Stromal-2 signature (Figure S3). This result indicated that our immune/mesenchymal DZ/LZ differential signature probed a different level of microenvironment heterogeneity when compared with that of consolidated DLBCL stromal features.
Discussion
B cells undergoing malignant transformation along lymphomagenesis receive an imprint from the transforming environment in the form of genetic lesions and phenotypical skewing. In the case of GC-related lymphomas, that imprint is traceable in gene rearrangement events promoted by AID mutational machinery activity, and by the replicative stress induced in the DZ, potentially affecting the genetic and molecular subtypes, that relate to the COO (Pasqualucci and Dalla-Favera, 2015). Within GC-DLBCL, the pathogenetic hypothesis that malignant B cell transformation may occur in either of the two functionally specialized compartments of the GC, DZ and LZ, has found experimental support through evidences pointing to differential expression of MHC I/II molecules, differential frequency of EZH2 mutations (Ennishi et al., 2019b), and differential expression of a subset of DZ/LZ-associated gene transcripts in different subsets of GCB DLBCL (Ennishi et al., 2019b; Victora et al., 2012). MHC classes I and II expression is transcriptionally modulated in the transition from the DZ to the LZ. Expression of MHC I/II cannot be exploited in the classification of GCB DLBCL subsets given the strong selection pressure imposed on the lymphoma cells by the immune system that causes recurrent extinction of these molecules from the surface of the malignant B cells because of mutational events (Ennishi et al., 2019b). Similarly, the attempt to link MHC negativity to the immunologically cold microenvironment of the DZ in GC-related DLBCL has been mostly based on the positive enrichment of signatures obtained by DZ B cells purified according to DZ/LZ surface markers (Ennishi et al., 2019b; Victora et al., 2012). Such B cell-centered approach, which has been successful in distinguishing clusters of DLBCL and in separating DLBCL from more aggressive entities, lacked the role of features representative of the in situ stromal and immune context that could affect the modulation/maintenance of programs resulting from the cross talk between B cells and stromal elements. We have applied DSP to non-neoplastic lymph node GCs to gain insight into the immune and stromal composition of the DZ and LZ, with the aim of deriving a spatially resolved microenvironment signature contributed by B cells and non-B stromal and immune cells, to probe DZ-/LZ-like microenvironment imprints in GC-related DLBCL and DH lymphomas. Along with a 53-genes signature including transcripts differentially expressed between DZ and LZ ROIs, we derived a 25-genes signature composed of those differential genes significantly more represented in the GC DZ and LZ stromal, non-B, sub-compartments and contributed mostly by transcripts expressed in adjacent stromal/immune cells that were asymmetrically distributed between DZ and LZ areas. The newly identified DZ/LZ microenvironmental signatures were exploited to question the origin of a highly heterogeneous set of GC-related aggressive B cell lymphomas. Our results establish the existence of two distinct tumor groups within GC-related aggressive B cell lymphomas, with those displaying a DZ-like microenvironment profile strikingly enriched for DH lymphomas. Consistently, this same DZ-like group showed strong positive enrichment for BL cases, indicating that MYC-driven lymphomagenesis in the GC (Basso and Dalla-Favera, 2015; Dominguez-Sola et al., 2012) is preferentially related with a DZ-like microenvironment.
Future studies will be needed to assess whether specific genetics of GC-type DLBCL may associate with LZ- and DZ-like microenvironment profiles, based on the preliminary observation that EZH2 mutations and BCL2 translocations, which have been demonstrated to display higher DH scores than other subtypes (Schmitz et al., 2018), showed a relative enrichment in DZ B cell signatures. Similar efforts should be placed to establish possible differences in the microenvironmental context of EZB-MYCneg lymphomas from that of EZB-MYC+ lymphomas, with the latter showing a worse prognosis (Wright et al., 2020).
The significant skewing in the distribution of DH cases in the DZ-like cluster provides preliminary insight into the unique immune and stromal microenvironment featuring these peculiar lymphomas, the biology and pathogenic trajectories of which are yet poorly understood. In summary, by applying Digital Spatial Profiling we provide first-time evidence for the existence of a GC DZ/LZ immune and stromal microenvironment signature able to cluster GC-related aggressive B cell lymphomas into two main groups with different response to standard chemo-immunotherapy regimens. In particular, our data provide experimental support for an origin from DZ GC B cells of highly aggressive DH lymphomas.
Limitations of the Study
A limitation of the experimental approach pursued in the analysis of segmented ROIs is represented by the level of resolution in the discrimination between different cellular components. Indeed, in the GC sub-compartments, B cells intermingle tightly with mesenchymal elements, such as FDCs or other immune cells including myeloid elements (e.g., GC macrophages). This tight interaction is responsible for the potential transfer of myeloid or stromal cell transcripts to B cell sub-compartments. To overcome this limitation, we attributed to B cell or stromal cell sub-compartments only those differentially expressed transcripts that were significantly enriched in that specific sub-compartment. New analytical tools are needed to overcome limitations imposed by the resolution of ROI segmentation in complex stromal-immune interfaces as the one we investigated.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Claudio Tripodo (claudio.tripodo@unipa.it).
Materials Availability
No unique reagents have been generated in this study and all the materials are commercially available.
Data and Code Availability
Gene expression data from published datasets are detailed in Transparent Methods, and normalized gene expression data generated in the Digital Spatial Profiling experiments are available in Table S4.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors wish to acknowledge Dr. Alessandro Gulino, Dr. Valeria Cancila (University of Palermo, Tumor Immunology Unit), and Dr. Yanfen Peng (Singapore University Hospital) for technical support and Dr. Michal Marek Hoppe (Singapore University Hospital) for helpful discussion. This study has been supported by the Italian Foundation for Cancer Research (AIRC) through the IG-2018 22145 Investigator Grant to C.T.; 5x1000 Grant 21198 to S.A.P.; by the Italian Ministry of Health Research Grant RF-2016-02364121 to M.P.C., C.C., and C.T.; and by the Italian Ministry of Education, University and Research (MIUR) grant 2017K7FSYB to C.T.
Author Contributions
C.T. conceived the study and wrote and reviewed the manuscript; F.I., F.Z., and S.M. analyzed the gene expression data and contributed to manuscript writing; M.V. contributed to human dataset analyses and interpretation of data; A.D.N., A.M., F.F., S.S., C.C., G.M., and A.D.J. contributed to the identification of markers, in the customization of the mRNA TAP panel, and in the interpretation of the results; A.V. provided assistance in panel customization and performed the digital spatial profiling; S.C. and M.P.C. contributed to the study conception, data interpretation, and manuscript writing; M.P. and S.A.P. supervised the study, contributed to manuscript writing and data interpretation, and reviewed the manuscript.
Declaration of Interests
A.V. works as Scientist for NanoString Technologies Inc. The remaining authors have nothing to disclose.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101562.
Supplemental Information
References
- Alizadeh A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Basso K., Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015;15:172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- Chapuy B., Stewart C., Dunford A.J., Kim J., Kamburov A., Redd R.A., Lawrence M.S., Roemer M.G.M., Li A.J., Ziepert M. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley I.D., Read K.A., Oestreich K.J. Trans-presentation of IL-15 modulates STAT5 activation and Bcl-6 expression in TH1 cells. Sci. Rep. 2015;5:15722. doi: 10.1038/srep15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Victora G.D., Ying C.Y., Phan R.T., Saito M., Nussenzweig M.C., Dalla-Favera R. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennishi D., Jiang A., Boyle M., Collinge B., Grande B.M., Ben-Neriah S., Rushton C., Tang J., Thomas N., Slack G.W. Double-hit gene expression signature defines a distinct subgroup of germinal center B-Cell-Like diffuse large B-cell lymphoma. J. Clin. Oncol. 2019;37:190–201. doi: 10.1200/JCO.18.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennishi D., Takata K., Beguelin W., Duns G., Mottok A., Farinha P., Bashashati A., Saberi S., Boyle M., Meissner B. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. 2019;9:546–563. doi: 10.1158/2159-8290.CD-18-1090. [DOI] [PubMed] [Google Scholar]
- Granai M., Mundo L., Akarca A.U., Siciliano M.C., Rizvi H., Mancini V., Onyango N., Nyagol J., Abinya N.O., Maha I. Immune landscape in Burkitt lymphoma reveals M2-macrophage polarization and correlation between PD-L1 expression and non-canonical EBV latency program. Infect. Agent. Cancer. 2020;15:28. doi: 10.1186/s13027-020-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremmidiotis G., Zola H. Changes in CD44 expression during B cell differentiation in the human tonsil. Cell. Immunol. 1995;161:147–157. doi: 10.1006/cimm.1995.1021. [DOI] [PubMed] [Google Scholar]
- Lenz G., Wright G., Dave S.S., Xiao W., Powell J., Zhao H., Xu W., Tan B., Goldschmidt N., Iqbal J. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C.R., Ong G.T., Church S.E., Barker K., Danaher P., Geiss G., Hoang M., Jung J., Liang Y., McKay-Fleisch J. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020;38:586–599. doi: 10.1038/s41587-020-0472-9. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin. Hematol. 2015;52:67–76. doi: 10.1053/j.seminhematol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Dalla-Favera R. Genetics of diffuse large B-cell lymphoma. Blood. 2018;131:2307–2319. doi: 10.1182/blood-2017-11-764332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R., Wright G.W., Huang D.W., Johnson C.A., Phelan J.D., Wang J.Q., Roulland S., Kasbekar M., Young R.M., Shaffer A.L. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D.W., Mottok A., Ennishi D., Wright G.W., Farinha P., Ben-Neriah S., Kridel R., Barry G.S., Hother C., Abrisqueta P. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J. Clin. Oncol. 2015;33:2848–2856. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha C., Barrans S., Cucco F., Bentley M.A., Care M.A., Cummin T., Kennedy H., Thompson J.S., Uddin R., Worrillow L. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J. Clin. Oncol. 2019;37:202–212. doi: 10.1200/JCO.18.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Dominguez-Sola D., Holmes A.B., Deroubaix S., Dalla-Favera R., Nussenzweig M.C. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.W., Huang D.W., Phelan J.D., Coulibaly Z.A., Roulland S., Young R.M., Wang J.Q., Schmitz R., Morin R.D., Tang J. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37:551–568.e14. doi: 10.1016/j.ccell.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data from published datasets are detailed in Transparent Methods, and normalized gene expression data generated in the Digital Spatial Profiling experiments are available in Table S4.