Abstract
In developed countries, colorectal cancer is the second cause of cancer-related mortality. Chemotherapy is considered a standard treatment for colorectal liver metastases (CLM). Among patients who develop CLM, the assessment of patient response to chemotherapy is often required to determine the need for second-line chemotherapy and eligibility for surgery. However, while FOLFOX-based regimens are typically used for CLM treatment, the identification of responsive patients remains elusive. Computer-aided diagnosis systems may provide insight in the classification of liver metastases identified on diagnostic images. In this paper, we propose a fully automated framework based on deep convolutional neural networks (DCNN) which first differentiates treated and untreated lesions to identify new lesions appearing on CT scans, followed by a fully connected neural networks to predict from untreated lesions in pre-treatment computed tomography (CT) for patients with CLM undergoing chemotherapy, their response to a FOLFOX with Bevacizumab regimen as first-line of treatment. The ground truth for assessment of treatment response was histopathology-determined tumor regression grade. Our DCNN approach trained on 444 lesions from 202 patients achieved accuracies of 91% for differentiating treated and untreated lesions, and 78% for predicting the response to FOLFOX-based chemotherapy regimen. Experimental results showed that our method outperformed traditional machine learning algorithms and may allow for the early detection of non-responsive patients.
Keywords: Deep convolutional neural network, Colorectal liver metastases, Chemotherapy, Prediction response, CT scans, FOLFOX-based regimen
Introduction
In the USA alone, colorectal cancer is the third main cause of cancer-related mortality in men and in women (second most common cause of cancer deaths when combined), causing approximately 51,020 deaths in 2019 [1]. Updated screening guidelines have decreased the incidence of colorectal cancer among older adults, but in recent years, incidence has increased by 3 to 4% among adults younger than 50, underscoring the need for novel approaches for diagnosis and treatment [2].
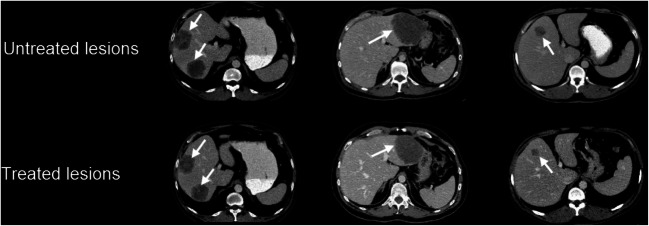
Chemotherapy is considered as an effective intravenous treatment option for colorectal liver metastases (CLM) [2]. However, the differentiation of treatment responsive vs. unresponsive untreated tumors from computed tomography (CT) remains a challenging problem, as this influences treatment options such as adding chemotherapy cycles or changing drug regimens. As illustrated in Fig. 1, the identification of new liver metastases appearing on axial CT images is difficult due to the similarity with previously treated tumors from past regimens, even to the trained eye of an experienced radiologist.
Fig. 1.
Case samples of untreated and treated colorectal liver metastases (white arrows) on axial CT images acquired in the same patient at baseline (top row) and follow-up examination 9 months later (bottom row), demonstrating the apparent similarity between lesions
The early assessment of tumor response to chemotherapy is critical to determine the need to change drug regimen and eligibility for surgery [3]. On contrast-enhanced CT, current evaluation paradigms compare tumor size in follow-up exams but remain very subjective and are based on substantial experience with the added difficulty of identifying new lesions on follow-up images from patients that were previously treated with chemotherapy [4].
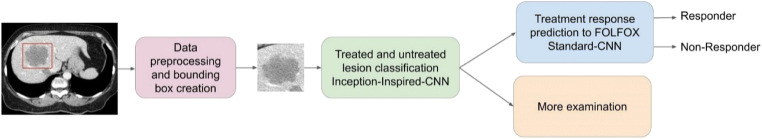
The pathological and histopathological responses for CLMs, which are derived from FOLFOX combined with a bevacizumab regimen, show good clinical result at the early stage of disease [5]. However, there is still no imaging evaluation criteria to enable early identification of response for patients. The purpose of this study is to develop an unsupervised image-based pipeline using a deep neural network to predict response to FOLFOX-based chemotherapy. Figure 2 shows the overall workflow of the proposed lesion classification pipeline, as well as the model for predicting the response to FOLFOX regimen.
Fig. 2.
Schematic illustration of the proposed workflow to classify untreated lesions from previously treated lesion in baseline CT scans, which are then fed to a predictive model to forecast response to a FOLFOX-based chemotherapy regimen
Related Work
Chemotherapy is a viable alternative to cease the growth of tumors and achieve regression [6]. Applications have used texture for assessing pixel intensity changes with respect to levels of necrosis and fibrosis [7]. These changes in pixel intensity can be measured by statistical methods such as gray level co-occurrence matrices (GLCM) [8]. Textural information can also help to predict the overall survival for primary colorectal cancers (CRC) and gastric tumors [9], including for colorectal cancer liver metastases (CRM) from CT textural features [10–12].
Texture analysis (TA) is frequently used for image recognition and for localizing specific regions of interest (ROI), such as organs or diseased tissue. In medical image analysis, assessing lesion normality/abnormality, as well as the graininess of the texture, can be assessed by TA. To date, there are several studies that have demonstrated the strengths of TA and its effects on inferring prognostic information [13, 14]. Even though the number significantly meaningful feature descriptors based on GLCM is limited [6], including correlation, energy, entropy, contrast, homogeneity, and dissimilarity, these can be effectively used with classical machine learning approaches (random forests, SVM’s), where their performance remains encouraging.
Typically, the chosen process for data representation and feature extraction will greatly affect performance of the machine learning method. For this reason, data preprocessing and data transformation play an important role in translating of machine learning algorithm in different domain. In machine learning, the very first step features are extracted and then fed to the classifier for training purposes such that the backbone of the features remains unchanged [15].
In the past few years, deep learning approaches showed tremendous progress in image recognition tasks and, in most cases, have been shown to surpass traditional state-of-the-art classical machine learning [16]. Until now, machine learning and deep learning in medical imaging have been widely applied in several fields such as prediction, automatic detection, and classification. The advantage of deep learning methods in contrast to with machine learning is that, the hierarchal features are extracted automatically at different layers of a neural network. These features representing some information such as intensity from an image, and learned through a back propagation process, which can be used for tumor detection and also classification of images in medical image processing [17–21].
In summary, deep neural networks are proposed to cover some constraints of classical machine learning for automatic feature extraction and data representation classification [22, 23]. Deep learning includes several level of representations in which the representation of the first level is transferred to the next higher level and so forth [23]. Deep learning approaches have recently been used for therapy outcome prediction and to assess the odds ratios of recurrence from pre-treatment examinations [24].
This paper introduces an architecture based on variant to state-of-the-art CNNs that are shown to have important clinical applications for automatic classification of treated and untreated lesion. Moreover, we propose a novel prediction model that may be used to select chemotherapy regimens. The purpose of this work was to develop a deep convolutional neural network (DCNN) to classify treated from untreated lesions and to predict response to a FOLFOX with Bevacizumab regimen as first-line of treatment.
Material and Methods
Patient Dataset
This retrospective‚ IRB-approved HIPAA compliant study included CT images of liver cancer patients from our tertiary referral center between June 2009 and June 2016 from an institutional biobank. Clinical parameters are acquired from the institutions clinical registry and laboratory system. Radiological response (assessment based on clinical images) is evaluated in consensus by two board certified radiologists experienced in hepatic imaging. For the proposed approaches, two dataset have been provided.
The clinical dataset included 202 colorectal cancer liver metastases patients, which yielded a total of 444 lesions, which are distributed in 230 treated and 214 untreated lesions. Treated patients received either a single line of therapy with a drug regimen based on FOLFOX with Bevacizumab, or a combination FOLFIRI drugs as a second line of treatment. Both treated and untreated lesions were segmented on contrast-enhanced CT scans (arterial phase). The lesions were automatically segmented using a joint liver/liver-lesion segmentation model build from two fully convolutional networks, connected in tandem, and trained together end-to-end [25], which yielded a Dice score over 90% from the 2017 MICCAI LITS challenge. The previous segmentation helps to minimize the size of the processed image, thus decreasing the computational cost, and helps the convolutional network from isolating several lesions contained in the same axial slice. The rationale for using an automatic segmentation of lesions over manual delineations was to ensure homogeneity and consistency in the segmentations. All lesion segmentation masks were then validated by an experienced radiologist with over 10 years of experience in hepatic imaging and liver cancer diagnosis.
From this radiological dataset, a total of 120 patients with CLM were used for treatment response prediction to FOLFOX chemotherapy. All these patients received as a first line of therapy a drug regimen based on FOLFOX with Bevacizumab, and treatment response following chemotherapy determined by histopathology using the tumor regression grade (TRG) [26], with 22 samples of TRG = 1, 67 samples with TRG = 2, 124 samples with TRG = 3, 149 samples with TRG = 4, and 19 samples with TRG = 5, with a TRG of 4 or 5 considered as non-responsive [27]. Pre-treatment lesions were segmented on baseline contrast-enhanced CT images in portal venous phase, yielding 381 segmented CLMs (213 responsive and 168 non-responsive).
Classification of Treated and Untreated Lesions
Data Preparation and Preprocessing
From our institutional liver biobank which included annotated CT images, all patient datasets included both a baseline and follow-up CT examination, all with validated tumor segmentation masks. The volume size was 512 × 512 cross-sectional, and the number of slices varied between 128 and 152. The original resolutions varied between 0.5 × 0.5 × 1 mm and 2.5 × 2.5 × 2 mm. Exposition was of 0.8 ms, while the tube current was current (25–300 reference mAs). In this experiment, the dataset was split in 80% for training and validation (to optimize the hyper-parameters) sets, with the remaining 20% of images kept as a separate test set for the final evaluation. Since the CT images used in this study originates from different CT scan devices, the images in this dataset possessed different spatial resolutions and voxel resolutions (voxel spacing). Therefore, all images were resampled to 0.8 × 0.8 × 1.0 mm by using cubic spline interpolation to have the same pixel spacing and slice thickness for the entire dataset. This allowed to provide consistency in resolution for the inputs of the neural network. Moreover, all CT images were processed as 2D slices in the training and testing phases. During preprocessing, bounding boxes were centered around each lesion (based on lesion size) and processed by a DCNN as training data. To select the size of each bounding box, the slice with the largest segmented lesion volume was chosen, providing the width and depth of the bounding box, while the height of the bounding box was determined from the number of slices covering the segmented lesion. Since the dataset included multiple lesions, the patch size for the bounding box is dependent on the lesion size which will vary from case to case. Finally, these bounding boxes were applied over the original CT images to extract the ROI. No data augmentation was used to increase the size of the dataset, as this was found to have little effect on the model performance. Prediction was done per lesion-based, where all the 2D slices were combined into a single input vector to the neural network, in order to generate a single outcome prediction.
Deep Learning Model
In this study, we propose to use DCNN based on a variation of Inception-Net and trained over the data samples to classify treated and untreated lesions. The key advantage of deep learning in comparison with classical machine learning is that these features descriptions at all layers are done without any human manipulation [20].
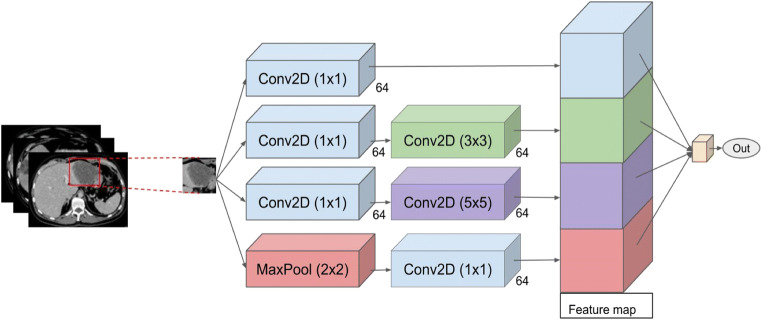
In order to implement our DCNN, Keras API [28] was used to design architecture of the layers and optimize the parameters. In a DCNN model, we use a stack of 3 × 3 or 5 × 5 kernel sizes, in combination with max pooling layers to extract predominant features from the images. As shown in Fig. 3, an Inception architecture is designed to combine these kernel outputs, such that all 1 × 1, 3 × 3, 5 × 5 filters which are fused together and performs the convolution on output from previous layers. Moreover, due to the importance of the pooling layer for the CNNs, the Inception architecture includes an additional pooling path. All lesion segmentation masks and bounding boxes for a region of interest were resized and normalized to yield an input vector with the same dimensionality for each training and testing sample. This handles cropped tumor images having different size. Finally, the output of all filters was concatenated and passed through the network as input of the next layer. Table 1 presents the detailed architecture of the network.
Fig. 3.
Proposed DCNN inspired by the Inception-Net architecture for classification of TACE treated and untreated lesions from CT image segmentation of CLM
Table 1.
Details of the network architecture for treated lesion classification
| Block type | Block name | Kernel size | Block name | Kernel size | Block name | Kernel size | Block name | Kernel size | No. of filters |
|---|---|---|---|---|---|---|---|---|---|
| Feature extraction | Conv2D | 1 × 1 | Conv2D | 1 × 1 | Conv2d | 1 × 1 | Maxpool | 2 × 2 | 64 |
| Feature extraction | Conv2D | 3 × 3 | Conv2D | 5 × 5 | Conv2D | 1 × 1 | - | - | 64 |
| Feature extraction | Concatenation | - | - | - | - | - | - | - | - |
| Classification | Fully connected | - | - | - | - | 2 | - | - | - |
| Classification | Softmax | - | - | - | - | - | - | - | - |
Baseline Machine Learning Model
In this study, standard feature extraction techniques, including gray level co-occurrence matrix (GLCM), were used as a basis of comparison to obtain high-level representation such as texture and shape, such that the spatial distribution of gray values. These were analyzed by calculating local features at each image point and extracting a set of statistics from the local feature distributions. By applying GLCM matrices, 14 textural features based on statistical theory were computed. All these features were presented as Haralick textural feature vectors and used as input for the classification methods. The Haralick textural features were computed with the mahotas library in Python, which is frequently used for image processing and computer vision applications [29]. Afterwards, in the learning phase, different classification algorithms were trained to classify treated and untreated lesions, including (1) decision tree (DT), (2) support vector machines (SVM) using an RBF kernel, and (3) artificial neural network (ANN). Implementation of these algorithms was done based on “Scikit-learn” as an open source tools for data analysis which is one of the python library packages [30], while hyperparameters of each method were chosen by empirically using a separate validation set. Comparison with these machine learning techniques helped to compare the performance of the proposed neural network with methods that are still used in recent studies to predict outcomes and grades for CLMs.
Treatment Response Prediction to FOLFOX-Based Chemotherapy Regimen
Data Preparation and Preprocessing
In the following experiment, the data preprocessing and bounding box extraction steps around each ROI were identical to the untreated lesion classification task described in the previous section. The dataset was split into 70%, 10%, and 20% for training, validation, and testing, respectively, as for the previous experiment. The CNN with fivefold cross-validation was trained for prediction purposes. The following four experiments were performed:
Bounding box on the largest untreated lesion per patient;
Segmentation of the largest untreated lesion per patient;
Bounding box on all untreated lesions per patient;
Segmentation of all untreated lesions per patient.
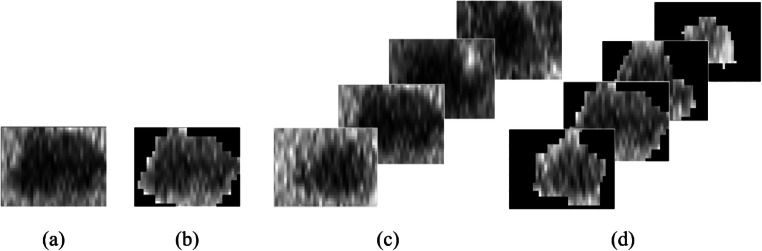
Figure 4 illustrates the details between all these proposed experiments. The output of the model yields a probabilistic prediction of response to FOLFOX with Bevacizumab chemotherapy regimen.
Fig. 4.
Schematic illustration of the set of experiments for treatment response prediction to Folfox regimen: a bounding box on the largest untreated lesion, b segmentation of the largest untreated lesion, c bounding box on all untreated lesions, and d segmentation of all untreated lesions
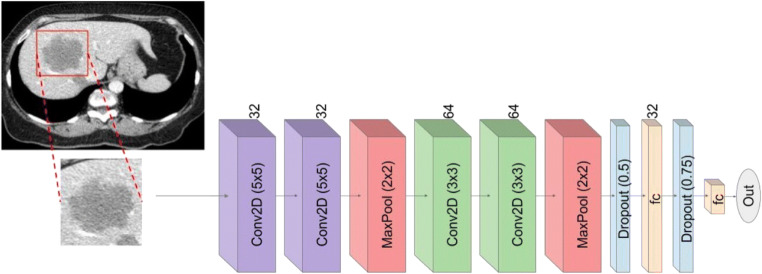
Network architecture
For the response prediction module, Keras API [27] was used for implementing the network layer architecture and for the parameter optimization. As shown in Fig. 5, the layer architecture that has been used for implementing the CNN classifier includes four convolutional 2D layers followed by a rectified linear unit (ReLU), two maxpooling 2D layers, two dropout, and a flatten layer. The final dense layer was followed by a ReLU activation function. The details of each block derived from the deep neural network and the details of framework architecture are shown in Table 2. This particular architecture was adapted from the previous module to reduce overfitting of data, as well handle the complexity and limited features for treatment response.
Fig. 5.
Proposed deep neural network inspired by the Inception-Net architecture for response prediction of FOLFOX with Bevacizumab chemotherapy regimen from untreated lesions in baseline CT images
Table 2.
Details of the model architecture used for response prediction
| Block type | Block name | Kernel size | No. of filters |
|---|---|---|---|
| Feature extraction | Conv2D | 5 × 5 | 32 |
| Feature extraction | Conv2D | 5 × 5 | 32 |
| Feature extraction | Maxpooling | 2 × 2 | - |
| Feature extraction | Conv2D | 3 × 3 | 64 |
| Feature extraction | Conv2D | 3 × 3 | 64 |
| Feature extraction | Maxpooling | 2 × 2 | - |
| Classification | Dropout (0.5) | - | - |
| Classification | Fully connect | - | 32 |
| Classification | Dropout (0.75) | - | - |
| Classification | Fully connected | - | 2 |
| Classification | Softmax | - | - |
Statistical Analysis
Statistical analysis of classification and prediction results was performed using paired t tests (IBM SPSS Statistics v20, Armonk, NY, USA), with a p value < 0.05 considered as statistically significant. The significance of systematic bias was evaluated with a paired t test on proportional difference in classification accuracy.
Results
Classification of Treated vs. Non-treated Lesions
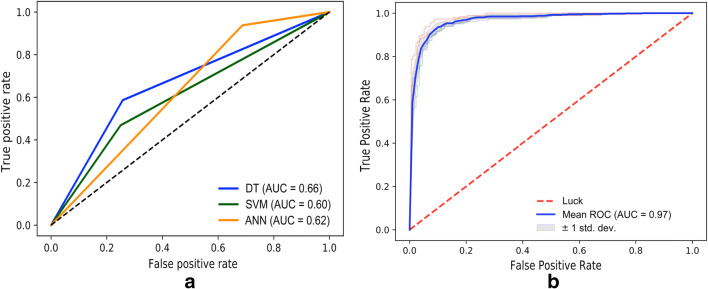
Using a fivefold cross-validation‚ the proposed deep neural network based on Inception-Inspired-CNN showed high classification performance discriminating treated vs untreated lesions in CT with an AUC of 0.97 which is statistically significant improvement as compared with AUC of 0.66, 0.60, and 0.62 for DT, SVM, and ANN classifiers respectively (see the “Related Work” section for details). In addition, for Inception-Inspired CNN-based model, a sensitivity of 90% (95% confidence interval, 86–93)‚ a specificity of 91% (95% CI, 85–94), and an overall accuracy of 0.91 (95% CI, 88–93) were obtained based on a per lesion evaluation. Figure 6 presents the receiver operating characteristics (ROC) curves that were used to measure the performance of classification tasks to define the ideal thresholds. In addition, the area under the curve (AUC) was utilized to evaluate how well the model is able to generalize the classification problem between treated and untreated lesions. The model showed better performance in comparison with traditional classification methods such as SVM, DT, and ANN, with respective accuracies of 59%, 67%, and 60% using local features based on Haralick textural features vector. Classification results between treated and untreated lesion through the CNN model are shown in Fig. 7.
Fig. 6.
ROC curve for classification performance using classical machine learning techniques (a) (decision trees (DT), support vector machine (SVM), and artificial neural network (ANN)), and using the proposed CNN framework (b)
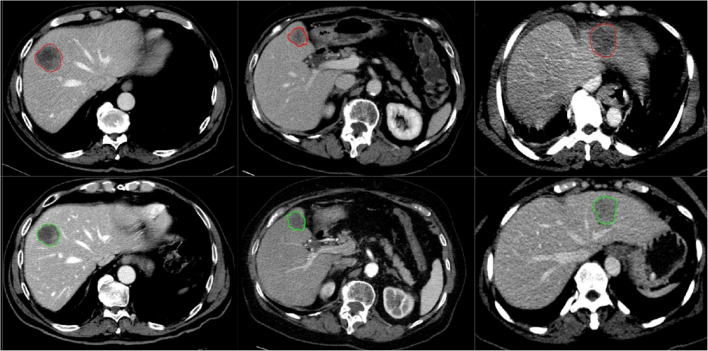
Fig. 7.
Classified untreated and treated based on CNN model, lesions delineated in red were identified as untreated and lesions delineated in green as treated lesions
Treatment Response Prediction
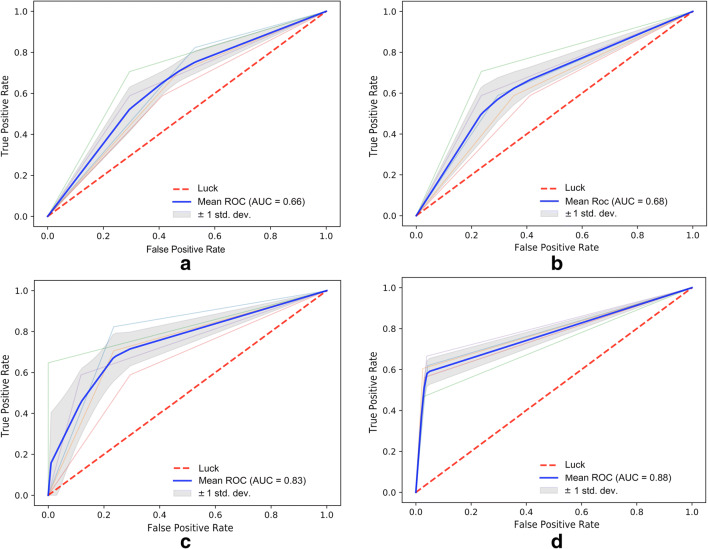
All the evaluation metrics for the experiments regarding the first-line regimen response prediction are shown in Table 3. The CNN-based framework was re-trained and re-tested using four different scenarios, as explained in the “Related Work” section. The first experiment based on the bounding box over the largest untreated lesion achieved an AUC of 0.66 (95% CI, 0.61–0.75), accuracy of 61% (95% CI, 54–73), sensitivity of 59% (95% CI, 53–68), and a specificity of 65% (95% CI, 62–77). The second experiment based on the segmentation of the largest untreated lesion achieved AUC of 0.68 (95% CI, 0.63–0.78), accuracy of 66% (95% CI, 62–75), sensitivity of 69% (95% CI, 62–80), and specificity of 60% (95% CI, 54–71). For the third experiment, bounding boxes on all lesions yielded an AUC of 0.83 (95% CI, 0.78–0.87), accuracy of 78% (95% CI, 74–83), sensitivity of 97% (95% CI, 94–99), and specificity of 59% (95% CI, 52–68). Finally, an AUC of 0.88 (95% CI, 0.85–0.94), accuracy of 76% (95% CI, 71–82), sensitivity of 98% (95% CI, 96–99), and specificity of 54% (95% CI, 50–60) were obtained with the fourth configuration, based on segmentations of all lesions. Results demonstrate improved performance with the third and fourth experiments where all lesions have been applied for training the CNN. From the 34 poor responders in the test set, the proposed model was able to predict response correctly in 27 cases. The descriptive ROC curves for each experiment are illustrated in Fig. 8.
Table 3.
Results of the treatment response prediction to FOLFOX chemotherapy from the proposed deep learning framework. Performance is reported for four different configurations experiments, each using the same number of images
| Experiments | AUC | Sensitivity (%) | Specificity (%) | Acc. (%) |
|---|---|---|---|---|
| Setup 1—Bounding box largest untreated lesion | 0.66 | 59.3 | 65.4 | 61.7 |
| Setup 2—Segmentation largest untreated lesion | 0.68 | 69.6 | 60.6 | 66.1 |
| Setup 3—Bounding box on all untreated lesions | 0.83 | 97.2 | 59.5 | 78.9 |
| Setup 4—Segmentation of all untreated lesions | 0.88 | 98.1 | 54.3 | 76.6 |
Fig. 8.
ROC curves for the different testing scenarios with the proposed DNN framework, including a bounding boxes on the largest lesion per patient, b segmentation of the largest lesion per patient, c bounding box on all untreated lesions per patient, and d segmentation of all untreated lesions per patient
Discussion and Conclusion
Unsupervised image classification is one of the fields of research which has been drastically changed by deep learning and which may assist radiologists to improve the yield for accurate diagnosis and improve patient follow-up. To the best of our knowledge, this is the first unsupervised deep learning framework which enables discriminating between treated and untreated lesions to identify potentially new lesions on baseline CT images and to provide a probability of response to chemotherapy with respect to all untreated lesions. Once established‚ such novel prediction models could easily be deployed in clinical routine to confirm which CLMs were previously treated with TACE using CT images in patients with incomplete clinical history and help in determining optimal chemotherapy regimen. On the other hand, a precise and efficient prediction model for recognizing the patients who will respond to a specific line of chemotherapy (FOLFOX plus bevacizumab) regimen according to baseline CT images (pretreatment) has several clinical benefits. By enabling early diagnosis of desired chemotherapy regimen for the recipients, we can prevent the negative side effects of this initial chemotherapy on liver toxicities [5, 31].
The envisioned integration in clinical practice would be to provide a tool to have an early indicator if untreated lesions are in fact resistant to FOLFOX therapy. In our preliminary analysis of the retrospective imaging exams, response prediction on previously treated lesions provided little value as chemotherapy tended to homogenize the intensity of voxels, significantly impacting the classification performance. Relying solely on patient history may not be fully reliable if new lesions appear on follow-up exams. Therefore, it is difficult to assume that all segmented lesions where either treated with chemotherapy or not. In order to facilitate the pipeline and clinical adoption, a consensus among hepatic radiologists was to first distinguish which lesions were already treated from those untreated in order to perform outcome prediction on these lesions. This would allow to streamline the integration of such a prediction tool.
With regard to classification between treated and untreated lesions, results show that while texture feature analysis is a reliable method to express the heterogeneity of tumors, these are based on a set of optimal handcrafted metrics which need to be pre-selected and do not necessarily reflect the high-dimensional features captured with neural networks. This study used different techniques such as GLCM and Haralick textural features descriptors to train classical machine learning algorithms (SVM, DT, ANN), and compared their performance to a CNN-based model. Fully connected neural networks prove superior in predicting highly heterogeneous CLM treated with TACE compared with traditional classification methods based on hand-crafted texture feature analysis methods for conventional machine learning. The results related to treatment response prediction show that deep neural networks might provide good accuracy for predicting response to FOLFOX on baseline CT in patients with CLMs. Once established‚ such a novel prediction model could easily be deployed in clinical routine and help in assessing other protocols, such as regimens based on FOLFIRI.
While the results presented in this paper are encouraging, there are some limitations. First, all of the images used in the model training included lesions, with no healthy subjects included. Furthermore, results show that the top-performing model is based on the segmentations of the untreated lesions. This requires highly accurate automatic segmentations to be produced, demanding a segmentation model which was trained on an important set of a lesions boundaries each validated by a trained radiologist. Finally, the training set is comprised of only 444 lesions from 202 patient CT scans, which is an order of magnitude less than in other deep learning applications in radiology, which includes thousands of images. The study was also performed in a single center and does not include a multi-center analysis. Further studies with larger sample sizes and independent datasets will be required to validate results. Other planned works include an evaluation of the predictive model on other chemotherapy regimens, such as with FOLFIRI compounds, as well as extending the validation in a multi-centric study, where the effect of acquisition and drug-regimen parameters will be evaluated.
Acknowledgments
We would like to express our appreciation to Imagia Cybernetics for providing the desired hardware for doing our experiments.
Funding Information
This study was financially supported by MEDTEQ and IVADO grants, as well as the MITACS organization. The clinical and radiological data was provided by Centre de recherche du Centre hospitalier de l’Université de Montréal (CR-CHUM). The Fonds de recherche du Québec en Santé and Fondation de l’association des radiologistes du Québec (FRQ-S and FARQ no. 34939) financially supported An Tang.
Compliance with Ethical Standards
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (include name of committee + reference number) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Footnotes
This work was accepted and presented as an abstract for SIIM 2019, in Denver, CO.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.American Cancer Society Key Statistics for Colorectal Cancer. Available at https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html. Accessed 24 January 2019.
- 2.Massmann A, Rodt T, Marquardt S, Seidel R, Thomas K, Wacker F, Richter GM, Kauczor HU, Bücker A, Pereira PL, Sommer CM. Transarterial chemoembolization (TACE) for colorectal liver metastases—current status and critical review. Langenbeck's Arch Surg. 2015;400:641–659. doi: 10.1007/s00423-015-1308-9. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau-Antonacci A, Petitclerc L, Gilbert G, Bilodeau L, Olivié D, Cerny M, Castel H, Turcotte S, Huet C, Perreault P, Soulez G, Chagnon M, Kadoury S, Tang A: Dynamic contrast-enhanced MRI to assess hepatocellular carcinoma response to Transarterial chemoembolization using LI-RADS criteria: A pilot study. Magn Reson Imaging, 2019. 10.1016/j.mri.2019.06.017 [DOI] [PubMed]
- 4.Bonanni L, Carino NDL, Deshpande R, Ammori BJ, Sherlock DJ, Valle JW, Tam E, O’Reilly DA. A comparison of diagnostic imaging modalities for colorectal liver metastases. Eur J Surg Oncol. 2014;40(5):545–550. doi: 10.1016/j.ejso.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 5.Loupakis F, Schirripa M, Caparello C, Funel N, Pollina L, Vasile E, Cremolini C, Salvatore L, Morvillo M, Antoniotti C, Marmorino F, Masi G, Falcone A. Histopathologic evaluation of liver metastases from colorectal cancer in patients treated with FOLFOXIRI plus bevacizumab. Br J Cancer. 2013;108:2549–2556. doi: 10.1038/bjc.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H. Brain tumor segmentation with Deep Neural Networks. Med Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Simpson AL, Doussot A, Creasy JM, Adams LB, Allen PJ, DeMatteo RP, Gönen M, Kemeny NE, Kingham TP, Shia J, Jarnagin WR, Do RKG, D’Angelica MI. Computed Tomography Image Texture: A Noninvasive Prognostic Marker of Hepatic Recurrence After Hepatectomy for Metastatic Colorectal Cancer. Ann Surg Oncol. 2017;24:2482–2490. doi: 10.1245/s10434-017-5896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern. 2007;SMC-3:610–621. doi: 10.1109/tsmc.1973.4309314. [DOI] [Google Scholar]
- 9.Ba-Ssalamah A, Muin D, Schernthaner R, Kulinna-Cosentini C, Bastati N, Stift J, Gore R, Mayerhoefer ME: Texture-based classification of different gastric tumors at contrast-enhanced CT. Eur J Radiol 82, 2013. 10.1016/j.ejrad.2013.06.024 [DOI] [PubMed]
- 10.Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V. Assessment of Primary Colorectal Cancer Heterogeneity by Using Whole-Tumor Texture Analysis: Contrast-enhanced CT Texture as a Biomarker of 5-year Survival. Radiology. 2012;266:177–184. doi: 10.1148/radiol.12120254. [DOI] [PubMed] [Google Scholar]
- 11.Hayano K, Yoshida H, Zhu AX, Sahani DV. Fractal analysis of contrast-enhanced CT images to predict survival of patients with hepatocellular carcinoma treated with sunitinib. Dig Dis Sci. 2014;59:1996–2003. doi: 10.1007/s10620-014-3064-z. [DOI] [PubMed] [Google Scholar]
- 12.Lubner MG, Stabo N, Lubner SJ, del Rio AM, Song C, Halberg RB, Pickhardt PJ. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40:2331–2337. doi: 10.1007/s00261-015-0438-4. [DOI] [PubMed] [Google Scholar]
- 13.Ganeshan B, Miles KA, Young RCD, Chatwin CR. In Search of Biologic Correlates for Liver Texture on Portal-Phase CT. Acad Radiol. 2007;14:1058–1068. doi: 10.1016/j.acra.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 14.Miles KA, Ganeshan B, Griffiths MR, Young RCD, Chatwin CR. Colorectal Cancer: Texture Analysis of Portal Phase Hepatic CT Images as a Potential Marker of Survival. Radiology. 2009;250:444–452. doi: 10.1148/radiol.2502071879. [DOI] [PubMed] [Google Scholar]
- 15.Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35:1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 16.Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E. Deep Learning for Computer Vision: A Brief Review. Comput Intell Neurosci. 2018;2018:1–13. doi: 10.1155/2018/7068349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gladis VP, Rathi P, Palani S. Brain Tumor Detection and Classification Using Deep Learning Classifier on MRI Images 1. Res J Appl Sci Eng Technol. 2015;10:177–187. [Google Scholar]
- 18.Paul R, Hawkins SH, Hall LO, Goldgof DB, Gillies RJ: Combining deep neural network and traditional image features to improve survival prediction accuracy for lung cancer patients from diagnostic CT. In: 2016 IEEE International Conference on Systems, Man, and Cybernetics, SMC 2016 - Conference Proceedings, 2017, pp 2570–2575
- 19.Nie D, Zhang H, Adeli E, Liu L, Shen D: 3D deep learning for multi-modal imaging-guided survival time prediction of brain tumor patients. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2016, pp 212–220. [DOI] [PMC free article] [PubMed]
- 20.Kumar N, Verma R, Arora A, Kumar A, Gupta S, Sethi A, Gann PH. Medical Imaging 2017: Digital Pathology. 2017. Convolutional neural networks for prostate cancer recurrence prediction; p. 101400H. [Google Scholar]
- 21.Chang C-C, Lin C-J. LIBSVM. ACM Trans Intell Syst Technol. 2011;2:1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 22.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 23.Schmidhuber J. Deep Learning in neural networks: An overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Bibault JE, Giraud P, Durdux C, Taieb J, Berger A, Coriat R, Chaussade S, Dousset B, Nordlinger B, Burgun A. Deep learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611. doi: 10.1038/s41598-018-30657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorontsov E, Tang A, Pal C, Kadoury S: Liver lesion segmentation informed by joint liver segmentation. In: Proceedings - International Symposium on Biomedical Imaging, 2018, pp 1332–1335
- 26.Rubbia-Brandt L, Giostra E, Brezault C, Roth AD, Andres A, Audard V, Sartoretti P, Dousset B, Majno PE, Soubrane O, Chaussade S, Mentha G, Terris B. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol. 2007;18:299–304. doi: 10.1093/annonc/mdl386. [DOI] [PubMed] [Google Scholar]
- 27.Rodel C, Martus P, Papadoupolos T, Füzesi L, Klimpfinger M, Fietkau R, Wittekind C. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23(34):8688–8696. doi: 10.1200/JCO.2005.02.1329. [DOI] [PubMed] [Google Scholar]
- 28.Chollet F, et al: Keras. 2015. Retrieved from https://github.com/fchollet/keras
- 29.Coelho LP. Mahotas: Open source software for scriptable computer vision. J Open Res Softw. 2013;1(1):e3. doi: 10.5334/jors.ac. [DOI] [Google Scholar]
- 30.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 31.Ravichandran K, Braman N, Janowczyk A, Madabhushi A. Medical Imaging 2018: Computer-Aided Diagnosis. 2018. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI; p. 105750C. [Google Scholar]