Abstract
Accurate and fully automatic brain tumor grading from volumetric 3D magnetic resonance imaging (MRI) is an essential procedure in the field of medical imaging analysis for full assistance of neuroradiology during clinical diagnosis. We propose, in this paper, an efficient and fully automatic deep multi-scale three-dimensional convolutional neural network (3D CNN) architecture for glioma brain tumor classification into low-grade gliomas (LGG) and high-grade gliomas (HGG) using the whole volumetric T1-Gado MRI sequence. Based on a 3D convolutional layer and a deep network, via small kernels, the proposed architecture has the potential to merge both the local and global contextual information with reduced weights. To overcome the data heterogeneity, we proposed a preprocessing technique based on intensity normalization and adaptive contrast enhancement of MRI data. Furthermore, for an effective training of such a deep 3D network, we used a data augmentation technique. The paper studied the impact of the proposed preprocessing and data augmentation on classification accuracy.
Quantitative evaluations, over the well-known benchmark (Brats-2018), attest that the proposed architecture generates the most discriminative feature map to distinguish between LG and HG gliomas compared with 2D CNN variant. The proposed approach offers promising results outperforming the recently supervised and unsupervised state-of-the-art approaches by achieving an overall accuracy of 96.49% using the validation dataset. The obtained experimental results confirm that adequate MRI’s preprocessing and data augmentation could lead to an accurate classification when exploiting CNN-based approaches.
Keywords: Classification, 3D convolutional neural network (CNN), Magnetic resonance imaging (MRI), Gliomas, Classification, Deep learning
Introduction
Early and accurate detection of brain tumor grade has a direct impact not only on the patient’s estimated survival but also on treatment planning and tumor growth evaluation. Among the central nervous system (CNS) primary brain tumors, gliomas could be considered as the most aggressive [1]. Recently, the World Health Organization (WHO), in its revised fourth edition published in 2016 [2], has considered two categories of glioma tumors: the low-grade (LG) and the high grade (HG) glioblastomas. The LG gliomas tend to exhibit benign tendencies. However, they have a uniform recurrence rate and could increase in grade over time. The HG gliomas are undifferentiated and carry a worse prognosis [3]. Among the recent sophisticated technologies, MRI could be considered as one of the main modalities used to image brain tumors for diagnosis and evaluation. Accurate identification of the tumor grade could be considered as a critical phase for various neuroimaging explorations [4]. Such an operation could be considered a time-consuming task. Consciously, several machine learning [5–7] and deep learning-based approaches [8, 9] have quickly evolved during the past few years illustrating that today’s medicine depends a lot on advanced information technologies. Several approaches have been proposed in the literature in order to classify brain tumors through MR imaging. They could be divided into two categories: supervised and unsupervised approaches. (1) The supervised approaches adopt a well “labeled” data. In fact, these approaches learn from labeled training data to predict results for unforeseen ones. (2) The unsupervised approaches could be defined as machine learning ones which do not need to supervise the model. Rather, we must permit the model to labor on its own to collect information from unlabeled data. The study [10] proved that the best classification accuracy of glioma brain tumors was achieved using 3D discrete wavelet transform (DWT) for feature extraction combined with random forest (RF) classifier. A comparative study with several classifiers such as multi-layer perceptron (MLP) [11], radial basis function (RBF) [12], or naive Bayes classifier [13] has been realized in order to attest the performance of the 3D DWT [10]. Support vector machine (SVM) has been also widely used for MR image classification [14, 15]. The study in [16] investigates a hybrid system based on genetic algorithm (GA) and SVM with Gaussian RBF kernel. GA optimization has been used to select the most relevant features. Experimental results illustrate that the use of GA improves the SVM’s classification accuracy.
Despite the significant potential of the supervised approaches in glioma brain tumor classification, it required specific expertise for optimal features extraction and selection techniques [17]. Over the past several years, unsupervised approaches [18] have gained researchers’ interest not only for their great performances but also because of the automatically generated features which could reduce the error rate. Recently, deep learning (DL)-based methods have emerged as one of the most prominent methods for medical image analysis such as classification [19], reconstruction [20], and even segmentation [21]. Recently, Iram et al. [22] discussed the use of a pre-trained VGG-16 model [23] for feature extraction. Feature map has been feed to long short-term memory (LSTM) recurrent neural network [24] to classify brain tumors into high/low grade. Authors assume that the pre-trained CNN models for feature extraction present better performance when cascaded with LSTM and achieve higher accuracy compared with GoogleNet [25], ResNet [26], and AlexNet [27]. Apparent 2D CNN limitations for brain tumors MRI classification have been discussed in recent work [28]. As a solution, a voxelwise residual network (VoxResNet) based on 3D CNN-based architecture for the identification of white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) has been proposed [28]. Experimental results confirm the efficiency of brain tissues’ classification from volumetric 3D MR images. This algorithm has been ranked first in the MR brain challenge in 2017, outperforming 2D CNN’s state-of-the-art methods. Authors in [29] have proposed an end-to-end 3-dimensional convolutional neural network (3D CNN) with gated multi-modal unit (GMU) fusion to integrate the information both in three dimensions and in multiple modalities. The whole MRI images have been used in order to be applied directly to 3D convolutional kernels using different MRI directions (sagittal, axial, and coronal). Inspired by the potential success of such architecture, we were motivated to implement a 3D CNN for brain glioma tumor classification.
The accurate glioma brain tumor classification has been considered as a harmful task due to highly inhomogeneous tumor regions composition. In fact, the tumor region includes edema, necrosis, and enhancing/non-enhancing tumor. Furthermore, some tumor sub-region intensities’ profiles may overlap with healthy tissues.
We propose, in this paper, a novel glioblastomas’ brain tumor grade classification approach based on deep three-dimensional convolutional neural network (3D CNN) in order to distinguish between HG and LG tumors. The principal research contributions of this paper are mainly:
The proposed approach presents an automated multi-scale 3D CNN-based architecture for brain MRI glioma tumor classification on the basis of the World Health Organization (WHO) standards.
A preprocessing method [30] for volumetric MR images has been used to improve the performance of CNN to overcome the major problem in MR images such data heterogeneity and low contrast.
Using deep architecture through small 3D kernels size, the proposed architecture has the potential to extract more local and global contextual features with highly discriminative power for glioma classification with reduced computational and memory requirements.
We have applied a data augmentation technique to generate new patches from the original ones to overcome the lack of data and to tackle the large variation of brain tumors heterogeneity.
Comparison results over MICAII challenge “BRATS-2018” prove that the proposed approach could yield the best performance by presenting the highest classification accuracy compared with supervised/unsupervised recent state-of-art methods.
The remainder of this paper is organized as follows: Proposed approach details the proposed approach. Results explores the experimental results and discusses the obtained results. Finally, conclusions are drawn in Discussion.
Proposed Approach
The proposed approach investigated a real 3D deep CNN architecture for automatic MRI glioma brain tumor grading. For instance, a 2D deep learning model learns increasingly complex features’ hierarchy, by implementing many trainable filters’ layers and optional pooling operations. The majority of these methods do not entirely examine the volumetric information in MR images but explore only two-dimensional slices. These slices could be considered independently or by using three orthogonal 2D patches to merge the contextual information [21]. Hence, our proposed approach, based on employing 3D convolutional filters, takes advantage of generating more powerful contextual features that deal with large brain tissues’ variations [13].
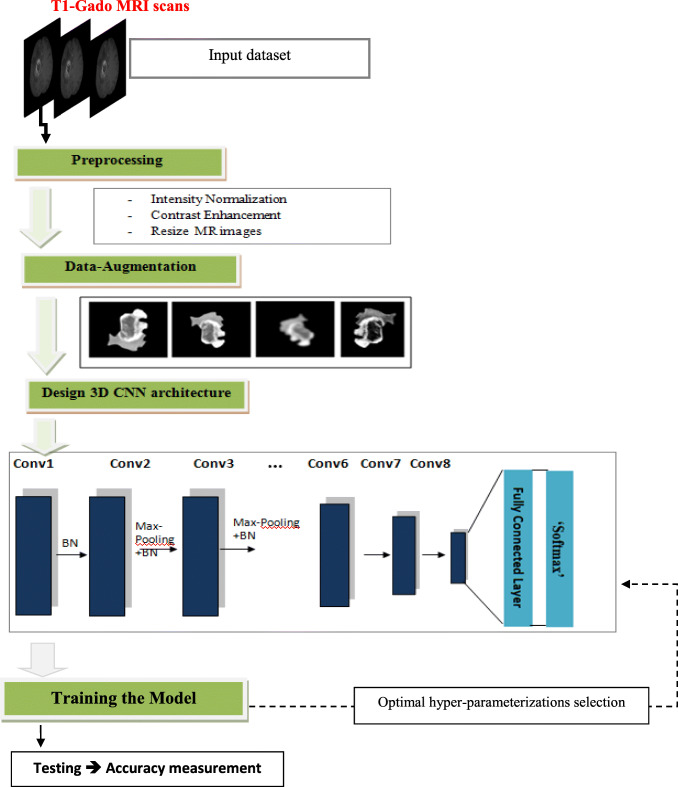
Furthermore, to boost the proposed model performance, we adopted a preprocessing approach based on an intensity normalization followed by a contrast enhancement technique for MR images. Such a process could be considered as not conventional (typical) in CNN-based classification approaches. The proposed approach flowchart is illustrated in Fig. 1.
Fig. 1.
Flowchart of the proposed approach for glioma brain tumor classification
The proposed approach includes essentially the following steps:
Data preprocessing: Normalization and contrast enhancement, through T1-Gado MR scans, have been applied in order to enhance the images’ quality, followed by a resizing step of the input images to optimize the required memory.
Data augmentation: A simple flipping method is performed in order to fill the gap of data’s lack to ensure an efficient CNN training.
3D CNN architecture design and optimization: The hyper-parameters, such as the number of convolution layers, pooling layers, and fully connected layers (FCLs) have been settled.
Model training: Training the proposed model using the augmented dataset and enhanced MR images.
Preprocessing
One of the major difficulties in MRI analysis is to deal with the thermal noise and the artifacts caused by the magnetic field and the small motions produced by the patient during the scanning process. In fact, existing noise in the acquired MRI scans could corrupt the fine details, blur tumor edges, and even decrease the images’ spatial resolution [31]. Thus, it could seriously degrade the performance of CNN-based methods by making the feature extraction more complicated [32]. For this reason, denoising and contrast enhancement [33] techniques for MR images [4] have gained recently a lot of interest and have been widely investigated by researchers to ameliorate the quality of the data before engaging in MRI brain tumors exploration such as classification and/or segmentation [28, 34, 35].
Since the MRI scans could be collected from multiple institutions, the MRI scan intensities may vary significantly. Therefore, the intensity normalization [21] based on linear transformation in the range [0, 1] through the min-max normalization technique is used to reduce intensities inhomogeneity.
In this work, our preprocessing consists of 3 steps as illustrated in Fig.2: First, we apply intensity normalization of the whole T1-Gado MRI scans followed by an MRI contrast enhancement method, previously developed in a previous work [30]. Finally, we resize the input MR images for memory optimization purposes. In fact, the size of the MR images in the BraTS database is 250 × 250 × 155. The considered size is then 112 × 112 × 94. The adopted image resizing is the cubic B-spline method [36].
Fig. 2.
Preprocessing steps
Data Augmentation
In computer vision, the data augmentation could be considered as an important key factor that is very effective in training highly deep learning based-methods [37]. A variety of data augmentation strategies have been proposed in the literature [38] for deep learning in medical imaging such as random crops, rotation, shears, and flipping. Recent studies [39] prove that some augmentation strategies could capture medical image features more effectively than others leading to better accuracy. This study demonstrates that the flipping technique is the optimal data augmentation strategy for medical imaging classification that leads to more discriminative feature maps compared with other techniques. Based on this study, we are encouraged to adopt the only horizontal and vertical flipping technique to generate new patches for each image in the training dataset.
Proposed Architecture: Deep Multi-scale 3D Convolutional Neural Network
The proposed architecture, illustrated in Table 1, is built with eight convolutional layers and three fully connected (FC) layers. For accurate brain glioma tumor classification, our proposed CNN model is based essentially on two principal components:
Unlike 2D-CNN architecture, which does not entirely examine the volumetric information in MR images but explore only two-dimensional slices, we adopt a 3D convolutional layer that provides a detailed feature map exploiting the entire volumetric spatial information to incorporate both the local and global contextual information.
Deep network architecture that produces a better quality of local optima. The additional nonlinearity, in such architecture, could yield highly discriminative power [40]. As a result of the richer structures captured by the deeper models, the deeper architectures have previously shown its effectiveness for natural images’ classification. On 3D networks, its impact could be considered as more drastic [40]
Table 1.
Proposed architecture
| Type | Filter size | # Number of filters | Stride | # FC units | |
|---|---|---|---|---|---|
| Layer 1 | Conv1 + BN | 3 × 3 × 3 | 32 | (2, 2, 2) | - |
| Layer 2 | Conv2 + Max Pooling + BN | 3 × 3 × 3 | 64 | (1, 1, 1) | - |
| Layer 3 | Conv3 + Max Pooling + BN | 3 × 3 × 3 | 128 | (2, 2, 2) | - |
| Layer 4 | Conv4 + Max Pooling + BN | 3 × 3 × 3 | 256 | (1, 1, 1) | - |
| Layer 5 | Conv5+ BN | 3 × 3 × 3 | 256 | (2, 2, 2) | - |
| Layer 6 | Conv6 + Up-sampling + BN | 3 × 3 × 3 | 128 | (1, 1, 1) | - |
| Layer 7 | Conv7 + Up-sampling + BN | 3 × 3 × 3 | 64 | (2, 2, 2) | - |
| Layer 8 | Conv8 | 3 × 3 × 3 | 32 | (1, 1, 1) | - |
| Layer 9 | FC-1 | - | - | - | 256 |
| Layer 10 | FC-2 | - | - | - | 256 |
| Layer 11 | FC-3(Softmax) | - | - | – | 2 |
Conv refers to convolutional layer, BN refers to Bach normalization, FC refers to fully connected (FC) layers, Up-sampling layer: it concatenates a repetition layer with a Conv + Max Pooling + BN layer with kernel size 3 × 3
However, 3D CNN is computationally and memory exhausted as an increased number of trainable parameters are required when compared with the 2D CNN variant. Thus, as a solution, we proposed the exclusive use of 3 × 3 kernels at each convolutional layer which could be considered as faster to convolve allowing stacking more layers with reduced weight. Meanwhile, the pooling layers were used in order to reduce the size of the intermediate layer. Another adopted solution to deal with memory constraints is the use of reduced filters’ number per layer especially in the first two layers of the network where the features have higher dimensionality (only32 filters in the first layer and 64 in the second layer).
Only eight convolutional layers were stacked to avoid that the extracted features become more abstract with the network’s increased depth. The next subsections detail the proposed CNN architecture and the adopted hyper-parameters.
Activation function
The activation function could be considered as the responsible of the nonlinearity which transforms the data. The rectifier linear unit (ReLU) is deployed as an activation function in the proposed model, defined in Eq. (1), where f (i) represents the function of neuron’s output of an input called “i.”
| 1 |
We adopt “ReLU” to achieve better performance considering its ability to faster train deep CNN alternately to classical “sigmoid” or “hyperbolic tangent” functions given by Eq. 2.
| 2 |
Pooling
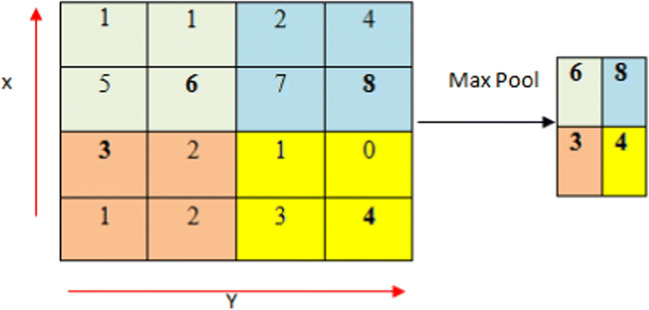
Pooling is a down-sampling strategy on CNN. We could specify essentially two conventional forms for pooling such as max pooling [41] and average pooling [42]. The average pooling is characterized by the consideration of all elements in a pooling region, even the parts which have low magnitude. The combination between the average pooling and the (ReLU) activation function leads to the creation of down-weighting strong activations’ effect as a result of the average computation takes into account many zero elements. Even with hyperbolic tangent activation functions, which could be considered as a worse case, the strong positive and the negative activation could cancel each other out, which could engender then smaller pooled responses [43]. Fortunately, max pooling does not present such drawbacks. For this reason, max pooling has been used, in this work, since it extracts the most relevant features for classification like tumor edges [21]. A max filter has been applied to the max pooling process to non-overlapping the initial representation’s sub-regions. Fig. 3
Fig. 3.
Max pooling concept with 2 × 2 filters and stride 2
Regularization
For the fully connected (FC) layers, we have used the dropout [44] as a regularization to boost the generalization ability and to prevent overfitting. Dropout removes stochastically the network’s nodes with probability for each region during training. Therefore, all FC layers’ nodes have been forced to learn better the data’s representations while preventing nodes from co-adapting to each other. All nodes have been used at the test time. Dropout could be then considered as a different networks’ set and a banging’s form because each network has been improved by a training data portion.
Training Step
For efficient data training, the owing parameters was discussed: optimizer, loss function, and initialization
Optimizer
Adam is an optimization algorithm that could be used as a substitute for the classical stochastic gradient descent (SGD) procedure to refresh network weights. Such optimizer combined two extensions of stochastic gradient descent’s advantages specifically: adaptive gradient algorithm (AdaGrad) and Root Mean Square Propagation (RMSProp) [45].
Loss Function
In this research, categorical cross-entropy is employed as a loss function. This function is used to compare the predictions’ distribution with the true one according to Eq. (3) where ŷ and y represent respectively the predicted and the target values.
| 3 |
Initialization
The Glorot normal called also Xavier normal has been used since it could be considered as one of the most recommended common initialization schemes for deep CNN architectures [21]. We ensure then to maintain in control of the activations and the gradients. Samples have been drawn from uniform distribution within [− limit, limit] given by Eq. 4 where fin and fout represent respectively the input’s number and the output units in the weight tensor.
| 4 |
Results
This section is dedicated to the presentation of the experiment’s results in order to justify the hyper-parameters choice and to validate the real impact of the proposed approach’s main contributions. Python 3.4 environment has been used to construct the proposed 3D-CNN using the KERAS and Tensorflow backend library on a workstation Intel-i7 2.60 GHz CPU, 19.5Go RAM equipped with NVIDIA GPU Geforce GTX 1080 Ti 11Go RAM.
Dataset
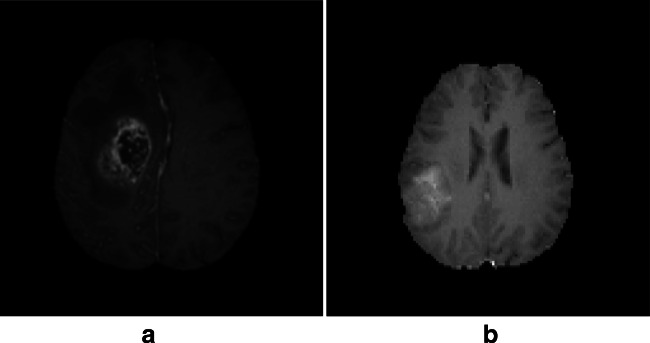
The evaluation was carried out on the multi-modal Brain Tumor Segmentation Challenge (BraTS 2018) [46] in conjunction with the MICCAI conference. This challenge has essentially been taken in order to compare among the current state-of-the-art methods for the multi-modal segmentation task. Nerveless, provided annotation into HG and LG glioma tumors, approved by experienced neuroradiologists, inspired us to use such database for the classification purpose. Each subject case in BraTS-2018 has four volumetric MRI scans: the native (T1), the post-contrast T1-weighted (T1-Gado), the T2-weighted (T2), and the fluid-attenuated inversion recovery (FLAIR). All data have been previously skull-stripped, co-registered to the same anatomical template, and interpolated to the same resolution (1 mm3).The BraTS-2018 training dataset comprises 284 subjects that include 209 HG and 75 LG glioma tumors. The validation data comprise 67 mixed grades glioma tumors. The neuroradiologists have assessed radiologically the complete original TCIA glioma collections, and the dataset has been updated with more routine clinically-acquired 3T MRI scans. The BraTS-2018 is available through the Image Processing Portal of the CBICA@UPenn (IPP, ipp.cbica.upenn.edu). Figure 4 illustrates a HG and LG subject case from the BraTS dataset.
Fig. 4.
a High-grade (HG) glioma subject case. b Low-grade (LG) glioma subject case
Evaluation Metric
The accuracy has been used as an evaluation metric to assess the proposed approach’s efficiency. The classification accuracy is defined as follows:
| 5 |
where the true positives (TP) represent the high-labeled data that are correctly classified, while the true negatives (TN) are the correctly predicted, although the false positives (FP) are MR images that are classified wrongly and the false negatives (FN) represent data that are correctly classified.
Validation of the Proposed Approach
In this section, we will firstly assess each processing step’s effects on the final classification accuracy. Then, we will validate the use of a deep architecture by making a comparative study with shallower networks architecture.
Preprocessing’s Validation
The training dataset quality has a direct impact on CNN performance. The publically available dataset (BraTS) could be considered as highly heterogeneous since it has been collected from multiple sites with different scanner technology and acquisition parameter settings. Thus, it may affect the MR scan qualities that could seriously limit the classification performance of the CNN model. To reduce the data heterogeneity, several researchers, such as Pereira et al. [21], found the main gain (4.6%) in the overall accuracy of CNN-based architecture after applying an intensity normalization using the same dataset for brain tumors segmentation. Insipid by the positive effect of intensity normalization on optimizing the performance of CNN for the segmentation [47], we applied the min-max normalization technique for voxels’ intensity normalization.
For contrast enhancement, we have applied an adaptive contrast stretching technique based on the original image statistical information [30].This technique preserves the tumor’s edge as well as the original image significant features. The applied technique has achieved encouraging results in MR images’ region of interest (ROI) contrast enhancing without an over noise amplification of the entire image. The final step of our proposed preprocessing is resizing the brain MR input image in order to overcome the computational complexity.
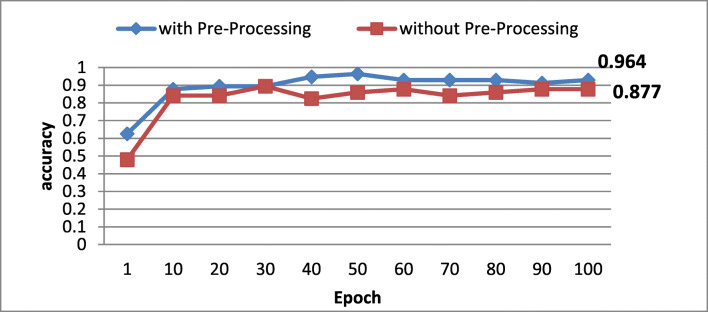
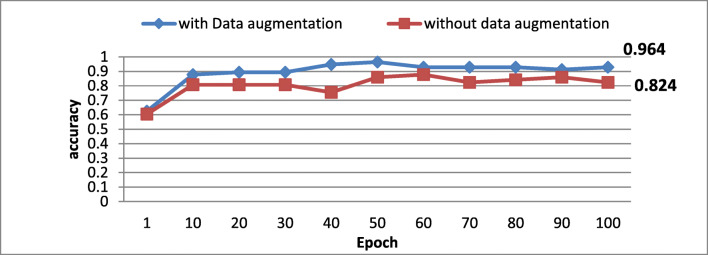
To gauge the suggested preprocessing’s impact on MRI glioma grading, we evaluate the accuracy with/without the preprocessing step. Figure 5 illustrates the obtained accuracy in the function of epoch’s number.
Fig. 5.
Preprocessing impact on classification’s accuracy
According to obtained results, one could notice that the accuracy is clearly better when applying the preprocessing confirming the effectiveness of the applied data preprocessing and the efficient model training.
Data Augmentation’s Validation
Medical imaging benchmarks are often imbalanced which could be considered as a serious problem especially when deep CNN is established for a fully automatic classification causing erroneous diagnosis guidance for the tumor grade diagnosis [39]. For instance, in the used dataset, the number of LG glioma brain tumor subject cases is much lower compared with the number of HG glioma subject cases. To balance the number of each class samples, data augmentation techniques are used. Moreover, the data augmentation is a common solution to alleviate the deeper networks’ overfitting. To evaluate such process impact on the proposed model performance, we have computed the classification’s accuracy with/without using data augmentation techniques. Figure 6 illustrates the effect of the data augmentation on the classification accuracy in terms of number of epoch.
Fig. 6.
Data augmentation technique’ impact on classification’ accuracy
As illustrated in Fig.6, one could notice that the use of the data augmentation’s techniques ameliorates clearly the classification’s accuracy from 0.8246 to 0.964. For this reason, we have adopted this amelioration on the proposed classification approach.
Deep Network Architecture’s Validation
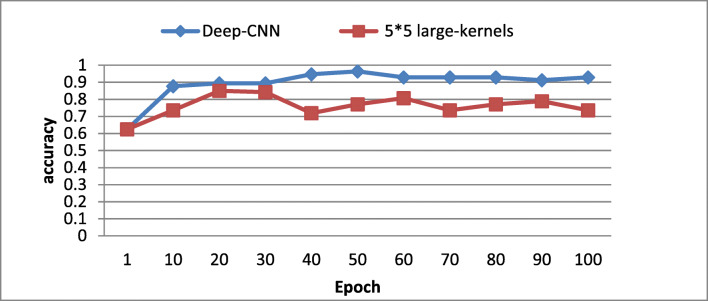
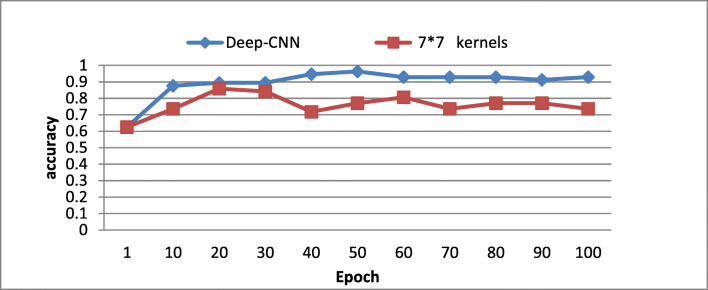
To validate the real effects of the proposed deep CNN architecture on the classification accuracy, we changed each convolutional layer of the proposed model with larger kernels which have the equivalent effective receptive field. Two variants of kernels size have been experimented using the proposed architecture:
(5 × 5) kernels size with maintaining the feature maps’ number as for the proposed architecture.
(7 × 7) kernels size where we increased the CNN’s capacity by augmenting the feature maps, namely, 64 in the first convolutional layer, 64 in the second, and 128 in the third and the fourth layers.
As illustrated in Figs. 7 and 8, one could notice that the proposed architecture yields higher accuracy value on testing and validation dataset compared with the shallower networks of (5 × 5) and to a (7 × 7) kernels size. These results confirm that our proposed architecture, using small (3 × 3), could capture more details compared with large kernels size even when increasing the feature maps. One could conclude that the proposed architecture has the advantage to maintain the effective receptive fields of bigger kernels while reducing the number of weights and allowing more non-linear transformations on the data. For this reason, we have adopted the use of (3 × 3) kernels size deep CNN.
Fig. 7.
Comparison of deep versus large kernels (5 × 5) based CNN architecture
Fig. 8.
Comparison of deep versus (7 × 7) large kernels with augmented features maps
Hyper-parameters’ Validation
In this section, we propose to study hyper-parameters’ effects on the classification’s accuracy and more specifically the effects of the pooling, the activation function, the optimizer, and the initializer.
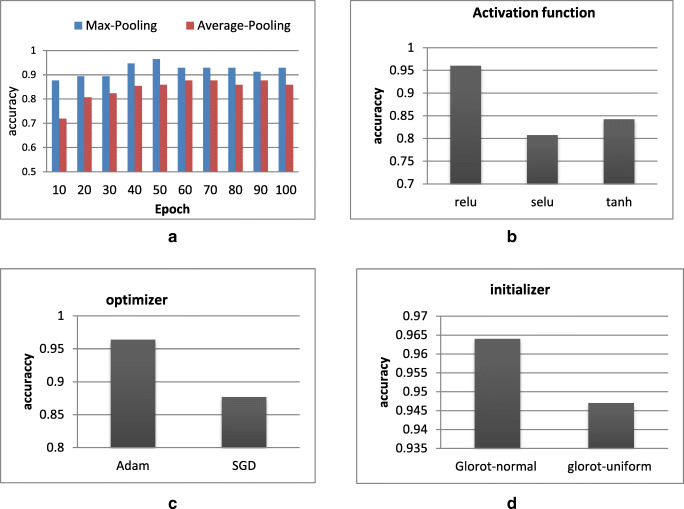
Pooling
We investigate average pooling versus max pooling. As illustrated in Fig. 9a, the max pooling has shown efficient performance compared with average one. For this reason, we have adopted the max pooling in the proposed CNN architecture.
Fig. 9.
Hyper-parameters’ validation. a Max pooling validation. b Relu activation function validation. c Adam optimize validation. d Glorot normal validation
Activation function
A comparative study between three activation function’s technique ReLu, selu, and tanh has been performed. As shown in Fig. 9b, one could notice that the ReLu activation function outperforms the two other activation function.
Optimizers
The Adam optimizer has been used to learn our network weights. Moreover, a second optimizer, the stochastic gradient descent (SGD), has been also tested in order to assess the classification performance. During the experiments, the initial learning rate for both optimizers was set to 0.001. As illustrated in Fig. 9c, the Adam optimizer provides much better performance compared with SGD optimizer.
Initializer
In order to justify our initializer’s choice, we have compared the performances of two different techniques: the Glorot normal and the Glorot uniform. As shown in Fig. 9d, the Glorot normal presents higher accuracy value that is why this initializer will be chosen in the proposed approach.
Discussion
In order to assess the proposed approach’s performances, a comparative study has been performed with both hand-crafted and deep learning-based approach from state of the art. Table 2 reports the obtained classification accuracy with the proposed approach as well as with the supervised approaches when applied to the BraTS dataset challenge for brain glioma classification.
Table 2.
Comparative study with supervised-based methods for brain MRI glioma classification (supervised method versus proposed approach)
| Method/year | Preprocessing | Feature extraction | Dim | Classifier | Accuracy (%) |
|---|---|---|---|---|---|
|
[48] (Hwan-ho, 2017) |
- | Histogram, shape/gray-level co-occurrence matrix (GLCM) | 2D | LASSO coefficient | 89.81% |
|
[49] (CHO et al., 2018) |
- | Histogram-based features GLCM intensity size-zone [ISZ] | 2D | Random Forest (RF) | 88% |
|
[10] (Ghazanfar et al., 2017) |
Intensity normalization filtering/histogram matching | 3D DWT | 3D | Random Forest | 89% |
|
[49] (CHO et al., 2018) |
- | Histogram-based features GLCM intensity size-zone [ISZ] | 2D | SVM | 87% |
| Proposed | Intensity normalization/contrast enhancement | Automatically | 3D | Deep CNN | 96.49% |
The approach proposed by [48] aims to classify glioma tumors into HG and LG. The features have been extracted by a fusion process between three modalities (MRI T1, T1-contrast, and FLAIR) based on the histogram, the shape, and the gray-level co-occurrence matrix (GLCM), and only forty-five significant features have been selected using LASSO method. The final classification is done by logistic regression (LR) using the LASSO score. The method achieved an accuracy of 89.81%.
The algorithm proposed by Cho et al. [49] investigated two types of classifiers mainly SVM and RF to distinguish between HG and LG through brain MRI. Qualitative evaluations could attest that the RF classifier showed the best performance (89%) compared with the SVM classifier (87%). For feature extraction, the authors have used shape and textural features. The study in [10] investigates brain glioma tumor’s classification into four classes (necrosis, edema, enhancing, and non-enhancing tumors). A 3D DWT has been used for feature extraction. A comparative study has been performed in order to evaluate different classifiers’ performances such as naive Bayes (NB), MLP with one hidden layer, and MLP with backpropagation, and the obtained accuracy was 60%, 70%, 76%, 80%, and 88%, respectively. RF classifier achieved then the better performance.
Table 3 illustrates a second comparative study between the proposed approach and the state-of-the-art CNN-based methods using the same dataset (BraTS).
Table 3.
Comparative study with CNN-based approaches
| Method/year | Preprocessing | Auto/semi-auto | Dimension | Method | Accuracy |
|---|---|---|---|---|---|
|
[50] (Ge et al. 2018) |
- | Auto | 2D | CNN | 90.4% |
|
[22] (Iram et al.2018) |
- | Semi-auto | 3D | AlexNet-LSTM | 71% |
|
[22] (Iram et al.2018) |
- | Semi-auto | 3D | ResNet-LSTM | 71% |
|
[22] (Iram et al.2018) |
- | Semi-auto | 3D | VGGNet-LSTM | 84% |
|
[51] (Pan,et al.,2015) |
Intensity normalization | Auto | 2D | CNN | 60% |
| Proposed approach | Intensity normalization/contrast enhancement | Auto | 3D | Deep CNN | 96.49% |
Entries in bold indicate the proposed method
The 3D CNN is rarely explored in MRI processing. To the best of our knowledge, only Iram et al. [22] have developed a 3D-based CNN model for MRI glioma grading. The feature’s maps have been extracted from the volumetric MR images and then are fed into the long short-term memory’s (LSTM) temporal direction network to classify brain tumors into HG and LG gliomas. In fact, this method is semi-automatic and does not explore sufficiently the 3D volumetric contextual information. However, automatic classification is highly desired in neurology’s practice.
In 2D CNN approaches, we have investigated two 2D CNN-based architectures. The first one has been proposed by Pan et al. in 2015 [51] which explore a pre-trained CNN model mainly the LeNet-5. Nerveless, this approach suffers from limited representation using shallower CNN networks. On the other hand, the approach, proposed by Ge et al. in 2018 [50], offers competing results for high- versus low-grade glioma classification. To enhance the obtained performances, they deployed a deep architecture exploiting multi-modality (multi-stream) fusion using six convolutional layers followed by three FC layers and data augmentation. However, the authors do not provide any comparative study due to datasets heterogeneity.
We could conclude that the present study offers a powerful approach for accurate glioma tumor classification outperforming several recent CNN architectures. Based on a fully 3D automatic deep CNN, it could harness the complimentary volumetric contextual information and offers then better results.
Conclusion
We presented in this study a multi-scale 3D CNN framework for automatic gliomas’ tumor grade classification in which, instead of patching the MR image, the whole 3D volumetric MRI sequences are passed to the network. Evaluation analysis shows that proposed architecture can learn high distinctive features to separate between LG and HG subject cases compared with the competitors using either variants of 2D CNNs or relatively shallower networks. Furthermore, the use of a preprocessing step has reduced significantly the dataset heterogeneity created by multi-scanner technologies and acquisitions protocols; meanwhile, the intensity normalization has a positive effect against to correct gliomas’ tissues large in-homogeneity. We found that data augmentation, through only a flipping technique, could improve significantly the overall accuracy especially when the dataset does not provide a satisfactory MR scans to train a deep CNN.
The comparative study with supervised and unsupervised state-of-the-art methods, using the same dataset, could attest that the proposed approach outperforms several well-known CNN-based architectures for glioma MRI classification. For future works, we proposed to investigate the newly technique of “capsule networks (CapsNet)” for MRI brain tumor classification.
Acknowledgments
The authors would to thank professors and doctors “Chokri Mhiri” and “Kheireddine Ben Mahfoudh” from the University Hospital Habib Bourguiba for their cooperation to achieve this work
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.M. L. Goodenberger, R. B. Jenkins, Genetics of adult glioma, Cancer genetics 205 (12) (2012) 613-621. [DOI] [PubMed]
- 2.Louis D, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee W, Ohgaki H, Wiestler O, Kleihues P, Ellison D. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Actaneuropathol (berl). 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.F. B. Mesfin, M. A. Al-Dhahir, Cancer, brain, gliomas, in: StatPearls [Internet], StatPearls Publishing, 2018.
- 4.H. Mzoughi, I. Njeh, M. B. Slima, A. B. Hamida, Histogram equalization-based techniques for contrast enhancement of MRI brain glioma tumor images: comparative study, in: 2018 4th International Conference on Advanced Technologies for Signal and Image Processing (ATSIP), IEEE, 2018,pp. 1. 6.510
- 5.X. Bi, J. G. Liu, Y. S. Cao, Classification of low-grade and high-grade glioma using multiparametric radiomics model, in: 2019 IEEE 3rd Information Technology, Networking, Electronic and Automation Control Conference (ITNEC), IEEE, 2019, pp. 574-577.
- 6.N. Marshkole, B. K. Singh, A. Thoke, Texture and shape based classification of brain tumors using back-propagation algorithm, International Journal of Computer Science and Information Technologies 2 (5) (2011) 2340-2342.
- 7.S. S. Nazeer, A. Saraswathy, A. K. Gupta, R. S. Jayasree, Fluorescence spectroscopy to discriminate neoplastic human brain lesions: a study using the spectral intensity ratio and multivariate linear discriminant analysis,Laser Physics 24 (2) (2014) 025602.
- 8.E. I. Zacharaki, S. Wang, S. Chawla, D. SooYoo, R. Wolf, E. R. Melhem, C. Davatzikos, Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magnetic Resonance in Medicine: An Official Journal of the International Society for MagneticResonance in Medicine 62 (6) (2009) 1609-1618. [DOI] [PMC free article] [PubMed]
- 9.V. Alex, Mohammed Safwan KP, S. S. Chennamsetty, G. Krishnamurthi, Generative adversarial networks for brain lesion detection, in: Medical Imaging 2017:Image Processing, Vol. 10133, International Society for Optics and Photonics, 2017, p. 101330G.
- 10.G. Latif, M. M. Butt, A. H. Khan, O. Butt, D. A. Iskandar, Multiclass brain glioma tumor classification using block-based 3d wavelet features of MRI mages, in: 2017 4th International Conference on Electrical and Electronic Engineering (ICEEE), IEEE, 2017, pp. 333-337.
- 11.S. M. Reza, K. M. Iftekharuddin, Glioma grading using cell nuclei morphologic features in digital pathology images, in: Medical Imaging 2016:Computer-Aided Diagnosis, Vol. 9785, International Society for Optics and Photonics, 2016, p. 97852U.20 [DOI] [PMC free article] [PubMed]
- 12.Chandra B, Babu KN. Classification of gene expression data using spiking wavelet radial basis neural network. Expert systems with applications. 2014;41(4):1326–1330. doi: 10.1016/j.eswa.2013.08.030. [DOI] [Google Scholar]
- 13.Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ. R. A.525 Gatenby, Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. Journal of Magnetic Resonance Imaging. 2017;46(1):115–123. doi: 10.1002/jmri.25497. [DOI] [PubMed] [Google Scholar]
- 14.V. Wasule, P. Sonar, Classification of brain MRI using SVM and KNN classifier,in: 2017 Third International Conference on Sensing, Signal Processing andSecurity (ICSSS), IEEE, 2017, pp. 218-223.
- 15.H. Mohsen, E. El-Dahshan, E. El-Horbaty, A. Salem, Brain tumor type classification based on support vector machine in magnetic resonance images, Annals Of Dunarea De Jos" University Of Galati, Mathematics,Physics, Theoretical mechanics, Fascicle II, Year IX (XL) (1).
- 16.J. Sachdeva, V. Kumar, I. Gupta, N. Khandelwal, C. K. Ahuja, Multi- class brain tumor classification using GA-SVM, in: 2011 Developments inE-systems Engineering, IEEE, 2011, pp. 182-187.
- 17.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. Journal of digital imaging. 2017;30(4):449–459. doi: 10.1007/s10278-017-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.H. Mohsen, E.-S. A. El-Dahshan, E.-S. M. El-Horbaty, A.-B. M. Salem, Classification using deep learning neural networks for brain tumors, Future Computing and Informatics Journal 3 (1) (2018) 68-71.
- 19.J. Wang, Y. Yang, J. Mao, Z. Huang, C. Huang, W. Xu, CNN-RNN: A unified framework for multi-label image classification, in: Proceedings ofthe IEEE conference on computer vision and pattern recognition, 2016, pp.2285-2294.
- 20.Zhan Z, Cai J-F, Guo D, Liu Y, Chen Z, Qu X. Fast multiclass dictionaries learning with geometrical directions in MRI reconstruction. IEEETransactions on Biomedical Engineering. 2015;63(9):1850–1861. doi: 10.1109/TBME.2015.2503756. [DOI] [PubMed] [Google Scholar]
- 21.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE transactions on medical imaging. 2016;35(5):1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 22.I. Shahzadi, T. B. Tang, F. Meriadeau, A. Quyyum, CNN-LSTM: Cascadedframework for brain tumour classification, in: 2018 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), IEEE, 2018, pp.633-637.
- 23.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556.
- 24.Palangi H, Deng L, Shen Y, Gao J, He X, Chen J, Song X, Ward R. Deep sentence embedding using long short-term memory networks: Analysis and application to information retrieval, IEEE/ACM Transactions onAudio. Speech and Language Processing (TASLP) 2016;24(4):694–707. [Google Scholar]
- 25.Deepak S, Ameer P. Brain tumor classification using deep CNN features via transfer learning. Computers in biology and medicine. 2019;111:103345. doi: 10.1016/j.compbiomed.2019.103345. [DOI] [PubMed] [Google Scholar]
- 26.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770-778.
- 27.A. Krizhevsky, I. Sutskever, G. E. Hinton, Imagenet classification with deep convolutional neural networks, in: Advances in neural informationprocessing systems, 2012, pp. 1097-1105.
- 28.H. Chen, Q. Dou, L. Yu, J. Qin, P.-A.Heng, Voxresnet: Deep voxelwise residual networks for brain segmentation from 3D MR images, NeuroImage 170 (2018) 446-455. [DOI] [PubMed]
- 29.F. Ye, J. Pu, J. Wang, Y. Li and H. Zha, “Glioma grading based on 3D multimodal convolutional neural network and privileged learning,” 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, 2017, pp. 759-763.
- 30.Mzoughi H, Njeh I, Slima MB, Hamida AB, Mhiri C, Mahfoudh KB. Denoising and contrast-enhancement approach of magnetic resonance imaging glioblastoma brain tumors. Journal of Medical Imaging. 2019;6(4):044002. doi: 10.1117/1.JMI.6.4.044002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohan J, Krishnaveni V, Guo Y, et al. A survey on the magnetic resonance image denoising methods. Biomedical Signal Processing and Control. 2014;9:56–69. doi: 10.1016/j.bspc.2013.10.007. [DOI] [Google Scholar]
- 32.Amutha S, Babu RD, Shankar R, Kumar HN. MRI denoising and enhancement based on optimized single-stage principle component analysis. International Journal of Advances in Engineering & Technology. 2013;5(2):224. [Google Scholar]
- 33.S. M. Pizer, R. E. Johnston, J. P. Ericksen, B. C. Yankaskas, K. E. Muller,Contrast-limited adaptive histogram equalization: speed and effectiveness ,in: Proceedings of the First Conference on Visualization in Biomedical Computing, IEEE, 1990, pp. 337-345.
- 34.M. F. Stollenga, W. Byeon, M. Liwicki, J. Schmidhuber, Parallel multi-dimensional LSTM, with application to fast biomedical volumetric image segmentation, in: Advances in neural information processing systems, 2015,pp. 2998-3006.
- 35.Chen, Wei, et al. “Computer-aided grading of gliomas combining automatic segmentation and radiomics.” International journal of biomedical imaging 2018 (2018). [DOI] [PMC free article] [PubMed]
- 36.Parsania PS, Virparia PV. A comparative analysis of image interpolation algorithms. International Journal of Advanced Research in Computer and Communication Engineering. 2016;5(1):29–34. doi: 10.17148/IJARCCE.2016.5107. [DOI] [Google Scholar]
- 37. L. Perez, J. Wang, The effectiveness of data augmentation in image classification using deep learning, arXiv preprint arXiv:1712.04621.
- 38.M. Sajjad, S. Khan, K. Muhammad, W. Wu, A. Ullah, S. W. Baik, Multi-grade brain tumor classification using deep CNN with extensive data augmentation, Journal of computational science 30 (2019) 174-182.
- 39.Zhang L, Yang H, Jiang Z. Imbalanced biomedical data classification using self-adaptive multilayer ELM combined with dynamic GAN. Biomedical engineering online. 2018;17(1):181. doi: 10.1186/s12938-018-0604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical imageanalysis. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 41.J. Nagi, F. Ducatelle, G. A. Di Caro, D. Cire_san, U. Meier, A. Giusti,F. Nagi, J. Schmidhuber, L. M. Gambardella, Max-pooling convolutional neural networks for vision-based hand gesture recognition, in: 2011 IEEEInternational Conference on Signal and Image Processing Applications (IC-SIPA), IEEE, 2011, pp. 342-347.
- 42.Y. Cui, F. Zhou, J. Wang, X. Liu, Y. Lin, S. Belongie, Kernel pooling for convolutional neural networks, in: Proceedings of the IEEE conference oncomputer vision and pattern recognition, 2017, pp. 2921-2930.
- 43.M. D. Zeiler, R. Fergus, Stochastic pooling for regularization of deep convolutional neural networks, arXiv preprint arXiv:1301.3557.
- 44.G. E. Hinton, N. Srivastava, A. Krizhevsky, I. Sutskever, R. R. Salakhut-dinov, Improving neural networks by preventing co-adaptation of feature detectors, arXiv preprint arXiv:1207.0580.
- 45.T. Kurbiel, S. Khaleghian, Training of deep neural networks based on distance measures using RMSProp, arXiv preprint arXiv:1708.01911.
- 46.Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, et al. The multimodal brain tumor image segmentation benchmark (brats) IEEE transactions onmedical imaging. 2014;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. Shah, Y. Xiao, N. Subbanna, S. Francis, D. L. Arnold, D. L. Collins,T. Arbel, Evaluating intensity normalization on MRIs of human brain with multiple sclerosis, Medical image analysis 15 (2) (2011) 267-282. [DOI] [PubMed]
- 48.H.-h. Cho, H. Park, Classification of low-grade and high-grade glioma using multi-modal image radiomics features, in: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society(EMBC), IEEE, 2017, pp. 3081-3084. [DOI] [PubMed]
- 49.Cho H-h, Lee S-H, Kim J, Park H. Classification of the glioma grading using radiomics analysis. Peer J. 2018;6:e5982. doi: 10.7717/peerj.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. Ge, I. Y.-H. Gu, A. S. Jakola, J. Yang, Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks,in: 2018 40th Annual International Conference of the IEEE Engineering inMedicine and Biology Society (EMBC), IEEE, 2018, pp. 5894-5897. [DOI] [PubMed]
- 51.Y. Pan, W. Huang, Z. Lin, W. Zhu, J. Zhou, J.Wong, Z. Ding, Brain tumor grading based on neural networks and convolutional neural networks, in:2015 37th Annual International Conference of the IEEE Engineering inMedicine and Biology Society (EMBC), IEEE, 2015, pp. 699-702. [DOI] [PubMed]