Abstract
There are data to suggest that some ductal carcinoma in situ (DCIS) may evolve through an evolutionary bottleneck, where minor clones susceptible to the imposed selective pressure drive disease progression. Here, we tested the hypothesis that an impact of the inflammatory environment on DCIS evolution is HER2-dependent, conferring proliferative dominance of HER2-negative cells. In tissue samples, density of tumour-infiltrating immune cells (TIICs) was associated only with high tumour nuclear grade, but in 9% of predominantly HER2-negative cases, the presence of tumoral foci (‘hot-spots’) of basal-like cells with HIF1-α activity adjacent to the areas of dense stromal infiltration was noted. Results of in vitro analyses further demonstrated that IL-1β and TNF-α as well as macrophage-conditioned medium triggered phosphorylation of NF-κB and subsequent upregulation of COX2 and HIF1-α, exclusively in HER2-negative cells. Treatment with both IL-1β and TNF-α resulted in growth stimulation and inhibition of HER2-negative and HER2-positive cells, respectively. Moreover, ectopic overexpression of HIF1-α rescued HER2-positive cells from the negative effect of IL-1β and TNF-α on cell growth. Our data provide novel insight into the molecular basis of HER2-dependent proliferation of DCIS cells and indicate the NF-κB/COX2 → HIF1-α signalling axis as a dominant mechanism of DCIS evolution induced by inflammatory microenvironment. Presented findings also highlight the clinical significance of heterogeneity of DCIS tumours and suggest that HIF1-α might be considered as a predictive marker of disease progression.
Abbreviations: BC, breast cancer; CAIX, carbonic anhydrase IX; DCIS, ductal carcinoma in situ; IDC, invasive ductal carcinoma; TME, tumour microenvironment; TIIC, tumour-infiltrating immune cell; TIL, tumour-infiltrating lymphocyte; TAM, tumour-associated macrophage
Keywords: DCIS, Inflammatory microenvironment, HER2, HIF1-α
Introduction
Ductal carcinoma in situ (DCIS), termed the non-obligate precursor of invasive ductal breast carcinoma (IDC), is characterised by proliferation of neoplastic cells within the duct lumen. Molecular mechanisms governing DCIS evolution are still poorly understood and no useful prognostic biomarkers have been identified.
Increasing evidence supports the role of epithelial-stromal interactions in tumour progression [1]. DCIS is no exception. As molecular profiles of synchronous DCIS and IDC are remarkably similar [2], DCIS → IDC development is thought to be dependent not solely on specific genomic alterations in preinvasive cells. Stimuli derived from tumour microenvironment (TME) and, in particular, stromal tumour-infiltrating immune cells (TIICs), i.e. tumour-infiltrating lymphocytes (TILs) and tumour-associated macrophages (TAMs), may be critically involved in the process [3], [4], [5], [6], [7], [8]. Moreover, recent studies suggest that an impact of immune TME modulation on tumour biology and disease outcome is particularly strongly pronounced at the early stages of BC development [8], [9], [10] and is dependent on BC cell phenotype [8], [11], [12], [13], [14].
Similar to its invasive counterpart, DCIS is not one entity but represents a heterogeneous group of pre-invasive breast lesions with different phenotypes as well as variable proliferative and invasive potentials [15]. HER2 (ErbB2) oncogene, the dominant driver of BC pathophysiology, is found to be overexpressed at high frequency in DCIS. Counterintuitively, DCIS → IDC evolution is associated with loss of HER2 expression (approximately 50% of DCIS HER2(+) vs 20% of IDC HER2(+)) [16], [17], [18]. Available data on HER2 importance in DCIS are scarce and contradicting [19], [20], [21], [22], but it has been suggested that the status of HER2 might affect the type of disease recurrence. While HER2(+) DCIS tumours are more likely to relapse as new in situ lesions, HER2-negative DCIS is associated with a higher risk of recurrent invasive BC. In addition, HER2 positivity was found more often in patients with pure DCIS compared to those with microinvasive DCIS and DCIS with IDC [23], [24].
Taken together, the HER2 status might be critical for cell responsiveness to inflammatory stimuli. Proliferative dominance of the HER2-negative subclones would comply with the evolutionary bottleneck model of DCIS progression [25], as well as point to a hitherto unknown aspect of HER2 role in BC biology. To verify this hypothesis the study combined clinical and mechanistic analyses. Evaluation of the extent and type of inflammatory infiltration in specimens from DCIS patients was carried out using a panel of specific markers selected on the basis of available clinical follow-up database [7], [12], [14]. A functional role of HER2 in the proliferative response of mammary epithelial cells to inflammatory stimuli was explored in a culture system representative of human DCIS [16], [26], [27].
Materials and methods
Patient selection and samples
Specimens of treatment-naïve, pure DCIS (without invasive component) diagnosed according to the WHO 2012 Classification of Breast Tumours [28] were obtained from patients treated at the Copernicus Memorial Hospital in Lodz and the Holycross Cancer Center in Kielce, Poland, between 2004 and 2018 (Table S1). To exclude an impact of genomic alterations on HER2 function, specimens with no HER2 amplification (n = 75) were selected (confirmed by FISH). The study was approved by the Local Research Ethics Committee (No. RNN/284/13/KE).
Histopathological procedures
Serial 5-μm sections of formalin-fixed paraffin-embedded (FFPE) blocks were processed for haematoxylin/eosin staining (H&E) and immunohistochemistry (IHC) for a panel of markers enabling quantitative and qualitative analysis of subclasses of (i) TILs: CD4, CD8 and FOXP3 (helper, cytotoxic and regulatory T cells) and (ii) TAMs: CD68 and CD163 (M1 and M2 macrophages). DCIS tumours were phenotyped by IHC for: (i) oestrogen/progesterone receptors (ER/PR) and HER2, (ii) cytokeratin 5/6 and (iii) carbonic anhydrase IX (CAIX), as an indicator of HIF-1α activity, using the protocols according to the manufacturers’ recommendations. ER/PR status was determined using Allred scoring system [29] and HER2, using both IHC (Herceptest™) and FISH [30], [31]. All antibodies are described in Table S2.
Evaluation of tumour-infiltrating lymphocytes and tumour-associated macrophages
Morphological and semi-quantitative analysis of stromal TIICs was carried out on H&E and IHC preparations using an UltraFast Scanner (Philips) with DigiPath™ software (Xerox), following the International Guidelines on TIL Assessment in Breast Cancer [32], [33], [34], [35], [36]. Infiltration was defined as the percentage of the tumour stroma area that is occupied by TIICs. In addition, the ‘hot-spots’ defined as areas rich in TIICs adjacent to tumour cells displaying high-grade characteristics, were selected for further phenotypical analysis, i.e. display of basal features (IHC for CK5/6) and activity of HIF1-α (IHC for CAIX). Cells positive for CD4, CD8, CD68, CD163, FOXP3 were counted for each tumour in four representative areas of 0.25 mm2 under magnification of 200×. Additionally, CD4/CD8 and CD68/CD163 ratios were calculated. Counting was conducted independently by two researchers (DP and LK) and supervised by the pathologist (MB). In case of significant disparities between the scores (20 or 5 cells/mm2 for TILs or TAMs, respectively, or difference >20% of the mean value), the case was additionally reassessed by another pathologist (HMR or RK).
Cell lines and reagents
HB2 cells were purchased from ECACC, MCF10A and THP-1 cell lines were from ATCC. Cells were passaged maximum for 3–4 months post-resuscitation and routinely tested for mycoplasma contamination. Cells were grown: (i) HB2 in DMEM supplemented with 10% FBS, 5 μg/ml insulin, 5 μg/ml hydrocortisone and penicillin/streptomycin solution (Pen-Strep); (ii) MCF10A in DMEM F12 supplemented with 5% horse serum, 20 ng/ml EGF, 0.5 mg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin and Pen-Strep; and (iii) THP-1 in RPMI-1640 with 10% FBS. Cell culture media and supplements were from Sigma-Aldrich, all growth factors and cytokines from PeproTech, celecoxib, magnolol and lapatinib from Selleckchem.
Treatment with cytokines
For analysis of the cytokine-induced activation of NF-κB/COX2 → HIF1-α, cells were starved overnight in serum-free media followed by stimulation with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for indicated times. Celecoxib (25 µM) and magnolol (10 µM) were applied for COX2 and NF-κB inhibition, respectively. For analysis of NF-κB/COX2 → HIF1-α involvement in 3D growth, cells were cultured in matrigel in the presence of IL-1β (10 ng/ml) or TNF-α (10 ng/ml) with/without celecoxib (10 µM) or magnolol (10 µM).
HER2 and HIF1-α overexpression
HB2(HER2+) and MCF10A(HER2+) cell lines were established with retroviral vector pBABEpuro-ERBB2 (Addgene #40978) [37]. HB2(HER2+/HIF1-α+) and MCF10A(HER2+/HIF1-α+) cells were generated with retroviral vector HA-HIF1α-wt-pBabe-puro (Addgene #19365) [38]. In all experiments involving the above cell variants, parental cells (wild type HB2 or MCF10A) were transfected with pBABEpuro vector and used as a control.
Analysis of cell growth in three-dimensional culture
Cells were cultured in 3D matrigel as previously described [39]. Briefly, 1.5 × 103 cells were resuspended in 40 μl of growth factor-reduced matrigel (mixed 1:1 with medium) and cultured for 8 (HB2) or 10 days (MCF10A). To evaluate cell growth, at least 50 colonies were measured for each condition using ImageJ software. For HER2-positive/HER2-negative co-cultures, GFP-expressing HB2(HER2+) or MCF10A(HER2+) cells were mixed with their wild-type counterparts in a ratio of 1:1 and incubated for 30–60 min on a slowly rocking platform to induce cell aggregation [40], [41]. Aggregates were gently suspended in matrigel and cultured for 7–10 days. For all 3D culture experiments, representative images were taken using ZEISS PrimoVert microscope.
THP-1 differentiation into M1- and M2-like macrophages
THP-1 cells were differentiated according to the published protocol [42], [43]. Briefly, cells were treated with 150 nM PMA (Sigma-Aldrich) for 24 hours, next day the medium was replaced and supplemented with INFγ (10 ng/ml) and LPS (20 ng/ml), for M1 or IL-13 (10 ng/ml) and IL-4 (10 ng/ml), for M2-differentiation. M1- and M2-like phenotypes were verified by immunostaining for CD68, iNOS and CD163, respectively. The concentration of IL-1β and TNF-α in macrophage-derived conditioned medium was evaluated using ELISA kit (R&D Systems).
Western blotting
Cells grown to 80–90% confluence were scraped in ice-cold PBS and lysed in the presence of Laemmli buffer (2× concentrated) supplemented with 2 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 5 mM EGTA, 1 mM EDTA, 2 mM Na4P2O7, 5 mM NaF and 5 mM Na3VO4. An equal amount of protein (∼20 μg) per lane was loaded, resolved in SDS-PAGE and transferred onto a nitrocellulose membrane. The membranes were blocked in 5% skimmed milk and probed overnight with primary antibodies (described in Table S2) at 4 °C. Secondary antibodies conjugated with IRDye 680RD or IRDye 800CW (from Jackson ImmunoResearch) and Odyssey Clx system or secondary antibodies conjugated with horseradish peroxidase (Sigma-Aldrich) and Western Lightning Plus-ECL (PerkinElmer) were used for visualisation.
Immunofluorescence
Cells were seeded onto 8-well chamber slide at 3 × 104 cells per well. For analysis of NF-κB localization, cells were serum-starved overnight and then stimulated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 30 min. In all immunostainings specimens were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 1 min, blocked in 5% serum and 3% BSA in PBS and incubated for 1 h with primary antibodies. After washing and subsequent 1-h incubation with AffiniPure DyLight 549 and AffiniPure DyLight 488 secondary antibodies (Jackson ImmunoResearch) individual proteins/markers were visualised using the fluorescent microscope Olympus IX83.
Statistical analysis
Continuous data were presented as medians with interquartile ranges (IQR), while nominal data as numbers with percentages. The Shapiro–Wilk test was used to assess the normality of distribution. For comparisons of continuous variables, following tests were used: Mann Whitney U-test; Student’s t-test; one-way or two-way block ANOVA (with post hoc Tukey’s multiple comparisons test); or Kruskal–Wallis test (with post hoc all-pairwise comparisons Conover–Inman test). Categorical variables were compared using the chi2 or two-tailed Fisher’s exact tests. For correlations, Spearman’s rank correlations were calculated. The analysis was conducted in Statistica 12.5 PL package (Statsoft, Tulsa, OK, USA). P values ≤0.05 were considered statistically significant.
Results
Clinical and pathological characteristics of the group
All 75 patients were diagnosed with pure DCIS (detailed characteristics in Table S1). HER2 expression (score by Herceptest™) ranged from HER2/0 (24% of cases), through HER2/1+ (15% of cases), HER2/2+ (52% of cases), to HER2/3+ (9% of cases).
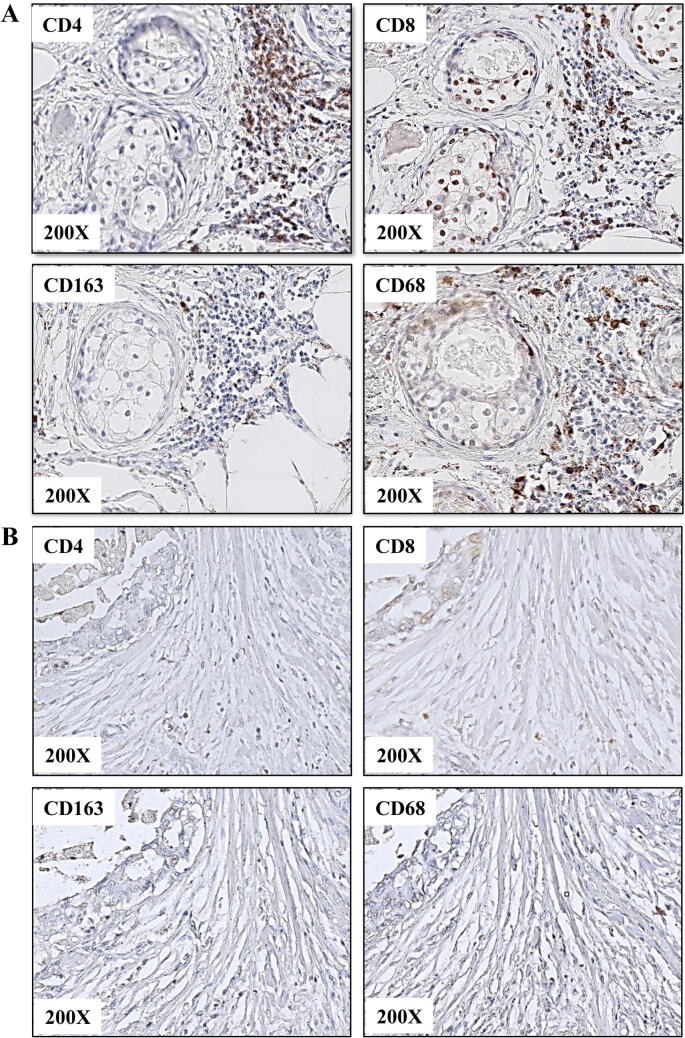
TIICs infiltration, as assessed on H&E and IHC preparations, was: (i) dense (≥30%, Fig. 1A) in 8 (11%) cases, and (ii) weak to moderate (median of 1% (IQR: 1–5%), Fig. 1B) in the majority of cases (89%) (Table 1). TIICs infiltration (%) correlated positively only with numbers of both CD4- and CD8-positive TILs (R = 0.85, p < 0.0001 and R = 0.69, p < 0.0001, respectively). Poor differentiation of the lesions (high nuclear grade) was associated with higher percentages of TIICs and higher numbers of CD4-positive TILs (G3 vs G1: 5.0% (IQR: 5.0–10.0%) vs 1.0% (IQR: 1.0–1.0%), p < 0.001 with post-hoc p = 0.002; and 64/mm2 (IQR: 36–171) vs 30/mm2 (IQR: 11–46), p = 0.010 with post-hoc p = 0.012) (Table 1). Higher percentages of TIICs were detected in HER2/1+ patients compared to HER2/2+ patients (p = 0.004, post-hoc p = 0.026), lower numbers of CD68-positive TAMs in HER2/0 patients compared to HER2/2+ patients (p = 0.007, post-hoc p = 0.028) and higher numbers of FOXP3-positive TILs in HER2/1+ and HER2/2+ patients compared to HER2/3+ patients (p = 0.003 with post-hoc p = 0.009 and post-hoc p = 0.005, respectively) (Table 2). No significant associations were found between TIICs counts and morphological subtype, necrosis, ER/PR status, age, Van Nuys index or tumour size (data not shown). Small numbers of survival events precluded an assessment of the prognostic value of TIICs infiltration. The subgroup analyses of patients with: (i) no TIICs (<5%) and (ii) TIICs (≥5%), divided according to the HER2 status (HER2/0 and HER2/1+/2+/3+), gave similar results as described above.
Fig. 1.
Density and type of inflammatory infiltration vary between DCIS lesions. Immunohistochemical presentation of (A) dense and (B) sparse infiltration by TILs (positive for CD4 and CD8) and TAMs (positive for CD163 and CD68).
Table 1.
Characteristics of tumour-infiltrating immune cells (TIICs) in the study group regarding nuclear grade of DCIS. Data presented as medians, interquartile ranges in brackets. Units for each variable are included in square brackets. Groups were compared using Kruskal–Wallis test. Bold values denote statistical significance.
| Feature | Whole group (n = 75) | Nuclear Grade 1 (n = 24) | Nuclear Grade 2 (n = 34) | Nuclear Grade 3 (n = 17) | p-Value |
|---|---|---|---|---|---|
| TIICs infiltration quantified in H&E [%] | 1.0 (1.0–5.0) |
1.0 (1.0–1.0) |
5.0 (1.0–5.0) |
5.0 (5.0–10.0) |
<0.001 |
| CD4 [number/mm2] | 43.0 (21.0–106.0) |
30.0 (11.0–46.5) |
50.0 (26.0–120.0) |
63.5 (36.0–171.0) |
0.010 |
| CD8 [number/mm2] | 49.0 (28.0–91.0) |
34.0 (21.0–65.0) |
54.0 (31.0–102.0) |
89.5 (25.5–144.0) |
0.062 |
| CD4/CD8 ratio | 0.9 (0.6–1.5) |
0.8 (0.3–1.2) |
1.0 (0.7–1.6) |
1.0 (0.7–1.7) |
0.157 |
| CD68 [number/mm2] | 20.0 (9.0–34.0) |
18.0 (6.5–36.0) |
25.5 (11.0–38.0) |
15.0 (8.0–20.5) |
0.397 |
| CD163 [number/mm2] | 9.0 (3.0–16.0) |
5.0 (1.0–13.0) |
11.0 (3.5–22.0) |
7.5 (2.5–14.0) |
0.123 |
| CD68/CD163 ratio | 2.3 (1.2–5.2) |
3.2 (1.9–6.3) |
2.3 (1.1–4.3) |
1.7 (1.1–7.0) |
0.225 |
| FOXP3 [number/mm2] | 4.0 (1.0–15.0) |
4.0 (1.0–11.0) |
8.0 (1.0–22.5) |
1.0 (0.0–10.0) |
0.089 |
Table 2.
Characteristics of tumour-infiltrating immune cells (TIICs) in the study group divided by HER2 expression. Data presented as medians and interquartile ranges in brackets. Units for each variable are included in square brackets. Groups were compared using Kruskal–Wallis test. Bold values denote statistical significance.
| Feature | Whole group (n = 75) | HER2-0 (n = 18) |
HER2-1 (n = 11) |
HER2-2 (n = 39) |
HER2-3 (n = 7) |
p-Value |
|---|---|---|---|---|---|---|
| TIICs infiltration quantified in H&E [%] | 1.0 (1.0–5.0) |
5.0 (1.0–10.0) |
5.0 (5.0–30.0) |
1.0 (1.0–5.0) |
5.0 (1.0–10.0) |
0.004 |
| CD4 [number/mm2] | 43.0 (21.0–106.0) |
38.8 (17.5–105.5) |
53.5 (27.0–594.0) |
34.3 (20.0–74.0) |
107.0 (45.5–171.0) |
0.161 |
| CD8 [number/mm2] | 49.0 (28.0–91.0) |
55.3 (28.0–104.0) |
47.0 (30.0–400.5) |
41.0 (26.0–66.0) |
94.5 (25.5–144.0) |
0.277 |
| CD4/CD8 ratio | 0.9 (0.6–1.5) |
0.8 (0.4–1.2) |
1.0 (0.7–1.8) |
1.0 (0.6–1.4) |
0.9 (0.7–1.7) |
0.525 |
| CD68 [number/mm2] | 20.0 (9.0–34.0) |
12.5 (5.0–20.0) |
20.5 (9.5–38.5) |
29.5 (12.0–42.0) |
8.8 (6.0–16.0) |
0.007 |
| CD163 [number/mm2] | 9.0 (3.0–16.0) |
5.3 (1.5–10.5) |
7.5 (2.0–16.5) |
11.8 (4.5–17.5) |
6.5 (1.0–8.5) |
0.107 |
| CD68/CD163 ratio | 2.3 (1.2–5.2) |
2.5 (1.1–5.7) |
2.3 (1.6–6.0) |
2.4 (1.2–4.3) |
2.4 (1.5–6.0) |
0.924 |
| FOXP3 [number/mm2] | 4.0 (1.0–15.0) |
2.0 (0.0–11.0) |
4.5 (1.5–44.5) |
8.3 (1.0–17.0) |
0.0 (0.0–1.0) |
0.003 |
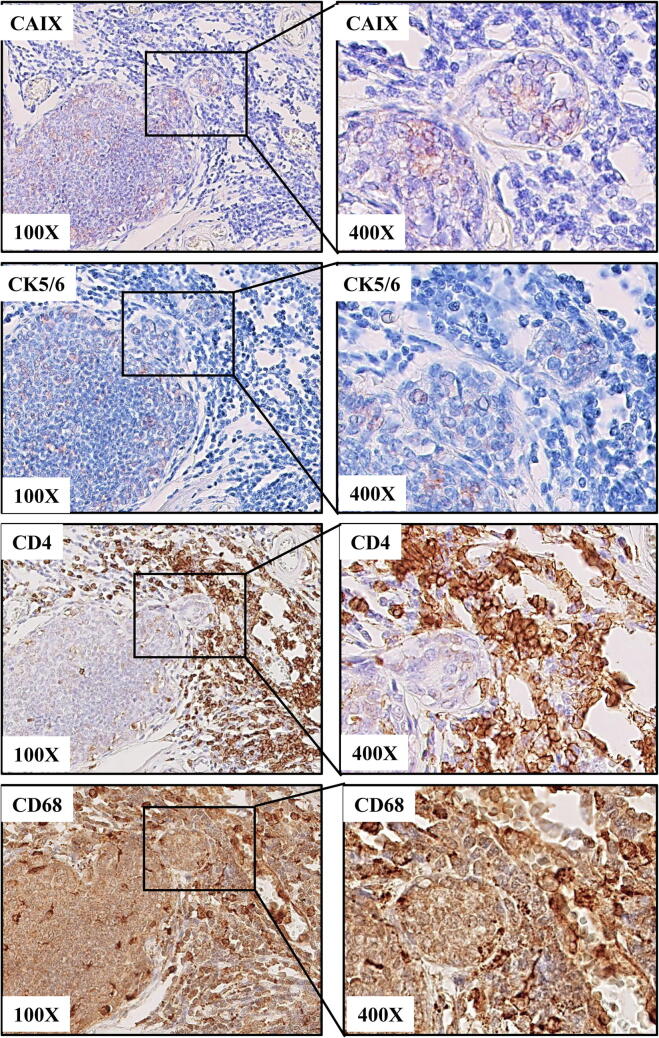
High-grade HER2-negative ‘hot-spots’ adjoin CD4+/CD68+-rich inflammatory foci
Most of the tumours, especially those with a prominent TIICs infiltration displayed a noticeable morphological heterogeneity regarding both tumour cells and numbers/type of infiltrates. Interestingly, in 7 (9%) cases we found tumoral foci of more aggressive morphological and immunophenotypic features, i.e. high nuclear grade, bland demarcation and phenotypic basal-like characteristics (Fig. 2). The epithelial cells of these ‘hot-spots’, found almost exclusively in HER2-negative DCIS lesions, displayed focal immunoreactivity for CK5/6 along with positivity for CAIX. These foci were tightly adjacent to dense TIICs infiltration dominated by CD4-positive TILs and CD68-positive TAMs (Fig. 2). A possible impact of HER2 on response of DCIS cells to the inflammatory TME was further evaluated using an in vitro model.
Fig. 2.
‘Hot-spots’ of basal-like cells with HIF1-α activity are adjacent to the areas of dense stromal pro-tumorigenic infiltration. Tumour cells displaying focal immunoreactivity for CAIX (histochemical marker of activated HIF1-α) and CK 5/6 border with CD4-positive TILs and CD68-positive TAMs.
IL-1β and TNF-α promote proliferative dominance of HER2-negative mammary epithelial cells
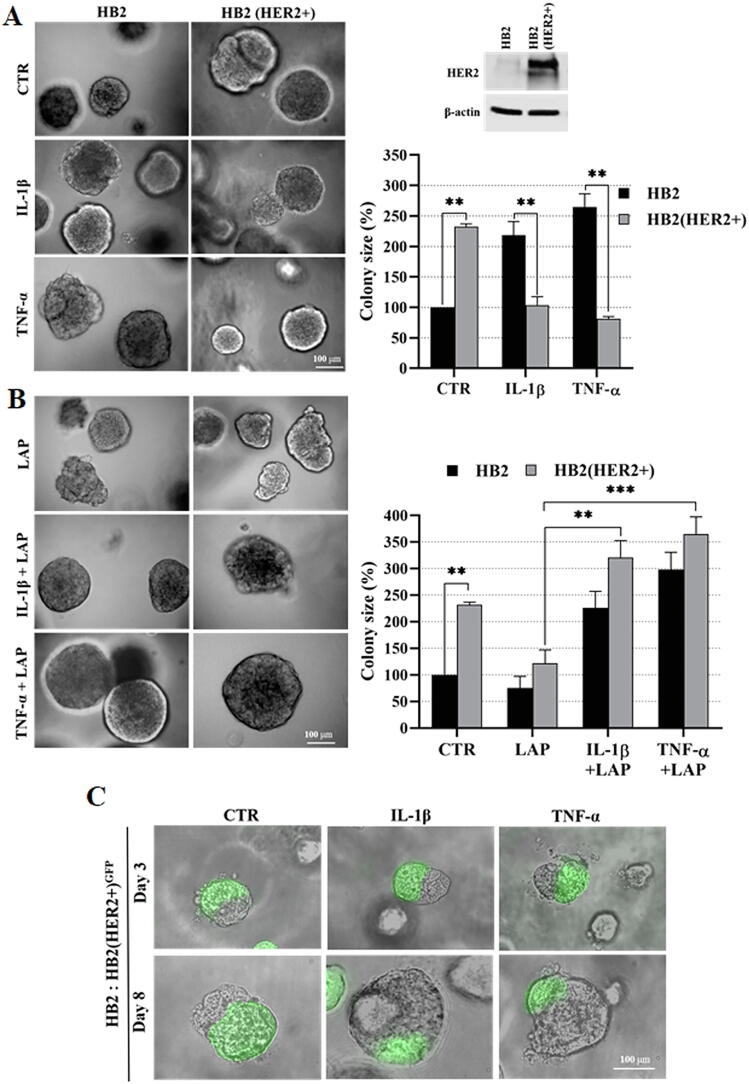
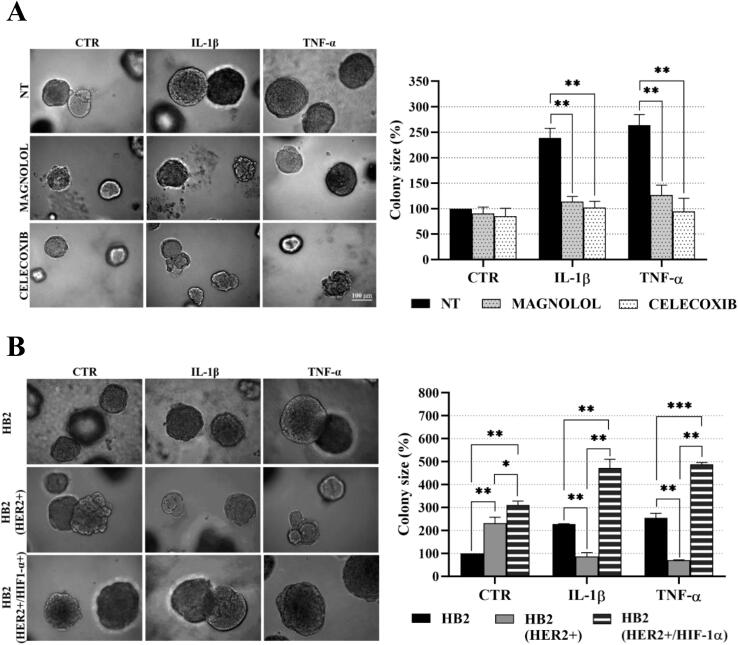
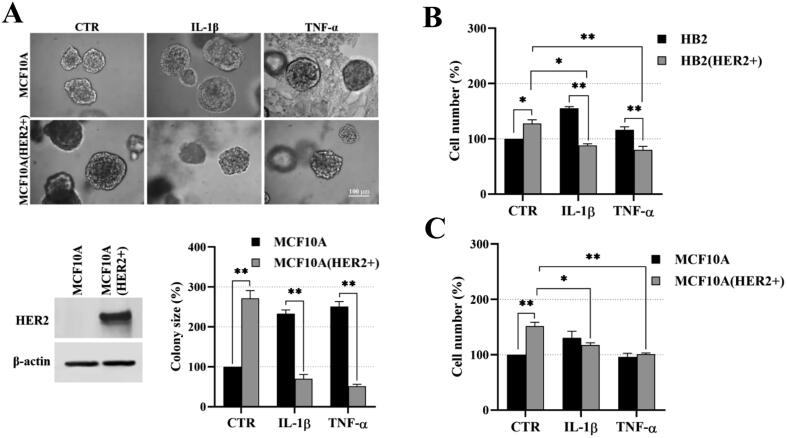
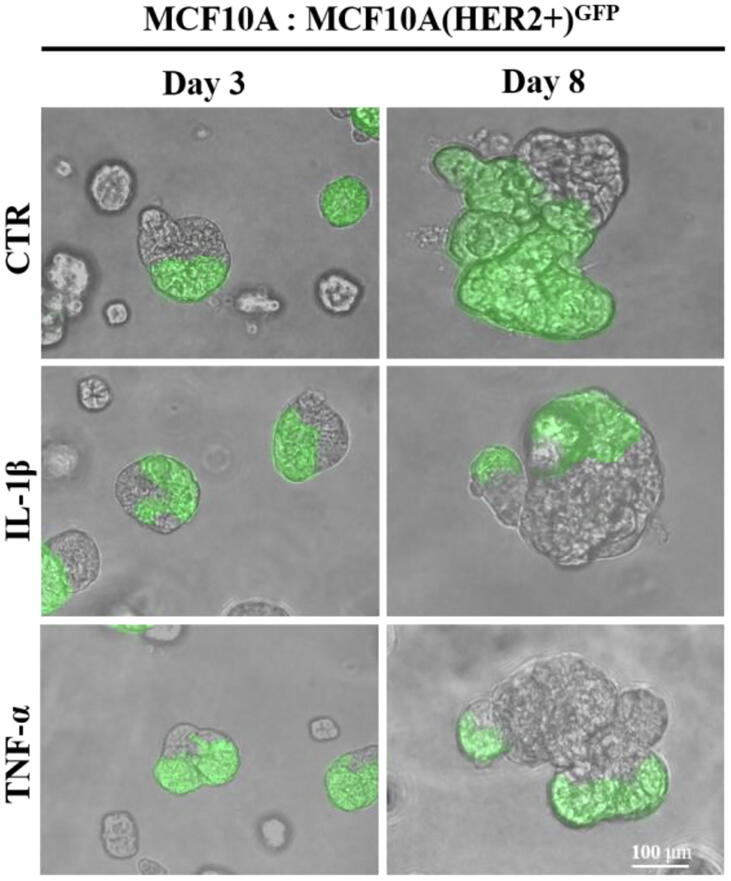
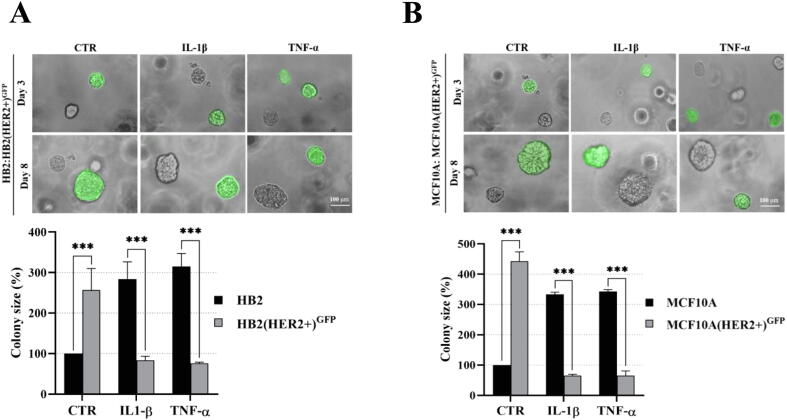
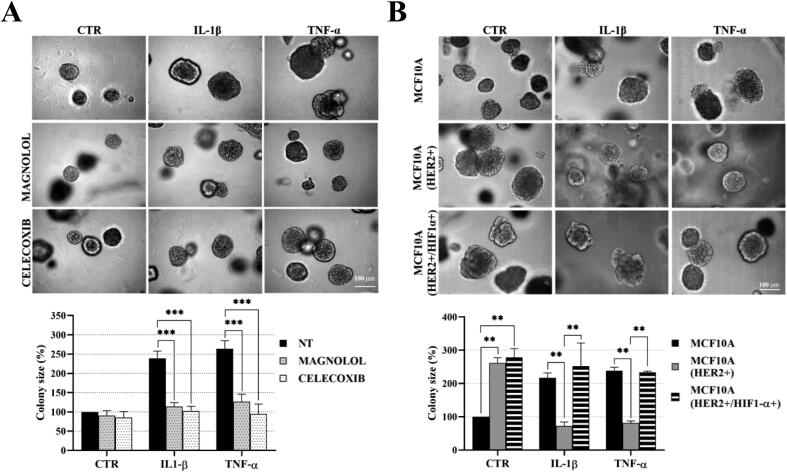
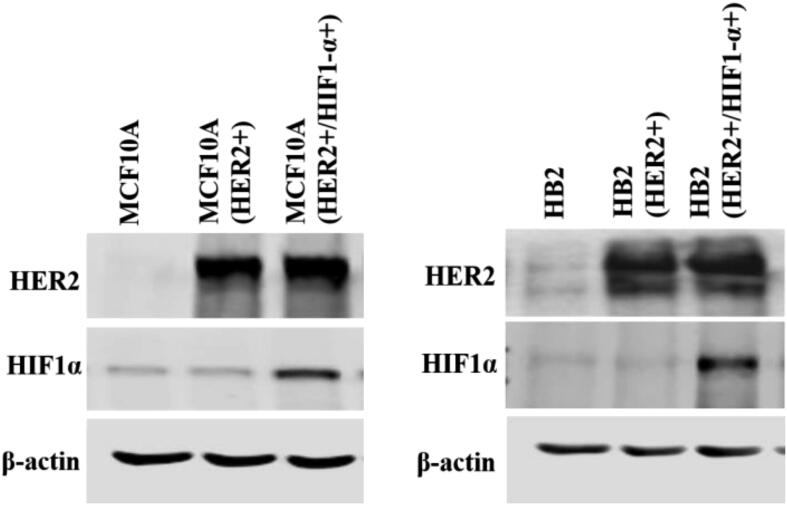
The experimental design was based on previous reports demonstrating that DCIS features could be recapitulated by overexpression of HER2 in HB2 and MCF10A mammary epithelial cells grown in three-dimensional (3D) cultures [16], [27]. Moreover, HB2 cells have also been shown to form DCIS-like lesion when xenotransplanted into nude mice [26]. As increased proliferation is the principal functional characteristic of DCIS cells, HB2 and MCF10A cells and their stable HER2-overexpressing variants (Fig. 3A and Fig. S1A) were used to assess HER2-dependent proliferative response to inflammatory stimuli. To relate in vitro model to histological findings (foci of tumoral cells positive for CAIX), IL-1β and TNF-α, two inflammatory cytokines known to activate HIF1-α under hypoxic and normoxic conditions [44], were used in our further analyses. As expected, overexpression of HER2 resulted in growth stimulation of HB2 and MCF10A cells (2.32- and 2.7-fold, respectively; in relation to the ‘wild-type’ control) in 3D matrigel (Fig. 3A and Fig. S1A). IL-1β or TNF-α treatment had a strong negative effect on proliferation (reflected by colony size) of HB2(HER2+) and MCF10A(HER2+) cells, which was opposite to the effect exerted on their HER2-negative counterparts. These findings were verified in classical 2D proliferation assays (Fig. S1B–C). To confirm that the influence of cytokines on cell growth was HER2-dependent, cells were cultured in the presence of lapatinib (HER2 and EGFR inhibitor [45]). We found that lapatinib strongly attenuated HER2-induced growth (Fig. 3B) and counteracted the negative impact of both cytokines on proliferation of HER2-overexpressing cells. These results suggest that inflammatory stimuli affect cell proliferation in a HER2-dependent manner. To further confirm these findings, 3D co-cultures of HB2 or MCF10A cells and their HER2-overexpressing variants expressing GFP, embedded in matrigel, as either pre-formed aggregates (Fig. 3C and Fig. S2) or single cells (1:1 ratio, Fig. S3A–B), were set-up. Similar to monocultures, stimulation with IL-1β and TNF-α led to a significant decrease and increase of size of HER2-positive and HER2-negative colonies, respectively, but this differential, HER2-dependent effect was aggravated (Fig. S3A–B vs Fig. 3A and Fig. S1A). These data further indicate that inflammatory milieu may induce proliferative dominance of HER2-negative clones.
Fig. 3.
IL-1β and TNF-α promote proliferative dominance of mammary epithelial cells in a HER2-dependent manner. (A) HER2-overexpression in HB2 cells was evaluated by Western blotting. HB2 and HB2(HER2+) cells were cultured in 3D matrigel for 8 days with IL-1β (10 ng/ml), TNF-α (10 ng/ml) and/or (B) lapatinib (1 µM). Representative images were taken and colony size was analysed using ImageJ software. Data are expressed as means ± SD (n = 3), **p < 0.005, ***p < 0.001. (C) HB2 and GFP-expressing HB2(HER2+) cells were mixed in a 1:1 ratio. Preformed cell aggregates were carefully embedded in matrigel and grown with IL-1β (10 ng/ml) or TNF-α (10 ng/ml). Representative images were taken after 8 days.
IL-1β and TNF-α activate the NF-κB/COX2 → HIF1-α axis in HER2-negative mammary epithelial cells
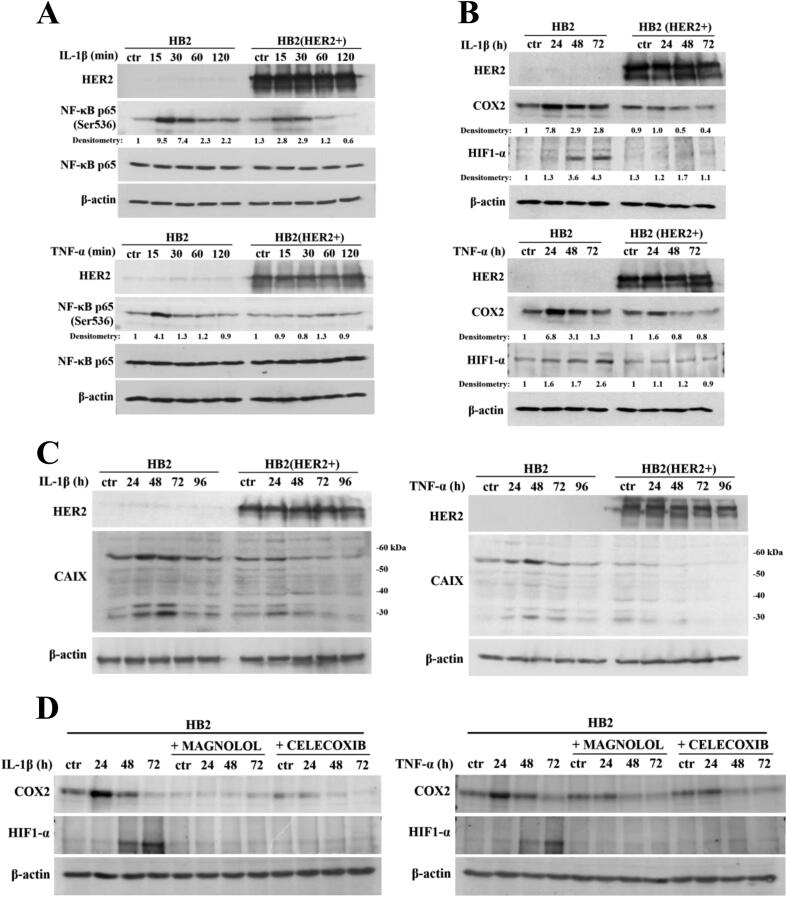
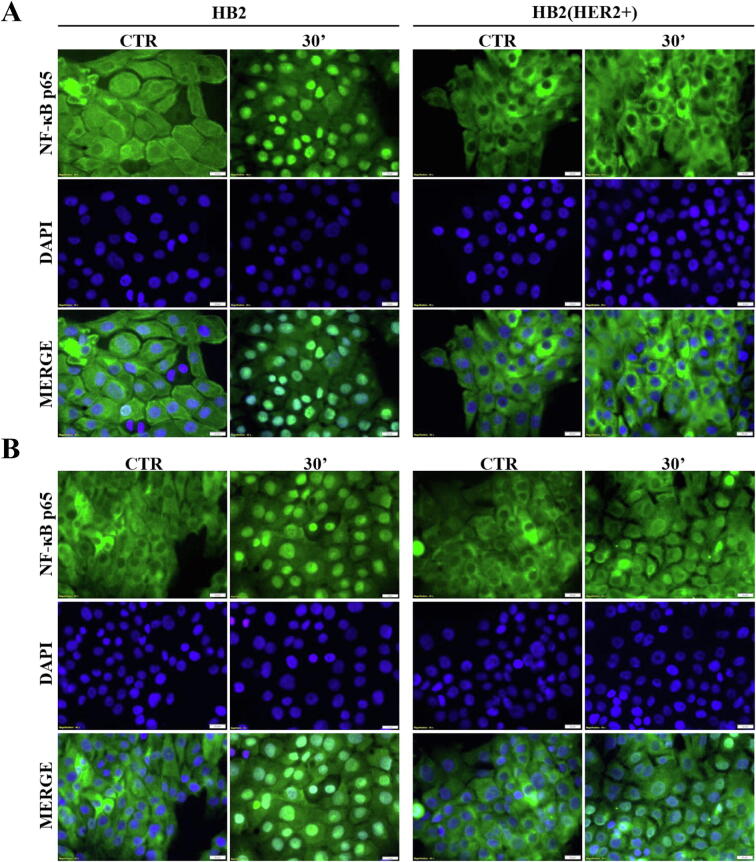
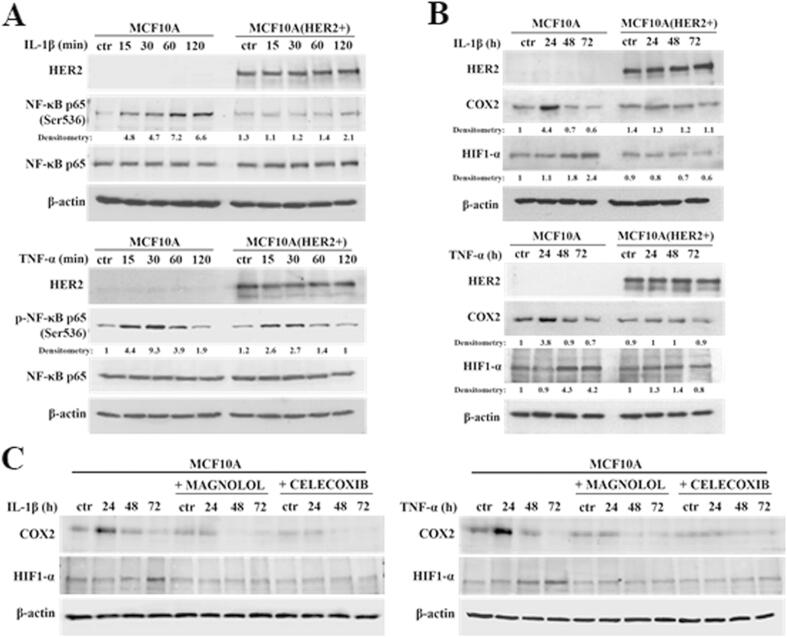
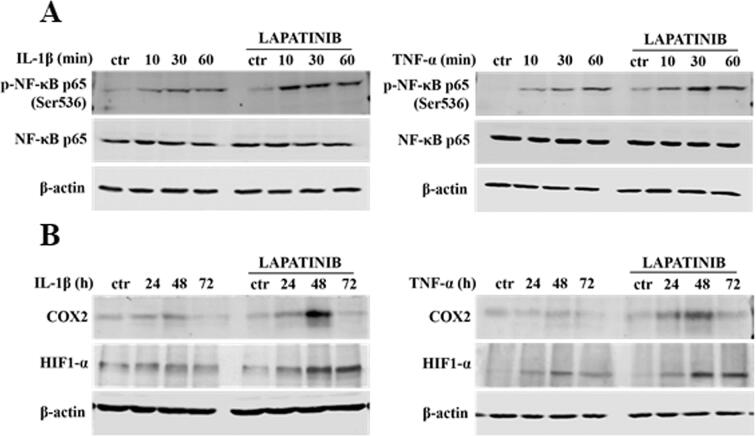
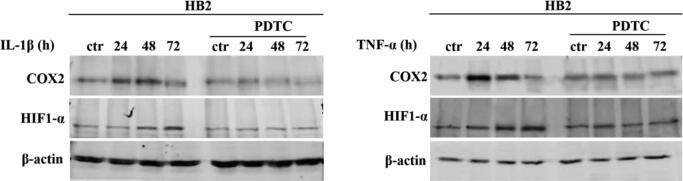
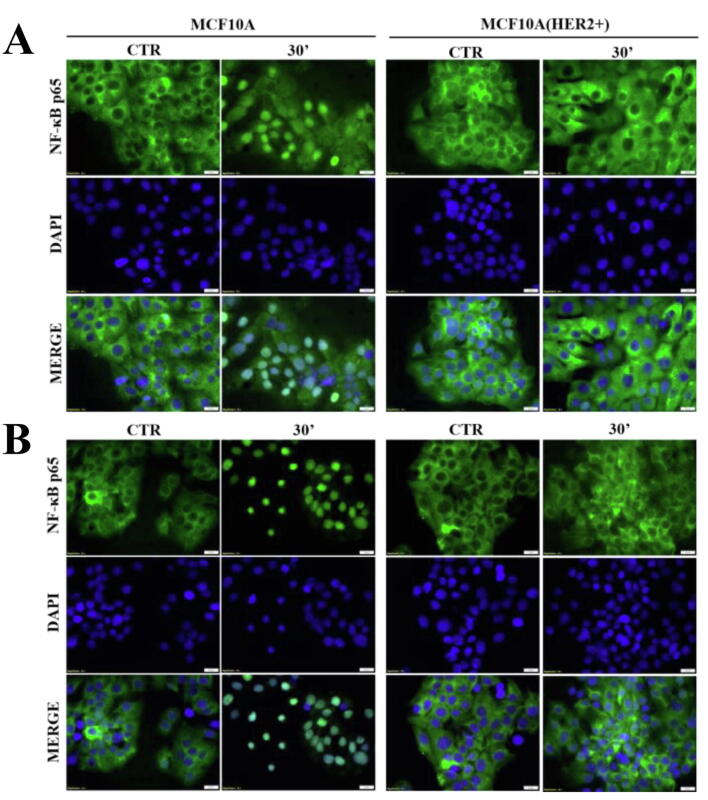
There is ample evidence to suggest that, in various biological set-ups, cytokines make use of the hypoxia signalling system under normoxia. Upregulation of HIF1-α through classical inflammatory signalling involving NF-κB and COX2 has been identified as a link between inflammatory and oncogenic pathways [46], [47]. To examine whether HIF1-α contributes to development of proliferative dominance of HER2-negative clones, HB2 and MCF10A cells and their HER2-overexpressing counterparts were stimulated with IL-1β and TNF-α and subjected for analyses of activation of the NF-κB/COX2 → HIF1-α axis. Using Western blotting analysis, we demonstrated that in both cell lines, treatment with IL-1β or TNF-α of HER2-negative cells resulted in strong phosphorylation of NF-κB p65-Ser536 as well as upregulation of COX2 (maximum after 24 h) and HIF1-α (maximum after 48–72 h) (Fig. 4A-B and Fig. S4A-B). In HER2-positive cells, phosphorylation of NF-κB was weaker and delayed, and no or only modest increase of COX2 and HIF1-α expression could be detected (Fig. 4A-B and Fig. S4A-B). Importantly, overexpression of HER2 did not affect the level of either NF-κB, COX2 or HIF1-α. Stimulation with cytokines of HER2-overexpressing HB2 cells, pre-treated with lapatinib, resulted in phosphorylation of NF-κB p65-Ser536 as well as an increase of COX2 and HIF1-α to the levels observed in HER2-negative cells (Fig. S5), confirming that the above effects were HER2-dependent. IL-1β and TNF-α treatment led also to upregulation of expression of carbonic anhydrase IX (CAIX), a HIF1-α-dependent gene, indicating HIF1-α activation [48]. This was seen only in HER2-negative cells (Fig. 4C). Pre-treatment of HB2 and MCF10A cells with magnolol, PDTC (NF-κB inhibitors [49], [50]) or celecoxib (COX2 inhibitor [51]) abolished IL-1β- and TNF-α-induced upregulation of COX2 and HIF1-α, respectively (Fig. 4D, Fig. S4C and Fig. S6), thus confirming the role of both NF-κB and COX2 in mediation of the process. HER2-dependenent specificity of NF-κB activation was additionally confirmed in experiment in which NF-κB p65 translocation to the nucleus upon IL-1β or TNF-α treatment was observed mostly in HER2-negative DCIS cells (Fig. 5A-B and Fig. S7A-B).
Fig. 4.
IL-1β and TNF-α activate NF-κB/COX2 → HIF1-α axis in HER2-negative mammary epithelial cells. (A) HB2 cells as well as their HER2-overexpressing variants were serum-starved and treated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 15, 30, 60 and 120 minutes, or (B) 24, 48 and 72 h. NF-κB activation, COX2 and HIF1-α expression were analysed by Western blotting. Densitometry was done with ImageJ software. (C) HB2 and HB2(HER2+) cells were serum-starved and treated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 24, 48, 72 and 96 h. CAIX (downstream target of HIF1-α activity) expression was evaluated via Western blotting. (D) HB2 cells were grown with magnolol (10 µM, NF-κB inhibitor) or celecoxib (25 µM, COX2 inhibitor), ± IL-1β (10 ng/ml), ± TNF-α (10 ng/ml) and analysed for COX2 and HIF1-α expression.
Fig. 5.
IL-1β and TNF-α promote NF-κB p65 translocation to the nucleus of HER2-negative DCIS cells. HB2 and HB2(HER2+) cells were serum starved overnight and treated with (A) IL-1β (10 ng/ml) or (B) TNF-α (10 ng/ml) for 30 min. NF-κB p65 translocation to the nucleus was assessed with fluorescent microscopy, scale bar is indicative of 20 µm.
IL-1β- and TNF-α-promoted growth of HER2-negative mammary epithelial cells is mediated by NF-κB/COX2 → HIF1-α pathway
We next sought to verify the contribution of NF-κB/COX2 → HIF1-α pathway to the cytokine-induced promotion of cell growth. In both analysed HER2-negative cell lines, magnolol or celecoxib almost completely abrogated stimulation of growth triggered by IL-1β or TNF-α (Fig. 6A, Fig. S8A), confirming involvement of both NF-κB and COX2. As the growth advantage conferred by cytokines was found to be associated with HIF1-α upregulation only in HER2-negative cells (Fig. 4B and Fig. S4B), we further assessed whether ectopic overexpression of HIF1-α might reverse cytokines-induced growth inhibition of HER2-positive counterparts. We found that, while HIF1-α overexpression did not affect the level of HER2 (Fig. S9), it enhanced growth of HB2(HER2+) cells and attenuated the negative effect of IL-1β and TNF-α on growth of both HB2(HER2+) and MCF10A(HER2+) cells (Fig. 6B and Fig. S8B). Taken together, these results suggest that NF-κB/COX2 → HIF1-α axis mediates HER2-dependent response of mammary epithelial cells to cytokine stimulation.
Fig. 6.
Proliferation of mammary epithelial cells in response to IL-1β and TNF-α is mediated by the NFκB/COX2 axis and relies on upregulation HIF1-α. (A) HB2 cells were cultured in 3D matrigel with IL-1β (10 ng/ml), TNF-α (10 ng/ml) and/or magnolol (10 µM) or celecoxib (10 µM). B) HB2 cells, their HER2- and HER2/HIF1-α-overexpressing variants were grown with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) in 3D matrigel. Representative images were taken after 8 days. Colony size was analysed using ImageJ software. Data are expressed as means ± SD (n = 3), **p < 0.005, ***p < 0.001.
Macrophages enhance growth of HER2-negative mammary epithelial cells
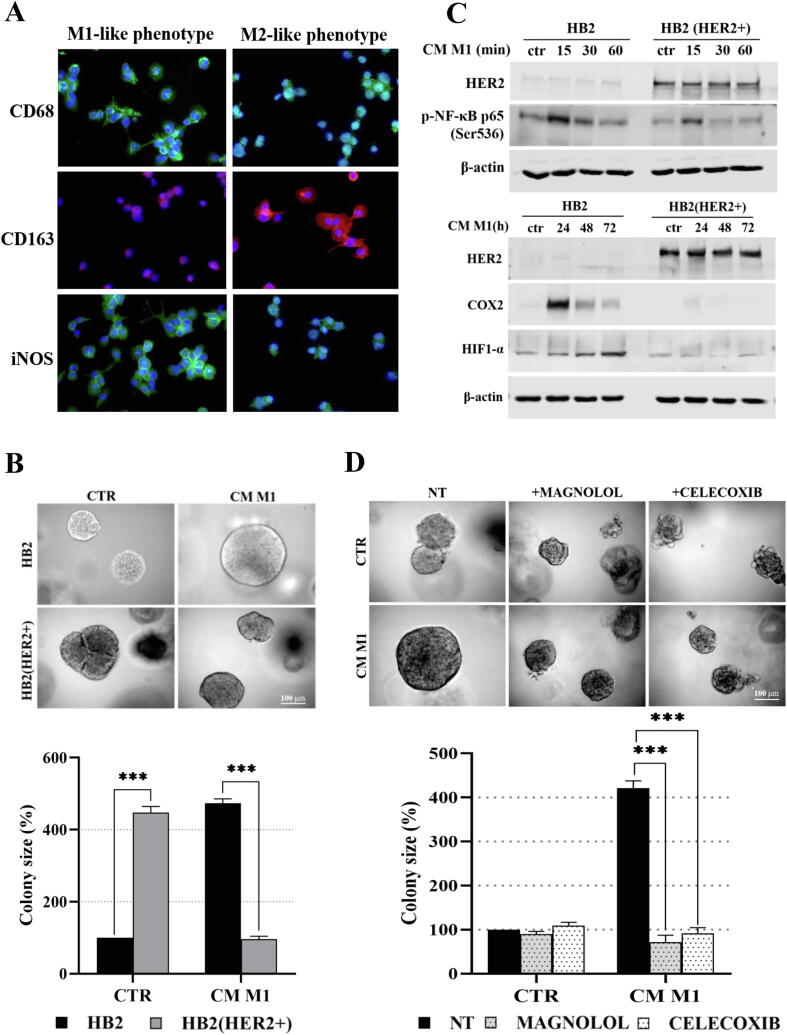
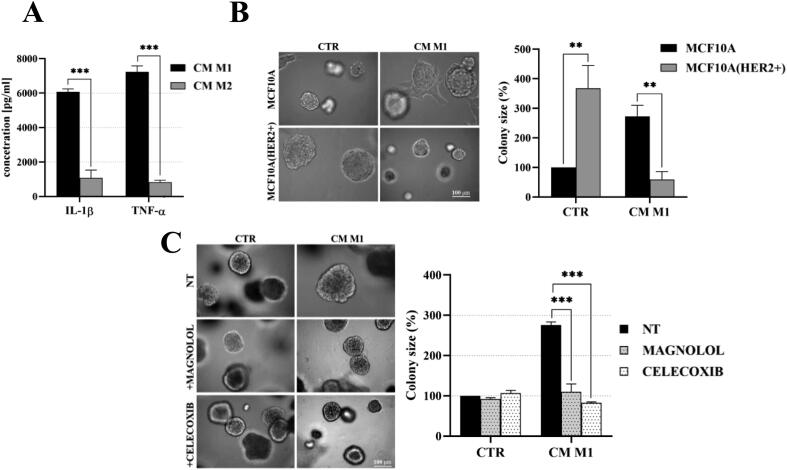
The in vivo relevance of cytokine-induced activation of NF-κB/COX2 → HIF1-α was assessed in experiments, where synthetic cytokines were replaced by macrophages-conditioned medium. The THP-1 cell line, widely exploited as a model of differentiation into macrophages, was used [42], [43]. Differentiation of THP-1 cells was verified by analyses of expression of specific markers i.e. iNOS and CD68 for M1- and CD163 for M2-like macrophages (Fig. 7A). Secretion of IL-1β and TNF-α by M1-like macrophages was found to be ∼6- and ∼13-fold higher, respectively, than that by M2-like cells (Fig. S10A), verifying predominance of the foreseen phenotypes in THP-1-derived subsets. We found that M1-like macrophages-conditioned medium (hereafter called M1-CM), similarly to the synthetic IL-1β and TNF-α (Fig. 3), promoted and inhibited growth of HER2-negative and HER2-overexpressing HB2 and MCF10A cells, respectively (Fig. 7B and Fig. S10B). Moreover, M1-CM induced stronger phosphorylation of NF-κB p65-Ser536 and upregulation of both COX2 and HIF1-α in HER2-negative than in HER2-positive HB2 cells (Fig. 7C). An involvement of NF-κB and COX2 activity in M1-CM-induced HER2-dependent growth of HB2 and MCF10A cells was verified with magnolol and celecoxib, which reversed the impact of M1-CM on cells growth (Fig. 7D and Fig. S10C). These results indicate that stromal inflammatory microenvironment may favour growth of HER2-negative clones via activation of the NF-κB/COX2 → HIF1-α pathway.
Fig. 7.
Macrophages-conditioned medium (M1-CM) promotes proliferation of HER2-negative mammary epithelial cells via activation of the NF-κB/COX2 → HIF1-α axis. (A) THP-1 cells were differentiated into M1- and M2-like macrophages as described in material and methods. Verification of THP-1 differentiation, i.e., expression of specific markers CD68 and iNOS for M1-like macrophages, CD163 for M2-like macrophages, was done by fluorescent microscopy. (B) HB2 and their HER2-overexpressing counterparts were cultured in 3D matrigel for 8 days in the presence of M1-like-conditioned medium (CM-M1). (C) HB2 cells were serum-starved and treated with CM-M1. NF-κB activation, COX2 and HIF1-α expression were analysed by Western blotting. (D) HB2 cells were grown in 3D matrigel for 8 days and treated with M1-like-conditioned medium ± magnolol (10 µM) or celecoxib (10 µM). Representative images were taken, an average colony size was determined with ImageJ software. Data are expressed as means ± SD (n = 3), ***p < 0.001.
Discussion
Evidence begins to accumulate that an impact of immune microenvironment on progression of BC is both cell type-specific [8], [12], [13], [14] and most prominent at the earliest stages of disease development [8], [9]. Given the importance of HER2 in BC biology, decreasing frequency of its expression as the disease evolves [16], [17], [18], here, we combined evaluation of clinical value of TIICs infiltration in DCIS with a mechanistic analysis of a role of HER2 in a proliferative cell response to inflammatory stimuli. Having focused on the effect of inflammation on HER2-related development of incipient BC, the analysed cohort was limited to patients with ‘pure’ DCIS with no HER2 amplification.
There were multiple attempts to assess the clinical value of the characteristics of the immune microenvironment in various types and at different stages of BC development. Most of these studies demonstrate an association between TIICs infiltration with poor disease outcome [8], [52], [53]. In DCIS, several reports support TILs infiltration as a poor prognostic factor for tumour progression and recurrence [54], [55], [56]. Our result linking TIICs with tumour grade is in line with these findings. The clinical value of density, types and ratios of TIICs vary between the studies [32], [54], [55], which may be due not only to the variability of sample sizes, inclusion criteria, methodologies and a lack of consistency in data interpretation but also may reflect the complexity of the immune system. Its interaction with the tumour is highly dynamic and, in reality, executed by distinct arms of its adaptive and innate components. It is also influenced by a variety of host factors, which can, at least partially, explain the reported differences between the clinical studies. What is consistently demonstrated, though, is an association between the density of immune infiltrates and the HER2-enriched and TNBC molecular subtypes, in both DCIS and IDC [8], [55], [57], [58]. The explanation for such a link is still unavailable. In our study, the size of the cohort precluded such an analysis, and we were not able to show an association between TIICs and the HER2 status. However, in 9% cases, histological examination revealed tumoral foci (‘hot-spots’) of basal-like cells with HIF1-α activity adjacent to the areas of dense stromal infiltration, which were almost exclusively present in HER2-negative DCIS lesions. Given the prevalence of HER2-negative IDCs, detection of even a small subset of potentially aggressive cells could have important biological implications. These findings also highlight the clinical significance of heterogeneity of DCIS tumours and value of identification of biological traits that could be used as predictors of progression.
A potential molecular mechanism governing the behaviour of such a subset was evaluated in further analyses using an in vitro DCIS model. Here, we show for the first time that the proliferative response of mammary epithelial cells to the inflammatory stimuli was HER2-dependent. Moreover, IL-1β and TNF-α promoted and inhibited growth of HER2-negative and HER2-positive clones, respectively. Our observation is in line with a recently reported association between IL-6 family of cytokines and decreased survival of patients with HER2-negative, but not HER2-positive, invasive breast carcinoma [59], however the molecular mechanism of this differential HER2-dependent cell response awaits to be revealed. Proliferative dominance of HER2-negative cells was demonstrated in two 3D experimental setups, i.e. mono- and co-cultures of HER2-positive and HER2-negative cells. In both, IL-1β and TNF-α exerted a similar effect but, surprisingly, in co-cultures the difference in response between HER2-negative and HER2-positive cells was exaggerated. This might be due to the addictive effects of synthetic cytokines and paracrine activity of HER2-overexpressing cells, known to be able to trigger a pro-inflammatory circuit in BC [60].
As demonstrated in many cancers, including IDC, a link between inflammatory and oncogenic pathways involves activation of the NF-κB/COX2 → HIF1-α axis [46], [47]. Our results support this observation and demonstrate that both IL-1β and TNF-α led to phosphorylation of NF-κB and subsequent upregulation of COX2 and HIF1-α, but the effect was highly dependent on the HER2 status. Moreover, while promoting proliferation of HER2-negative cells, inflammatory stimuli caused severe growth inhibition of their HER2-positive counterparts. Increasing evidence indicates that, although HIF1-α stabilisation primarily occurs in response to hypoxia, stimulation with cytokines, growth factors as well as oncogenic activation, involving PI3K, MAPK and/or NF-κB signalling, can lead to synthesis of HIF1-α under normoxic conditions [44]. A functional cross-talk between HIF1-α and HER2 has been demonstrated by several studies. HER2-overexpressing BC cells have been shown to stabilise HIF1-α under normoxia and require HIF1-α for HER2-mediated 3D in vitro growth [61]. HIF1-α has been identified as an upstream regulator of MAPK activity through direct and indirect regulation of dual-specificity phosphatases (DUSP) transcription. Expression of a stable form of HIF1-α in HER2-expressing BC cells was sufficient to induce lapatinib resistance via activation of the ERK pathway [62]. Thus, it is conceivable, that in the inflammatory microenvironment, activation of the HIF1-α-mediated pathway becomes the main driver of cell growth and allows the cells to overcome addiction to HER2. Induced in HER2-negative cells, it promotes proliferation, while an inability of HER2-positive cells to maintain expression and activation of HIF1-α results in suppression of 3D growth.
Conclusion
Our study showed for the first time that the proliferative response of mammary epithelial cells to inflammatory stimuli, mediated by the NF-κB/COX2 → HIF1-α axis, is HER2-dependent. As our in vitro experiments were carried out under normoxic conditions, the data also indicate that activation of HIF1-α, a protein commonly perceived to be a classical oxygen sensor, may be induced by several alternative means, including inflammatory cytokines. In tissue ‘inflammatory foci’, upregulation of HIF1-α may play a dominant role, conferring growth advantage to HER2-negative cells. In the context of DCIS heterogeneity and predominance of HER2-negative IDCs, this mechanism might significantly contribute to the evolution of the disease.
Conflict of interest
All authors have read the journal’s policy on disclosure of potential conflicts of interest. The authors have no financial disclosures to make and no conflicts of interest to disclose.
CRediT authorship contribution statement
Dominika Piasecka: Formal analysis, Investigation, Investigation, Resources, Writing - original draft, Validation, Funding acquisition. Marcin Braun: Formal analysis, Investigation, Resources, Writing - original draft, Validation. Magdalena Mieszkowska: Investigation. Lukasz Kowalczyk: Investigation. Janusz Kopczynski: Resources. Radzislaw Kordek: Resources. Rafal Sadej: Conceptualization, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Validation, Supervision. Hanna M. Romanska: Conceptualization, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Validation, Supervision, Project administration, Funding acquisition.
Acknowledgments
Acknowledgments
pBABEpuro-ERBB2 and HA-HIF1-alpha-wt-o-pBABE-puro were a gift from Matthew Meyerson and William Kaelin (Dana-Farber Cancer Institute). We thank Synevo Central Lab (Lodz, Poland) for providing access to UltraFast Scanner and DigiPath™ Professional Production Software.
The study was funded by Polish National Science Centre grants: OPUS No. UMO-2013/09/NZ4/02512 (to HMR) and PRELUDIUM no. UMO-2018/29/N/NZ3/02407 (to DP).
Ethical approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Medical University of Lodz (No. RNN/284/13/KE). Informed consent was not required for this study. The research was only retrospective and did not require any intervention on patients.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.09.003.
Contributor Information
Rafal Sadej, Email: rafal.sadej@gumed.edu.pl.
Hanna M. Romanska, Email: hanna.romanska@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
IL-1β and TNF-α promote proliferative dominance of mammary epithelial cells in a HER2-dependent manner. (A) HER2-overexpressing MCF10A cell line was generated by retroviral transduction of appropriate plasmid and verified by Western blotting. MCF10A and MCF10A(HER2+) cells were cultured in 3D matrigel for 10 days in the presence of IL-1β (10 ng/ml) or TNF-α (10 ng/ml). Representative images were taken, colony size was analysed using ImageJ software. (B) HB2 vs HB2(HER2+), and (C) MCF10A vs MCF10A(HER2+) cells, treated with of IL-1β (10 ng/ml) or TNF-α (10 ng/ml), were analysed for 2D proliferation. Cells were counted manually after 96 hours of growth. Data are expressed as means ± SD (n = 3), *p < 0.05, **p < 0.005 and ***p < 0.001.
Supplementary Fig. S2.
IL-1β and TNF-α promote proliferative dominance of mammary epithelial cells in a HER2-dependent manner. MCF10A and GFP-expressing MCF10A(HER2+)GFP cells were mixed in a 1:1 ratio. Preformed cell aggregates were embedded in matrigel and grown with IL-1β (10 ng/ml) or TNF-α (10 ng/ml). Representative images were taken after 8 days.
Supplementary Fig. S3.
IL-1β and TNF-α promote proliferative dominance of mammary epithelial cells in a HER2-dependent manner. (A) HB2 and (B) MCF10A cell lines and their GFP-expressing (HER2+)GFP variants were mixed in a ratio of 1:1 and embedded in 3D matrigel. After 8 days of growth in the presence of IL-1β (10 ng/ml) or TNF-α (10 ng/ml) representative images were taken and colony size analysed using ImageJ software. Data are expressed as means ± SD (n = 3), ***p < 0.001.
Supplementary Fig. S4.
IL-1β and TNF-α activate NF-κB/COX2 → HIF1-α axis in HER2-negative mammary epithelial cells. A) MCF10A cells, as well as their HER2-overexpressing variants, were serum-starved and treated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 15, 30, 60 and 120 minutes, or (B) 24, 48 and 72 hours. NF-κB activation, COX2 and HIF1-α expression were analysed by Western blotting. Densitometry was done with ImageJ software. C) MCF10A cells were grown with magnolol (10 µM) or celecoxib (25 µM), stimulated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 24, 48 and 72 h and analysed for COX2 and HIF1-α expression.
Supplementary Fig. S5.
HER2-inhibition restores IL-1β- and TNF-α-triggered activation of NF-κB/COX2 → HIF1-α axis. MCF10A(HER2+) cells were serum-starved, pre-treated with lapatinib (1 µM) for 3 h and incubated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 10, 30, 60 and 120 min, or 24, 48 and 72 h. (A) NF-κB activation and (B) COX2, HIF1-α expression were analysed by Western blotting.
Supplementary Fig. S6.
IL-1β and TNF-α activate NF-κB/COX2 → HIF1-α axis in HER2-negative mammary epithelial cells. HB2 cells were grown with PDTC (10 µM), stimulated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for 24, 48 and 72 h and analysed for COX2 and HIF1-α expression.
Supplementary Fig. S7.
IL-1β and TNF-α promote NF-κB p65 translocation to the nucleus of HER2-negative DCIS cells. MCF10A and MCF10A(HER2+) cells were serum starved overnight and treated with A) IL-1β (10 ng/ml) or B) TNF-α (10 ng/ml) for 30 min. NF-κB p65 translocation to the nucleus was assessed with fluorescent microscopy, scale bar represents 20 µm.
Supplementary Fig. S8.
IL-1β and TNF-α promote growth of HER2-negative DCIS cells via activation of the NF-κB/COX2 → HIF1-α axis. (A) MCF10A cells were cultured in 3D matrigel with IL-1β (10 ng/ml), TNF-α (10 ng/ml) and/or magnolol (10 µM) or celecoxib (10 µM). (B) MCF10A cells, their HER2- and HER2/HIF1-α-overexpressing variants were grown at the presence of IL-1β (10 ng/ml) or TNF-α (10 ng/ml) in 3D matrigel. Representative images were taken after 10 days, colony size was analysed using ImageJ software. Data are expressed as means ± SD (n = 3), **p < 0.005 and ***p < 0.001.
Supplementary Fig. S9.
Ectopic overexpression of HIF1-α in HB2 and MCF10A cell lines. HIF1-α overexpression in HB2(HER2+) and MCF10A(HER2+) cell lines was established via retroviral transduction of appropriate plasmid (described in material and methods) and confirmed via Western blotting.
Supplementary Fig. S10.
Macrophages enhance growth of HER2-negative mammary epithelial cells. (A) IL-1β and TNF-α concentration in macrophage-derived conditioned medium (CM) was determined by ELISA. Data are expressed as means ± SD (n = 3), ***p < 0.001. (B) MCF10A and MCF10A(HER2+) cells were cultured in 3D matrigel in the presence of M1-like macrophages-conditioned medium (CM-M1). (C) MCF10A cells were grown with M1-like macrophages-conditioned medium and/or magnolol (10 µM) or celecoxib (10 µM). Representative images were taken after 10 days of growth, average colony size was determined with ImageJ software. Data are expressed as means ± SD (n = 3), **p < 0.005, ***p < 0.001.
References
- 1.Hu M., Peluffo G., Chen H., Gelman R., Schnitt S., Polyak K. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc Natl Acad Sci U S A. 2009;106:3372–3377. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010;2010:210–213. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F., Zhuang X., Lin L., Yu P., Wang Y., Shi Y., Hu G., Sun Y. New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med. 2015;13:45. doi: 10.1186/s12916-015-0278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pages F., Galon J., Dieu-Nosjean M.C., Tartour E., Sautes-Fridman C., Fridman W.H. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene. 2010;29:1093–1102. doi: 10.1038/onc.2009.416. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Esquivel-Velazquez M., Ostoa-Saloma P., Palacios-Arreola M.I., Nava-Castro K.E., Castro J.I., Morales-Montor J. The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res. 2015;35:1–16. doi: 10.1089/jir.2014.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxevanis C.N., Sofopoulos M., Fortis S.P., Perez S.A. The role of immune infiltrates as prognostic biomarkers in patients with breast cancer. Cancer Immunol Immunother. 2019;68:1671–1680. doi: 10.1007/s00262-019-02327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil Del Alcazar C.R., Aleckovic M., Polyak K. Immune escape during breast tumor progression. Cancer Immunol Res. 2020;8:422–427. doi: 10.1158/2326-6066.CIR-19-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savas P., Loi S. Metastatic breast cancer: TIL it is too late. Clin Cancer Res. 2019 doi: 10.1158/1078-0432.CCR-19-3490. [DOI] [PubMed] [Google Scholar]
- 10.Szekely B., Bossuyt V., Li X., Wali V.B., Patwardhan G.A., Frederick C., Silber A., Park T., Harigopal M., Pelekanou V. Immunological differences between primary and metastatic breast cancer. Ann Oncol. 2018;29:2232–2239. doi: 10.1093/annonc/mdy399. [DOI] [PubMed] [Google Scholar]
- 11.Camp J.T., Elloumi F., Roman-Perez E., Rein J., Stewart D.A., Harrell J.C., Perou C.M., Troester M.A. Interactions with fibroblasts are distinct in Basal-like and luminal breast cancers. Mol Cancer Res. 2011;9:3–13. doi: 10.1158/1541-7786.MCR-10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng S., Li L., Zhou M., Jiang W., Niu H., Yang K. Distribution and prognostic value of tumorinfiltrating T cells in breast cancer. Mol Med Rep. 2018;18:4247–4258. doi: 10.3892/mmr.2018.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart D.A., Yang Y., Makowski L., Troester M.A. Basal-like breast cancer cells induce phenotypic and genomic changes in macrophages. Mol Cancer Res. 2012;10:727–738. doi: 10.1158/1541-7786.MCR-11-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Droeser R., Zlobec I., Kilic E., Guth U., Heberer M., Spagnoli G., Oertli D., Tapia C. Differential pattern and prognostic significance of CD4+, FOXP3+ and IL-17+ tumor infiltrating lymphocytes in ductal and lobular breast cancers. BMC Cancer. 2012;12:134. doi: 10.1186/1471-2407-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allred D.C., Wu Y., Mao S., Nagtegaal I.D., Lee S., Perou C.M., Mohsin S.K., O'Connell P., Tsimelzon A., Medina D. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 16.Pradeep C.R., Kostler W.J., Lauriola M., Granit R.Z., Zhang F., Jacob-Hirsch J., Rechavi G., Nair H.B., Hennessy B.T., Gonzalez-Angulo A.M. Modeling ductal carcinoma in situ: a HER2-Notch3 collaboration enables luminal filling. Oncogene. 2012;31:907–917. doi: 10.1038/onc.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Vijver M.J., Peterse J.L., Mooi W.J., Wisman P., Lomans J., Dalesio O., Nusse R. Neu-protein overexpression in breast cancer. Association with comedo-type ductal carcinoma in situ and limited prognostic value in stage II breast cancer. N Engl J Med. 1988;319:1239–1245. doi: 10.1056/NEJM198811103191902. [DOI] [PubMed] [Google Scholar]
- 18.Park K., Han S., Kim H.J., Kim J., Shin E. HER2 status in pure ductal carcinoma in situ and in the intraductal and invasive components of invasive ductal carcinoma determined by fluorescence in situ hybridization and immunohistochemistry. Histopathology. 2006;48:702–707. doi: 10.1111/j.1365-2559.2006.02403.x. [DOI] [PubMed] [Google Scholar]
- 19.Provenzano E., Hopper J.L., Giles G.G., Marr G., Venter D.J., Armes J.E. Histological markers that predict clinical recurrence in ductal carcinoma in situ of the breast: an Australian population-based study. Pathology. 2004;36:221–229. doi: 10.1080/00313020410001692558. [DOI] [PubMed] [Google Scholar]
- 20.Ringberg A., Anagnostaki L., Anderson H., Idvall I., Ferno M., South Sweden Breast Cancer G Cell biological factors in ductal carcinoma in situ (DCIS) of the breast-relationship to ipsilateral local recurrence and histopathological characteristics. Eur J Cancer. 2001;37:1514–1522. doi: 10.1016/s0959-8049(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 21.Roses R.E., Paulson E.C., Sharma A., Schueller J.E., Nisenbaum H., Weinstein S., Fox K.R., Zhang P.J., Czerniecki B.J. HER-2/neu overexpression as a predictor for the transition from in situ to invasive breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:1386–1389. doi: 10.1158/1055-9965.EPI-08-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latta E.K., Tjan S., Parkes R.K., O'Malley F.P. The role of HER2/neu overexpression/amplification in the progression of ductal carcinoma in situ to invasive carcinoma of the breast. Mod Pathol. 2002;15:1318–1325. doi: 10.1097/01.MP.0000038462.62634.B1. [DOI] [PubMed] [Google Scholar]
- 23.Zhou W., Jirstrom K., Johansson C., Amini R.M., Blomqvist C., Agbaje O., Warnberg F. Long-term survival of women with basal-like ductal carcinoma in situ of the breast: a population-based cohort study. BMC Cancer. 2010;10:653. doi: 10.1186/1471-2407-10-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borgquist S., Zhou W., Jirstrom K., Amini R.M., Sollie T., Sorlie T., Blomqvist C., Butt S., Warnberg F. The prognostic role of HER2 expression in ductal breast carcinoma in situ (DCIS); a population-based cohort study. BMC Cancer. 2015;15:468. doi: 10.1186/s12885-015-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casasent A.K., Edgerton M., Navin N.E. Genome evolution in ductal carcinoma in situ: invasion of the clones. J Pathol. 2017;241:208–218. doi: 10.1002/path.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novitskaya V., Romanska H., Dawoud M., Jones J.L., Berditchevski F. Tetraspanin CD151 regulates growth of mammary epithelial cells in three-dimensional extracellular matrix: implication for mammary ductal carcinoma in situ. Cancer Res. 2010;70:4698–4708. doi: 10.1158/0008-5472.CAN-09-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nash C.E., Mavria G., Baxter E.W., Holliday D.L., Tomlinson D.C., Treanor D., Novitskaya V., Berditchevski F., Hanby A.M., Speirs V. Development and characterisation of a 3D multi-cellular in vitro model of normal human breast: a tool for cancer initiation studies. Oncotarget. 2015;6:13731–13741. doi: 10.18632/oncotarget.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinn HP, Kreipe H (2013). A brief overview of the WHO classification of breast tumors, 4th Edition, Focusing on issues and updates from the 3rd Edition, Breast Care (Basel) 8, 149-154. [DOI] [PMC free article] [PubMed]
- 29.Allred D.C., Harvey J.M., Berardo M., Clark G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–168. [PubMed] [Google Scholar]
- 30.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W. Human epidermal growth factor receptor 2 testing in breast cancer: American Society Of Clinical Oncology/College Of American Pathologists Clinical Practice Guideline focused update. Arch Pathol Lab Med. 2018;142:1364–1382. doi: 10.5858/arpa.2018-0902-SA. [DOI] [PubMed] [Google Scholar]
- 31.Lal P., Salazar P.A., Hudis C.A., Ladanyi M., Chen B. HER-2 testing in breast cancer using immunohistochemical analysis and fluorescence in situ hybridization: a single-institution experience of 2,279 cases and comparison of dual-color and single-color scoring. Am J Clin Pathol. 2004;121:631–636. doi: 10.1309/VE78-62V2-646B-R6EX. [DOI] [PubMed] [Google Scholar]
- 32.Salgado R., Denkert C., Demaria S., Sirtaine N., Klauschen F., Pruneri G., Wienert S., Van den Eynden G., Baehner F.L., Penault-Llorca F. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dieci M.V., Radosevic-Robin N., Fineberg S., van den Eynden G., Ternes N., Penault-Llorca F., Pruneri G., D'Alfonso T.M., Demaria S., Castaneda C. Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: A report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin Cancer Biol. 2018;52:16–25. doi: 10.1016/j.semcancer.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Hendry S., Salgado R., Gevaert T., Russell P.A., John T., Thapa B., Christie M., van de Vijver K., Estrada M.V., Gonzalez-Ericsson P.I. Assessing tumor infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the International Immuno-Oncology Biomarkers Working Group: Part 1: Assessing the host immune response. TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv Anat Pathol. 2017;24:235–251. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denkert C., von Minckwitz G., Darb-Esfahani S., Lederer B., Heppner B.I., Weber K.E., Budczies J., Huober J., Klauschen F., Furlanetto J. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 36.Mao Y., Qu Q., Chen X., Huang O., Wu J., Shen K. The prognostic value of tumor-infiltrating lymphocytes in breast cancer: a systematic review and meta-analysis. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0152500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greulich H., Kaplan B., Mertins P., Chen T.H., Tanaka K.E., Yun C.H., Zhang X., Lee S.H., Cho J., Ambrogio L. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–14481. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Q., Bartz S., Mao M., Li L., Kaelin W.G., Jr. The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol. 2007;27:2092–2102. doi: 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turczyk L., Kitowska K., Mieszkowska M., Mieczkowski K., Czaplinska D., Piasecka D., Kordek R., Skladanowski A.C., Potemski P., Romanska H.M. FGFR2-driven signaling counteracts tamoxifen effect on ERα-positive breast cancer cells. Neoplasia. 2017;19:791–804. doi: 10.1016/j.neo.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadej R., Romanska H., Kavanagh D., Baldwin G., Takahashi T., Kalia N., Berditchevski F. Tetraspanin CD151 regulates transforming growth factor beta signaling: implication in tumor metastasis. Cancer Res. 2010;70:6059–6070. doi: 10.1158/0008-5472.CAN-09-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mieszkowska M., Piasecka D., Potemski P., Debska-Szmich S., Rychlowski M., Kordek R., Sadej R., Romanska H.M. Tetraspanin CD151 impairs heterodimerization of ErbB2/ErbB3 in breast cancer cells. Transl Res. 2019;207:44–55. doi: 10.1016/j.trsl.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Genin M., Clement F., Fattaccioli A., Raes M., Michiels C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 2015;15:577. doi: 10.1186/s12885-015-1546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park E.K., Jung H.S., Yang H.I., Yoo M.C., Kim C., Kim K.S. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res. 2007;56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 44.Zhou J., Brune B. Cytokines and hormones in the regulation of hypoxia inducible factor-1alpha (HIF-1alpha) Cardiovasc Hematol Agents Med Chem. 2006;4:189–197. doi: 10.2174/187152506777698344. [DOI] [PubMed] [Google Scholar]
- 45.Albanell J., Gascon P. Small molecules with EGFR-TK inhibitor activity. Curr Drug Targets. 2005;6:259–274. doi: 10.2174/1389450053765888. [DOI] [PubMed] [Google Scholar]
- 46.Jung Y.J., Isaacs J.S., Lee S., Trepel J., Neckers L. IL-1beta-mediated up-regulation of HIF-1alpha via an NFkappaB/COX-2 pathway identifies HIF-1 as a critical link between inflammation and oncogenesis. FASEB J. 2003;17:2115–2117. doi: 10.1096/fj.03-0329fje. [DOI] [PubMed] [Google Scholar]
- 47.Stasinopoulos I., O'Brien D.R., Bhujwalla Z.M. Inflammation, but not hypoxia, mediated HIF-1alpha activation depends on COX-2. Cancer Biol Ther. 2009;8:31–35. doi: 10.4161/cbt.8.1.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiche J., Ilc K., Laferriere J., Trottier E., Dayan F., Mazure N.M., Brahimi-Horn M.C., Pouyssegur J. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009;69:358–368. doi: 10.1158/0008-5472.CAN-08-2470. [DOI] [PubMed] [Google Scholar]
- 49.Tse A.K., Wan C.K., Zhu G.Y., Shen X.L., Cheung H.Y., Yang M., Fong W.F. Magnolol suppresses NF-kappaB activation and NF-kappaB regulated gene expression through inhibition of IkappaB kinase activation. Mol Immunol. 2007;44:2647–2658. doi: 10.1016/j.molimm.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Zhou J., Zhang H., Gu P., Bai J., Margolick J.B., Zhang Y. NF-kappaB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res Treat. 2008;111:419–427. doi: 10.1007/s10549-007-9798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basu G.D., Pathangey L.B., Tinder T.L., Lagioia M., Gendler S.J., Mukherjee P. Cyclooxygenase-2 inhibitor induces apoptosis in breast cancer cells in an in vivo model of spontaneous metastatic breast cancer. Mol Cancer Res. 2004;2:632–642. [PubMed] [Google Scholar]
- 52.Jones L.M., Broz M.L., Ranger J.J., Ozcelik J., Ahn R., Zuo D., Ursini-Siegel J., Hallett M.T., Krummel M., Muller W.J. STAT3 establishes an immunosuppressive microenvironment during the early stages of breast carcinogenesis to promote tumor growth and metastasis. Cancer Res. 2016;76:1416–1428. doi: 10.1158/0008-5472.CAN-15-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeNardo D.G., Barreto J.B., Andreu P., Vasquez L., Tawfik D., Kolhatkar N., Coussens L.M. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campbell M.J., Baehner F., O'Meara T., Ojukwu E., Han B., Mukhtar R., Tandon V., Endicott M., Zhu Z., Wong J. Characterizing the immune microenvironment in high-risk ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2017;161:17–28. doi: 10.1007/s10549-016-4036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruneri G., Lazzeroni M., Bagnardi V., Tiburzio G.B., Rotmensz N., DeCensi A., Guerrieri-Gonzaga A., Vingiani A., Curigliano G., Zurrida S. The prevalence and clinical relevance of tumor-infiltrating lymphocytes (TILs) in ductal carcinoma in situ of the breast. Ann Oncol. 2017;28:321–328. doi: 10.1093/annonc/mdw623. [DOI] [PubMed] [Google Scholar]
- 56.Toss M.S., Miligy I., Al-Kawaz A., Alsleem M., Khout H., Rida P.C., Aneja R., Green A.R., Ellis I.O., Rakha E.A. Prognostic significance of tumor-infiltrating lymphocytes in ductal carcinoma in situ of the breast. Mod Pathol. 2018;31:1226–1236. doi: 10.1038/s41379-018-0040-8. [DOI] [PubMed] [Google Scholar]
- 57.Kristensen V.N., Vaske C.J., Ursini-Siegel J., Van Loo P., Nordgard S.H., Sachidanandam R., Sorlie T., Warnberg F., Haakensen V.D., Helland A. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci U S A. 2012;109:2802–2807. doi: 10.1073/pnas.1108781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asano Y., Kashiwagi S., Goto W., Kurata K., Noda S., Takashima T., Onoda N., Tanaka S., Ohsawa M., Hirakawa K. Tumour-infiltrating CD8 to FOXP3 lymphocyte ratio in predicting treatment responses to neoadjuvant chemotherapy of aggressive breast cancer. Br J Surg. 2016;103:845–854. doi: 10.1002/bjs.10127. [DOI] [PubMed] [Google Scholar]
- 59.Tawara K, Scott H, Emathinger J, Ide A, Fox R, Greiner D, LaJoie D, Hedeen D, Nandakumar M, Oler AJ, et al. (2019). Co-Expression of VEGF and IL-6 Family Cytokines is Associated with Decreased Survival in HER2 Negative Breast Cancer Patients: Subtype-Specific IL-6 Family Cytokine-Mediated VEGF Secretion12, Vol. 12, pp. 245–255. [DOI] [PMC free article] [PubMed]
- 60.Liu S., Lee J.S., Jie C., Park M.H., Iwakura Y., Patel Y., Soni M., Reisman D., Chen H. HER2 overexpression triggers an IL-1α pro-inflammatory circuit to drive tumorigenesis and promote chemotherapy resistance. Cancer Res. 2018;78:2040–2051. doi: 10.1158/0008-5472.CAN-17-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whelan K.A., Schwab L.P., Karakashev S.V., Franchetti L., Johannes G.J., Seagroves T.N., Reginato M.J. The oncogene HER2/neu (ERBB2) requires the hypoxia-inducible factor HIF-1 for mammary tumor growth and anoikis resistance. J Biol Chem. 2013;288:15865–15877. doi: 10.1074/jbc.M112.426999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karakashev S.V., Reginato M.J. Hypoxia/HIF1alpha induces lapatinib resistance in ERBB2-positive breast cancer cells via regulation of DUSP2. Oncotarget. 2015;6:1967–1980. doi: 10.18632/oncotarget.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.