Abstract
The tumor microenvironment (TME) chiefly consists of tumor cells and tumor-infiltrating immune cells admixed with the stromal component. A recent clinical trial has shown that the tumor immune cell infiltration (ICI) is correlated with the sensitivity to immunotherapy and the head and neck squamous cell carcinoma (HNSC) prognosis. However, to date, the immune infiltrative landscape of HNSC has not yet been elucidated. Herein, we proposed two computational algorithms to unravel the ICI landscape of 1,029 HNSC patients. Three ICI patterns were defined, and the ICI scores were determined by using principal-component analysis. A high ICI score was characterized by an increased tumor mutation burden (TMB) and the immune-activating signaling pathways. Activation of transforming growth factor-β (TGF-β) and WNT signaling pathways were observed in low ICI score subtypes, indicating T cell suppression, and may be responsible for poor prognosis. Two immunotherapy cohorts confirmed patients with higher ICI scores demonstrated significant therapeutic advantages and clinical benefits. This study demonstrated that the ICI scores serve as an effective prognostic biomarker and predictive indicator for immunotherapy. Evaluating the ICI patterns of a larger cohort of samples will extend our understanding of TME, and it may provide directions to the current research investigations on immunotherapeutic strategies for HNSC.
Keywords: head and neck squamous cell carcinoma, immune cell infiltration, tumor microenvironment, immunotherapy
Graphical Abstract
Understanding the tumor-immune microenvironment is critical for improving the efficacy of current immunotherapies. Zhang et al. comprehensively evaluated the cellular, molecular, and genetic factors associated with immune cell infiltration patterns. These have important implications for how tumors respond to immunotherapies and may guide more effective immunotherapy strategies.
Introduction
Head and neck cancer ranks as the sixth most common malignancy worldwide and claims around 3,50,000 lives per year.1 The squamous cell carcinoma is the most common pathological type of HNSC.2 Local recurrence, cervical node metastasis, and treatment failure caused by resistance to conventional chemotherapy are the leading causes of death in patients with advanced head and neck squamous cell carcinoma (HNSC).
Immunotherapy activates the host’s natural defense system, which identifies and eliminates the tumor cells. It has emerged as an effective treatment with unparalleled and synergistic survival benefits in multiple cancers.3, 4, 5, 6 However, a major limitation of this treatment is that it benefits only a minority of patients. Thus, novel therapeutic markers demand urgent investigation so that the ideal HNSC subgroups for immunotherapy could be identified.3,7
The tumor microenvironment (TME) in HNSC contains transformed cells admixed with immune cells and stromal cellular elements.8 Extensive research on TME has revealed a crucial role of the tumor-infiltrating immune cells in tumor dissemination, relapse, metastasis, and therapeutic response to immunotherapy.9,10 For instance, tumor-associated macrophages (TAMs) exert multiple pro-tumor effects by secreting immunosuppressive cytokines, such as interleukin-10 (IL-10) and transforming growth factor-β (TGF-β), and enhancing the TAM density, which is associated with unfavorable prognosis.11, 12, 13 In contrast, the escalated levels of tumor-infiltrating lymphocytes (TLSs), such as CD4+ T cells and CD8+ T cells, have been associated with improved survival rate and response to immunotherapy.14 Because the immune checkpoint blockade activates the pre-existing TLS cells, which recognize and eliminate the dysplastic and neoplastic cells, TLS cells play crucial roles in response to immunotherapy.15,16 However, the identification of TLS cells is not sufficient to characterize the complex tumor immune milieu. Besides, resistance to immunotherapy can also be observed in patients with higher TLSs.13,17,18 The immunosuppressive cytokines secreted by TAMs can impair the anti-tumor effects of TLSs. Moreover, the excessive infiltration of stromal components in tumor tissue can decrease the TLS trafficking into tumors.16,17 It elucidates that the intercellular relationships in TME are more critical than the single-cell population. So far, the expansive landscape of immune cells infiltrating the TME of HNSC has not been elucidated.
Over the past decades, advances in the next-generation sequencing (NGS) technology, specifically the NGS algorithm technology, have unveiled massive biological information about HNSC tumorigenesis and metastasis.19 In this study, we used two computational algorithms, CIBERSORT and ESTIMATE, to analyze the gene-expression profiles of bulk tumor sample and acquire a comprehensive outlook about the intra-tumoral immune landscape.20,21 Besides, we classified the HNSC into three discrete subtypes as per the immune cell-infiltration patterns. Conclusively, in this study, we established the ICI scores to characterize the various immune landscapes, which could precisely predict patient outcome and response to immunotherapy.
Results
The Landscape of Immuno-cell Infiltration in the TME of HNSC
First, we performed the CIBERSORT and ESTIMATE algorithms to quantify the activity or enrichment levels of immune cells in HNSC tumor tissues (Table S1).20,21 Based on 1,029 tumor samples with matched immune cell infiltration (ICI) profiles from the meta-cohort (Array express databse: GSE41613, GSE42743, E-MTAB-1328 and GSE65858; The Cancer Genome Atlas [TCGA]-HNSC; ), unsupervised clustering was performed using the ConsesusClusterPlus package of R software to classify the HNSC patients into distinct subtypes.
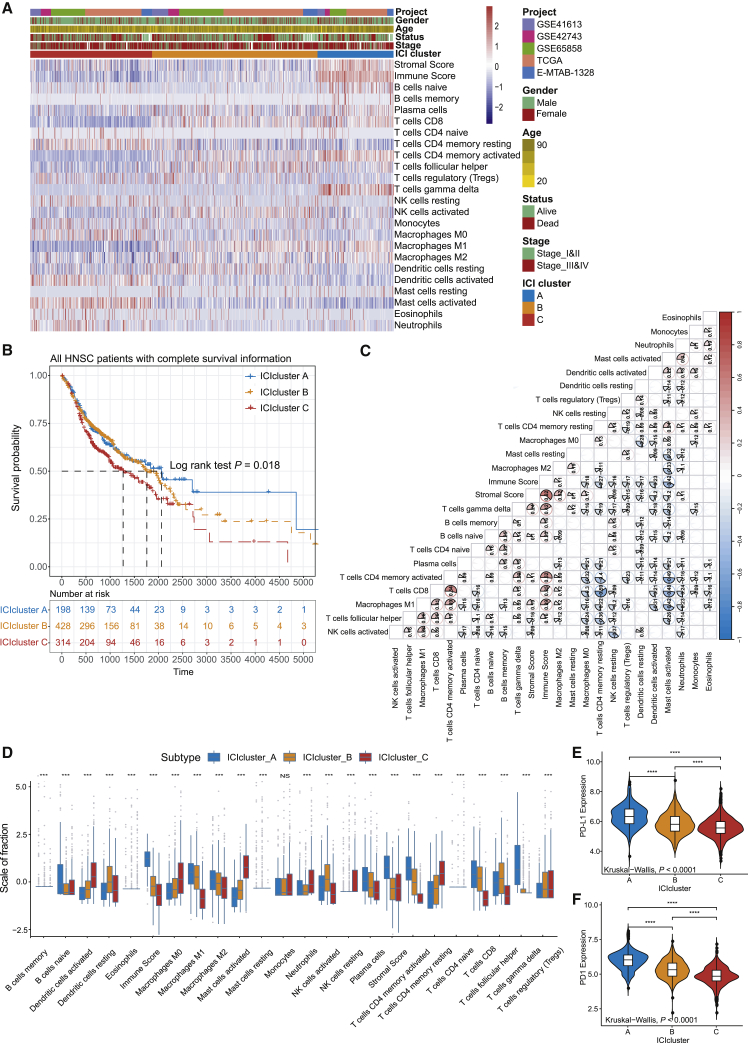
We identified three independent ICI subtypes with significant survival differences (log rank test, p = 0.018; Figure S1B; Figures 1A and 1B). To further clarify the intrinsic biological differences that led to distinct clinical phenotypes, we compared the immune cell composition of the TME. Among the three main immune subtypes, the ICI cluster A was associated with a favorable prognosis with a median survival of 2,064 days. It was marked by high naive B cells, M1 and M2 macrophages, plasma cells, memory CD4 T cells, CD8 T cells, and gamma delta T cells infiltration. The median survival time of ICI cluster B was 1,762 days, and patients in ICI cluster B were characterized by a significantly higher density of resting dendritic cells (DCs), activated natural killer (NK) cells, and follicular helper T cells. The subjects in the ICI cluster C witnessed a shorter overall survival (OS; median survival 1,281 days) and exhibited a significant increase in the infiltration of activated DCs, activated mast cells, neutrophils, resting NK cells, memory resting CD4+ T cells, and T regulatory cells. Additionally, the correlation coefficient heatmap was generated to visualize the universal landscape of immune cell interaction in TME (Figure 1C). We also analyzed the two vital immune checkpoints, i.e., PD1 and PD-L1 in each ICI subtype. ICI cluster A was characterized by a significantly higher PD1/PD-L1 expression level and ICI cluster C with a lower PD1/PD-L1 expression level. The Kruskal-Wallis test was used to detect the significant differences between the immune cells and the PD1/PD-L1 expression level in three distinct ICI subtypes (Figures 1D–1F).
Figure 1.
The Landscape of Immuno-cell Infiltration in the TME of HNSC
(A) Unsupervised clustering of tumor-infiltrating immune cells in five independent HNSC cohorts. Rows represent tumor-infiltrating immune cells, and columns represent samples. (B) Kaplan-Meier curves for overall survival (OS) of all HNSC patients with immune cell-infiltrating classes. Log rank test showed an overall p = 0.018. (C) The fraction of tumor-infiltrating immune cells in three ICI clusters. We also plotted the immune score and stromal score of three ICI clusters. The statistical difference of three ICI clusters was compared through the Kruskal-Wallis test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. (D) Cellular interaction of the tumor-infiltrating immune cell types. (E and F) Difference in PD-L1 (E) and PD1 (F) expression among distinct ICI clusters (Kruskal-Wallis test, p < 0.0001).
Identified Immune Gene Subtype
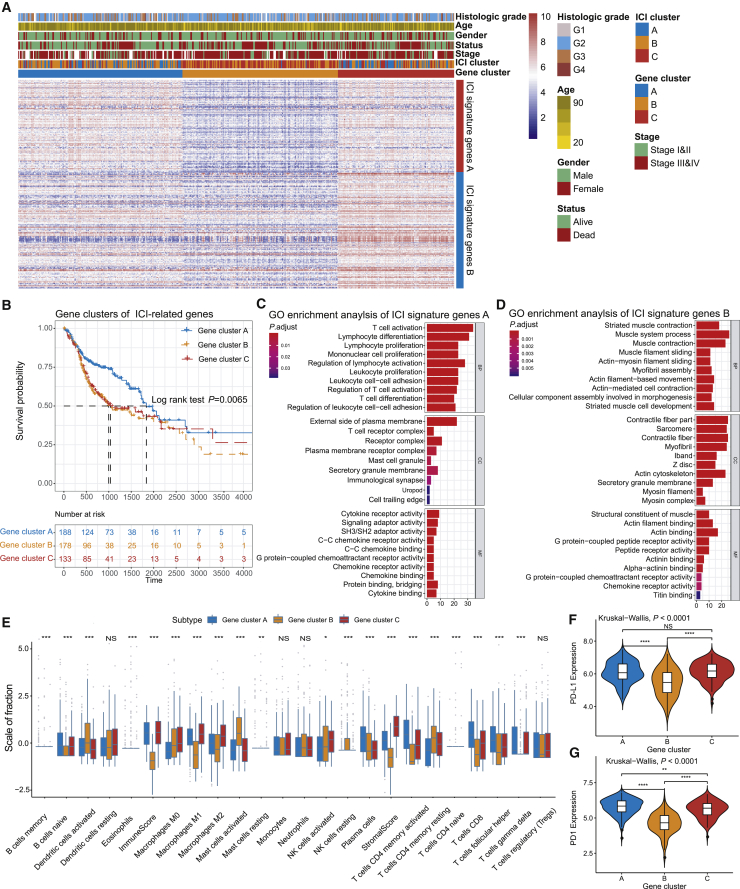
To unravel the underlying biological characteristics of distinct immunophenotypes, we performed differential analyses to determine the transcriptome variations among these subtypes by limma packages of R software. In the subsequent analysis, primary emphasis was laid on the TCGA-HNSC cohort, which had the most exhaustive clinical information. We performed the unsupervised clustering of 1,089 differentially expressed genes (DEGs) (Table S2), obtained by previous differential analyses, which classified the TCGA cohort into three genomic clusters, namely, gene clusters A–C (Figures S2A and S2B). The 588 gene signatures that were positively correlated with the gene cluster were termed as the ICI gene signature A, and the rest of the DEGs were named as the ICI gene signature B (Table S3). Concurrently, in order to reduce the noise or redundant genes, we used the Boruta algorithm to perform dimension reduction in the ICI gene signatures A and B.22 The heatmap delineated the transcriptomic profile of the 276 most abundant DEGs identified across the genomic clusters (Figure 2A), which were annotated by the “clusterProfiler” R package.23 The significantly enriched biological processes are summarized in Figures 2C and 2D, and a detailed description is provided in Table S4. The correlation coefficient between the ICI signature gene and PD1/PDL1 was determined through correlation analysis, and we observed that a majority of ICI signatures genes were significantly correlated to the PD1/PD-L1 expression level (Table S5).
Figure 2.
Identification of Immunogenic Gene Subtypes
(A) Unsupervised clustering of common DEGs among three ICI cluster groups to classify patients into three groups: gene clusters A–C. (B) Kaplan-Meier curves for the three groups of patients. The log rank test showed an overall p = 0.0065. (C and D) Gene Ontology (GO) enrichment analysis of the two ICI-relevant signature genes: ICI signature genes A (C) and B (D). The x axis indicates the number of genes within each GO term. (E) The fraction of tumor-infiltrating immune cells in three gene clusters. We also plotted the immune and stromal scores of three ICI clusters. The statistical difference of three ICI clusters was compared through the Kruskal-Wallis test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. (F and G) Difference in PD-L1 (F) and PD1 (G) expression among distinct ICI gene clusters (Kruskal-Wallis test, p < 0.0001).
Furthermore, we explored the prognostic implications of the ICI gene clusters by integrating them with survival information. The analysis was performed by using the Kaplan-Meier plotter, and we found that patients in gene cluster A had a better prognosis, whereas patients in gene clusters B and C had the unfavorable outcomes (log rank test, p = 0.0065; Figure 2B). Interestingly, we found that gene clusters A and C were associated with significantly high immune scores. Multiple studies have shown that the immune system may have favorable and adverse outcomes, which could be exhibited in the form of pro-tumor or anti-tumor activity, as observed in our analysis.13,16 As depicted in Figure 2E, gene cluster C showed an escalated stromal component infiltration and immunosuppression-associated M2 macrophages and decreased DC infiltration, which was characterized as the “immune exhausted phenotype” by the previous studies.13,24 The gene cluster A exhibited the highest CD8+ T cell infiltration. The active immune phenotypes were characterized by the presence of plasma cells and memory activated CD4+ T cells. Additionally, the three genomic clusters showed significant differences in PD1/PD-L1 expression levels (Kruskal-Wallis, p < 0.0001; Figures 2F and 2G). The ICI gene clusters A and C were associated with relatively higher PD1/PD-L1 expression levels, whereas ICI gene cluster B with the lowest PD1/PD-L1 expression level. Taken together, the consistency between the immune profile and prognostic profile in different gene clusters implied that our classification method is scientific and reasonable.
Construction of the ICI Score
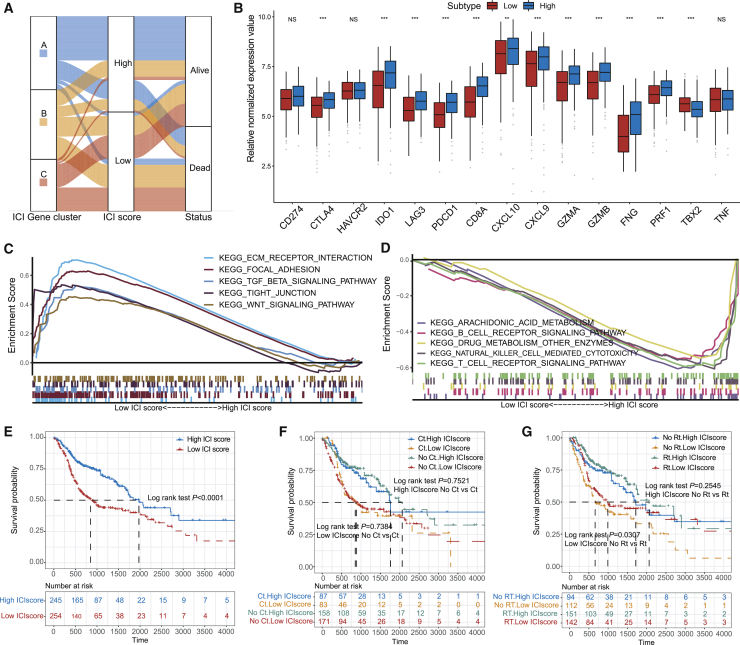
To obtain quantitative indicators of ICI landscape in HNSC patients, we used principal-component analysis (PCA) to compute two aggregate scores: (1) the ICI score A from ICI signature gene A, and (2) the ICI score B from ICI signature gene B. The ISA and ISB of each patient in this investigation were computed as the sum of individual relevant individual scores. Finally, we acquired the prognostic signature score that was defined as the ICI score. The patients in the TCGA cohort were stratified into two groups as the high or low ICI scores by using the optimal cutoff value acquired by the X-tile software. The distribution of patients in three gene clusters is represented in Figure 3A. We analyzed the immune activity and tolerance condition of each group in the TCGA cohort before determining the prognostic value of the ICI score in the TCGA cohort and other independent datasets. To evaluate this, first, we selected CD274, CTLA4, HAVCR2, IDO1, LAG3, and PDCD1 as immune-checkpoint-relevant signatures, and CD8A, CXCL10, CXCL9, GZMA, GZMB, IFNG, PRF1, TBX2, and TNF as immune-activity-related signatures.18,25 We observed that most of the immune-checkpoint-relevant and immune-activity-relevant genes except CD274, TBX2, HAVCR2, and TNF were significantly overexpressed in the high ICI group as demonstrated by the Wilcoxon test (Figures 3B and 3C). Besides, the gene set enrichment analysis (GSEA) revealed that the WNT and TGF-β signaling pathways were significantly enriched in the low ICI score group, whereas NK cell-mediated cytotoxicity, B cell receptor, and T cell receptor signaling pathways were enriched in the high ICI group (Figures 3D and 3E; Table S6).
Figure 3.
Construction of the ICI Scores
(A) Alluvial diagram of ICI gene cluster distribution in groups with different ICI clusters, ICI scores, and survival outcomes. (B) Immune-checkpoint-relevant genes (IDO1, CD274, HAVCR2, PDCD1, CTLA4, and LAG3) and immune-activation-relevant genes (CD8A, CXCL10, CXCL9, GZMA, GZMB, PRF1, IFNG, TBX2, and TNF) expressed in high and low ICI score subgroups. (C) Enrichment plots showing ECM receptor interaction, focal adhesion, TGF-β, tight junction, and Wnt signaling pathways in the low ICI score subgroup. (D) Enrichment plots showing T cell receptor signaling pathway, B cell receptor signaling pathway, natural killer cell-mediated cytotoxicity, drug metabolism other enzymes, and arachidonic acid metabolism in the high ICI score subgroup. (E) Kaplan-Meier curves for high and low ICI score groups in the TCGA-HNSC cohort. Log rank test, p < 0.001. (F) Kaplan-Meier curves for patients in the TCGA-HNSC cohort stratified by both receipt of adjuvant chemotherapy (Ct) and ICI score. Log rank test, p = 0.7521, high ICI score no Ct versus Ct; log rank test, p = 0.7384, low ICI score no Ct versus Ct. (G) Kaplan-Meier curves for patients in the TCGA-HNSC cohort stratified by both receipt of adjuvant radiotherapy (Rt) and ICI scores. Log rank test, p = 0.2545, high ICI score no Rt versus Rt; log rank test, p = 0.0307, low ICI score no Rt versus Rt.
The subsequent analysis involved the evaluation of the prognostic implications of the ICI scores. ICI score subgroups analysis done using the Kaplan-Meier plotter showed that the patients in the high ICI score group (median survival time 1,972 days) had significantly better OS rate than the low ICI score group (median survival time 1,972 days; log rank test, p < 0.0001; Figure 3F). Moreover, the prognostic efficiency of the ICI score was validated in GenBank: GSE41613 (n = 97), GenBank: GSE65858 (n = 270), and the total HNSC cohort (n = 940; log rank test; Figures S3A–S3D). However, because of inadequate clinical data, we were not able to establish a statistically significant correlation in GenBank: GSE42743 (n = 79). We also evaluated the effect of adjuvant therapy on the prognosis of each ICI subgroup. We found that the survival advantage in the low ICI score group was evident in the patients who received adjuvant radiotherapy (log rank test; Figures 3G and 3H).
The Correlation between the ICI Scores and Somatic Variants
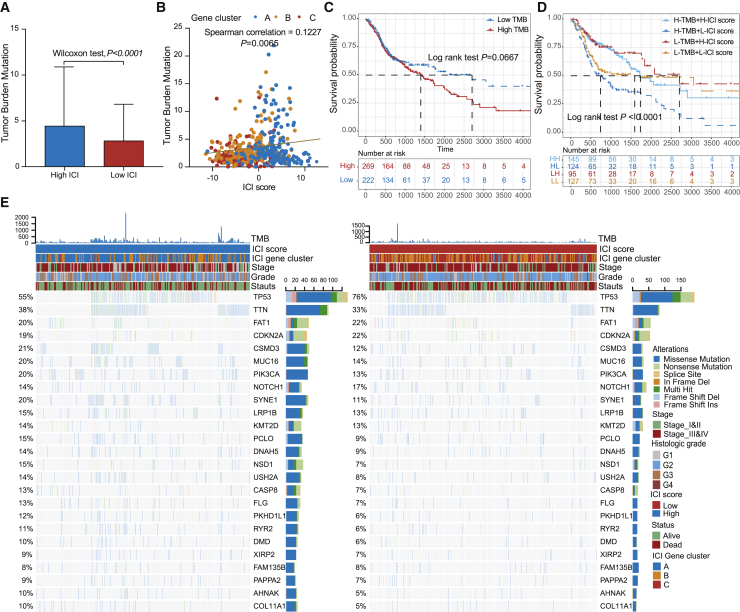
A myriad of evidence has demonstrated that tumors harboring high mutation burden (non-synonymous variants) were associated with increased CD8+ T cell infiltration in tumor tissue, which recognizes and eliminates these tumors. It implies that tumor burden mutation (TMB) might determine the individual’s response to cancer immunotherapy.26,27 An increased TMB has been correlated to an improved response to PD-1 blockades and prolonged progressive free survival in the KEYNOTE 012 clinical trial.3,28 Considering the significant clinical implications of TMB, we sought to explore the intrinsic correlation between the TMB and ICI scores to elucidate the genetic imprints of each ICI subgroup. To execute this, we first compared the TMB of patients with high ICI and low ICI score groups. As shown in Figure 4A, patients in the high ICI score subgroup showed a significantly higher TMB than patients in the low ICI score subgroup (Wilcoxon test p < 0.001). Further correlation analyses confirmed that the ICI score was significantly and positively correlated with the TMB (Spearman coefficient: R = 0.1227, p = 0.0065; Figure 4B). Next, we categorized the patients into discrete subgroups based on the immune set point of TMB, as described previously.28 As demonstrated in Figure 4A, we found that patients with low TMB showed better OS than the high TMB (log rank test, p = 0.0667). Taking into account the contraindicatory prognostic value of TMB and ICI scores, we next evaluated the synergistic effect of these scores in prognostic stratification of HNSC. Stratified survival analysis revealed that the TMB status did not interfere with ICI scores-based predictions. ICI score subtypes showed significant survival differences in both high and low TMB subgroups (log rank test, High TMB & High ICI score (HH) versus High TMB & Low ICI score (HL), p < 0.0001; Low TMB & Low ICI score (LH) versus Low TMB & Low ICI score (LL), p = 0.0203; Figure 4D). Collectively, these findings indicate that the ICI score might serve as an underlying predictive indicator that is independent of TMB and effectively measures the response to immunotherapy.
Figure 4.
The Correlation between the ICI Score and Somatic Variants
(A) TMB difference in the high and low ICI score subgroups. Wilcoxon test, p < 0.0001. (B) Scatterplots depicting the positive correlation between ICI scores and mutation load in the TCGA-HNSC cohort. The Spearman correlation between ICI scores and mutation load is shown (p = 0.0058). (C) Kaplan-Meier curves for high and low TMB groups of the TCGA-HNSC cohort. Log rank test, p = 0.0067. (D) Kaplan-Meier curves for patients in the TCGA-HNSC cohort stratified by both TMB and ICI scores. Log rank test, p < 0.001. (E) The oncoPrint was constructed using high ICI scores on the left (red) and low ICI scores on the right (blue). Individual patients are represented in each column.
Furthermore, we assessed the distribution of somatic variants in HNSC driver genes between the low and high ICI subgroups. The HNSC driver genes were accessed by using the maftools.29 The top 25 driver genes with the highest alteration frequency were further analyzed (Figure 4E). Analysis of the mutation annotation files of the TCGA cohort revealed that the alteration frequency of TP53, NSD1, CSMD3, SYNE1, PKHD1L1, USH2A, PIK3CA, CASP8, FLG, PCLO, AHNAK, COL11A1, and RYR2 was significantly different between the low and high ICI score groups (chi-square test; Table 1). These outcomes might provide novel ideas for investigating the mechanism of tumor ICI composition and gene mutation in immune checkpoint blockade therapy.
Table 1.
Association of ICI Scores with Somatic Variants
| Gene Symbol | High ICI Score (%) | Low ICI Score (%) | p Value |
|---|---|---|---|
| TP53 | 132 (55) | 191 (76) | <0.0001 |
| NSD1 | 36 (15) | 18 (7) | 0.0040 |
| CSMD3 | 50 (21) | 30 (12) | 0.0051 |
| SYNE1 | 48 (20) | 28 (11) | 0.0058 |
| PKHD1L1 | 29 (12) | 15 (6) | 0.0171 |
| USH2A | 34 (14) | 20 (8) | 0.0216 |
| PIK3CA | 48 (20) | 33 (13) | 0.0293 |
| CASP8 | 31 (13) | 18 (7) | 0.0342 |
| FLG | 31 (13) | 18 (7) | 0.0342 |
| PCLO | 36 (15) | 23 (9) | 0.0378 |
| AHNAK | 24 (10) | 13 (5) | 0.0400 |
| COL11A1 | 24 (10) | 13 (5) | 0.0400 |
| RYR2 | 26 (11) | 15 (6) | 0.0498 |
| MUC16 | 48 (20) | 35 (14) | 0.0550 |
| DNAH5 | 34 (14) | 23 (9) | 0.0683 |
| DMD | 24 (10) | 15 (6) | 0.0953 |
| TTN | 91 (38) | 83 (33) | 0.1833 |
| XIRP2 | 22 (9) | 18 (7) | 0.4110 |
| PAPPA2 | 22 (9) | 18 (7) | 0.4110 |
| NOTCH1 | 34 (14) | 43 (17) | 0.4601 |
| LRP1B | 36 (15) | 33 (13) | 0.5171 |
| CDKN2A | 46 (19) | 55 (23) | 0.5791 |
| KMT2D | 34 (14) | 33 (14) | 0.6941 |
| FAT1 | 48 (20) | 55 (22) | 0.7413 |
| FAM135B | 19 (8) | 20 (8) | >0.9999 |
p value was obtained from the chi-square test between different ICI score subtypes.
The Role of ICI Scores in the Prediction of Immunotherapeutic Benefits
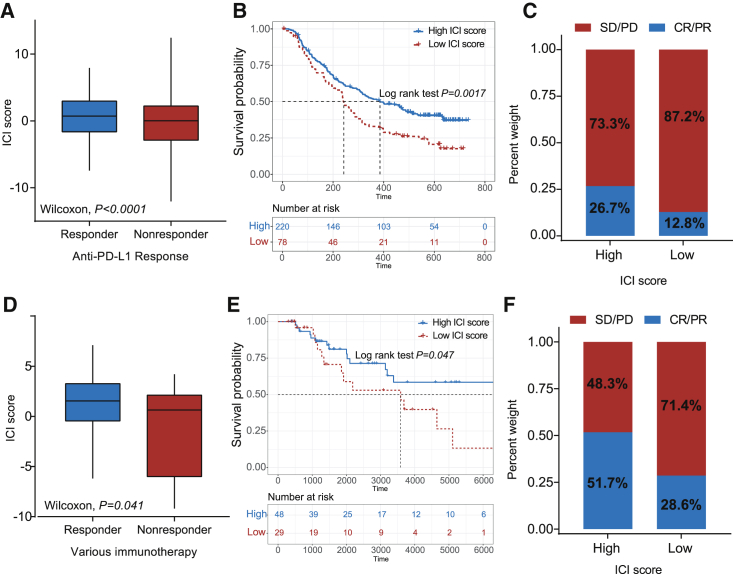
Emerging immune checkpoint blocking therapy, which is used in cancer treatment to block the T cell inhibitory molecules in cancer treatment, has shown astounding outcomes with the potential in improving diseased conditions in advanced cancers, but it is not effective in all patients.3,4 In the subsequent analysis, we examined the utility of the ICI score in speculating the therapeutic benefit in patients. To conduct this, the patients who received anti-PD-L1 immunotherapy in the IMvigor210 cohort were assigned high or low ICI scores. Notably, patients with high ICI scores significantly outlived patients with low ICI scores in the IMvigor210 cohort (log rank test, p = 0.0017; Figure 5A). The objective response rate of anti-PD-L1 therapy was higher in the high ICI score group than in the low ICI group in the IMvigor210 cohort (chi-square test, p = 0.0143; Figure 5B). We also found that higher ICI scores are correlated with objective response to anti-PD-L1 therapy in the IMvigor210 cohort (Wilcoxon test, p < 0.0001; Figure 5C). A similar outcome was observed in the TCGA-SKCM cohort, which received distinct immunotherapies, such as cytokines, vaccines, and checkpoint blockers (log rank test, p = 0.0470; Figure 5D; chi-square test, p = 0.0143, Figure 5E; chi-square test, p = 0.1520, Figure 5F). Collectively, these data indicate that ICI scores could be correlated to response to immunotherapy.
Figure 5.
The Role of ICI Scores in the Prediction of Immunotherapeutic Benefits
(A) ICI scores in groups with a different anti-PD-1 clinical response status. Wilcoxon test, p < 0.0001. (B) Kaplan-Meier curves for patients with high and low ICI scores in the IMvigor210 cohort. Log rank test, p = 0.0017. (C) Rate of clinical response (complete response [CR]/partial response [PR] and stable disease [SD]/progressive disease [PD]) to anti-PD-L1 immunotherapy in high or low ICI score groups in the IMvigor210 cohort. (D) Distribution of ICI score in different response status to immunotherapy in the TCGA-SKCM cohort. Wilcoxon test, p = 0.041. (E) Kaplan-Meier curves for patients with high and low ICI scores in the TCGA-SKCM cohort. Log rank test, p = 0.047. (F) Rate of clinical response (CR/PR and SD/PD) to various immunotherapies in high or low ICI score groups in the TCGA-SKCM cohort.
Discussion
The early clinical trials of immunotherapy have demonstrated its high efficacy in tumorigenic growth eradication and improvement in the quality of life in patients with advanced HNSC. Due to these factors, the US Food Drug Administration and the European Medicines Agency have approved pembrolizumab as the front-line treatment in subjects with unresectable recurrent/metastatic HNSC.3 A significant limitation of immunotherapy is that only a minority of patients have benefitted from it. Even the Society for Immunotherapy of Cancer, who issued the first guidelines on immunotherapy for the treatment of HNSC, has emphasized that appropriate patients for immunotherapy should be identified.30 In this study, we established a methodology to quantify the comprehensive tumor immune milieu in HNSC. The outcome of our study revealed that the ICI score is an efficient prognostic biomarker and predictive indicator in assessing the response to immunotherapy.
Mounting evidence has identified that the immune cell dysfunction within the HNSC-TME promotes immunosuppression and thus the associated tumor survival and progression. It later necessitates the therapeutic intervention.31,32 In this study, we analyzed the ICI in a meta-cohort of 1,029 HNSC samples and categorized the HNSC into three distinct immune subtypes. Our analysis indicated that densities of CD4+ T cells, CD8+ T cells, plasma cells, and M1 macrophages and a high immune score were correlated to the patient’s prognosis, which is in line with the previous studies.33,34 This emphasizes the fact that the pre-existing immune response has an anti-tumor effect and positively affects the response to immunotherapy. A few pioneering clinical and genomic studies17,20,35 reported that HNSC is one of the tumors that is highly infiltrated with immune cells. However, less than 20% of HNSC patients responded to immunotherapy as compared with patients with other tumor types with lower immune infiltration.3,36 This insinuates that even the immune phenotypes in the tumor cannot absolutely predict the response to immunotherapy. The molecular analysis of HNSC has identified a series of cytokines, chemokines, and other TME components that determine the host’s ability to exercise anti-tumor immune response. These molecular alterations during tumorigenesis may disturb the intercellular communication between the infiltrating immune cells, and thus tip the balance between immunity tolerance and activity.16
In the current study, we hypothesized that the comprehensive characterization of the ICI profiles and immune-related gene expression pattern would be a novel approach in strategizing the patient-specific tailored therapy. Our primary concern was the molecular characterization of HNSC-TME that modulates the immune system, and so we first fetched the immune-related genes based on previous and present novel ICI gene clusters. Within these gene clusters, we found that ICI gene cluster B had the lowest immune score, stromal score, and other immune-response-related cells, which suggests an immune-cold phenotype. Conversely, the ICI gene clusters A and C exhibited relatively high immune scores and inflammatory cell density. Also, we observed that a high stromal score was associated with increased infiltration of TAMs and resting DCs in ICI gene cluster C, implying a humoral immune response in this cluster.24,37 Additionally, the ICI gene cluster A had a more favorable immune-activated phenotype with the highest density of CD8+ T cells, activated CD4+ T cells, and plasma cells.38,39 On the other hand, the impact of the TME on patients’ OS was well documented by the previous studies.13,40 In line with these findings, our results revealed that the humoral immune response in the ICI gene cluster C and immune-cold phenotype in ICI gene cluster B were associated with a poor prognosis that could trigger tumor cell evasion from the immune system and impart resistance to immunotherapy. The anti-tumor immune response in the ICI gene cluster A was associated with a favorable prognosis, and we speculated that the patients in ICI gene cluster A might benefit from immunotherapy. The outcome of our analysis is in line with the previous studies, which indicate that the gene clusters in the current study might lead to the development of more precise immunotherapy.
Considering the individual heterogeneity of immune milieu, it was urgently demanded to quantify the ICI patterns of individual tumors. The individual-based model derived from tumor subtype-specific biomarkers has been well established in breast and colorectal cancers to improve outcome prediction.41,42 In the current study, with the help of the Boruta algorithm, we obtained potential “subtypes biomarkers” and established an ICI score to quantify the ICI pattern. Through GSEA, we found genes that were involved in the immunosuppressive pathways, such as TGF-β and WNT signaling pathways, and these genes were enriched explicitly in the low ICI score groups. Recently, the preclinical reports have identified an association between the gene mutations and response or tolerance to immunotherapy.43,44 An integrated ICI score at the genome level revealed the significant variant frequency differences in multiple genes between the high and low ICI scores, and few of these genes were explicitly associated with sensitivity or resistance. Besides, we also detected that the TMB, which is more sensitive to immunotherapy, was significantly reduced in patients with low ICI scores. The correlation between the ICI score and TMB was found to be 0.1227. The stratified analysis revealed that the prognosis value of ICI scores was independent of TMB in HNSC. The lack of correlation, coupled with the observed individual predictive values and GSEA outcome, implies that the ICI score and TMB represent distinct aspects of tumor immunobiology and can predict patient response to immunotherapy independently of TMB.
The patients receiving immunotherapy were assessed by the IMvigor210 and TCGA-SKCM, and we found that the ICI score was significantly elevated in patients responding to immunotherapy, which validated its predictive value. Overall, this suggests that single-agent immunotherapy might be beneficial for the patients with high ICI scores. Taking into account the activity of the TGF-β signaling pathway in low ICI score subtypes, TGF-β inhibition coupled with immune checkpoint blockade might be beneficial for patients with low ICI scores.45,46 In this context, previous preclinical studies have confirmed that the synergistic therapeutic effect of the TGF-β inhibitor and immune checkpoint inhibitor were more efficient than single-agent immunotherapy for solid tumors. Moreover, there is an ongoing phase 1b/2 clinical trial (ClinicalTrials.gov: NCT02423343) to test the therapeutic effects of the combined application of TGF-β and nivolumab in advanced solid tumors. Furthermore, the findings of the current investigation demand clinical trial-based validation in a larger HNSC cohort receiving immunotherapy.
In summary, we comprehensively analyzed the ICI landscape of HNSC, providing a clear picture of the anti-/pro-tumor immune response regulation in HNSC. The difference in ICI patterns was found to be correlated to tumor heterogeneity and treatment complexity. Thus, systematic evaluation of tumor ICI patterns carried out by the current study has crucial clinical implications. Also, it can facilitate the identification of ideal candidates for tailoring optimal immunotherapeutic strategies.
Materials and Methods
HNSC Datasets and Samples
A total of 1,029 HNSC samples datasets (TCGA-HNSC was derived from TCGA database GSE41613, GSE65858, GSE42743 and E-MTAB-1328 were derived from Array express database) were procured from five publicly available datasets. The detailed information of 1,029 HNSC patients was shown in Table S7. The RNA sequencing (RNA-seq; Fragments Per Kilobase Million [FPKM] value) data of TCGA-HNSC datasets were downloaded from the University of California Santa Cruz (UCSC) Xena browser (Genomic Data Commons [GDC] hub: https://xenabrowser.net/datapages/?hub=https://gdc.xenahubs.net:443, accessed June 15, 2019). The microarray datasets (GenBank: GSE65858, GSE41613, and GSE42743 and E-MTAB-1328) were downloaded from the Array Express database (https://www.ebi.ac.uk/arrayexpress, accessed June 15, 2019). Out of these five datasets, OS data for TCGA-HNSC and GenBank: GSE65858, GSE42743, and GSE41613 were available. As described previously, the expression profile (FPKM values) of TCGA-HNSC datasets was transformed into TPMs (transcripts per kilobase million), which was identical to those resulting from microarrays.47 The “ComBat” algorithm was applied to reduce the likelihood of batch effects from non-biological technical biases between different datasets.48
Consensus Clustering for Tumor-Infiltrating Immune Cells
Infiltration levels for distinct immune cells in HNSC were quantified by using “CIBERSORT” R package21 and employing the LM22 signature and 1,000 permutations. ESTIMATE evaluated the immune and stromal contents (immune and stromal score) for each HNSC sample.20 The hierarchical agglomerative clustering of HNSC was executed as per the ICI pattern of each sample. The unsupervised clustering “Pam” method based on Euclidean and Ward’s linkage was used in this analysis, executed by using the “ConsensuClusterPlus” R package,23 and repeated 1,000 times to ensure the classification stability.
DEGs Associated with the ICI Phenotype
Patients were grouped into the ICI clusters based on immune-cell infiltration to identify genes associated with the ICI patterns. DEGs among ICI subtypes were determined by setting significance cutoff criteria to p < 0.05 (adjusted) and absolute fold-change > 1.4, which was implemented by employing the limma R package.
Dimension Reduction and Generation of ICI Score
First, unsupervised clustering was employed to categorize the patients in TCGA as per the DEG values. Furthermore, DEG values that were positively and negatively correlated with the clusters signature were termed as the ICI gene signatures A and B, respectively. Furthermore, the Boruta algorithm was employed for the dimension reduction of the ICI gene signatures A and B,22 and principal component 1 was extracted as the signature score by employing the PCA. Lastly, we applied a method similar to Gene expression grade index49 to define the ICI score of each patient:
Collection of Somatic Alteration Data
The corresponding mutation data of patients in the TCGA-HNSC cohort were downloaded from TCGA data portal (https://www.cancer.gov/tcga/). To determine the mutational burden of HNSC, we counted the total number of non-synonymous mutations in HNSC. The somatic alterations in HNSC driver genes were evaluated for the high or low ICI scores. The HNSC driver genes were identified using “maftool” R package.29 The top 25 driver genes with the highest alteration frequency were further analyzed.
Gene Expression Data with Immunotherapy
Two independent datasets, IMvigor210 and TCGA-SKCM, were downloaded and analyzed to determine the predictive value of the ICI scores. The IMvigor210 dataset was downloaded from a freely available, fully documented software and data package, under the Creative Commons 3.0 license that can be downloaded from http://research-pub.gene.com/IMvigor210CoreBiologies. The TCGA-SKCM dataset, the expression profiles, and related-clinical information were downloaded from the UCSC Xena browser (GDC hub: https://xenabrowser.net/datapages/?hub=https://gdc.xenahubs.net:443). A total of 298 urothelial cancer cases with complete clinical information and 80 skin melanoma cases who received immunotherapy were analyzed to determine the ICI scores.
Statistical Analyses
All statistical analyses were conducted using GraphPad Prism version 7.0 or SPSS version 21.0 (IBM Corporation, Armonk, NY, USA) software. The Kruskal-Wallis test was used to compare more than two groups, and the Wilcoxon test was used to compare two groups. The X-tile software, which iteratively tests possible cut points to select the one with the maximum rank statistic, was used to classify patients into two subtypes in each dataset and to reduce the computational batch effect.50 The Kaplan-Meier plotter was employed to generate survival curves for the subgroups in each dataset. The log rank test evaluated the statistically significant differences. The chi-square test analyzed the correlation between the ICI score subgroups and somatic mutation frequency, and the Spearman analysis computed the correlation coefficient. Two-tailed p < 0.05 was considered as statistically significant.
Author Contributions
B.Z. conceived and designed the whole project and drafted the manuscript. X.Z. and M.S. analyzed the data and wrote the manuscript. X.Z. carried out data interpretations and helped data discussion. T.C. provided specialized expertise and collaboration in data analysis. M.S. revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.08.030.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Shield K.D., Ferlay J., Jemal A., Sankaranarayanan R., Chaturvedi A.K., Bray F., Soerjomataram I. The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J. Clin. 2017;67:51–64. doi: 10.3322/caac.21384. [DOI] [PubMed] [Google Scholar]
- 3.Seiwert T.Y., Burtness B., Mehra R., Weiss J., Berger R., Eder J.P., Heath K., McClanahan T., Lunceford J., Gause C. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 4.Curran M.A., Montalvo W., Yagita H., Allison J.P. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc. Natl. Acad. Sci. USA. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garassino M.C., Gadgeel S., Esteban E., Felip E., Speranza G., Domine M., Hochmair M.J., Powell S., Cheng S.Y., Bischoff H.G. Patient-reported outcomes following pembrolizumab or placebo plus pemetrexed and platinum in patients with previously untreated, metastatic, non-squamous non-small-cell lung cancer (KEYNOTE-189): a multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21:387–397. doi: 10.1016/S1470-2045(19)30801-0. [DOI] [PubMed] [Google Scholar]
- 6.Robert C., Ribas A., Wolchok J.D., Hodi F.S., Hamid O., Kefford R., Weber J.S., Joshua A.M., Hwu W.J., Gangadhar T.C. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet. 2014;384:1109–1117. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 7.Hamid O., Robert C., Daud A., Hodi F.S., Hwu W.J., Kefford R., Wolchok J.D., Hersey P., Joseph R.W., Weber J.S. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng D., Zhou R., Yu Y., Luo Y., Zhang J., Sun H., Bin J., Liao Y., Rao J., Zhang Y., Liao W. Gene expression profiles for a prognostic immunoscore in gastric cancer. Br. J. Surg. 2018;105:1338–1348. doi: 10.1002/bjs.10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y., Zhang Q., Hu Y., Li T., Yu J., Zhao L., Ye G., Deng H., Mou T., Cai S. ImmunoScore Signature: A Prognostic and Predictive Tool in Gastric Cancer. Ann. Surg. 2018;267:504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 11.Noy R., Pollard J.W. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Palma M., Lewis C.E. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y.P., Wang Y.Q., Lv J.W., Li Y.Q., Chua M.L.K., Le Q.T., Lee N., Colevas A.D., Seiwert T., Hayes D.N. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann. Oncol. 2019;30:68–75. doi: 10.1093/annonc/mdy470. [DOI] [PubMed] [Google Scholar]
- 14.Vassilakopoulou M., Avgeris M., Velcheti V., Kotoula V., Rampias T., Chatzopoulos K., Perisanidis C., Kontos C.K., Giotakis A.I., Scorilas A. Evaluation of PD-L1 Expression and Associated Tumor-Infiltrating Lymphocytes in Laryngeal Squamous Cell Carcinoma. Clin. Cancer Res. 2016;22:704–713. doi: 10.1158/1078-0432.CCR-15-1543. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg J.E., Hoffman-Censits J., Powles T., van der Heijden M.S., Balar A.V., Necchi A., Dawson N., O’Donnell P.H., Balmanoukian A., Loriot Y. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 17.Şenbabaoğlu Y., Gejman R.S., Winer A.G., Liu M., Van Allen E.M., de Velasco G., Miao D., Ostrovnaya I., Drill E., Luna A. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R., Albright A., Cheng J.D., Kang S.P., Shankaran V. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedberg M.L., Goh G., Chiosea S.I., Bauman J.E., Freilino M.L., Zeng Y., Wang L., Diergaarde B.B., Gooding W.E., Lui V.W. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J. Clin. Invest. 2016;126:169–180. doi: 10.1172/JCI82066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kursa M.B., Rudnicki W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010;36:1–13. [Google Scholar]
- 23.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas S.K., Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 25.Hugo W., Zaretsky J.M., Sun L., Song C., Moreno B.H., Hu-Lieskovan S., Berent-Maoz B., Pang J., Chmielowski B., Cherry G. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGranahan N., Furness A.J., Rosenthal R., Ramskov S., Lyngaa R., Saini S.K., Jamal-Hanjani M., Wilson G.A., Birkbak N.J., Hiley C.T. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J., Sher X., Liu X.Q., Lu H., Nebozhyn M. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen E.E.W., Bell R.B., Bifulco C.B., Burtness B., Gillison M.L., Harrington K.J., Le Q.T., Lee N.Y., Leidner R., Lewis R.L. The Society for Immunotherapy of Cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J. Immunother. Cancer. 2019;7:184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czystowska M., Gooding W., Szczepanski M.J., Lopez-Abaitero A., Ferris R.L., Johnson J.T., Whiteside T.L. The immune signature of CD8(+)CCR7(+) T cells in the peripheral circulation associates with disease recurrence in patients with HNSCC. Clin. Cancer Res. 2013;19:889–899. doi: 10.1158/1078-0432.CCR-12-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis R.J., Van Waes C., Allen C.T. Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and Tregs as mediators of the immunosuppressive microenvironment in head and neck cancer. Oral Oncol. 2016;58:59–70. doi: 10.1016/j.oraloncology.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y., Jiang Z., Chen C., Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J. Exp. Clin. Cancer Res. 2018;37:327. doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandal R., Şenbabaoğlu Y., Desrichard A., Havel J.J., Dalin M.G., Riaz N., Lee K.W., Ganly I., Hakimi A.A., Chan T.A., Morris L.G. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829. doi: 10.1172/jci.insight.89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarchoan M., Hopkins A., Jaffee E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Probst H.C., McCoy K., Okazaki T., Honjo T., van den Broek M. Resting dendritic cells induce peripheral CD8+ T cell tolerance through PD-1 and CTLA-4. Nat. Immunol. 2005;6:280–286. doi: 10.1038/ni1165. [DOI] [PubMed] [Google Scholar]
- 38.Hamanishi J., Mandai M., Iwasaki M., Okazaki T., Tanaka Y., Yamaguchi K., Higuchi T., Yagi H., Takakura K., Minato N. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc. Natl. Acad. Sci. USA. 2007;104:3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang M.L., Lukens J.R., Bullock T.N. Cognate memory CD4+ T cells generated with dendritic cell priming influence the expansion, trafficking, and differentiation of secondary CD8+ T cells and enhance tumor control. J. Immunol. 2007;179:5829–5838. doi: 10.4049/jimmunol.179.9.5829. [DOI] [PubMed] [Google Scholar]
- 40.Li B., Cui Y., Nambiar D.K., Sunwoo J.B., Li R. The Immune Subtypes and Landscape of Squamous Cell Carcinoma. Clin. Cancer Res. 2019;25:3528–3537. doi: 10.1158/1078-0432.CCR-18-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callari M., Cappelletti V., D’Aiuto F., Musella V., Lembo A., Petel F., Karn T., Iwamoto T., Provero P., Daidone M.G. Subtype-Specific Metagene-Based Prediction of Outcome after Neoadjuvant and Adjuvant Treatment in Breast Cancer. Clin. Cancer Res. 2016;22:337–345. doi: 10.1158/1078-0432.CCR-15-0757. [DOI] [PubMed] [Google Scholar]
- 42.Bramsen J.B., Rasmussen M.H., Ongen H., Mattesen T.B., Ørntoft M.W., Árnadóttir S.S., Sandoval J., Laguna T., Vang S., Øster B. Molecular-Subtype-Specific Biomarkers Improve Prediction of Prognosis in Colorectal Cancer. Cell Rep. 2017;19:1268–1280. doi: 10.1016/j.celrep.2017.04.045. [DOI] [PubMed] [Google Scholar]
- 43.George S., Miao D., Demetri G.D., Adeegbe D., Rodig S.J., Shukla S., Lipschitz M., Amin-Mansour A., Raut C.P., Carter S.L. Loss of PTEN Is Associated with Resistance to Anti-PD-1 Checkpoint Blockade Therapy in Metastatic Uterine Leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burr M.L., Sparbier C.E., Chan Y.C., Williamson J.C., Woods K., Beavis P.A., Lam E.Y.N., Henderson M.A., Bell C.C., Stolzenburg S. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravi R., Noonan K.A., Pham V., Bedi R., Zhavoronkov A., Ozerov I.V., Makarev E., V Artemov A., Wysocki P.T., Mehra R. Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat. Commun. 2018;9:741. doi: 10.1038/s41467-017-02696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan Y., Zhang D., Xu C., Hance K.W., Marelli B., Qi J., Yu H., Qin G., Sircar A., Hernández V.M. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci. Transl. Med. 2018;10:eaan5488. doi: 10.1126/scitranslmed.aan5488. [DOI] [PubMed] [Google Scholar]
- 47.Wagner G.P., Kin K., Lynch V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 48.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 49.Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 50.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.