Abstract
Background
Hepatocellular carcinoma (HCC) is a common lethal malignant tumor worldwide. Circular RNAs (circRNAs) have been reported to affect the development of human cancers, including HCC. In this project, we aim to clarify the functional effect of circular CDR1as (circ_CDR1as) on HCC progression.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot is implemented to detect the expression of circ_CDR1as, microRNA (miR)-1287 and Raf-1 proto-oncogene, serine/threonine kinase (Raf1). Cell proliferation is assessed via colony formation and 3-(4, 5)-dimethylthiazole-2-y1)-2, 5-biphenyl tetrazolium bromide (MTT) assays. Cell migration and invasion are measured by Transwell assay. The target relationship between miR-1287 and circ_CDR1as or Raf1 is validated through dual-luciferase reporter assay. The levels of epithelia–mesenchymal transition (EMT) markers and the MEK/ERK signal pathway-related proteins are examined by Western blot. Model in nude mice is constructed to determine the role of circ_CDR1as in vivo.
Results
Expression of circ_CDR1as and Raf1 is elevated, while miR-1287 expression is decreased in HCC. Depletion of circ_CDR1as or Raf1 could inhibit proliferation and metastasis of HCC cells. Besides, circ_CDR1as regulates Raf1 expression by targeting miR-1287. MiR-1287 upregulation or Raf1 depletion could partially counterbalance circ_CDR1as depletion-mediated inhibitory effects on HCC cell behaviors. Moreover, circ_CDR1as depletion represses HCC progression through inactivating MEK/ERK pathway. In addition, circ_CDR1as depletion suppresses tumor growth in vivo via regulating miR-1287/Raf1 pathway.
Conclusion
Circ_CDR1as depletion inhibits HCC cell proliferation and metastasis by miR-1287/Raf1 and MEK/ERK pathways, highlighting a promising molecular target for HCC treatment.
Keywords: HCC, circ_CDR1as, Raf1, miR-1287, proliferation, metastasis, MEK/ERK pathway
Introduction
With a high mortality, hepatocellular carcinoma (HCC) ranks the most prevalent human malignancy derived from liver.1 Nowadays, operative excision, liver transplantation and radiotherapy are conventional therapeutic options, while increasing cognition about hepatocarcinogenesis highlights the potential of targeted therapy.2,3 Therefore, clarifying the molecular mechanism in HCC progression is meaningful for the treatment of HCC.
Circular RNAs (circRNAs) are a group of circular, closed, non-coding RNA molecules without 5ʹ end cap and 3ʹ end poly(A) tail.4 Numerous studies have reported the dysregulated expression of circRNAs in many human diseases, including cancers.5,6 CircRNAs are reported to be closely relevant to tumor occurrence and development, functioning as potential biomarkers.7 Certain circRNAs have been proved to take part in the pathogenesis of HCC, such as circMTO1,8 circC3P19 and circFBLIM1.10 As for circ_CDR1as, it has been identified to be an oncogene to accelerate HCC progression.11 However, the specific actin mechanism of circ_CDR1as in HCC development has not been fully illuminated yet.
Similar to circRNA, microRNAs (miRNAs) are also non-coding RNAs, with 22 nucleotides in length, which could function as key modulators in animals and plants via binding to mRNAs.12 MiRNAs are said to be involved in the development of many solid tumors, such as HCC.13 MiRNA-1287 (miR-1287) has been documented to play as a tumor suppressor in HCC.14 Having known that miR-1287 is a potential target of circ_CDR1as (predicted utilizing starBase), we intend to explore the impact of miR-1287 on the circ_CDR1as-mediated HCC progression, so as to better understand the role of circ_CDR1as in HCC.
Raf-1 proto-oncogene, serine/threonine kinase (Raf1) is a vital signaling molecule, involved in the modulation of cellular proliferation and differentiation.15,16 In HCC, Raf1 is related to hepatocarcinogenesis, and could repress its progression.17 In addition, Raf1 is an important ingredient of RAS/RAF/MEK/ERK signal pathway.16 Here, we analyze the role of Raf1 (a target of miR-1287 forecasted via starBase) in the circ_CDR1as-mediated HCC development.
In our work, the dysregulation of circ_CDR1as in HCC tissues and cells is detected. Additionally, functional role of circ_CDR1as in HCC cell proliferation, metastasis and tumorigenesis, and the potential mechanistic pathway are explored.
Materials and Methods
HCC Samples
One hundred and five pairs of HCC tissues (Tumor) and matched adjacent healthy tissues (Normal) are postoperatively procured from Affiliated Dongguan People’s Hospital, Southern Medical University of Dongguan. After resection, all tissues are preserved in a −80°C refrigerator. All participators involved in this assay submit written informed consent, and none of them have received any form of anti-tumor therapy before surgery. The current assay has obtained the authorization of the Ethics Committee of Affiliated Dongguan People’s Hospital, Southern Medical University of Dongguan.
Cell Culture
Human Liver Epithelial-2 (THLE-2, ATCC® CRL-2706) and HCC cell line Hep3B (ATCC® HB-8064) are purchased from American Type Culture Collection (ATCC; Manassas, VA, USA), HCC cell line Huh7 (BNCC337690) is bought from BeNa culture collection (Beijing, China). Above cells are maintained in Dulbecco’s Modified Eagle Medium (DMEM; Solarbio, Shanghai, China) supplemented with 10% (v/v) fetal bovine serum (FBS; Beyotime, Nantong, China) and 1% penicillin/streptomycin in a 5% CO2/95% air humidified incubator at 37°C.
Cell Transfection
To silence circ_CDR1as or Raf1, small interference RNA (siRNA) targeting circ_CDR1as (si-circ_CDR1as) or Raf1 (si-Raf1) and the negative control (si-NC) are synthesized by KeyGEN Biotech (Nanjing, China). For overexpression, overexpression vector of circ_CDR1as (circ_CDR1as) and Raf1 (Raf1) and their corresponding negative control Vector and pcDNA are supplied by Hanbio Biotechnology Co., ltd (Shanghai, China). Furthermore, miR-1287 mimic (miR-1287) and the negative control (miR-NC), miR-1287 inhibitor (anti-miR-1287) and the negative control (anti-miR-NC) are constructed by GeneCopoeia (Guangzhou, China). These oligonucleotides or plasmids are introduced into Hep3B and Huh7 cells using Lipofectamine 3000 (Solarbio) according to the recommended protocols.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA is extracted from cultured cells, HCC tissues and adjacent healthy tissues, as well as from formed tumors in nude mice using the RNA Isolation Kit (Beyotime) referring to the guideline supplied by the manufacturer. After reverse-transcription utilizing BeyoRT™ III M-MLV reverse transcriptase (Beyotime) or miRNA First-Strand Synthesis Kit (Clontech, Mountain View, CA, USA), qPCR is performed with SYBR Premix ExTaq kit (TaKaRa, Otsu, Japan) or miRNA qRT-PCR TB Green Kit (Clontech). The relative expression levels of miR-1287 (normalized to U6), circ_CDR1as and Raf1 (normalized to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) are calculated using the 2−ΔΔCt method. Sequences of all primers are: circ_CDR1as-forward primer, 5ʹ-TCAACTGGCTCAATATCCATGTC-3ʹ and circ_CDR1as-reverse primer, 5ʹ-ACCTTGACACAGGTGCCAT-3ʹ; Raf1-forward primer, 5ʹ-GGGAGCTTGGAAGACGATCAG-3ʹ and Raf1-reverse primer, 5ʹ-ACACGGATAGTGTTGCTTGTC-3ʹ; GAPDH-forward primer, 5ʹ-TCGACAGTCAGCCGCATCTTCTTT-3ʹ, and GAPDH-reverse primer, 5ʹ-ACCAAATCCGTTGACTCCGACCTT-3ʹ; miR-1287-forward primer, 5ʹ-GTGCTGGATCAGTGGTTC-3ʹ, and miR-1287-reverse primer 5ʹ-GTCCAGTTTTTTTTTTTTTTTGACTC-3ʹ; U6-forward primer, 5ʹ-CGCTTCGGCAGCACATATACTA-3ʹ and U6-reverse primer 5ʹ-CGCTTCACGAATTTGCGTGTCA-3ʹ.
Western Blot
Total protein is isolated from cultured cells, HCC tissues and adjacent healthy tissues, as well as formed tumors in nude mice utilizing the protein extraction kit (Solarbio) and quantified using bicinchoninic acid assay Kit (Beyotime). After separation through sodium dodecyl sulfate (SDS)-polyacrylamide gels (PAGE), protein samples are subjected to transfer onto polyvinylidene difluoride membranes (Pall Corporation, East Hills, NY, USA). Later, the membranes are blocked in defatted milk, incubated with special primary antibody against Raf1 (ab173539, 1:1500), snail (ab216347, 1:1000), E-cadherin (ab11512, 1:1000), MEK1 (ab32091, 1:1000), phosphorylated MEK1 (p-MEK1; ab96379, 1:1500), ERKs (ab218017, 1:1000), phosphorylated ERKs (p-ERKs; ab223500, 1:1000), or GAPDH (internal control; ab181602, 1:2000), then incubated with horseradish peroxidase-conjugated secondary antibody (ab205718, 1:5000). Visualization of protein blots is achieved using the enhanced chemiluminescence kit (Beyotime).
RNase R Digestion and Actinomycin D Treatment
For the validation of the stability of circ_CDR1as, 5 μg RNA isolated from Hep3B and Huh7 cells is disposed with RNase R (3 U/μg, Epicentre Biotechnologies, Shanghai, China) or not (Mock) at 37°C for 30 min. Expression of circ_CDR1as and GAPDH mRNA is examined by qRT-PCR.
Moreover, Actinomycin D (2 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) or dimethyl sulfoxide solution (DMSO; Sigma-Aldrich) is added to DMEM. After routine culture at 37°C for 4 h, 8 h, 12 h and 24 h, total RNA is extracted and the levels of circ_CDR1as and GAPDH are detected via qRT-PCR.
Nuclear-Cytoplasmic Fractionation Assay
To determine the subcellular location of circ_CDR1as, the PARIS™ Kit (Thermo Fisher Scientific, Carlsbad, CA, USA) is applied based on the user’s manual, with GAPDH (for cytoplasmic fraction) and U6 (for nuclear fraction) as internal controls. Eventually, the ratio of circ_CDR1as in the cytoplasmic or nuclear fraction of Hep3B and Huh7 cells is determined by qRT-PCR.
Colony Formation Assay and 3-(4, 5)-Dimethylthiazole-2-Y1)-2, 5-Biphenyl Tetrazolium Bromide (MTT) Assay
For colony formation assay, 200 cells per well are seeded into 6-well plate containing complete medium (DMEM plus 10% FBS) after digestion with trypsin. After culture for 12 days, formed visible colonies (cell mass harboring more than 50 cells) are fixed with methanol, dyed by crystal violet, and counted.
For MTT assay, treated Hep3B and Huh7 cells are plated onto 96-well plate at a density of 2 × 103 cells and maintained for 0 day, 1 day, 2 days or 3 days. These cells are cultured for another 4 h after the addition with 5 mg/mL MTT solution. The optimal density (OD) value of each well at 490 nm is measured utilizing a microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
Transwell Assay
This assay is applied to evaluate the metastasis ability of treated Hep3B and Huh7 cells. For invasion detection, insert chambers are enveloped with Matrigel (Solarbio). About 1×105 treated cells suspended in serum-free DMEM are plated onto the upper chambers, while the lower ones contain complete medium. 24 h later, cells invaded to the bottom of the insert are immobilized with polyoxymethylene and stained with crystal violet, then counted under a light microscope (×200 magnification).
As for migration examination, the experimental procedures are similar, except for the Matrigel-free insert chambers.
Bioinformatics and Dual-Luciferase Reporter Assay
Bioinformatic analyses for the miRNA directly interacted with circ_CDR1as and the downstream mRNA of the predicted miRNA are conducted utilizing online software starBase 3.0 (http://starbase.sysu.edu.cn/), a huge database usually used for predicting the target miRNAs of circRNAs and the target mRNAs of miRNAs.
To perform dual-luciferase reporter assay, luciferase reporters, including circ_CDR1as-wt, circ_CDR1as-mut, Raf1-wt and Raf1-mut, are constructed. Among them, circ_CDR1as-mut and Raf1-mut are severally derived from circ_CDR1as-wt and Raf1-wt through mutating the binding sites to miR-1287 using Quick-Change Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA). Hep3B and Huh7 cells are co-transfected with above luciferase reporter and miR-1287 or miR-NC utilizing Lipofectamine 3000 for 48 h. The luciferase activity is measured by the employment of Dual-Luciferase reporter system (Beyotime), following the recommended instructions.
Xenograft Tumor Assay
The lentiviral small hairpin RNA (shRNA) targeting circ_CDR1as (sh-circ_CDR1as) and the negative control (sh-NC) are synthesized by GenePharma Co. Ltd. (Shanghai, China). Then, Hep3B cells stably expressing sh-circ_CDR1as or sh-NC are constructed.
Approximately 2 × 106 Hep3B cells with stable transfection are subjected to hypodermic injection into the right flank of male nude mice (BALB/c, 5 weeks old, Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China)), called sh-circ_CDR1as group (n=4) and sh-NC group (n=4). At 7 days post injection, volume of generated tumors is monitored every 5 days, utilizing the formula: 0.5 × length × width2. Thirty-two days later, all mice are killed and generated tumors are resected for weighing and photographing. Subsequently, the expression levels of circ_CDR1as, miR-1287 and Raf1 are examined. The animal experiment is ratified by the Ethics Committee of Affiliated Dongguan People’s Hospital, Southern Medical University of Dongguan and performed according to the Guide for the Care and Use of Laboratory Animals (GB/T 35892–2018).
Statistical Analysis
All data in this project come from three independent experiments, and are shown as mean ± standard deviation. The statistical analysis is accomplished by Student’s t-test or one-way analysis of variance on SPSS 20.0 software (IBM Corp., Armonk, NY, USA). The significant difference is defined as a P value less than 0.05. The correlation analysis among the levels of circ_CDR1as, miR-1287 and Raf1 in HCC tissues is executed by Pearson correlation analysis.
Results
Circ_CDR1as and Raf1 are Highly Enriched in HCC Tissues
For the determination of the expression of circ_CDR1as and Raf1 in HCC, qRT-PCR is implemented. As shown in Figure 1A and B, circ_CDR1as and Raf1 mRNA are significantly upregulated in HCC tissues compared to adjacent normal tissues. Additionally, the upregulation of Raf1 in HCC tissues is also validated at protein level by Western blot analysis (Figure 1D). Moreover, the expression of circ_CDR1as and Raf1 is positively correlated in HCC tissues (Figure 1C). Collectively, circ_CDR1as and Raf1 embrace potential to function in HCC progression.
Figure 1.
Circ_CDR1as and Raf1 are highly enriched in HCC tissues. (A and B) QRT-PCR assay for the relative expression of circ_CDR1as and Raf1 mRNA in HCC tissues and adjacent normal tissues (n=105). (C) Pearson correlation analysis for the correlation between the levels of circ_CDR1as and Raf1 in HCC tissues, r=0.4849, P < 0.0001. (D) Western blot analysis for the level of Raf1 protein in HCC tissues and adjacent normal tissues. *P < 0.05.
Circ_CDR1as is a Stable circRNA
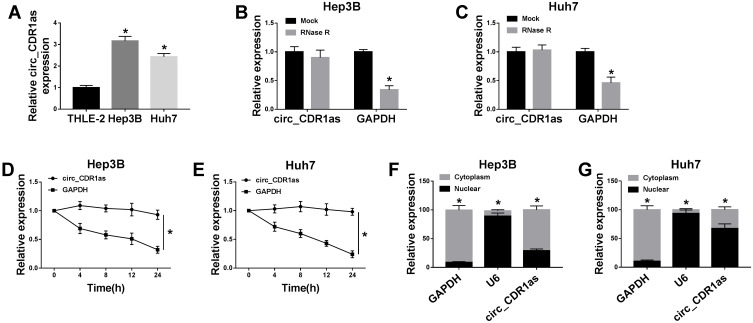
By preforming qRT-PCR assay, we find that circ_CDR1as expression is higher in Hep3B and Huh7 cells in contrast to THLE-2 cells (Figure 2A). In fact, circRNAs harbor covalently closed, single-stranded circular structure, making them resistant to the digestion of ribonucleases, like RNase R and conferring them a longer half-life than that of linear mRNAs.18 Obviously, the relative expression of GAPDH is declined in Hep3B and Huh7 cells treated with RNase R, when compared with cells in Mock group, but circ_CDR1as expression changes little, indicating that circ_CDR1as is resistant to RNase R digestion (Figure 2B and C). Analogously, circ_CDR1as is tolerant of Actinomycin D treatment, and is more stable than linear gene GAPDH (Figure 2D and E). CircRNAs are widely distributed in organisms, and the majority of circRNAs are located in the cytoplasm.19 As determined by Nuclear-cytoplasmic fractionation assay and qRT-PCR assay, circ_CDR1as is mainly located in the cytoplasmic fractionation of Hep3B and Huh7 cells (Figure 2F and G). Altogether, circ_CDR1as is a stable circRNA.
Figure 2.
Circ_CDR1as is a stable circRNA. (A) QRT-PCR assay for the relative expression of circ_CDR1as in THLE-2, Hep3B and Huh7 cells. (B and C) QRT-PCR assay for the relative expression of circ_CDR1as and GAPDH mRNA in Hep3B and Huh7 cells treated with RNase R or not. (D and E) QRT-PCR assay for the relative expression of circ_CDR1as and GAPDH mRNA in Hep3B and Huh7 cells disposed with Actinomycin D or not. (F and G) Nuclear-cytoplasmic fractionation assay for the relative expression of GAPDH, U6 and circ_CDR1as in the cytoplasmic and nuclear fractionations of Hep3B and Huh7 cells. *P < 0.05.
Circ_CDR1as Positively Regulates HCC Cell Proliferation and Metastasis
Having known that circ_CDR1as is dysregulated in HCC, we subsequently explore the functional role of circ_CDR1as in the cellular behaviors of HCC Hep3B and Huh7 cells. After transient transfection, gain-of-function and loss-of-function assays are performed. As validated in Figure 3A, the knockdown efficiency of si-circ_CDR1as and the overexpression efficiency of circ_CDR1as in Hep3B and Huh7 cells are all-right, with si-NC and Vector as controls, respectively. Following colony formation assay demonstrates that depletion of circ_CDR1as reduces the number of the colonies (Figure 3B), while introduction of circ_CDR1as increases the number of the colonies (Figure 3E). Besides, MTT assay discloses that circ_CDR1as depletion inhibits the OD value of transfected Hep3B and Huh7 cells at 490 nm (Figure 3C and D), while gain of circ_CDR1as triggers reverse result (Figure 3F and G). Meanwhile, the migration and invasion of transfected Hep3B and Huh7 cells are declined due to circ_CDR1as depletion (Figure 3H and I), while promoted by circ_CDR1as upregulation (Figure 3J and K), which are manifested via Transwell assay. Western blot analysis witnesses a lower level of snail and a higher level of E-cadherin in Hep3B and Huh7 cells with circ_CDR1as depletion (Figure 3L), as well as a higher level of snail and a lower level of E-cadherin in cells with circ_CDR1as overexpression (Figure 3M). From the above data, we conclude that depletion of circ_CDR1as suppresses HCC cell proliferation and metastasis; otherwise, circ_CDR1as upregulation contributes to HCC cell proliferation and metastasis.
Figure 3.
Circ_CDR1as positively regulates HCC cell proliferation and metastasis. Hep3B and Huh7 cells are transfected with si-NC, si-circ_CDR1as, Vector or circ_CDR1as. (A) QRT-PCR assay for the relative expression of circ_CDR1as in transfected cells. (B and E) Colony formation assay for the number of the colonies formed by transfected cells. (C, D, F and G) MTT assay for the OD value of transfected cells at 490 nm. (H–K) Transwell assay for the migration and invasion abilities of transfected cells. (L and M) Western blot analysis for the protein levels of snail and E-cadherin in transfected cells. *P < 0.05.
Raf1 Depletion Inhibits HCC Cell Proliferation and Metastasis
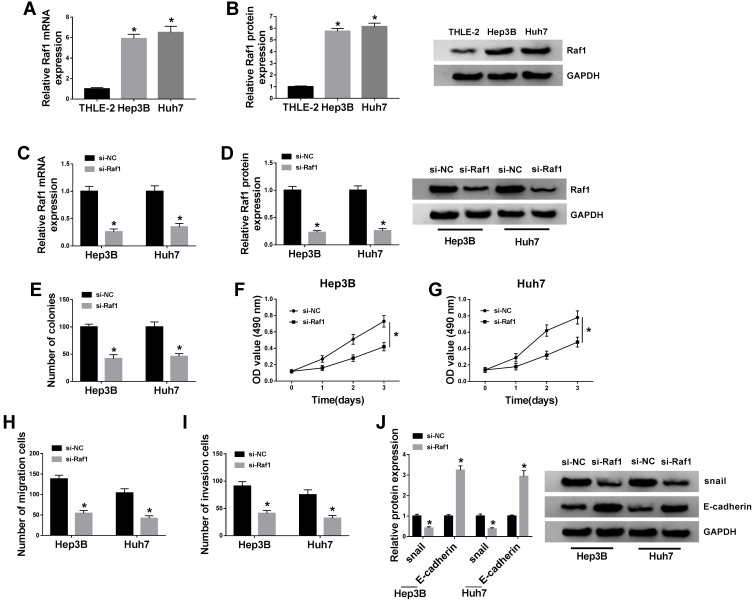
To figure out the enrichment of Raf1 in HCC cells, qRT-PCR and Western blot assays are performed, and confirm the upregulation of Raf1 in Hep3B and Huh7 cells, at both mRNA and protein levels (Figure 4A and B). Next, we conduct loss-of-function assay to clarify the function of Raf1 in HCC cell proliferation and metastasis. Hep3B and Huh7 cells are transfected with si-NC or si-Raf1, and following qRT-PCR and Western blot assays also prove the knockdown efficiency of si-Raf1 (Figure 4C and D). As illustrated in Figure 4E, deficiency of Raf1 weakens the colony formation ability of HCC cells. Moreover, MTT assay uncovers the inhibitory impact of si-Raf1 on the OD value of Hep3B and Huh7 cells (Figure 4F and G). Loss of Raf1 also hinders the migration and invasion abilities of transfected HCC cells (Figure 4H and I). Data from Western blot suggest that Raf1 depletion lowers the protein level of snail, while elevates E-cadherin level (Figure 4J). These results imply that Raf1 depletion represses HCC cell proliferation and metastasis.
Figure 4.
Raf1 depletion inhibits HCC cell proliferation and metastasis. (A and B) QRT-PCR and Western blot assays for the relative expression of Raf1 in THLE-2, Hep3B and Huh7 cells. (C–J) Hep3B and Huh7 cells are introduced with si-NC or si-Raf1. (C and D) QRT-PCR and Western blot assays for the relative expression of Raf1 in treated cells. (E) Colony formation assay for the number of the colonies formed by treated cells. (F and G) MTT assay for the OD value of treated cells at 490 nm. (H and I) Transwell assay for the migration and invasion abilities of treated cells. (J) Western blot analysis for the protein levels of snail and E-cadherin in treated cells. *P < 0.05.
Circ_CDR1as Targets miR-1287, an Upstream miRNA of Raf1
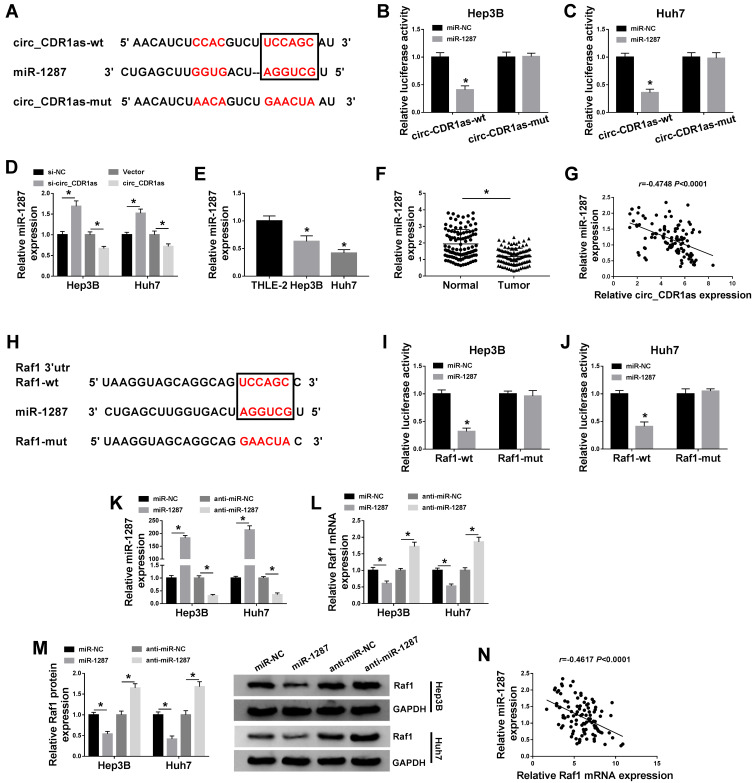
Based on the above results, we intend to figure out the mechanistic pathway of circ_CDR1as in HCC progression. To begin, we identify a target miRNA of circ_CDR1as utilizing starBase, miR-1287 is able to bind to circ_CDR1as. The binding region is exhibited in Figure 5A. Dual-luciferase reporter assay reveals that miR-1287 rather than miR-NC evidently reduces the luciferase activity of circ_CDR1as-wt in Hep3B and Huh7 cells, while no obvious change emerges in the luciferase activity of circ_CDR1as-mut of both the two cells (Figure 5B and C). Furthermore, depletion of circ_CDR1as elevates miR-1287 expression, while circ_CDR1as accumulation downregulates miR-1287 in Hep3B and Huh7 cells (Figure 5D). The downregulation of miR-1287 is observed in Hep3B and Huh7 cells, as well as in HCC tissues, relative to THLE-2 cells and adjacent healthy tissues, respectively (Figure 5E and F). We find that miR-1287 expression in HCC tissues is inversely correlated to circ_CDR1as expression (Figure 5G).
Figure 5.
Circ_CDR1as targets miR-1287, an upstream miRNA of Raf1. (A) The binding sites of miR-1287 on circ_CDR1as, as well as the mutant sites based on starBase. (B and C) Dual-luciferase reporter assay for the luciferase activities of circ_CDR1as-wt and circ_CDR1as-mut in Hep3B and Huh7 cells. (D) QRT-PCR assay for the relative expression of miR-1287 in Hep3B and Huh7 cells introduced with si-NC, si-circ_CDR1as, Vector or circ_CDR1as. (E) QRT-PCR assay the relative expression of miR-1287 in THLE-2, Hep3B and Huh7 cells. (F) QRT-PCR assay for the relative expression of miR-1287 in HCC tissues and adjacent normal tissues (n=105). (G) Pearson correlation analysis for the correlation between the levels of miR-1287 and circ_CDR1as in HCC tissues, r=−0.4748, P < 0.0001. (H) The binding sites of miR-1287 on the 3ʹUTR of Raf1, as well as the mutant sites based on starBase. (I and J) Dual-luciferase reporter assay for the luciferase activities of Raf1-wt and Raf1-mut in Hep3B and Huh7 cells. (K–M) Hep3B and Huh7 cells are transfected with miR-NC, miR-1287, anti-miR-NC or anti-miR-1287. (K and L) QRT-PCR assay the relative expression levels of miR-1287 and Raf1 in transfected cells. (M) Western blot analysis for the protein level of Raf1 in transfected cells. (N) Pearson correlation analysis for the correlation between the levels of miR-1287 and Raf1 in HCC tissues, r=−0.5270, P < 0.0001. *P < 0.05.
We also search the target mRNA of miR-1287 utilizing starBase, and 3ʹUTR of Raf1 contains the binding sites for miR-1287 (Figure 5H). After cotransfection with miR-1287, the luciferase activity of Raf1-wt in Hep3B and Huh7 cells is conspicuously declined, but it has no significant impact on the luciferase activity of Raf1-mut (Figure 5I and J). To examine the effects of miR-1287 on the Raf1 expression in HCC cells, Hep3B and Huh7 cells with miR-1287 upregulation or miR-1287 interference are successfully constructed, which is confirmed by qRT-PCR assay (Figure 5K). As depicted in Figure 5L and M, miR-1287 inversely regulates the mRNA and protein levels of Raf1 in Hep3B and Huh7 cells. Additionally, miR-1287 expression in HCC tissues is negatively correlated to Raf1 mRNA (Figure 5N). To sum up, circ_CDR1as targets miR-1287, and miR-1287 targets Raf1 in HCC cells.
Circ_CDR1as Depletion Represses HCC Cell Proliferation and Metastasis by Sponging miR-1287 to Downregulate Raf1
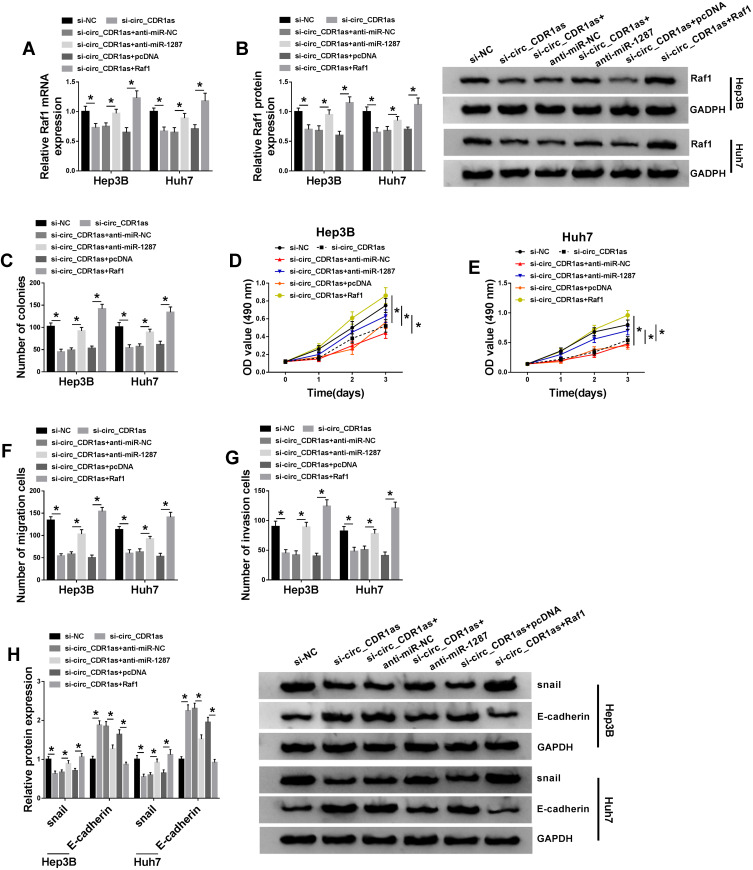
On the account of the target relationship among circ_CDR1as, miR-1287 and Raf1 in HCC cells, we infer that miR-1287 and Raf1 are involved in circ_CDR1as depletion-mediated HCC cell proliferation and metastasis inhibition. To validate our guess, a series of rescue experiments are implemented. As illustrated in Figure 6A and B, circ_CDR1as depletion downregulates the mRNA and protein expression of Raf1 in Hep3B and Huh7 cells, which is ameliorated by miR-1287 depletion or Raf1. What is more, circ_CDR1as depletion-mediated the declined colony formation ability (Figure 6C), OD value at 490 nm (Figure 6D and E), migration and invasion capacities (Figure 6F and G), snail level as well as the elevated E-cadherin level (Figure 6H) are all alleviated by cotransfection with miR-1287 inhibitor or Raf1. Herewith, circ_CDR1as modulates HCC cell proliferation and metastasis by regulating miR-1287/Raf1 axis.
Figure 6.
Circ_CDR1as depletion represses HCC cell proliferation and metastasis by sponging miR-1287 to downregulate Raf1. Hep3B and Huh7 cells are introduced with si-NC, si-circ_CDR1as, si-circ_CDR1as+anti-miR-NC, si-circ_CDR1as+anti-miR-1287, si-circ_CDR1as+pcDNA or si-circ_CDR1as+Raf1. (A and B) QRT-PCR and Western blot assays for the relative expression of Raf1 in treated cells. (C) Colony formation assay for the number of the colonies formed by treated cells. (D and E) MTT assay for the OD value of treated cells at 490 nm. (F and G) Transwell assay for the migration and invasion abilities of treated cells. (H) Western blot analysis for the protein levels of snail and E-cadherin in treated cells. *P < 0.05.
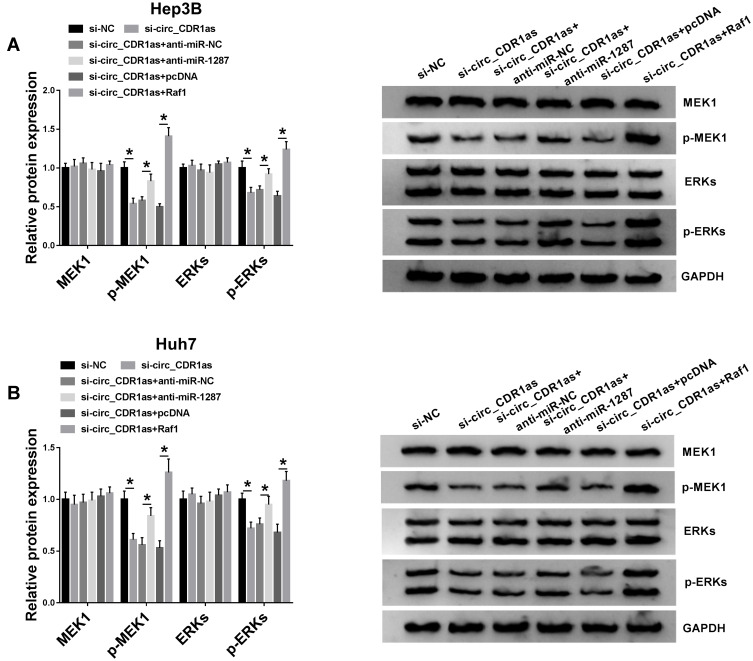
Circ_CDR1as Depletion Inactivates MEK/ERK Pathway by Regulating miR-1287/Raf1 Axis
The influence of circ_CDR1as depletion on the MEK/ERK pathway in Hep3B and Huh7 cells is analyzed. Western blot assay validates that circ_CDR1as depletion represses the levels of p-MEK1 and p-ERK, leading to the inactivation of MEK/ERK pathway, while cotransfection of miR-1287 inhibitor or Raf1 relieves the inactivated impact (Figure 7A and B). Therefore, circ_CDR1as depletion functions as a MEK/ERK pathway inactivator in Hep3B and Huh7 cells through regulating miR-1287/Raf1 axis.
Figure 7.
Circ_CDR1as depletion inactivates MEK/ERK pathway by regulating miR-1287/Raf1 axis. Hep3B and Huh7 cells are introduced with si-NC, si-circ_CDR1as, si-circ_CDR1as+anti-miR-NC, si-circ_CDR1as+anti-miR-1287, si-circ_CDR1as+pcDNA or si-circ_CDR1as+Raf1. (A and B) Western blot analysis for the protein levels of MEK1, p-MEK1, ERKs and p-ERKs in treated cells. *P < 0.05.
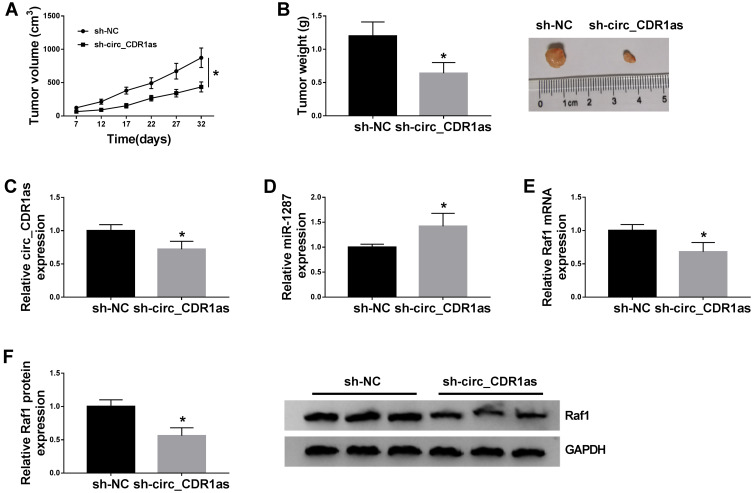
Circ_CDR1as Depletion Suppresses HCC Tumor Growth in vivo
For the confirmation of the role of circ_CDR1as in HCC tumorigenesis in vivo, BALB/c nude mice are treated with hypodermic injection with Hep3B cells stably expressing sh-circ_CDR1as or sh-NC. The volume of formed tumors in sh-circ_CDR1as group is smaller than those in sh-NC group (Figure 8A). Besides, at 32 days post injection, all formed tumors are resected, and we find that generated tumors in sh-circ_CDR1as group exhibit a lower weight in reference to sh-NC group (Figure 8B). Furthermore, we discover the upregulation of circ_CDR1as (Figure 8C) and Raf1 (Figure 8E and F), as well as the downregulation of miR-1287 (Figure 8D) in sh-circ_CDR1as group relative to sh-NC group. In conclusion, circ_CDR1as depletion represses HCC tumor growth in vivo.
Figure 8.
Circ_CDR1as depletion suppresses HCC tumor growth in vivo. Hep3B cells stably transfected with sh-circ_CDR1as or sh-NC are subcutaneously injected into BALB/c nude mice. (A) The volume of formed tumors. (B) The weight of formed tumors. (C–E) QRT-PCR assay for the relative expression of circ_CDR1as, miR-1287 and Raf1 in formed tumors. (F) Western blot analysis for the protein level of Raf1 in formed tumors. *P < 0.05.
Discussion
Nowadays, more and more circRNAs have emerged as key regulators, involved in progression of HCC, participating in many cellular processes.20 Here, in HCC, we find that circRNA circ_CDR1as is highly expressed. We also observe its positive effects on HCC cellular behaviors, including proliferation, migration and invasion, epithelia–mesenchymal transition (EMT) in vitro, and on tumor growth in vivo, as well as its action mechanistic pathway by acting as miR-1287 sponge.
CircRNAs are hot researched non-coding RNAs, and dysregulated expression of circRNAs is manifested to be relevant to HCC initiation, invasion, and metastasis.21 What is important, circRNAs are also potential molecules, functioning in the diagnosis, therapy, and prognosis of patients with HCC.22 It is a fact that numerous circRNAs often exhibit cell type-specific, tissue-specific or developmental stage-specific expression.23,24 Situated at chromosome X: 139865339-139866824, circ_CDR1as (also named as hsa_circ_0001946 or ciRS-7), is a risk factor for HCC; Besides, circ_CDR1as is downregulated in HCC cells and tissues, affecting physiological function of HCC cells by targeting miR-7 and regulating EGFR signaling.25 Inversely, Su et al pronounce that circ_CDR1as is highly expressed in HCC cells and tissues, and gain of circ_CDR1as largely promotes HCC cells to proliferate and metastasize, as well as accelerates HCC progression, highlighting its oncogenic role.11 What is more, Xu et al point out that the expression level of circ_CDR1as in HCC and matched non-tumor tissues is comparable.26 From our data, we also discover that circ_CDR1as expression is significantly upregulated in HCC tissues and cells. Besides, we determine that circ_CDR1as is a stable circRNA, suggesting that circ_CDR1as might be a diagnostic biomarker for HCC patients. Besides, circ_CDR1as could also promote the development of non-small-cell lung cancer (NSCLC),27 nasopharyngeal carcinoma,28 while inhibit ovarian cancer progression,29 suggesting its oncogenic role. The altered expression of circ_CDR1as in HCC prompts us to investigate the function of circ_CDR1as in vitro and in vivo. In this work, we observe that deficiency of circ_CDR1as represses HCC cell proliferation and metastasis in vitro, as well as tumor growth in vivo, in consistent with a former paper.11
According to the competing endogenous RNA (ceRNA) hypothesis, circRNA could act as ceRNA to sponge miRNA to exert its regulatory potency in human cancer development. A former work points out that depletion of Cdr1as acts tumor suppressor role in HCC development by serving as a ceRNA of miR-7.30 Here, we utilize starBase to seek the potential target miRNA of circ_CDR1as for better understanding its mechanism. Luckily, miR-1287 is forecasted to be a candidate, which is further validated by dual-luciferase reporter assay. As reported, miR-1287 embraces important diagnostic and prognostic value in colorectal cancer.27 In breast cancer, miR-1287 could be sponged by circ-UBE2D2 to affect tumor progression.28 And, in NSCLC, miR-1287 functions as the target of circ_0026134 to regulate cell proliferation and invasion of A549 and H1299 cells.29 Lu et al declare that upregulation of miR-1287 decreases the tumorigenesis phenotypes of HCC cells in vitro, including cell proliferation, cell cycle, invasion and migration in vitro model.14 In this project, the downregulation of miR-1287 is detected in HCC tissues and cells. Next, we find that depletion of miR-1287 reverses the circ_CDR1as depletion-mediated the reduced tumorigenesis phenotypes of HCC cells. So, we conclude that circ_CDR1as deficiency inhibits HCC progression via upregulating miR-1287 expression.
Additionally, we also search starBase to forecast the downstream mRNA of miR-1287 to clarify the regulatory axis. Later, we observe the binding sites between the miR-1287 and 3ʹUTR of Raf1. We utilize dual-luciferase reporter assay to validate this binding. Based on our data, we find that Raf1 depletion inhibits the tumorigenesis phenotypes of Hep3B and Huh7 cells, suggesting the oncogenic function of Raf1 in vitro, which is similar to the investigation of Ghousein et al.17 Raf1 is a well-investigated oncogene in various human cancers, such like lung cancer,30 papillary thyroid cancer,31 colorectal cancer32 and cervical cancer.33 Tian and his partners prove that Raf1 could function as an important prognostic factor of the development of the patients with NSCLC after radiotherapy.15 And Raf1 is identified to be the target of miR-195, which reduces the growth of thyroid cancer.34 In this project, gain of Raf1 also rescues the circ_CDR1as depletion-mediated the repressed proliferation and metastasis of HCC cells. Thus, circ_CDR1as depletion inhibits HCC development by regulating miR-1287/Raf1 pathway.
Furthermore, we try to seek the possible signal pathway by which circ_CDR1as affecting HCC progression. The Raf/MEK/ERK pathway has vital effects on cell growth, cell cycle and drug resistance, whose inhibitor could reduce tumor development.35,36 In our work, circ_CDR1as depletion could break the activation of MEK/ERK pathway through the miR-1287/Raf1 axis. Therefore, we confirm that circ_CDR1as/miR-1287/Raf1 axis is involved in the development of HCC in a MEK/ERK pathway-dependent manner, so as to affect the HCC cell proliferation, metastasis and tumorigenesis phenotypes.
Due to the circRNAs are expressed in cell-, tissue- and developmental stage-dependent manners,23,24 the expression pattern and regulatory function of circ_CDR1as in other HCC cell lines will be our future research direction. Furthermore, the correlation between the circ_CDR1as expression and the clinicopathological characteristics of the above-mentioned 105 HCC patients, including gender, TNM stage, histological grade, etc. will be examined.
To sum up, the upregulation of circ_CDR1as and Raf1, and the downregulation of miR-1287 are observed in HCC. Circ_CDR1as sponges miR-1287 to upregulate Raf1, leading to HCC cell proliferation and metastasis inhibition in vitro and tumor growth repression in vivo, through modulating the MEK/ERK pathway. Our study identifies a novel action mechanism of circ_CDR1as functioning in HCC progression, implying its application potential in HCC targeted treatment.
Funding Statement
There is no funding to report.
Abbreviations
HCC, Hepatocellular carcinoma; circRNA, circular RNA; miRNA, microRNAs; Raf1, Raf-1 proto-oncogene, serine/threonine kinase; EMT, epithelia–mesenchymal transition; qRT-PCR, Quantitative real-time polymerase chain reaction; MTT, 3-(4, 5)-dimethylthiazole-2-y1)-2, 5-biphenyl tetrazolium bromide; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Slotta JE, Kollmar O, Ellenrieder V, Ghadimi BM, Homayounfar K. Hepatocellular carcinoma: surgeon’s view on latest findings and future perspectives. World J Hepatol. 2015;7(9):1168–1183. doi: 10.4254/wjh.v7.i9.1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giuliani F, Colucci G. Treatment of hepatocellular carcinoma. Oncology. 2009;77(Suppl 1):43–49. doi: 10.1159/000258495 [DOI] [PubMed] [Google Scholar]
- 3.Worns MA, Galle PR. Future perspectives in hepatocellular carcinoma. Dig Liver Dis. 2010;42(Suppl 3):S302–S309. doi: 10.1016/s1590-8658(10)60521-x [DOI] [PubMed] [Google Scholar]
- 4.Dragomir M, Calin GA. Circular RNAs in cancer – lessons learned from microRNAs. Front Oncol. 2018;8:179. doi: 10.3389/fonc.2018.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Z, Yan Y, Zeng S, et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2017;9(1):1444–1460. doi: 10.18632/oncotarget.22846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology (Baltimore, Md). 2017;66(4):1151–1164. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 9.Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499(4):1044–1049. doi: 10.1016/j.bbrc.2018.03.221 [DOI] [PubMed] [Google Scholar]
- 10.Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37(1):172. doi: 10.1186/s13046-018-0838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging. 2019;11(19):8182–8203. doi: 10.18632/aging.102312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 13.Wu W, Sun M, Zou G-M, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. 2007;120(5):953–960. doi: 10.1002/ijc.22454 [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Tang L, Xu Y, et al. Mir-1287 suppresses the proliferation, invasion, and migration in hepatocellular carcinoma by targeting PIK3R3. J Cell Biochem. 2018;119(11):9229–9238. doi: 10.1002/jcb.27190 [DOI] [PubMed] [Google Scholar]
- 15.Tian H, Yin L, Ding K, et al. Raf1 is a prognostic factor for progression in patients with non‑small cell lung cancer after radiotherapy. Oncol Rep. 2018;39(4):1966–1974. doi: 10.3892/or.2018.6277 [DOI] [PubMed] [Google Scholar]
- 16.Cobb MH, Hepler JE, Cheng M, Robbins D. The mitogen-activated protein kinases, ERK1 and ERK2. Semin Cancer Biol. 1994;5(4):261–268. [PubMed] [Google Scholar]
- 17.Ghousein A, Mosca N, Cartier F, et al. miR-4510 blocks hepatocellular carcinoma development through RAF1 targeting and RAS/RAF/MEK/ERK signalling inactivation. Liver Int. 2020;40(1):240–251. doi: 10.1111/liv.14276 [DOI] [PubMed] [Google Scholar]
- 18.Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102(4):837–852. [DOI] [PubMed] [Google Scholar]
- 19.Qian L, Yu S, Chen Z, Meng Z, Huang S, Wang P. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870(2):247–260. doi: 10.1016/j.bbcan.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 20.Qiu L, Xu H, Ji M, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. doi: 10.1016/j.lfs.2019.116660 [DOI] [PubMed] [Google Scholar]
- 21.Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. 2018;7(7):3101–3109. doi: 10.1002/cam4.1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332. doi: 10.3389/fonc.2018.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. doi: 10.1242/dev.12807424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi: 10.1080/15476286.2016.1220473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res. 2017;16(10):3891–3902. doi: 10.1021/acs.jproteome.7b00519 [DOI] [PubMed] [Google Scholar]
- 26.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS‑7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Zhang J, Pan S, Zhou J, Diao X, Liu S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac Cancer. 2020. doi: 10.1111/1759-7714.13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong Q, Huang J, Wei J, Circular WR. RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer Cell Int. 2019;19:252. doi: 10.1186/s12935-019-0959-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Mao M, Jiang J, Zhu D, Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019;12:3869–3879. doi: 10.2147/OTT.S207938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347. doi: 10.1371/journal.pone.0158347 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Fateh A, Feizi MAH, Safaralizadeh R, Azarbarzin S, Ravanbakhsh R. Diagnostic and prognostic value of miR-1287 in colorectal cancer. J Gastrointest Cancer. 2016;47(4):399–403. doi: 10.1007/s12029-016-9833-5 [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Li J, Du C, et al. Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl Oncol. 2019;12(10):1305–1313. doi: 10.1016/j.tranon.2019.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother. 2019;112:108743. doi: 10.1016/j.biopha.2019.108743 [DOI] [PubMed] [Google Scholar]
- 34.Jiang Q, Cheng L, Ma D, Zhao Y. FBXL19-AS1 exerts oncogenic function by sponging miR-431-5p to regulate RAF1 expression in lung cancer. Biosci Rep. 2019;39(1):BSR20181804. doi: 10.1042/BSR20181804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Kong Q. LncRNA LINC00460 promotes the papillary thyroid cancer progression by regulating the LINC00460/miR-485-5p/Raf1 axis. Biol Res. 2019;52(1):61. doi: 10.1186/s40659-019-0269-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borovski T, Vellinga TT, Laoukili J, et al. Inhibition of RAF1 kinase activity restores apicobasal polarity and impairs tumour growth in human colorectal cancer. Gut. 2017;66(6):1106–1115. doi: 10.1136/gutjnl-2016-311547 [DOI] [PubMed] [Google Scholar]
- 37.Wu F, Zhou J. CircAGFG1 promotes cervical cancer progression via miR-370-3p/RAF1 signaling. BMC Cancer. 2019;19(1):1067. doi: 10.1186/s12885-019-6269-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, Jiang C, Sun Q, et al. miR-195 is a key regulator of Raf1 in thyroid cancer. Onco Targets Ther. 2015;8:3021–3028. doi: 10.2147/OTT.S90710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773(8):1263–1284. doi: 10.1016/j.bbamcr.2006.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: structural and pharmacological perspectives. Eur J Med Chem. 2016;109:314–341. doi: 10.1016/j.ejmech.2016.01.012 [DOI] [PubMed] [Google Scholar]