Abstract
Non-enzymatic glycation of proteins is believed to be the root cause of high dietary sugar associated pathophysiological maladies. We investigated the structural changes in protein during progression of glycation using ribosylated Bovine Serum Albumin (BSA). Non enzymatic attachment of about 45 ribose molecules to BSA resulted in gradual reduction of hydrophobicity and aggregation as indicated by red-shifted tryptophan fluorescence, reduced ANS binding and lower anisotropy of FITC-conjugated protein. Parallely, there was a significant decrease of alpha helicity as revealed by Circular Dichroism (CD) and Fourier transformed-Infra Red (FT-IR) spectra. The glycated proteins assumed compact globular structures with enhanced Thioflavin-T binding resembling amyloids. The gross structural transition affected by ribosylation led to enhanced thermostability as indicated by melting temperature and Transmission Electron Microscopy. At a later stage of glycation, the glycated proteins developed non-specific aggregates with increase in size and loss of amyloidogenic behaviour. A parallel non-glycated control incubated under similar conditions indicated that amyloid formation and associated changes were specific for ribosylation and not driven by thermal denaturation due to incubation at 37 °C. Functionality of the glycated protein was significantly altered as probed by Isothermal Titration Calorimetry using polyphenols as substrates. The studies demonstrated that glycation driven globular amyloids form and persist as transient intermediates during formation of misfolded glycated adducts. To the best of our knowledge, the present study is the first systematic attempt to understand glycation associated changes in a protein and provides important insights towards designing therapeutics for arresting dietary sugar induced amyloid formation.
Keywords: Chemistry, Biochemistry, Ribosylated BSA, Globular amyloid, Thermostability of glycated protein, Glycation driven vs. heat induced protein aggregation, Dietary sugar induced amyloid formation
Chemistry; Biochemistry; Ribosylated BSA; Globular amyloid; Thermostability of glycated protein; Glycation driven vs. heat induced protein aggregation; Dietary sugar induced amyloid formation
1. Introduction
Dietary sugars are an inevitable part of the metabolic load of our body since carbohydrates constitute the primary source of energy. However, the detrimental effects associated with dietary sugars are have been complicated in modern urban lifestyle due to consumption of high colrie overcooked or junked food. One of the major cellular complicacies arising out of consistent persistence of reducing sugars is posed by non-enzymatic glycation of proteins leading to their subsequent aggregation. Protein aggregation is implicated in many detrimental conditions ranging from neurodegenerative diseases to degeneration of therapeutically important proteins [1, 2]. Despite protein folding and protein aggregation being distinct processes, both intrinsic and extrinsic factors govern whether a folding pathway will lead to a native structure or to misfolded aggregate [3]. In recent years, many unnatural post-translational modifications have been discovered which can significantly alter the native structure and function of proteins [4]. One of these is the non-enzymatic glycation or attachment of reducing sugars to the lysine and arginine residues and free thiols of cysteine side chains via Schiff base formation [5]. Glycation of proteins eventualize in a complex series of reactions known as Maillard reactions [5] which terminate in the form of stable irreversibly misfolded adducts - the Advanced Glycation End (AGE) products [6]. Glycation has been associated with severely altered structure and functionality of native proteins [7] including formation of covalent cross-links with nearby proteins [8]. AGE formation and accumulation has a diverse spectrum of diseases encompassing several physiological maladies such as arteriosclerosis, renal failure, diabetic complications and Alzheimer disease [9]. Cellular effects of AGEs are exerted by binding to specific receptors termed RAGE (Receptor for AGE) with subsequent initiation of downstream signaling cascade leading ROS generation, oxidative stress and NF-κβ activation [8, 10]. More significantly, glycated proteins have been found to resemble the behaviour of other misfolded proteins associated with amyloid deposits such as beta-amyloid, tau, prions etc. [11, 12, 13]. Glycation has the potential to induce unfolding and refolding of globular proteins into cross-β structure thus automatically making it eligible as a candidate amyloidogenic protein [14, 15]. However, contrasting reports have been obtained thus far regarding the role of glycation in creation of amyloidogenic aggregates. While many reports suggest that glycation selectively makes the protein amyloidogenic [16, 17, 18], few others indicate that glycation of amyloidogenic precursor protein inhibits fibril formation [19, 20]. Additionally, the glycatedproteins have also been reported to be more thermostable than their native counterparts [21, 22]. In vitro glycation of whey proteins is a well-known practice to improve its thermostability for industrial applications [23].
Bovine Serum albumin (BSA), the most abundant serum protein with versatile applications both in vivo and in vitro, has been a particularly popular model for understanding the effects of glycation [18, 24, 25, 26, 27]. However, the majority of reports in this regard have only concentrated on the structural characterization of the final glycated form instead of probing the changes that occur progressively throughout the entire period of glycation starting from initial formation of sugar-protein adduct to generation of the terminally misfolded Advanced Glycated End Product. Additionally, the methodology used universally to glycate proteins involves incubation of the protein with the respective sugars at 37 °C. Sustained incubation at such elevated temperatures is known to cause structural alterations in the protein [2, 26] including formation of aggregates. However, most of the studies have not systematically accounted for the changes which occur in the control non glycated protein incubated under similar conditions. The present studies report the formation of a globular amyloid like aggregate as an intermediate in the generation of terminally misfolded protein aggregate resulting from in vitro ribosylation of BSA and characterizes its physico-chemical attributes in terms of thermostability and substrate binding efficiency. The structural and functional changes entailed by the protein due to glycation are also compared with a parallel non glycated control maintained under similar conditions.
2. Material and methodology
2.1. Chemicals
Bovine Serum Albumin (highest available purity), ANS (8-anilino-1-naphthalenesulfonic acid), NPN (N-Phenyl-1-naphthylamine), Thioflavin-T, FITC (fluorescein isothiocyanate)were purchased from Sigma Chemicals. Other chemicals and reagents needed of Analytical grade were purchased locally from SRL.
2.2. Glycation of BSA
Bovine Serum Albumin (0.15 mM) was incubated separately with 0.5 M of different sugars (glucose, galactose, fructose, ribose and mannose) in presence of 0.05 M sodium phosphate buffer, pH 7.4 at 37 °C. Reaction mixtures were kept in this condition for 10 days. It was kept into sterile glass tubes in presence of 3 mM sodium azide to prevent bacterial contamination. Aliquots were taken under sterile conditions at desired times and frozen at -20 °C until further assayed. The samples were then diluted five times with 0.2 M sodium acetate buffer, pH 5 to reduce the glycated protein and incubated at 37 °C for 2 h to release glycosylamine adducts. In order to remove free ribose molecules, this treated protein was then dialyzed overnight against 0.1 M sodium phosphate buffer, pH 7.4, at 4 °C. After that the dialyzed protein was reduced by adding a 50-fold molar excess of sodium borohydride in presence of 0.1 M sodium hydroxide. The solution was then kept at room temperature for 4 h after which the reaction was terminated by the adding 1 N HCI slowly to eliminate excess sodium borohydride. Finally, the samples were dialyzed overnight at 4 °C against double distilled water [28]. Parallely, the control incubated samples (without ribosylation) were also treated in a similar manner and protein concentrations were measured by Bradford assay.
2.3. Fluorescence spectroscopy
2.3.1. AGE fluorescence
The formation of Advanced Glycated End products (AGE) was monitored by measuring the emitted fluorescence spectra of 1.5 μM glycated BSA with excitation at 370 nm. The emission spectra were obtained in the range of 390–500 nm with both excitation and emission bandwidths at 5 nm [29]. All these measurements were performed on Perkin-Elmer LS55 spectrofluorimeter.
2.3.2. Intrinsic tryptophan fluorescence
Steady state fluorescence was recorded with 3 μM protein dissolved in 50 mM phosphate buffer on a Perkin-Elmer LS55 spectrofluorimeter. Intrinsic tryptophan fluorescence spectra were carried out by exciting the samples at 295 nm with excitation and emission slit widths set at 5 nm. The emission spectra were recorded between 315 to 500 nm. Baseline corrections were performed with buffer in all cases [30, 31].
2.3.3. ANS fluorescence
Steady-state fluorescence of ANS was measured on the same Perkin-Elmer LS55 spectrofluorimeter as of intrinsic tryptophan fluorescence measurements. ANS concentration was 0.1mM and bandwidth for excitation and emission monochromators were 5 nm. Fluorescence of ANS was measured by excitation at 360 nm with protein concentration of 0.2 μM in a 50 mM sodium phosphate buffer (pH 7.2). The emission spectra for the samples were recorded in the range of 380 and 600 nm [32].
2.3.4. NPN fluorescence
NPN fluorescence of BSA samples was carried out on the Perkin-Elmer LS55 spectrofluorimeter using 60 μM aqueous solution of NPN fluorophore [33]. Fluorescence spectra were recorded with 1.5 μM of protein samples in the range of 370–600 nm. The emission spectra were measured by exciting the samples at 350 nm and bandwidths of both excitation and emission spectra at 5 nm.
2.4. MALDI – mass spectroscopy analysis
Extents of glycation in BSA samples were obtained by Mass spectroscopy analysis of the samples from Bruker spectrophotometer (model: Ultra FiexTreme). Samples were prepared in microcrystalline matrix surfaces that were made on the probe tips of the mass spectrometer. Protein samples were spotted by mixing the samples in a 50% aqueous acetonitrile solution (0.6 μL) saturated with sinapinic acid and containing 0.05% TFA. Sample analysis was carried out in linear TOF mode with 550 ns delay. Further data processing and calibrations were done using the computer program attached to the spectrophotometer [34]. The number (N) of ribose residues conjugated with BSA molecule was calculated from the following formula:
| N = (MWBSA-ribose− MWBSA)/MWRibose |
2.5. DTNB assay
The free thiol contents in BSA, incubated and glycated BSA samples were determined using Ellman's assay [35]. 0.1mM DTNB (5,5′-dithiobis-2-nitrobenzoic acid) or Ellman's reagent in presence of 0.1mMTris buffer (pH 8) were mixed with different concentrations of cysteine (10 μM–100 μM) to obtain a standard curve. The samples were then incubated at room temperature for 5 min and the absorbance was measured at 412 nm in Jasco V-630 spectrophotometer. The assay was carried out with the protein samples as well and from the standard curve of cysteine, the free thiol contents of the proteins were obtained.
2.6. FITC- BSA conjugation
BSA samples were labeled with fluorescein isothiocyanate (Sigma) in 0.1 M carbonate buffer, pH 9. The samples were kept at room temperature and in the dark for 4 h. The labelling ratio (F:P) for control and incubated BSA were 1:1 and for ribosylated BSA it was 0.5:1. Then the solutions were dialyzed four times in dark at 4 °C against 0.01 M phosphate buffered saline (PBS), pH 7.4 in order to remove any unbound FITC. The F:P ratio range was selected depending on the sensitivity and photophysics of the dye conjugated to the protein [36].
2.7. Fluorescence anisotropy
Fluorescence anisotropy decays of FITC conjugated BSA samples were recorded in a Varian-Cary-UV Eclipse spectrofluorimeter at 25 °C [37]. Polarized decays were collected at the vertical (IVV) and horizontal (IVH) positions of the emission polarizer. The samples were excited at 490 nm and the bandwidth for excitation and emission monochromator were set at 5 nm. The emission intensities were measured at 522 nm. Fluorescence spectra were corrected for the emission of the control solution containing buffer. Both decays were collected in the same conditions and for the same amount of time. A high number of counts were essential to increase the signal-to-noise ratio for anisotropy decays. G represents a correction factor for the sensitivity of the optics to light with different polarizations. The G factor was determined using the instruments software for the samples in the absence of protein, for which the rotational correlation time was short and the anisotropy decayed to zero within the time scale for the measurement. This procedure is necessary since small changes in the G factor can bring about changes in the baseline for the anisotropy of the samples. Then the G factor determined in the absence of protein was used for anisotropy decays in the presence of the proteins.
2.8. DLS
Dynamic light scattering measurements were performed with BSA in the concentration of 0.1 mg/ml [38]. The concentration range was selected according to the response and sensitivity of the instrument. Data were obtained in a Nano zeta-sizer (Malvern Instruments) and a Beckman Coultier DLS instrument at a fixed 90ο scattering angle using Nd-doped solid state laser of 632.5 nm with 100 s of integration time; the intensity correlation function was obtained by acquiring data between 5 and 1,000 ms, in 200 channels.
2.9. Native gel electrophoresis
Native-polyacrylamide gel electrophoresis (7%) under reducing condition was carried out according to the methods described earlier [39, 40]. The protein samples (10 μg) were incubated at different temperatures between 25 °C and 85 °C for 1 h and subsequently were dissolved in the sample buffer. The gel was run at 20 mA till the tracking dye reached its lower edge. Thereafter, it was immediately subjected to silver staining using ProteoSilverTM silver stain kit (Sigma) according to the manufacturer's instructions.
2.10. Circular Dichroism
Circular dichroic (CD) spectra of different preparations of BSA (1.5 μM) in the far-UV wavelength range (190–250 nm) were recorded at 25 °C on a JASCO J-720 spectropolarimeter (calibrated with d-10-camphorsulfonic acid) using a cylindrical quartz cuvette of path length 1 mm. The following scan parameters were used: 1 nm bandwidth, 2 s response time, 0.1 nm step resolution and 20 nm/min scan speed [40, 41, 42]. Each spectrum had an average of four continuous scans. The final spectra were acquired by subtracting blank runs on appropriate protein-free buffer (10 mM-phosphate buffer, pH 7.0) and subjected to a moderate degree of noise-reduction analysis. Alpha helix content was obtained using the formula:
| % α-helix = ((MRE222nm – 2340) / 30300) x 100 |
Where, Mean Residual Ellipticity (MRE) = MRE222 = θobs(mdeg)/(10∗n∗c∗l), n = number of peptide bonds, c = concentration of the sample in M, l = pathlength = 0.1 cm.
2.11. FT-IR analysis
Fourier Transform-Infra Red (FT-IR) spectra of BSA samples were recorded on a Perkin-Elmer FTIR instrument. The samples were prepared by grinding 1 mg of the protein sample with 100 mg of KBr together previously kept in dessicator [43, 44, 45]. After mixing properly pellets were made by pressing the samples at 10 tons for 1 min. The spectral data were obtained within the range of 4000 cm−1 - 450 cm−1 with a spectral resolution of 1 cm−1. The spectrum of the background was recorded initially and it was further subtracted from the spectra of samples.
2.12. Melting point analysis from spectral studies
Melting points of glycated and incubated BSA were acquired from absorption measurements of the samples [46]. In brief, absorbance (280 nm) of BSA samples (7.5 μM aliquots) were taken corresponding to the temperature range of 25–90 °C in a Jasco V-630 spectrophotometer fitted with a peltier. Baseline corrections for each sample were performed with buffer in absence of the protein.
2.13. Thioflavin T binding
A ThT stock solution was prepared by dissolving 2.5 mM Thioflavin T (Sigma) in phosphate buffer (10 mM phosphate, 150 mM NaCl, pH 7.0) and then filtering it through a 0.2 μm syringe filter [47]. The working solution was generated by diluting the stock solution 50 fold in the phosphate buffer on the day of analysis. BSA samples at concentration of 1.5 μM were mixed with the working solution and were allowed to bind the ThT for 1min. The fluorescence intensities of the samples were measured on Perkin-Elmer LS55 spectrofluorimeter by excitation at 440 nm (slit width 5 nm) and emission 482 nm (slit width 10 nm) at medium speed.
2.14. Transmission Electron Microscopy (TEM)
TEM was carried out as reported earlier [48], with few modifications. Briefly, twenty five square mesh copper grids with continuous carbon film (Electron Microscopy Sciences) were taken to incubate the samples. 10 μL of each BSA samples (control BSA, non-glycated and glycated BSA) were applied onto the grids and allowed to dry in air before analysis. Glycated and non-glycated BSA samples were kept at 60 °C for 1hour before applying onto the grids. TEM images were collected using JEOL-JEM 2100 (Japan) electron microscope operated at 200 kV and size of the proteins were acquired using the associated software.
2.15. Isothermal titration calorimetric measurements (ITC)
The isothermal titration calorimetric experiments were performed on a VP-ITC titration microcalorimeter (MicroCal Inc., Northampton, MA). Protein and ligand concentrations were adjusted at 12 μM and 250 μM, respectively, to maintain a high ligand:protein ratio. After that multiple injections of 60 μl each from curcumin and resveratrol (250 μM) were made into the sample cell maintaining the temperature of the system at 25 °C. The principle method of ITC necessitates presence of 30–40 times ligand concentration from that of protein [49]. To correct the heat effect due to mixing and dilution, control experiments were performed in which ligands were injected into the buffer solution without BSA.
2.16. Statistical analyses
Standard deviations in all fluorescence measurements and DLS were performed with instrument associated software. Statistical analyses of ITC data was carried out in Origin ver 9.0.
3. Results and discussion
3.1. Monitoring the formation of AGE through characteristic fluorescence emission
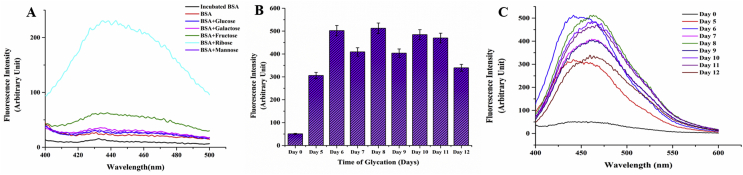
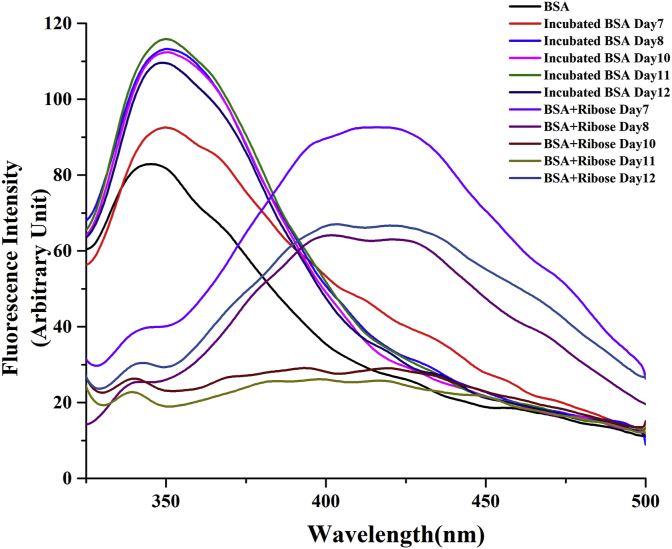
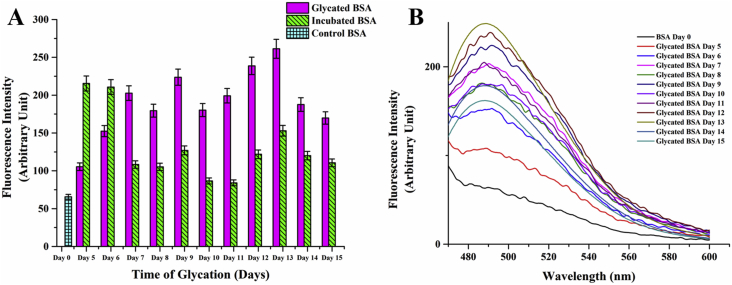
In order to generate a working model of glycated protein, in vitro glycation of Bovine Serum Albumin was carried out individually with the monosaccharides, ribose, glucose and fructose, galactose and mannose. The reaction was monitored by the characteristic fluorescence emission of Advanced Glycation End product at around 440 nm. AGE fluorescence was highest with ribose followed by fructose whereas no significant increase in fluorescence was noted for BSA glycated with the other sugars (Figure 1A). It may be mentioned here that although glucose is the most commonly encountered sugar in the intracellular milieu, it needs a significantly long time to glycate serum albumins [50, 51]. Since the secondary structure of the protein was abolished almost completely beyond twelve days of sustained incubation at 37 °C (as elaborated later), ribose was selected for generation of a glycated BSA model. Advancement of glycation was followed by observing daywise fluorescence of the samples. Fluorescence intensity was highest on the 8th day of incubation (Figure 1B). It decreased subsequently with a significant red shift in emission indicating that the AGE fluorescence was shifted to a more polar environment due to a gross structural transition of the protein (Figure 1C). This was investigated at a later part of the studies.
Figure 1.
Fluorescence spectrum of Advanced Glycated End Product A) AGE formation with different sugar-protein adducts on 10th day B) Progression of AGE formation in ribosylated BSA. Error bars represent the deviations observed in triplicate data sets C) Corresponding fluorescence spectra of B acquired with 0.15 mM BSA (with 0.5 M ribose) in 0.05 M sodium phosphate buffer, pH 7.4 at 37 °C.
3.2. BSA gets optimally ribosylated within 10 days of glycation reaction
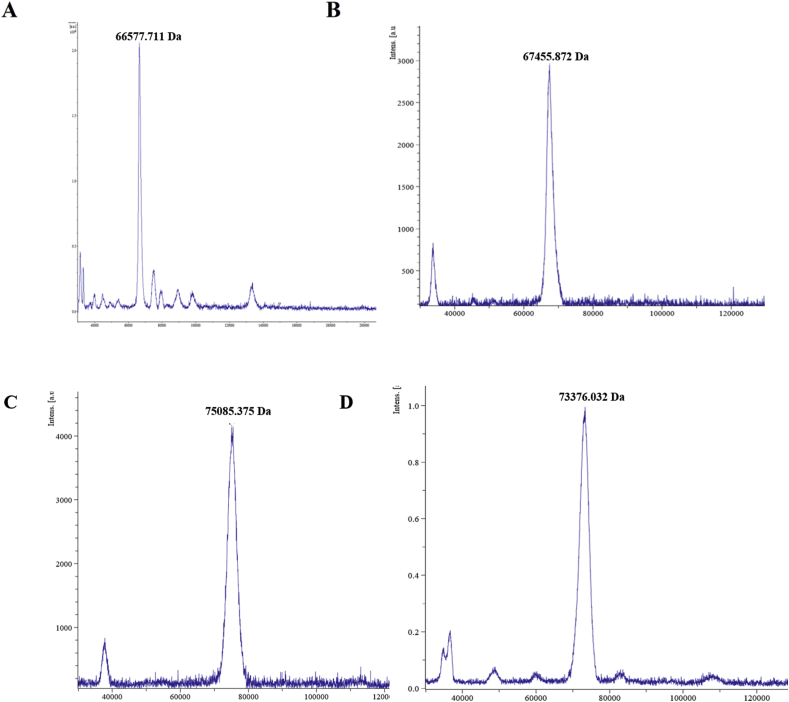
Extent of glycation of the 10th day ribosylated sample was measured quantitatively using MALDI-MS (Figure 2) after a round of purification though HPGPLC to separate any glycated peptide from the commercial protein preparation (data not shown). Number of ribose residues attached was ascertained at different time intervals during the course of glycation. Molecular weight of 4th day sample was found to be 67.45 kD corresponding to attachment of about 5 sugar residues (Figure 2B). Maximum number of ribose residues was attached within 10 days as the molecular weight of the protein increased to 75.08 kD indicating attachment of as many as 56 sugar molecules (Figure 2C). During the later stages of glycation, molecular weight of BSA was found to be about 73.373 kD corresponding to about 45 molecules of ribose (Figure 2D). This can be attributable to partial degradation of the protein or loss of attached sugar or both occurring simultaneously under prolonged thermal incubation. The 10th day glycated protein model was used for some of the subsequent studies such as morphology investigation by TEM, temperature dependence of amyloid formation, FT-IR spectroscopy and evaluation of substrate binding capability by ITC. Since the in vitro glycation was performed by incubating the protein at 37 °C, a parallel control without the sugar was maintained in every case to account for the temperature induced structural changes which might have occurred in the protein.
Figure 2.
MALDI Spectra of glycated protein A) Control Non-glycated BSA B) BSA glycated with ribose for 4 days C) BSA glycated with ribose for 10 days D) BSA glycated with ribose for 15 days.
3.3. Glycation delays thermal aggregation of BSA
Bovine Serum Albumin is a naturally aggregation prone protein [52, 53] where intermolecular disulphide bonds are generally held responsible for mediating the aggregation [54]. Although BSA is not naturally amyloidogenic, intermolecular aggregation of BSA is implicated in formation of irreversible amyloid fibrils in the protein [55, 56]. Free thiol content in purified 8th day glycated protein decreased to more than 50% from 18.96 mM to 8.96 mM. However, the same was also noted in a parallel control where the protein was incubated at 37 °C without the sugar. Therefore it was evident that the decrease in number of free thiols (indicating disulphide mediated cross-linking of protein sub-units) was attributable to prolonged incubation at higher temperature and not due to glycation.
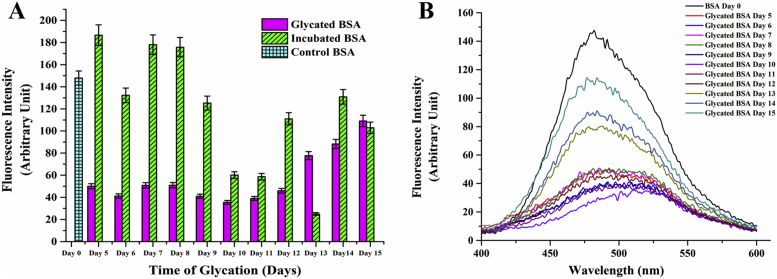
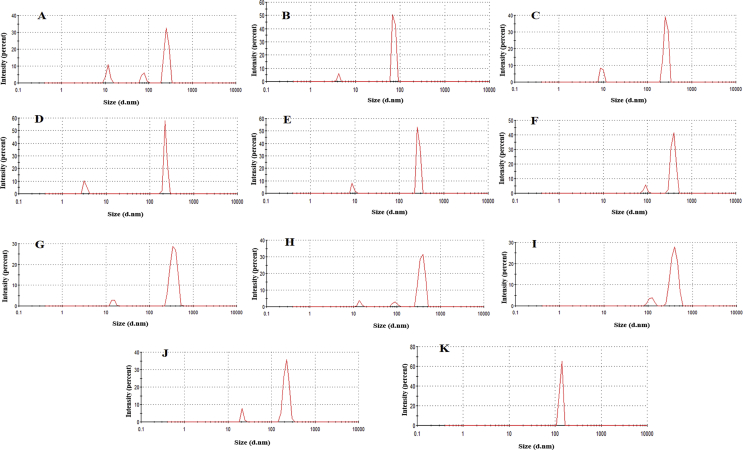
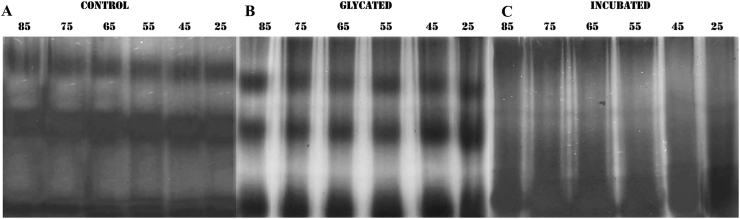
Apart from involvement of disulfide bonds, natural aggregation also occurs due to exposed hydrophobic interfaces [57, 58]. This is of commonplace occurrence for non-aggregation prone proteins due to solution induced deviation from native structure [59]. ANS is an extrinsic fluorophore known to bind to hydrophobic surfaces of proteins [60], where binding is mostly mediated by cataionic groups in protein side chains [61]. Therefore, it is widely used for probing the extent of accessible hydrophobic surface of a protein during its thermal unfolding. ANS binding at different stages of glycation was monitored and the resultant effect on aggregation was studied using a combination of Dynamic Light Scattering and Fluorescence Anisotropy. ANS fluorescence was reduced significantly in glycated samples on the 5th day of incubation and remained more or less constant thereafter till the 12th day (Figures 3A and 3B). Thereafter, it increased steeply again indicating conversion into a molten globule and hydrophobicity driven aggregation. In contrast, fluorescence from the incubated (control) protein samples was strikingly higher throughout the entire period indicating thermal denaturation of BSA due to prolonged incubation at 37 °C. Since glycation reaction consumes most of the free amino groups of protein side chains [62], ANS binding was also possibly reduced due to non-availability of free lysine and arginine groups. 1-N-Phenylnaphthylamine (NPN) is another fluorescent dye known to bind to hydrophobic patches in proteins [63]. Binding of NPN to glycated BSA was also significantly lesser than the control protein (Figure 4A and 4B). Therefore, it was plausible that glycation imparted thermostability to the protein during sustained incubation above ambient temperature. Increase or decrease in hydrophobicity has a direct bearing on the aggregation status of the protein [64]. BSA being an intrinsically self-association prone protein, it was imperative to investigate the effect on glycation on aggregation of the protein. Dynamic light scattering data (Table 1, Figure 5) of the control non-incubated protein gave three different peaks of the native protein at around 258 nm, 76 nm and 12 nm justifying its spontaneous aggregation. The glycated protein showed initial increase in hydrodynamic radius from 11.6 nm to 14.6 nm which was probably attributable to the increased solvent hydration shell around the protein. Accordingly, the diameters of the other two species were also increased to 90 nm and 370 nm respectively. However, the corresponding incubated protein revealed only two peaks. This can be probably attributable to the fact that the protein due to sustained incubation at an elevated temperature somewhat lost its natural ability to associate into a plethora of multimeric entities. In case of the glycated protein, three peaks were visible till the sixth day of incubation and thereafter the structural changes inflicted resulted in only one discernible peak on the 12th day. However, the average size of the entities in glycated proteins became significantly smaller than the incubated proteins after 6 days of incubation. This was at the same time indicative of formation of structured entity in the glycated sample and also was in unison to our assumption that prolonged incubation at elevated temperature brought about thermal aggregation of non-resulting into bigger entities. The aggregation propensity was further verified using anisotropy of FITC conjugated BSA and native PAGE. Through the entire period of incubation, the ribosylated BSA-FITC conjugates had significantly lower anisotropy values as compared to their non glycated counterparts incubated under same conditions (Table 2). This was again testimonial to the fact that ribosylation offered protection to the protein from thermal denaturation and resultant aggregation. The proteins were subsequently subjected to progressively higher temperatures (25 °C–85 °C) for 1 h and their migration patterns were analyzed by Native PAGE (Figure 6). The unincubated control protein was seen to form higher MW aggregates at higher temperatures (Figure 6A). In contrast, the positions of the bands for the glycated samples were more or less constant (Figure 6B). The incubated protein did not present any conclusive picture since most of the bands were rendered smeary due to thermal denaturation (Figure 6C). This was again an added testimonial to the integrity of the glycated protein in spite of being kept at elevated temperature (37 °C) for more than a week.
Figure 3.
A) ANS binding profile of ribosylated and incubated proteins during progression of glycation. Error bars represent the deviations observed in triplicate data sets B) Corresponding fluorescence spectra of A; the trend reveals reduced ANS binding between the period of 5–12 days.
Figure 4.
A) NPN Binding spectra of ribosylated and incubated proteins during progression of glycation. Error bars represent the deviations observed in triplicate data sets B) Corresponding fluorescence spectra of A; the trend shows reduced NPN binding indicating suppression of hydrophobicity between 8 to 12 days period.
Table 1.
Size distribution of ribosylated and incubated proteins during different times of glycation.
| Sample | Hydrodynamic radius (d.nm) | Polydispersity index (P.I) | |
|---|---|---|---|
| BSA | 258.6 ± 27.88 11.63 ± 0.9924 76.09 ± 7.286 |
1 | |
| Incubated | Day4 | 72.99 ± 5.361 4.157 ± 0.1266 |
1 |
| Day6 | 264.1 ± 26.81 9.373 ± 0.6883 |
0.986 | |
| Day8 | 915.2 ± 161.1 1.002 ± 0.114 |
1 | |
| Day10 | 271.6 ± 19.83 8.966 ± 0.5268 |
1 | |
| Day12 | 385.1 ± 43.02 89.85 ± 7.137 |
1 | |
| Glycated | Day4 | 360.7 ± 59.3 14.67 ± 1.186 |
0.726 |
| Day6 | 376.3 ± 52.06 89.95 ± 9.508 13.86 ± 1.037 |
1 | |
| Day8 | 393.3 ± 66.93 117.2 ± 15.18 |
0.858 | |
| Day10 | 218.7 ± 29.19 21.4 ± 1.037 |
0.870 | |
| Day12 | 135 ± 9.236 | 1 | |
Protein samples at a concentration of 0.1 mg/ml in 0.05 M phosphate buffer with 0.05 M Nacl were used for the studies.
Figure 5.
Size distribution of glycated (G–K) and incubated (B–F) BSA (0.1 mg/ml) A) BSA B) 4th day incubated BSA C) 6th day incubated BSA D)8th day incubated BSA E) 10th day incubated BSA F) 12th day incubated BSA G)4th day glycated BSA H) 6th day glycated BSA I) 8th day glycated BSA J)10th day glycated BSA G) 12th day glycated BSA.
Table 2.
Anisotropy values of glycated BSA incubated for different days along with their respective incubated BSA.
| Sample | R - value | G - factor | |
|---|---|---|---|
| BSA | 0.126 ± 0.008 | 1 | |
| Incubated | Day4 | 0.132 ± 0.007 | 1 |
| Day6 | 0.124 ± 0.01 | 1 | |
| Day8 | 0.108 ± 0.009 | 1 | |
| Day10 | 0.062 ± 0.007 | 1 | |
| Day12 | 0.093 ± 0.007 | 1 | |
| Glycated | Day4 | 0.062 ± 0.005 | 1 |
| Day6 | 0.064 ± 0.005 | 1 | |
| Day8 | 0.059 ± 0.006 | 1 | |
| Day10 | 0.065 ± 0.0048 | 1 | |
| Day12 | 0.062 ± 0.0053 | 1 | |
Experiments were performed thrice and mean R values have been reported.
Figure 6.
Native PAGE analysis of A) control, B) glycated and C) incubated proteins (10 μg each) after incubation at increasing temperatures. In each case lanes have been labelled with the respective temperature of incubation.
There have been contrasting reports regarding the effect of glycation on aggregation of the protein. While several studies have shown that glycation leads to aggregation of the protein [65, 66], it was found to inhibit/slow down aggregation and resultant plaque formation in most of the amyloidogenic proteins [19]. In the present studies, it was clearly shown that prolonged incubation at 37 °C brought about thermal aggregation of BSA mediated by increased contact of hydrophobic interfaces. Glycation delayed this thermal aggregation but promoted formation of globular amyloid aggregates (as studied later). Therefore, it was apparent that aggregation caused by thermal denaturation and glycation were mechanistically different from each other. Probably, it is safer to state that glycation induced aggregation is specific and dependent on both the nature of the protein molecules as well as the sugar attached.
3.4. Glycation causes gross structural alteration of BSA
Since there was a marked change in aggregation propensity of the native protein after ribosylation, a corresponding structural change was also imperative. BSA has two tryptophan residues at positions 134 and 212 respectively [67]. Trp-134 is fully solvent accessible being localized in the second helix of the first domain whereas Trp-212 is localized in a hydrophobic pocket of domain II. Therefore, BSA is an ideal model for probing structural transition due to polarity changes [30, 68, 69]. Especially, Trp-134 fluorescence is widely used to gain preliminary idea about the helix integrity of the protein [70]. There was a drastic decrease in fluorescence of ribosylated proteins together with a red shift of about 55 nm (Figure 7). However, for the control incubated protein samples, a higher fluorescence was observed without any significant change in emission maxima. The observations were in unison with our hypothesis that following ribosylation there was a gross increase in polarity around the tryptophan microenvironment due to partial denaturation of the glycated protein which accounted for the red shift. In case of the incubated proteins, there was only a slight red shift which was indicative of a perturbation in structural integrity of the protein. Additionally, there was a marked decrease in fluorescence intensity of the ribosylated protein from 8th to 11th day of incubation and it again increased on the 12th day. This could be accounted for by the fact that due to formation of a compact aggregate of ribosylated BSA, tryptophan fluorescence was quenched due to close proximity with peptide bonds [71].
Figure 7.
Intrinsic tryptophan fluorescence of glycated proteins in comparison to incubated protein samples during the period of progression of glycation.
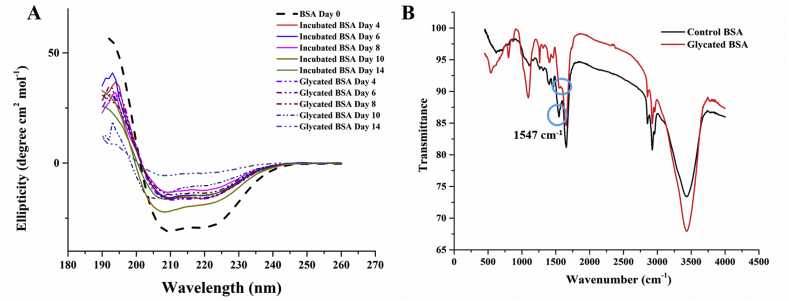
Since there was a gross indication in disruption of the helical structure of the protein from intrinsic tryptophan fluorescence, the structural changes following glycation were further investigated with CD and FT-IR spectroscopy (Figure 8A and Figure 8B). Alpha helicity decreased gradually in ribosylated sample by more than 7% on the 8th day of incubation (Table 3). However, the loss in helical structure was more drastic till the 10th day of incubation for the control incubated sample without glycation. This was again consistent with our observation that glycation provided resistance again thermal denaturation of the protein. However at later stages (beyond 12 days), the structure of the glycated protein collapsed completely. In order to further warrant the observations, FT-IR spectra of lyophilized protein samples (10th day) were obtained. Alongside CD, FT-IR is also a powerful tool to investigate the changes in secondary structure of proteins by through the study of amide bands [72]. The Amide I band (between 1600 and 1700 cm−1) is mainly associated with the peptide backbone C=O stretching vibration. The Amide II (between 1510 and 1580 cm−1) region is more complex arising out of in-plane N–H bending and C–N and C–C stretching vibrations (1700−1600 cm−1). Second derivative spectra of the protein revealed that the 1547 cm−1 amide II band was almost non-existent in the glycated sample which was indicative of substantial loss of alpha helicity of protein.
Figure 8.
A) CD spectra of ribosylated and incubated sample showing substantial loss of alpha helicity (retention of alpha helicity greater in ribosylated protein as compared to incubated protein) B) FT-IR spectra (of lyophilized protein samples) showing loss of alpha helicity as indicated by reduced 1547 cm−1 amide II band.
Table 3.
Alpha helix contents of glycated and incubated BSA samples during progression of glycation.
| Period of glycation (in day) | Glycated BSA | Incubated BSA |
|---|---|---|
| 4 | 53.39 | 52.77 |
| 6 | 51.95 | 49.61 |
| 8 | 46.69 | 42.59 |
| 10 | 36.308 | 34.49 |
| 14 | 20.43 | 28.41 |
| Control BSA | 49.67 | |
Alpha helicity was determined from the average of mean residual ellipticity values from three independent experiment sets.
3.5. Ribosylation imparts thermal stability
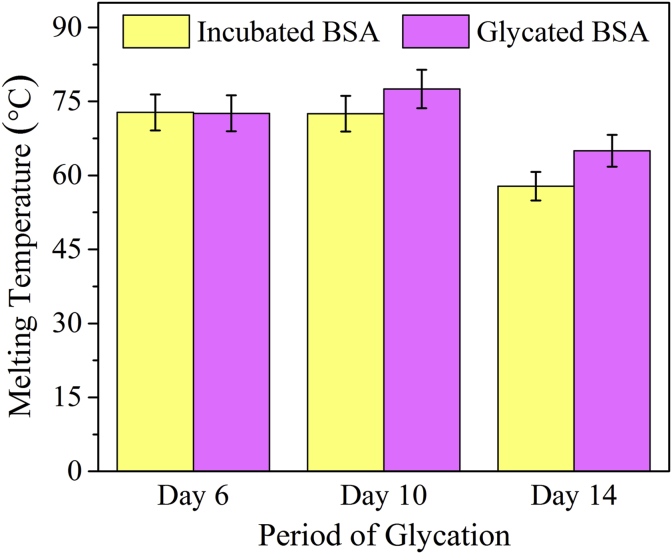
It was predicted from structural analysis that the glycated protein was more thermostable than its native counterpart. Melting points of the glycated and incubated protein samples were calculated from temperature dependent UV absorbance data at three different phases a) initial stage (4th-6th day), intermediate stage (8-12th day) and late stage (13th to 15th day) (Figure 9) to evaluate the thermal stability during the progression of glycation. Tm of incubated and glycated proteins from the 6th day sample (representative of the initial stage) were the same (72.5 °C) and similar to that of the control unincubated protein. However, in case of the 10th day sample (representative of the intermediate stage), it was raised by about 5 °C indicating enhanced thermostability of the glycated protein. At the same time point, Tm of the incubated protein was grossly unaltered. Thermostability of both glycated and incubated samples decreased drastically during the final stages of glycation as assessed from the 14th day sample. The data was in accordance with those acquired from assessment of hydrophobicity and CD spectrum and indicated that thermostability of the protein was visible only during the intermediate stage (day 8–12) and was lost subsequently during subsequent advanced stages of glycation.
Figure 9.
Absorbance based melting temperatures of different protein samples during initial (6 days), intermediate (10 days) and late stages (14 days) of glycation. Error bar represent the deviation from five individual data sets.
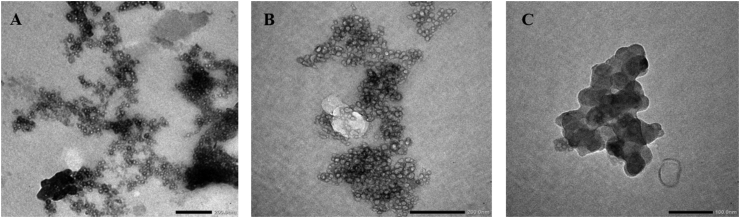
The effect of thermal denaturation on the glycated protein was also examined by TEM imaging after incubation at 60 °C for one hour (Figure 10). Globular morphology of glycated protein was found to be preserved in-spite of the severe heat treatment (Figure 10B), however, it was significantly more aggregated as compared the control non-incubated protein (Figure 10A). The non-glycated protein treated under similar conditions was grossly denatured and converted into a structureless entity (Figure 10C).
Figure 10.
TEM imaging of glycated and incubated BSA (10th day) acquired after subjecting to thermal incubation at 60 °C for one hour. A) Control non-incubated protein B) Glycated protein C) Non-glycated but incubated protein. Scale bar represents 200 nm in figures 9A and 9B and 100 nm in 9C.
3.6. Globular amyloid formation in BSA occurs due to glycation and does not owe to thermal aggregation
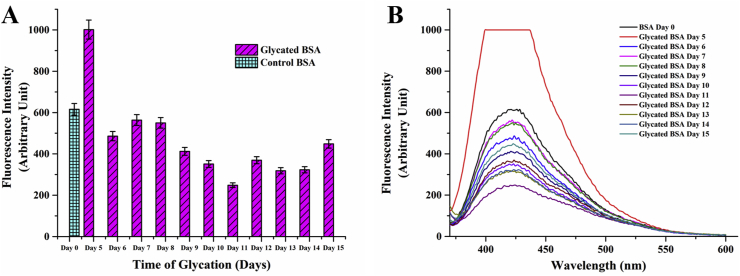
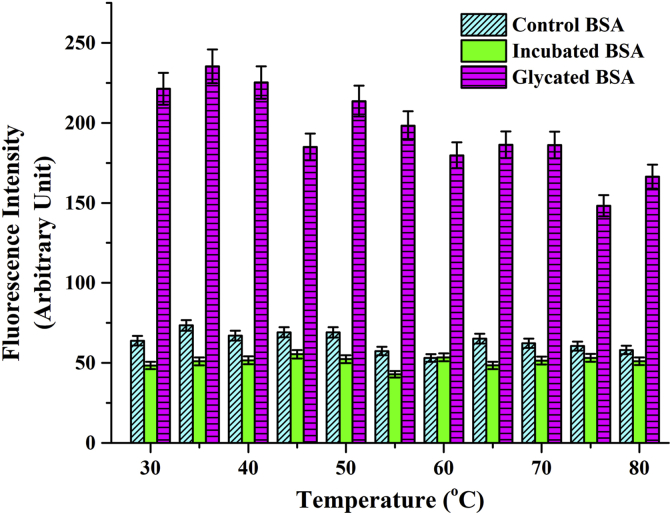
There has been several reports of Advanced Glycated End Products developing into amyloid like aggregates [16, 73] suggesting a direct link between type 2 diabetes and neurodegenerative disorders [74]. However, understanding the structural basis of this correlation has not been simple. Glycation has been found to both promote [17] and inhibit [19] amyloid fibril formation. Moreover, the amyloid like aggregates generated from non-amyloidogenic proteins like serum albumin have been either shown to be transformed into β-fibrils hallmark of classical amyloid plaques [11, 75] or retain their globular structure showing amyloid specific histochemistry [18] depending upon the nature of the attached sugar. In addition, thermal aggregation of serum albumin had also been shown to induce amyloid formation [76]. We subjected the ribosylated proteins obtained at different times of glycation to Thioflavin T binding analyses. Thio-T fluorescence was found to increase significantly from 9th day of incubation and was highest on the 12th day of glycation (Figure 11A and 11B). Subsequently, the fluorescence decreased gradually. This was testimonial to the fact that amyloid like behaviour of the protein was present only during the intermediate stages of glycation and wasn't necessarily a hallmark of the Advanced Glycation End Product arising out of the protein-ribose adduct. Heat induced fibrillation, as observed for the control non-glycated sample was also present during the initial stages of glycation (till 6th day) which then decreased to a basal level. This indicated that Thio-T binding property of the protein occurred distinctly for a) thermal aggregation, and b) for glycation. In order to further confirm the fact that ribosylation induced amyloidogenic property of the protein was independent of thermal fibrillation, an additional temperature dependent binding profile of Thio-T was carried out (Figure 12) with the 10th day ribosylated protein. Fluorescence intensity was highest at 35 °C and decreased gradually for higher temperatures. The incubated and unincubated controls subjected to similar conditions gave no noticeable Thioflavin T binding. This confirmed our hypothesis that Thio-T binding was specific for ribosylation and amyloidogenic transition of the protein simply could not be induced instantaneously by subjecting the protein to higher temperatures. Thermal aggregation can only be accounted for such structural transitions if the protein is subjected to elevated temperatures for much longer time periods. The glycated protein aggregates, inspite of showing Thio-T binding didn't reveal formation of classical fibrillar structure in TEM and retained their globular structures. Accordingly, they also didn't respond to the Congo red birefringence assay, another hallmark of the amyloid fibrils. Therefore, the amyloids arising out of glycation were non-classical globular in nature as has also been observed earlier for ribosylated proteins [18]. Deposition of non-canonical globular amyloid aggregates showing classical histochemistry like Thioflavin-T binding or Congo red birefringence have also been reported for other pathophysiological conditions [77, 78]. Therefore, amyloid formation is often more ubiquitous and versatile than is believed and drawing a structural consequence arising out of glycationis evidence based and cannot be generalized.
Figure 11.
A) Thioflavin T binding profiles of glycated vs incubated proteins during the entire period of glycation. Error bars represent the deviations observed in triplicate data sets B) Corresponding fluorescence spectra of A; the data shows amyloid like formation between 9th and 12th days.
Figure 12.
Thioflavin T binding to 10th day ribosylated BSA at different temperatures indicates that amyloid like formation is specific for the glycated protein and is not thermally induced.
3.7. Glycation causes significant change in affinity of BSA towards various substrates
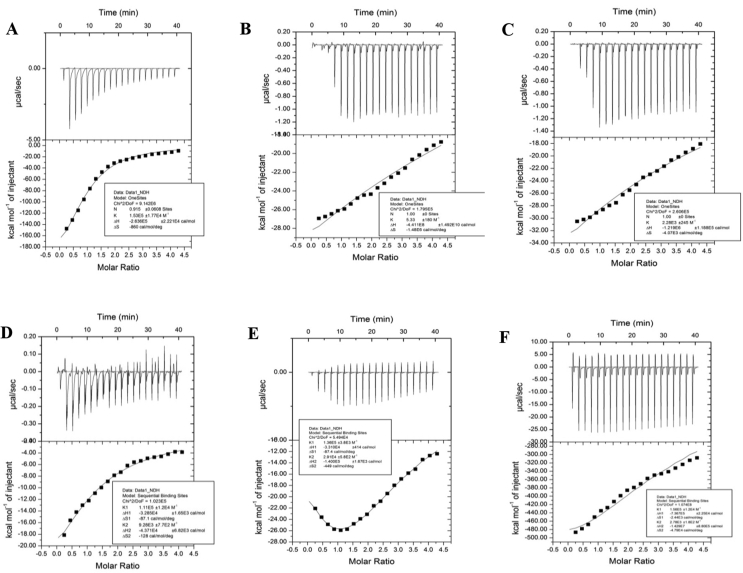
Residual substrate binding ability of BSA after ribosylation was checked with two standard polyphenols which are physiological substrates of the protein, curcumin and resveratrol. BSA has got a single binding site for curcumin [79] and two sites for resveratrol [80]. The observed titration curve (Figure 13 A-F) with a single binding site of curcumin indicated an enthalpy driven moderately strong binding of the substrate largely mediated by non-covalent interactions [81]. The Wiseman parameter “c” of 1.836 was a testimonial of this strong binding [82] (Table 4). However, the substrate binding was completely abolished for the 10th day glycated sample with an almost 100 fold decrease in binding constant. Consequently, the Wiseman parameter was decreased drastically to 0.027. It seemed that this drastic fall in activity of the native protein owed more to thermal denaturation since the non-glycated control protein subjected to incubation under similar conditions underwent even more drastic effects with binding constant being decreased more than 105 fold. Binding of resveratrol was also found to be exothermic and enthalpy driven in nature. However, completely dissimilar trends were noted, however, for the other substrate resveratrol having two known binding sites in BSA. For the first site, binding constant increased from 1.11 × 105to 1.56 × 105 mol−1. However, for the second site, resveratrol bound with greater affinity to control BSA than the glycated counterpart as binding constant declined from 9.28 × 103 to 2.78 × 105 mol−1. The fate of the incubated protein without glycation was an intermediate one as its binding affinity with respect to the first site was not improved as significantly as for the glycated protein but it suffered a lesser decline in affinity with its substrate as compared to the first site. In all cases, where the Wiseman parameter was evaluated to be much lesser than 1, the corresponding thermodynamic parameters of binding were not evaluated since those cases represented very poor ligand-protein interaction. It was evident that the gross structural transition entailed due to glycation brought about significant loss in functionality of the protein. In fact, this also explains the widespread toxicity caused by glycation induced changes in cellular proteome [83]. In addition to inducing structural perturbation, glycation can also interfere directly by denying physical access of the ligands to their respective binding pockets in the protein. However, this cannot be warranted without actual knowledge of the location of lysine and arginine residues which have been modified by glycation.
Figure 13.
Assessment of residual substrate binding capability of control (A,D), incubated (B,E) and glycated (C,F) BSA against curcumin (A,B,C) and resveratrol (D,E,F) by Isothermal Titration Calorimetry.
Table 4.
Evaluation of residual ligand binding capability of glycated BSA against resveratrol and curcumin.
| Ligand | Sample | Number of binding sites (n) | Binding constant (kd) (mol−1) |
Wiseman parameter(c) (binding constant x receptor concentration) | Free energy change (ΔG) (KJmol−1) |
Entropy change (ΔS) (Kcal/mol/deg) |
Enthalpy change (ΔH) (Kcal/mol) |
|---|---|---|---|---|---|---|---|
| Curcumin | BSA | 1 | 1.53 × 105 ± 1.77 × 104 | 1.836 | -6.72 | -0.86 | -2.64 × 102 ± 22.21 |
| Incubated BSA | 1 | 5.33 ± 180 | 0.639 × 10−4 | --- | --- | --- | |
| Glycated BSA | 1 | 2.28 × 103 ± 245 | 0.027 | --- | --- | --- | |
| Resveratrol | BSA | 2 | 1.11 × 105 ± 1.2 × 104 9.28 × 103 ± 7.7 × 102 |
1.332 0.11136 |
-6.874 -5.556 |
-0.087 -0.128 |
-32.8 ± 1.65 -43.7 ± 6.82 |
| Incubated BSA | 2 | 1.36 × 105 ± 3.8 × 103 2.91 × 104 ± 5.8 × 102 |
1.632 0.349 |
-7.055 -6.198 |
-0.0874 -0.449 |
-33.1 ± 0.414 -1.4 × 102 ± 1.67 |
|
| Glycated BSA | 2 | 1.56 × 105 ± 1.2 × 104 2.78 × 103 ± 1.8 × 102 |
1.872 0.033 |
-58.88 --- |
-2.44 --- |
-7.86 × 102 ± 22.5 --- |
4. Conclusion
Glycation of proteins and other macromolecules occurs as inevitable consequence of carbohydrate based regimen of our diet. Slow accumulation and attachment of sugar residues to proteins gradually bring about a structural transition in them which eventualize into terminally misfolded aggregates termed as Advanced Glycation End Products. The role of glycation in amyloid formation is well documented and many studies have reported thus far that glycation may accelerate or inhibit formation of amyloid plaques. Based upon the reports obtained thus far, glycation has been mostly associated with non-canonical amyloidogenic aggregates from non amyloidogenic proteins without the formation of classical cross β fibrillar structure. We have shown in this study that dietary sugar induced glycation and amyloid formation is not a generic property of the ribosylated BSA; rather it occurs transiently during a specific phase of progression of glycation, which finally eventualize into irreversibly misfolded aggregate termed as Advanced Glycation End Product. We have also shown that these globular amyloids possess enhanced thermostability, have minimal solvent accessible hydrophobic interface and therefore not prone to hydrophobic aggregation. In addition, we have been able to dissect the amyloidogenic behavior arising out of glycation from that caused by sheer thermal aggregation of proteins. The observation of enhanced thermostability of amyloids although doesn't come as a surprise because of stubborn recalcitrant nature of amyloid plaques or fibrils formed intracellularly. However, these globular amyloids do raise an important issue, are these clearable by the RAGE axis eventually? AGE molecules are recognized chiefly by their ability to bind to specific receptors on the surfaces of predatory immune cells such as macrophages and initiate a plethora of cellular defense mechanisms such as release of inflammatory cytokines and elicitation of oxidative stress response [8, 10]. Probably, the AGE-RAGE pathway has evolved as a defense mechanism to sequester these toxic protein aggregates from the cellular microenvironment, because presence of these aggregates can mediate aggregation and malfunctioning of other cellular proteins. However, whether the intermediates of glycation pathway can also bind to RAGE and initiate downstream signaling is yet to be deciphered. Under the circumstances, the present studies are believed to provide significant insights to predict glycation associated malfunctioning of cellular and extracellular proteins and design therapeutics to arrest the transformation of the Schiff base adducts to obnoxious amyloids and AGE molecules.
Declarations
Author contribution statement
S. Banik: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
M. Bhattacharyya: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
A. Das: Performed the experiments; Wrote the paper.
A. Ghosh and M. Guria: Performed the experiments.
P. Basak: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
A. Pramanik and R. Majumder: Performed the experiments; Contributed reagents, materials, analysis tools or data.
S. Hazra: Analyzed and interpreted the data.
Funding statement
This work was supported by STAR College Scheme, Department of Biotechnology, Govt. of India. A. Das was supported by a Fellowship provided by University Grants Commission, Govt. of India.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
TEM images were acquired from the facility at University Science Instrumentation Centre, University of Burdwan with technical supervision from Dr. Subhajit Karmakar and Mr. Debanjan Mukherjee.
Contributor Information
Maitree Bhattacharyya, Email: bmaitree@gmail.com.
Samudra Prosad Banik, Email: samudrapb@gmail.com.
References
- 1.Kumar V., Sami N., Kashav T., Islam A., Ahmad F., Hassan M.I. Protein aggregation and neurodegenerative diseases: from theory to therapy. Eur. J. Med. Chem. 2016;124:1105–1120. doi: 10.1016/j.ejmech.2016.07.054. [DOI] [PubMed] [Google Scholar]
- 2.Verdugo M., Encinar J.R., Costa-Fernández J.M., Menendez-Miranda M., Bouzas-Ramos D., Bravo M., Quiroz W. Study of conformational changes and protein aggregation of bovine serum albumin in presence of Sb (III) and Sb (V) PloS One. 2017;12(2) doi: 10.1371/journal.pone.0170869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2004;1739(1):5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 4.del Monte F., Agnetti G. Protein post-translational modifications and misfolding: new concepts in heart failure. Proteomics–Clin. Appl. 2014;8(7-8):534–542. doi: 10.1002/prca.201400037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q., Ames J.M., Smith R.D., Baynes J.W., Metz T.O. A perspective on the Maillard reaction and the analysis of protein glycation by mass spectrometry: probing the pathogenesis of chronic disease. J. Proteome Res. 2009;8(2):754–769. doi: 10.1021/pr800858h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gkogkolou P., Böhm M. Advanced glycation end products: key players in skin aging? Derm. Endocrinol. 2012;4(3):259–270. doi: 10.4161/derm.22028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luthra M., Balasubramanian D. Nonenzymatic glycation alters protein structure and stability. A study of two eye lens crystallins. J. Biol. Chem. 1993;268(24):18119–18127. [PubMed] [Google Scholar]
- 8.Ott C., Jacobs K., Haucke E., Santos A.N., Grune T., Simm A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014;2:411–429. doi: 10.1016/j.redox.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salahuddin P., Fatima M.T., Abdelhameed A.S., Nusrat S., Khan R.H. Structure of amyloid oligomers and their mechanisms of toxicities: targeting amyloid oligomers using novel therapeutic approaches. Eur. J. Med. Chem. 2016;114:41–58. doi: 10.1016/j.ejmech.2016.02.065. [DOI] [PubMed] [Google Scholar]
- 10.Ramasamy R., Yan S.F., Schmidt A.M. Receptor for AGE (RAGE): signaling mechanisms in the pathogenesis of diabetes and its complications. Ann. N. Y. Acad. Sci. 2011;1243:88. doi: 10.1111/j.1749-6632.2011.06320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouma B., Kroon-Batenburg L.M., Wu Y.-P., Brünjes B., Posthuma G., Kranenburg O., de Groot P.G., Voest E.E., Gebbink M.F. Glycation induces formation of amyloid cross-β structure in albumin. J. Biol. Chem. 2003;278(43):41810–41819. doi: 10.1074/jbc.M303925200. [DOI] [PubMed] [Google Scholar]
- 12.Ko S.-Y., Ko H.-A., Chu K.-H., Shieh T.-M., Chi T.-C., Chen H.-I., Chang W.-C., Chang S.-S. The possible mechanism of advanced glycation end products (AGEs) for Alzheimer’s disease. PloS One. 2015;10(11) doi: 10.1371/journal.pone.0143345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S.M., Rabbani N., Tabrez S., Ul Islam B., Malik A., Ahmed A., AAlsenaidy M., M Alsenaidy A. Glycation induced generation of amyloid fibril structures by glucose metabolites. Protein Pept. Lett. 2016;23(10):892–897. doi: 10.2174/0929866523666160831153858. [DOI] [PubMed] [Google Scholar]
- 14.Iannuzzi C., Irace G., Sirangelo I. Differential effects of glycation on protein aggregation and amyloid formation. Front. Mol. Biosci. 2014;1:9. doi: 10.3389/fmolb.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iannuzzi C., Maritato R., Irace G., Sirangelo I. Glycation accelerates fibrillization of the amyloidogenic W7FW14F apomyoglobin. PloS One. 2013;8(12) doi: 10.1371/journal.pone.0080768. e80768. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Arasteh A., Farahi S., Habibi-Rezaei M., Moosavi-Movahedi A.A. Glycated albumin: an overview of the in vitro models of an in vivo potential disease marker. J. Diabetes Metab. Disord. 2014;13(1):49. doi: 10.1186/2251-6581-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obrenovich M.E., Monnier V.M. Glycation stimulates amyloid formation. Sci. Aging Knowl. Environ. 2004;2:3. doi: 10.1126/sageke.2004.2.pe3. 3. [DOI] [PubMed] [Google Scholar]
- 18.Wei Y., Chen L., Chen J., Ge L., He R.Q. Rapid glycation with D-ribose induces globular amyloid-like aggregations of BSA with high cytotoxicity to SH-SY5Y cells. BMC Cell Biol. 2009;10(1):1–15. doi: 10.1186/1471-2121-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emendato A., Milordini G., Zacco E., Sicorello A., Dal Piaz F., Guerrini R., Thorogate R., Picone D., Pastore A. Glycation affects fibril formation of Aβ peptides. J. Biol. Chem. 2018;293(34):13100–13111. doi: 10.1074/jbc.RA118.002275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno-Gonzalez I., Soto C. Semin. Cell Dev. Biol. Elsevier; 2011. Misfolded protein aggregates: mechanisms, structures and potential for disease transmission; pp. 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G., Zhong Q. Glycation of whey protein to provide steric hindrance against thermal aggregation. J. Agric. Food Chem. 2012;60(38):9754–9762. doi: 10.1021/jf302883b. [DOI] [PubMed] [Google Scholar]
- 22.Trębacz H., Szczęsna A., Arczewska M. Thermal stability of collagen in naturally ageing and in vitro glycated rabbit tissues. J. Therm. Anal. Calorim. 2018;134(3):1903–1911. [Google Scholar]
- 23.Liu G., Zhong Q. High temperature-short time glycation to improve heat stability of whey protein and reduce color formation. Food Hydrocolloids. 2015;44:453–460. [Google Scholar]
- 24.Coussons P.J., Jacoby J., McKay A., Kelly S.M., Price N.C., Hunt J.V. Glucose modification of human serum albumin: a structural study. Free Radic. Biol. Med. 1997;22(7):1217–1227. doi: 10.1016/s0891-5849(96)00557-6. [DOI] [PubMed] [Google Scholar]
- 25.Mendez D.L., Jensen R.A., McElroy L.A., Pena J.M., Esquerra R.M. The effect of non-enzymatic glycation on the unfolding of human serum albumin. Arch. Biochem. Biophys. 2005;444(2):92–99. doi: 10.1016/j.abb.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Rondeau P., Armenta S., Caillens H., Chesne S., Bourdon E. Assessment of temperature effects on β-aggregation of native and glycated albumin by FTIR spectroscopy and PAGE: relations between structural changes and antioxidant properties. Arch. Biochem. Biophys. 2007;460(1):141–150. doi: 10.1016/j.abb.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Rondeau P., Navarra G., Cacciabaudo F., Leone M., Bourdon E., Militello V. Thermal aggregation of glycated bovine serum albumin. Biochim. Biophys. Acta Protein Proteonomics. 2010;1804(4):789–798. doi: 10.1016/j.bbapap.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Watkins N.G., Thorpe S.R., Baynes J.W. Glycation of amino groups in protein. Studies on the specificity of modification of RNase by glucose. J. Biol. Chem. 1985;260(19):10629–10636. [PubMed] [Google Scholar]
- 29.Sharifi E., Sattarahmady N., Habibi-Rezaei M., Farhadi M., Sheibani N., Ahmad F., Moosavi-Movahedi A.A. Inhibitory effects of β-cyclodextrin and trehalose on nanofibril and AGE formation during glycation of human serum albumin. Protein Pept. Lett. 2009;16(6):653–659. doi: 10.2174/092986609788490212. [DOI] [PubMed] [Google Scholar]
- 30.Das A., Basak P., Pattanayak R., Kar T., Majumder R., Pal D., Bhattacharya A., Bhattacharyya M., Banik S.P. Trehalose induced structural modulation of Bovine Serum Albumin at ambient temperature. Int. J. Biol. Macromol. 2017;105:645–655. doi: 10.1016/j.ijbiomac.2017.07.074. [DOI] [PubMed] [Google Scholar]
- 31.Das A., Basak P., Pramanick A., Majumder R., Pal D., Ghosh A., Guria M., Bhattacharyya M., Banik S.P. Trehalose mediated stabilisation of cellobiase aggregates from the filamentous fungus Penicillium chrysogenum. Int. J. Biol. Macromol. 2019;127:365–375. doi: 10.1016/j.ijbiomac.2019.01.062. [DOI] [PubMed] [Google Scholar]
- 32.Schönbrunn E., Eschenburg S., Luger K., Kabsch W., Amrhein N. Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. Proc. Natl. Acad. Sci. U.S.A. 2000;97(12):6345–6349. doi: 10.1073/pnas.120120397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H.-L., Zhang L.-Y., Zhuang S.-L., Ni C.-X., Shang H.-W. Fluorescence investigation on the interaction of a prevalent competitive fluorescent probe with entomic odorant binding protein. Spectrosc. Lett. 2013;46(7):527–534. [Google Scholar]
- 34.Liu W., Cohenford M.A., Frost L., Seneviratne C., Dain J.A. Inhibitory effect of gold nanoparticles on the D-ribose glycation of bovine serum albumin. Int. J. Nanomed. 2014;9:5461. doi: 10.2147/IJN.S70777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulaj G., Kortemme T., Goldenberg D.P. Ionization− reactivity relationships for cysteine thiols in polypeptides. Biochemistry. 1998;37(25):8965–8972. doi: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- 36.Hungerford G., Benesch J., Mano J.F., Reis R.L. Effect of the labelling ratio on the photophysics of fluorescein isothiocyanate (FITC) conjugated to bovine serum albumin. Photochem. Photobiol. Sci. 2007;6(2):152–158. doi: 10.1039/b612870j. [DOI] [PubMed] [Google Scholar]
- 37.Jain N., Mukhopadhyay S. Appl. Spectrosc. Sci. Nanomater. Springer; 2015. Applications of fluorescence anisotropy in understanding protein conformational disorder and aggregation; pp. 41–57. [Google Scholar]
- 38.Li S., Xing D., Li J. Dynamic light scattering application to study protein interactions in electrolyte solutions. J. Biol. Phys. 2004;30(4):313–324. doi: 10.1007/s10867-004-0997-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Majumder R., Banik S.P., Khowala S. AkP from mushroom Termitomyces clypeatus is a proteoglycan specific protease with apoptotic effect on HepG2. Int. J. Biol. Macromol. 2016;91:198–207. doi: 10.1016/j.ijbiomac.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 41.Majumder R., Banik S.P., Khowala S. Purification and characterisation of κ-casein specific milk-clotting metalloprotease from Termitomyces clypeatus MTCC 5091. Food Chem. 2015;173:441–448. doi: 10.1016/j.foodchem.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 42.Wilson C.J., Apiyo D., Wittung-Stafshede P. Role of cofactors in metalloprotein folding. Q. Rev. Biophys. 2004;37(3-4):285–314. doi: 10.1017/S003358350500404X. [DOI] [PubMed] [Google Scholar]
- 43.Majumder R., Sheikh L., Naskar A., Mukherjee M., Tripathy S. Depletion of Cr (VI) from aqueous solution by heat dried biomass of a newly isolated fungus Arthrinium malaysianum: a mechanistic approach. Sci. Rep. 2017;7(1):1–15. doi: 10.1038/s41598-017-10160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naskar A., Majumder R., Goswami M. Bioaccumulation of Ni (II) on growing cells of Bacillus sp.: response surface modeling and mechanistic insight. Environ. Technol. Innov. 2020:101057. [Google Scholar]
- 45.Naskar A., Majumder R., Goswami M., Mazumder S., Maiti S., Ray L. Implication of greener biocomposite bead for decontamination of nickel (II): column dynamics study. J. Polym. Environ. 2020;28(7):1985–1997. [Google Scholar]
- 46.Schechter A.N., Epstein C.J. Spectral studies on the denaturation of myoglobin. J. Mol. Biol. 1968;35(3):567–589. doi: 10.1016/s0022-2836(68)80015-4. [DOI] [PubMed] [Google Scholar]
- 47.Bolder S.G., Sagis L.M., Venema P., van der Linden E. Thioflavin T and birefringence assays to determine the conversion of proteins into fibrils. Langmuir. 2007;23(8):4144–4147. doi: 10.1021/la063048k. [DOI] [PubMed] [Google Scholar]
- 48.Banik S.P., Pal S., Ghorai S., Chowdhury S., Majumder R., Mukherjee S., Khowala S. In situ reversible aggregation of extracellular cellobiase in the filamentous fungus Termitomyces clypeatus. Biotechnol. Bioproc. Eng. 2012;17(5):925–936. [Google Scholar]
- 49.Zaidi N., Ahmad E., Rehan M., Rabbani G., Ajmal M.R., Zaidi Y., Subbarao N., Khan R.H. Biophysical insight into furosemide binding to human serum albumin: a study to unveil its impaired albumin binding in uremia. J. Phys. Chem. B. 2013;117(9):2595–2604. doi: 10.1021/jp3069877. [DOI] [PubMed] [Google Scholar]
- 50.Raghav A., Ahmad J., Alam K. Nonenzymatic glycosylation of human serum albumin and its effect on antibodies profile in patients with diabetes mellitus. PloS One. 2017;12(5) doi: 10.1371/journal.pone.0176970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubio-Ruiz M.E., Díaz-Díaz E., Cárdenas-León M., Argüelles-Medina R., Sánchez-Canales P., Larrea-Gallo F., Soria-Castro E., Guarner-Lans V. Glycation does not modify bovine serum albumin (BSA)-induced reduction of rat aortic relaxation: the response to glycated and nonglycated BSA is lost in metabolic syndrome. Glycobiology. 2008;18(7):517–525. doi: 10.1093/glycob/cwn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubey K., Anand B.G., Shekhawat D.S., Kar K. Eugenol prevents amyloid formation of proteins and inhibits amyloid-induced hemolysis. Sci. Rep. 2017;7:40744. doi: 10.1038/srep40744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polyanichko A., Mikhailov N., Romanov N., Baranova Y.G., Chikhirzhina E. Intermolecular interactions in solutions of serum albumin. Cell Tissue Biol. 2017;11(1):9–15. [PubMed] [Google Scholar]
- 54.Maruyama T., Katoh S., Nakajima M., Nabetani H. Mechanism of bovine serum albumin aggregation during ultrafiltration. Biotechnol. Bioeng. 2001;75(2):233–238. doi: 10.1002/bit.10001. [DOI] [PubMed] [Google Scholar]
- 55.Bhattacharya M., Jain N., Mukhopadhyay S. Insights into the mechanism of aggregation and fibril formation from bovine serum albumin. J. Phys. Chem. B. 2011;115(14):4195–4205. doi: 10.1021/jp111528c. [DOI] [PubMed] [Google Scholar]
- 56.Dasgupta M., Kishore N. Selective inhibition of aggregation/fibrillation of bovine serum albumin by osmolytes: mechanistic and energetics insights. PloS One. 2017;12(2) doi: 10.1371/journal.pone.0172208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J.-E., Sang J.C., Rodrigues M., Carr A.R., Horrocks M.H., De S., Bongiovanni M.N., Flagmeier P., Dobson C.M., Wales D.J. Mapping surface hydrophobicity of α-synuclein oligomers at the nanoscale. Nano Lett. 2018;18(12):7494–7501. doi: 10.1021/acs.nanolett.8b02916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z., Li Y., Jiang L., Qi B., Zhou L. Relationship between secondary structure and surface hydrophobicity of soybean protein isolate subjected to heat treatment. J. Chem. 2014;2014 [Google Scholar]
- 59.Tarar H.S., Roughton B.C., Camarda K.V. Comput. Aided Chem. Eng. Vol. 34. Elsevier; 2014. Designing optimum protein-excipient interactions using molecular docking simulations; pp. 441–446. [Google Scholar]
- 60.Cardamone M., Puri N. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992;282(2):589–593. doi: 10.1042/bj2820589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasymov O.K., Glasgow B.J. ANS fluorescence: potential to augment the identification of the external binding sites of proteins. Biochim. Biophys. Acta Protein Proteonomics. 2007;1774(3):403–411. doi: 10.1016/j.bbapap.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stefanowicz P., Kijewska M., Kluczyk A., Szewczuk Z. Detection of glycation sites in proteins by high-resolution mass spectrometry combined with isotopic labeling. Anal. Biochem. 2010;400(2):237–243. doi: 10.1016/j.ab.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Loh B., Grant C., Hancock R. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984;26(4):546–551. doi: 10.1128/aac.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weids A.J., Ibstedt S., Tamás M.J., Grant C.M. Distinct stress conditions result in aggregation of proteins with similar properties. Sci. Rep. 2016;6:24554. doi: 10.1038/srep24554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iram A., Alam T., Khan J.M., Khan T.A., Khan R.H., Naeem A. Molten globule of hemoglobin proceeds into aggregates and advanced glycated end products. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0072075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Linetsky M., Shipova E., Cheng R., Ortwerth B.J. Glycation by ascorbic acid oxidation products leads to the aggregation of lens proteins. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2008;1782(1):22–34. doi: 10.1016/j.bbadis.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarkar D. Probing the interaction of a globular protein with a small fluorescent probe in the presence of silver nanoparticles: spectroscopic characterization of its domain specific association and dissociation. RSC Adv. 2013;3(46):24389–24399. [Google Scholar]
- 68.Ghisaidoobe A.B., Chung S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: a focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014;15(12):22518–22538. doi: 10.3390/ijms151222518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ursache F.M., Aprodu I., Nistor O.V., Bratu M., Botez E., Stănciuc N. Probing the heat-induced structural changes in bovine serum albumin by fluorescence spectroscopy and molecular modelling. Int. J. Dairy Technol. 2017;70(3):424–431. [Google Scholar]
- 70.Ali M.F., Kaushik A., Gupta D., Ansari S., Jairajpuri M.A. Changes in strand 6B and helix B during neuroserpin inhibition: implication in severity of clinical phenotype. Biochim. Biophys. Acta Protein Proteonomics. 2020:140363. doi: 10.1016/j.bbapap.2020.140363. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y., Liu B., Yu H.-T., Barkley M.D. The peptide bond quenches indole fluorescence. J. Am. Chem. Soc. 1996;118(39):9271–9278. [Google Scholar]
- 72.Kong J., Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007;39(8):549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 73.Hsu Y.-H., Chen Y.-W., Wu M.-H., Tu L.-H. Protein glycation by glyoxal promotes amyloid formation by islet amyloid polypeptide. Biophys. J. 2019;116(12):2304–2313. doi: 10.1016/j.bpj.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vitek M.P., Bhattacharya K., Glendening J.M., Stopa E., Vlassara H., Bucala R., Manogue K., Cerami A. Advanced glycation end products contribute to amyloidosis in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 1994;91(11):4766–4770. doi: 10.1073/pnas.91.11.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiti F., Dobson C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009;5(1):15. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 76.Holm N.K., Jespersen S.K., Thomassen L.V., Wolff T.Y., Sehgal P., Thomsen L.A., Christiansen G., Andersen C.B., Knudsen A.D., Otzen D.E. Aggregation and fibrillation of bovine serum albumin. Biochim. Biophys. Acta Protein Proteonomics. 2007;1774(9):1128–1138. doi: 10.1016/j.bbapap.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 77.Demirhan B., Bilezikçi B., Kiyici H., Boyacioglu S. Globular amyloid deposits in the wall of the gastrointestinal tract: report of six cases. Amyloid. 2002;9(1):42–46. doi: 10.3109/13506120209072444. [DOI] [PubMed] [Google Scholar]
- 78.Makhlouf H.R., Goodman Z.D. Globular hepatic amyloid: an early stage in the pathway of amyloid formation: a study of 20 new cases. Am. J. Surg. Pathol. 2007;31(10):1615–1621. doi: 10.1097/PAS.0b013e318054e6b6. [DOI] [PubMed] [Google Scholar]
- 79.Bourassa P., Kanakis C., Tarantilis P., Pollissiou M., Tajmir-Riahi H. Resveratrol, genistein, and curcumin bind bovine serum albumin. J. Phys. Chem. B. 2010;114(9):3348–3354. doi: 10.1021/jp9115996. [DOI] [PubMed] [Google Scholar]
- 80.Latruffe N., Menzel M., Delmas D., Buchet R., Lançon A. Compared binding properties between resveratrol and other polyphenols to plasmatic albumin: consequences for the health protecting effect of dietary plant microcomponents. Molecules. 2014;19(11):17066–17077. doi: 10.3390/molecules191117066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Du X., Li Y., Xia Y.-L., Ai S.-M., Liang J., Sang P., Ji X.-L., Liu S.-Q. Insights into protein–ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17(2):144. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Turnbull W.B., Daranas A.H. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 2003;125(48):14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 83.Fournet M., Bonté F., Desmoulière A. Glycation damage: a possible hub for major pathophysiological disorders and aging. Aging Dis. 2018;9(5):880. doi: 10.14336/AD.2017.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]