Graphical abstract
Keywords: Antibiotic crisis, Bacterial predators, BALOs, Myxobacteria
Abbreviations: AR, antibiotic resistance; ARB, antibiotic-resistant bacteria; ARG, antibiotic-resistant gene; MDRB, multi-drug resistant bacteria; WHO, World Health Organization; HGT, horizontal gene transfer; SM, secondary metabolite; BALOs, Bdellovibrio and like organisms; OSMAC, one strain many compounds; BGC, biosynthetic gene cluster; NRPS, nonribosomal peptide synthetase; PKS, polyketide synthase; OMV, outer membrane vesicle
Summary
Discovery of antimicrobials in the past century represented one of the most important advances in public health. Unfortunately, the massive use of these compounds in medicine and other human activities has promoted the selection of pathogens that are resistant to one or several antibiotics. The current antibiotic crisis is creating an urgent need for research into new biological weapons with the ability to kill these superbugs. Although a proper solution requires this problem to be addressed in a variety of ways, the use of bacterial predators is emerging as an excellent strategy, especially when used as whole cell therapeutic agents, as a source of new antimicrobial agents by awakening silent metabolic pathways in axenic cultures, or as biocontrol agents. Moreover, studies on their prey are uncovering mechanisms of resistance that can be shared by pathogens, representing new targets for novel antimicrobial agents. In this review we discuss potential of the studies on predator-prey interaction to provide alternative solutions to the problem of antibiotic resistance.
1. Introduction
Antibiotics revolutionized medicine after the serendipitous discovery of penicillin by Fleming in the 1920s [1], the subsequent development of synthetic antimicrobials [2], the establishment of useful platforms for screening and isolation of antibiotic-producing microorganisms [3], and the development of the pharmaceutical industry. These antibacterial drugs have saved millions of lives, not only by combating fatal infectious diseases, but also by enabling physicians to make advances in surgery, in organ transplantation, in cancer chemotherapy, and in the use of artificial devices [4], [5]. For these reasons, the discovery and use of antibiotics in public health is one of the main pillars of modern medicine, considered one of the great achievements of the 20th century.
Antibiotics are natural products produced by microorganisms or their semisynthetic derivatives. These compounds have been a feature of the environment for a long time, so that bacteria have needed to evolve some forms of antibiotic resistance (AR) to survive. For instance, bacteria inactivate antibiotics by producing enzymes that modify them, but they can also alter their target of action, or prevent the antibiotics from accumulating, either because they are expelled using efflux pumps or by altering the permeability of the membrane [6], [7]. The antibiotic-resistant genes (ARGs) are favored in nature due to the selective pressure of natural production of antibiotics by bacteria. However, the use, overuse, or misapplication of these drugs, not only for therapeutic purposes, but also in animal feed and agriculture, have added an additional selective pressure and accelerated the ability of bacteria to evolve and, consequently, led to the inevitable emergence and spread of AR [8], [9], [10], [11]. Furthermore, bacteria can collect multiple resistance mechanisms, and this fact has promoted the appearance of the so-called superbugs or MDRB (multi-drug resistant bacteria), which are strains that have acquired resistance to a wide variety of antibiotics. The World Health Organization (WHO) has issued the priority pathogens list with thirteen human pathogens that have developed high levels of resistance across the world [12], [13]. Members of the so-called ESKAPE pathogens constitute an especially urgent threat to human health [14].
The first step in AR transmission is the transfer of ARGs of environmental bacteria through mobile genetic elements by horizontal gene transfer (HGT) to human or animal commensal bacteria or pathogens, generating resistant clones carrying ARGs that can then be spread in the environment [15], [16]. The interconnection between humans, animals and their ecosystems leads to the transmission of resistance. Although AR in hospitals or health centers represents a major concern, the dissemination is not restricted to clinical settings. Antibiotics eliminated by humans or animals are discharged into urban, agricultural or farm waste water, and can be incorporated by food crops through irrigation with those contaminated waters, increasing the selective pressure [17]. Moreover, waste water treatment plants, which receive a great variety of bacteria, including antibiotic-resistant bacteria (ARB) from various sources, act as reservoirs and offer convenient conditions for the interaction and exchange of ARGs by HGT, contributing to a broader dissemination [18]. Other types of industrial contaminants, such as metals, can contribute to AR, since many resistance mechanisms that protect against them, such as multidrug efflux pumps, also confer resistance to antibiotics [19], [20], [21]. Finally, physical forces contribute to their dissemination. The hyper-connection of the current world promotes the worldwide spread of AR. This has serious consequences, not only in the treatment of clinical and/or veterinary infections, but also in maintaining the balance of microbial communities across the biosphere.
Currently there are 700,000 deaths per year due to ARB. It is estimated that by 2025 many of the current antibiotics will be ineffective [22], and that by 2050 superbugs may cause 10 million deaths annually worldwide [23]. The problems derived from AR in healthcare could cost the world a trillion USD per year in healthcare, which would lead to a reduction of 2% to 3.5% in gross domestic product [23], [24].
Many public health agencies and economic and political institutions across the world recognize that AR is one of the great challenges for the 21st century. Many experts consider that we are initiating the post-antibiotic era, in which we will see the return of epidemics prevalent during the pre-antibiotic era [5]. But the antibiotic crisis that we have experienced in the last decade is caused not only by the appearance of MDRB, but also by the scarcity of resources dedicated to the search for new products, and, in this scenario, the pipeline for the discovery of new antibiotics is running dangerously low [4], [5], [12], [24], [25], [26], [27]. Since the development of resistance is inevitable, even if we make good and sparing use of antibiotics, the search for new antibiotics and alternative therapies is critical. The WHO, in a recent report, proposes several guidelines to effectively deal with AR to avoid drifting back to a pre-antibiotic era. These leading strategies, along with procedures to reduce antibiotic pressure and transmission of AR and ARGs, are: i) developing new ideas that question pre-existing forms of knowledge; ii) protecting research from purely commercial determinants; iii) creating new interpretations and strategies to address resistance; and iv) reorientating research to better understand the role of bacteria and human-microbe relationships [22]. In this context, laboratories around the world are developing new therapeutic approaches for reducing the AR burden, which include microbiota transplant procedures to displace ARB, genetic engineering to interrupt resistance genes (such as editing based on CRISPR/Cas), metagenomic engineering, phage cocktails against ARB, vaccines against pathogens, antimicrobial peptides, the use of metals (Cu, Ag, Ga), development of molecules that interfere with the ability of bacteria to communicate and group together in antibiotic-resistant biofilms, the search for new antimicrobial products, and the use of bacteria as therapeutic agents [16], [29].
Predatory bacteria play an essential role in these new lines of research [16], [28], [29], [30], [31]. The fact that these micropredators are designed to lyse and kill other bacteria, including MDRB and those pathogens that form biofilms, has led to them being proposed as a reasonable antibiotic alternative to be explored. The aim of this review is to collect and discuss the advances in the use of predatory bacteria in this fight against AR, either as biocontrol agents (“living antibiotics”) or as biological resources for innovative antimicrobial products (“microfactories”).
2. Predatory bacteria: what are they? How do they kill?
Predatory bacteria use other bacteria or yeasts as a food source; hence, they are able to actively hunt and kill their neighbors to later consume their macromolecules as nutrients. Various studies have shown that these predators are widely distributed in many natural and artificial environments (such as artificial microcosms, experiments in laboratory, waste waters treatment plants, or aquaculture plants) where they play an important role not only in maintaining microbial diversity, but also in shaping ecosystems [32], [33], [34], [35], [36], [37], [38], [39].
In the context of fighting AR, predatory bacteria in nature contribute to limiting the spread of ARGs in different environments, because they can prey upon ARB and degrade their DNA [40], [41], reducing the ARG pools and their spread to other bacteria by HGT via conjugation or transformation [42]. Moreover, they can also limit the possibility of the transfer of ARGs via transduction by preying on bacteria targeted by phage-resistant bacteria [43].
Bacterial hunting strategies can be categorized in two major groups: epibiotic and endobiotic predation [44]. In the epibiotic strategy, predators attack and consume the prey from the outside. Two different subgroups can be differentiated within this group: epibiotic “solitary hunters”, which, strongly attached to the prey cell envelope, suck their contents out before dividing into two daughter cells; and the epibiotic “cooperative hunters”, where individual cells cooperate within the community by sharing a combination of diffusible hydrolytic enzymes and secondary metabolites (SMs), including antibiotics, that kill and decompose the prey in a non-species-specific manner before consuming the macromolecules [44]. Whether hunting is truly cooperative or merely communal is not clear [45]. In the epibiotic lone-hunter subgroup the most studied genera are Micavibrio, Vampirococcus and Vampirovibrio, and the species Bdellovibrio exovorus, [32]. The cooperative (or communal) hunting strategy subgroup includes bacteria whose mode of attack is based on lysis at a distance that does not require contact with the prey, such as the genus Streptomyces [46], [47], and those that require close proximity to the prey cells such as myxobacteria [44], [48], [49], [50], [51], [52].
The endobiotic strategy is used by lone predatory bacteria that actively move to encounter their prey, stick to their outside, perforate and modify the prey cell wall by secreting hydrolytic enzymes, and penetrate the periplasmic or the cytoplasmic space, devouring it from within. Once the cycle is finished, predators move on to attack neighboring cells. A representative species of this group is Bdellovibrio bacteriovorus [32], [53], [54], [55].
Biochemical and microbiological studies carried out in several laboratories and analyses of the genomes and transcriptomes during predation (predatosomes) of obligate epibiotic predators such as Micavibrio aeruginosavorus, Vampirovibrio chlorellavorus, and B. exovorus [56], [57], [58], and endobiotic B. bacteriovorus and other BALOs (Bdellovibrio and like organisms), have revealed that they are able to synthesize and secrete an unusual number of hydrolytic proteins, such as lipases, glycanases, peptidases and proteases, which are probably involved in damaging and digesting prey cell structures [53], [58], [59], [60], [61]. They are, however, non-competent antibiotic producers [62], since their killing strategies are based on the active search and recognition of the prey, the specific binding to the external structures, and its degradation. This particular way of life has led researchers to mainly focus on its possible application in alternative live therapies in which the predators' whole cells can be used to kill or control the growth of other ARB; that is, the use of predators in bacterial therapy as biocontrol agents [53], [61].
However, the cooperative facultative epibiotic predators, which include genera of the two most important groups of bacterial SM and hydrolytic enzyme producers, Actinobacteria and myxobacteria, are by far the most promising resources for innovative bioactive natural products. Within Actinobacteria, the genus Streptomyces has been thoroughly investigated for many years for the isolation of antibiotics [63], [64], [65], but it has only recently been described as being a bacterial predator that kills other soil bacteria by secreting hydrolytic enzymes and SMs far away from the prey [46], [47]. Myxobacteria include broad prey range predators such as Myxococcus xanthus, Myxococcus flavescens, Myxococcus virescens, Myxococcus macrosporus, Corallococcus coralloides, Stigmatella aurantiaca, Chondromyces apiculatus and Chondromyces crocatus [44]. Compared with Bdellovibrio and BALOs, which exhibit a restricted prey spectrum, myxobacteria are able to kill a broad range of bacteria, including clinically relevant species [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78]. This is probably due to their capacity to secrete a plethora of lytic metabolites which, acting either in isolation or synergistically, are able to attack a wide variety of prey. They are able to produce cell wall-degrading enzymes, lipases, nucleases, polysaccharidases, proteases and, unlike many other antibiotic producers, various classes of antibiotics, some of which are hardly ever found as microbial SMs [79], [80], [81], [82]. Although they have been underestimated in the past, in the last decade they have emerged as excellent “microfactories”, with a high capacity to produce an unlimited number of previously unknown promising antimicrobials [82].
The use of epibiotic predators in alternative therapies in medicine or as biocontrol in different fields should be done in such a way that they contribute to the fight against MDBR, without increasing the accumulation of antibiotics in the environment that may generate AR.
3. BALOS as biocontrol agents
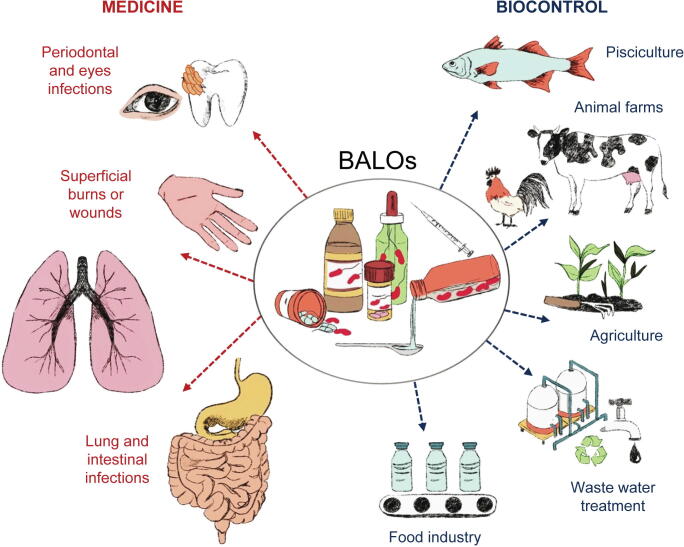
Endobiotic predators, such as B. bacteriovorus, other BALOs, and some lone hunters that follow an epibiotic strategy, such as M. aeruginosavorus, are generating interest for the treatment of intractable, antibiotic-resistant infections. Multiple studies, using human cells or serum, or animal models such as chicks, zebrafish, rabbits, guinea pigs, mice, and rats, and many in vitro studies have shown that predator whole cells or their enzymes can be used as therapeutic agents and as alternative or complementary applications in biological control [83], [84], [85], [86], [87], [88], [89] (Fig. 1 and Supplementary Table 1).
Fig. 1.
Applications of Bdellovibrio and BALOs in medicine and as biocontrol agents.
The potential of these predatory bacteria as a preferred or supplementary treatment of multidrug-resistant bacterial infections is supported by a range of evidence.
First, predators such as Bdellovibrio and Micavibrio have the ability to kill human Gram-negative pathogens such as Acinetobacter, Aeromonas, Bordetella, Burkholderia, Citrobacter, Enterobacter, Escherichia, Klebsiella, Listonella, Morganella, Proteus, Salmonella, Serratia, Shigella, Vibrio, Yersinia, Helicobacter pylori, and Legionella that have acquired, or are at risk of acquiring, resistance to antibiotics [86], [90], [91], [92], [93], [94], [95]. These predators also reduce the bacterial burden of drug-resistant members of the ESKAPE group such as K. pneumoniae, A. baumannii [86], [96], [97], [98] or P. aeruginosa [99], Stenotrophomonas maltophilia [100] and other MDRB Gram-negative clinical pathogens such as Shigella [101]. The ability to kill all these Gram-negative bacteria makes BALOS to postulate as an alternative to fight against most of the MDBR identified as a critical priority by WHO. However, there are also several Gram-positive bacteria in this list, what makes interesting to explore the capacity of BALOs to kill them. In fact, it has recently been reported that B. bacteriovorus seems to attack S. aureus cystic fibrosis isolates by using an unusual epibiotic strategy [99].
These predators may also alter the commensal microbiota, and it is known that they increase the number of Gram-positive species in the gut microbiome of chickens and rats [94], [102]. The consequences of this imbalance on animal health remain to be investigated. Iebba et al. [103] reported that Bdellovibrio is present and abundant in the human gut microbiota populations of healthy individuals and suggested that the use of predatory bacteria as probiotics could restore the balance in the intestinal ecosystem to control dysbiotic events. However, studies by Shatzkes et al. [102] on the effect on rat gut microbiota showed mixed effects, since they found that intrarectal inoculation of Bdellovibrio contributes to health, while M. aeruginosavorus has potential adverse consequences. Therefore, although further studies are required to decipher the consequences of changes in the microbiota due to the administration of predatory bacteria, it is worth bearing in mind that antibiotics also may have adverse effects on commensal bacteria.
Second, these predators are able to inhibit the formation of, or reduce, preformed Gram-negative and even Gram-positive biofilms through a plethora of secreted enzymes, particularly proteases and nucleases [86], [92], [104], [105], [106], [107], [108]. Biofilms can be found in many natural environments and on industrial equipment, waste water treatment plants, hospital surfaces, health care settings and instruments. Bacteria are able to persist as reservoirs within these multicellular structures, and biofilms inside the host allow pathogens to overcome innate immune defenses. This capacity to disrupt biofilms of relevant pathogens and to remove environmental and industrial biofilms opens the way to using these predators or their enzymes not only in medicine, but also in many biocontrol processes that will be discussed below [100], [109], [110].
Third, topical application, ingestion, injection, or intranasal inoculation of whole cells have no apparent cytotoxicity, neither in terms of pathological effects, nor in terms of diminution in cell viability in in vitro cell culture models [111], [112], [113] or in vivo animal models [94], [96], [101], [102], [113], [114], [115], [116].
Fourth, they do not incite a systemic or sustained immune response, most likely due to the special structure of their lipopolysaccharide, which lacks the typical negatively charged phosphate groups, resulting in only low binding affinity to the lipopolysaccharide receptors in human immune cell pathogens [101], [117]. They are passively engulfed by macrophages and are able to persist inside these cells over 24–48 h as non-replicative forms, although they retain their predatory competence. Although they do not affect host cell viability, they stimulate moderate cytokine responses [111], [112], [113], [115]. Predator persistence inside macrophages for sufficient time to prey on pathogens opens the way to using predatory bacteria to eliminate intracellular pathogens, such as Salmonella, Klebsiella or Francisella species. Moreover, the relatively benign occupancy of macrophages by Bdellovibrio could prevent other intracellular pathogens from entering. However, this passive uptake suggests that macrophages might present predatory bacteria to antibody-forming cells and, consequently, humans could develop immune reactions against them after repeated exposures [30]. In fact, Raghunathan et al. [118] detected, although at low levels, IgG and IgM antibodies against two B. bacteriovorus strains in more than 90% of human serum samples from a biobank, suggesting that the predatory treatment could be used only once. On the other hand, experiments in immune-compromised zebrafish inoculated with B. bacteriovorus HD100 demonstrated that cooperation between the host immune system and bacterial predation is important, and it must be taken into consideration in order to maximize antibacterial therapy benefits [101]. Although a lot of progress has been made in recent years, more research is needed to understand not only the interactions and fate of these predatory bacteria within human immune cells, but also to elucidate the processes involved in their uptake by phagocytic cells, how long bacteria survive in the host cells, and the mechanisms induced by the predatory bacteria.
Fifth, Bdellovibrio is an aerobic predator, conditioning it to be an effective treatment in aerobic environments such as superficial burns or wounds, eyes and lungs [85], [96], [119], [120]. Nevertheless, this predator is able to tolerate microaerophilic conditions, which widens its areas of use to treating gastrointestinal and periodontal infections [83], [85], [121], [122], [123], [124].
Sixth, one of the main advantages of using these predators as therapeutic agents is the failure of prey bacteria to develop induced resistance against predation. The main reason for this is that these predators are not antibiotic producers, and their killing mechanisms do not target specific receptor proteins that can evolve resistance. However, predation can weaken due to different adaptations arising in the prey bacteria. For instance, transient resistance has been observed in some prey similar to bacterial persistence [83], [125]. As a consequence, Bdellovibrio predation never completely kills off the attacked population. To resolve this difficulty, the combined use of B. bacteriovorus and antibiotics in a co-therapy has been proposed [85]. This combined therapy has the additional advantage of expanding the individual killing spectra of both the antibiotics and the predator. Im et al. [126] have confirmed that a combination of Bdellovibrio and violacein is effective against Gram-positive bacteria, multidrug-resistant pathogens, and Gram-positive and negative mixed species populations. However, for an efficient development of these co-therapies it is necessary to determine the correct predator-antibiotic combination to increase the antimicrobial efficiency, while avoiding development of drug resistance. With this in mind, researchers are making efforts to determine the sensitivity of Bdellovibrio against antibiotics [127], and more research must be carried out to determine its ability to grow on antibiotic-inactivated cells and the AR mechanisms developed by the predator. Recently, experiments using Bdellovibrio and prey-specific bacteriophages eradicated E. coli when preyed upon by both predators, demonstrating the potential of this innovative co-therapy as a future alternative antibacterial treatment that would reduce the selection for single predator-specific resistance [128].
And seventh, there is a lack of uptake of prey genetic material during predation, and it has been shown that, even after regular repeated high exposure to the prey genome, there is no incorporation of any major pathogenicity island(s) or of any potentially virulent genes from the prey [129]. The exchange of genetic material would have an additional negative impact because prey could also incorporate ARGs, which could contribute to the spread of resistance. The absence of DNA interchange further strengthens the possibility of using these bacteria in the future as a safe alternative in the fight against AR.
Besides clinical uses (Fig. 1, Supplementary Table 1), BALOs are also being explored as a feasible alternative to antibiotics as biocontrol agents in horticulture, aquaculture, livestock farming and food processing. In addition, they are being assayed for their application in other biotechnological processes such as waste water treatment, extraction of bioplastics, reduction of horizontal transfer of ARGs, and other environmentally-friendly applications in which the reduction of unwanted bacteria would be positive [43], [85], [130], [131], [132] (Fig. 1, Supplementary Table 1).
The use of epibiotic predators as biocontrol agents remains largely unexplored. However, the ability of myxobacteria (besides their predatory behaviors) to glide on solid surfaces, produce a wide variety of antibiotics, and form myxospores, make these bacteria excellent, commercially viable candidates for the biocontrol of phytopathogenic bacteria and fungi [69], [73], [133], [134], [135] (Supplementary Table 1).
4. Myxobacteria as sources of new antibiotics, bioactive products and lytic enzymes
Although Bdellovibrio and BALOs are mainly being explored within the concept of whole cell therapy, other investigations are being carried out that aim to make use of the large amount of bacterially destructive lytic enzymes that predators employ to kill their prey [59], [61], [130], [131], [136], [137]. Those applications include some interesting uses such as tools for antibody analysis or for controlling the spread of ARGs in mixed microbial communities through the degradation of cell-free DNA or inactivation of phage particles, thus limiting two of the genetic DNA transfer processes that dictate the outcome of spread: transformation and transduction [43].
For the discovery of new antimicrobial agents, cooperative predators are the most promising bacteria. In the past, the producers of antibiotics and other bioactive products were mainly isolated from soil and tested by traditional diffusion methods using culture filtrates or extracts [4], [138]. The advent of the post-genomic era, with the increase in genome sequencing and annotation, next-generation sequencing technologies, modern proteomic, transcriptomic and metabolomic tools, and scientific and technological advances in genetics, biology, robotics, chemistry, metabolic engineering, and bioinformatics, has amplified the range of methodologies used for the isolation and identification of producers and has opened new doors to the identification of their “antibiotic'omes”, defined as the specific subset of microbial natural products with antibiotic activity [65], [139], [140], [141].
Although isolation of new bacterial producers is one of the priority lines in antibiotic discovery, another approach is the use of new technologies to exploit the knowledge accumulated over the years about well recognized “microfactories”. The term “OSMAC” (one strain many compounds) was introduced at the beginning of this century [142], and it refers to the fact that a single bacterial strain is capable of producing a diverse collection of structurally different SMs. Bacteria never produce the full collection of compounds encoded by their genomes at the same time under a specific environmental condition, since this would be energetically costly. To avoid unnecessary costs, they modify their transcriptomes, proteomes, and metabolomes to survive under changing conditions. For that reason, biosynthesis pathways are firmly controlled by complex regulatory networks that respond to internal and/or external signals, many of which are still unknown. The OSMAC approach focuses on the systematic alteration of culturing parameters, such as medium components (salts, amino acids, carbon source), temperature, pH, culture aeration, type of culture vessel, etc., to imitate in the laboratory those natural environments that optimize the production of useful compounds [143]. The detailed study of every single step of the complex metabolic pathways for the biosynthesis of SMs, their regulative crosstalk, and the signals that activate the regulatory bacterial sensors is facilitating the discovery of novel products from one bacterium or fungus [144]. Myxobacteria are perfect candidates to be considered from the OSMAC point of view [82], [142]. The Helmholtz-Center for Infection Research in Germany has a collection of myxobacteria with more than 9,000 strains, and more than 1,000 biosynthetic gene clusters (BGCs) associated with myxobacteria are deposited in the antisMASH database (antismash.secondarymetabolites.org).
M. xanthus DK1622, besides being a predator, is also a model organism for studying prokaryotic development. Over many years, numerous groups have accumulated a deep knowledge about the production of SMs and lytic enzymes, multicellular behavior, intra- and extracellular signaling, movement mechanisms, adaptations to changing environments, etc. [145]. This predator holds a large genome with an unprecedentedly high number of regulatory mechanisms [146], [147], [148] and an outstanding biosynthetic capacity for degradative enzymes and SM production, including 18 nonribosomal peptide synthetases (NRPS), 22 polyketide synthases (PKS), and six mixed PKS/NRPS, which concords well with its predatory lifestyle [146], [149]. Analyses using the antisMASH server calculate that those clusters correspond to a total of 14.5% of its 9.1 Mb genome [150]. Several of these natural products have been isolated [80], [82], [151], [152], [153], [154], [155], [156], [157], [158] (Fig. 2) and even expressed in heterologous hosts [159]. However, many BGCs are not yet assigned to their corresponding hypothetical SMs, probably because some of these genes remain silenced (cryptic genes) or are expressed at low levels, yielding low quantities of the respective compounds in the growth conditions assayed [82], [149], [160]. The same situation is observed in Streptomyces, a genus notorious for its ability to produce a large quantity and variety of antibiotics and SMs [65], [161]. Streptomyces genomes are also large and typically contain more than 20 BGCs dedicated to specialized metabolism such as NRPSs and PKSs. In the case of Streptomyces coelicolor A3, these enzymes are found in up to 27 clusters, which represent 10.6% of its 8.7 Mb genome [162], [163]. Only a small fraction of them are transcriptionally active under laboratory conditions and most of their products have never been characterized [161], [164].
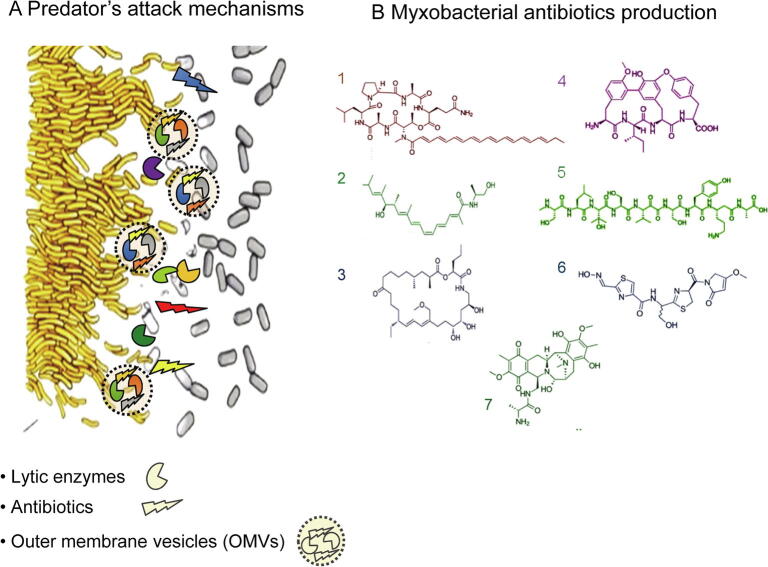
Fig. 2.
The cooperative predator M. xanthus as a source of antibiotics and new products. A. During predation, M. xanthus secretes lytic enzymes, antibiotics and other secondary metabolites, some of them included in OMVs. B. Antibiotics and bioactive products identified in M. xanthus species. 1: myxochromide; 2: myxalamid; 3: myxovirescin TA; 4: cittilin A; 5: myxoprincomide; 6: althiomycin; 7: saframycin Mx1.
This huge untapped biosynthetic potential makes the unlocking of the cryptic pathways of those cooperative predators another good strategy for antibiotic discovery. It is reasonable to think that one of the ecological roles of antibiotics in nature is to be used as weapons against susceptible bacteria during predatory processes and, in fact, some of them have been shown to be involved in predation [46], [165]. It has been recently found that although the genes responsible for SM biosynthesis in M. xanthus are expressed during growth, their expression increases during development [148]. These expression profiles suggest that SMs are used to protect developing cells from other microbes in the soil, to defend spores inside the fruiting bodies or to release nutrients from prey to promote germination [148]. It is also realistic to infer that production of such metabolites by predators might be regulated by contact with and/or proximity to the prey. Therefore, it would be useful to design efficient screening methods in the presence of a variety of pathogens for the induction of novel antibiotics against them.
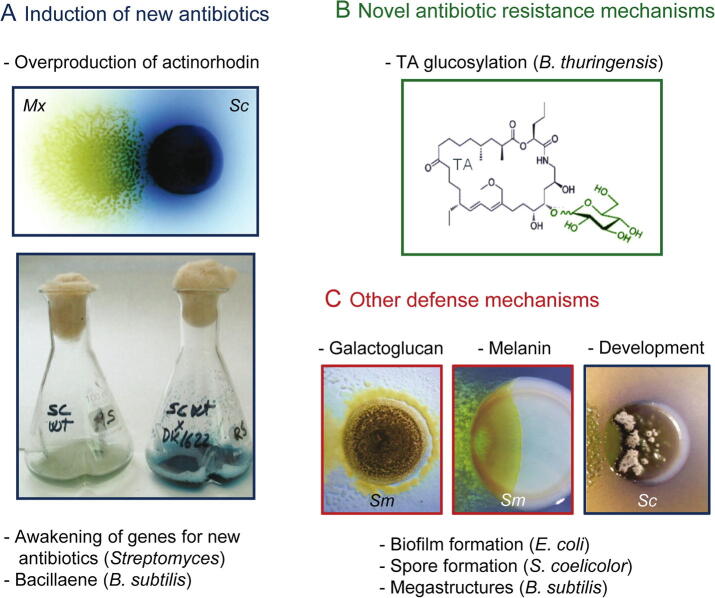
But interspecies cross talk can also trigger the production by prey of specific bioactive compounds for defensive purposes and other resistance mechanisms to counterattack such as biofilm formation [166], spore differentiation, or induction of new ARGs [167] (Fig. 3). Co-cultivation experiments, followed by deep analysis of the influence on the secondary metabolome, are gaining increasing attention and are already yielding exciting results. Some studies have demonstrated that the presence in cultures of other bacteria stimulates Streptomyces to produce some SMs [144], [164]. Most of these pairwise interactions have been assayed with other related Actinobacteria or Bacillus. Moreover, M. xanthus is able to induce the overproduction of the blue antibiotic actinorhodin while preying on S. coelicolor (Fig. 3A) [168]. Other results involving the production of SMs during predatory relationships are the induction of bacillaene by B. subtilis [76], [169], or the biosynthesis of melanin by S. meliloti to protect against predation by M. xanthus (Fig. 3) [77], [170].
Fig. 3.
Defense mechanisms in the prey with biotechnological applications. A. Silenced antibiotics are induced in the prey during the predatory process. In the pictures, M. xanthus (Mx, predator) induces in Streptomyces coelicolor (Sc, prey) the blue antibiotic actinorhodin, in solid and liquid media. B. Novel antibiotic resistance mechanism have been discovered in B. thuringensis: myxovirescin TA glucosylation. C. Other physical/chemical defenses mechanisms induced by M. xanthus predation in different bacteria. Galactoglucan (left picture) and melanin (middle picture) protect Sinorhizobium meliloti from predation. M. xanthus induces development in Streptomyces (right picture). Pictures from panel A and right picture from panel C are reproduced from Pérez et al. (2011) Microb Biotechnol 4: 175–183. Left picture in panel C is adapted from Muñoz-Dorado et al., (2016) Front Microbiol 7: 781. Middle picture in panel C is from Contreras-Moreno et al. (2020) Front Microbiol 11: 94. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The first transcriptomic study during a predatory process of M. xanthus against E. coli [171] indicated that M. xanthus does not perceive live prey as food, and that the prey shows extensive transcriptomic changes when co-cultured with the predator. Moreover, M. xanthus supernatants and secreted outer membrane vesicles (OMVs, see below) also induced changes in the expression of large numbers of prey genes. Another transcriptomic analysis confirmed the actinorhodin overproduction observed in Streptomyces by Pérez et al [168], and confirmed that it is iron competition during this interaction which leads to the activation of actinorhodin BGC plus 21 other SM BGCs [172] (Fig. 3A). This is a good example of how basic research on bacterial predator-prey interaction is helping to discover new ways to activate cryptic biosynthetic pathways and is improving our comprehension of awakening mechanisms.
Another aspect to be considered in the search for innovative applications for predators are the Gram-negative OMVs. OMVs are spherical portions of the bacterial outer membrane with a diameter of ~ 20–250 nm, which represent an alternative to general secretion systems. The myxobacterial OMVs contain a highly concentrated dose of hydrolytic enzymes and molecules associated with antibiotic activity, and are able to deliver these compounds to distant and inaccessible locations, enabling more efficient epibiotic cooperative killing of prey [51], [173], [174], [50]. The OMVs can be used as shuttles to deliver hydrophobic compounds with any desired properties (Fig. 2). For instance, they have been confirmed to be good adjuvants in vaccine delivery [175]. The myxobacterial OMVs have recently been explored as natural antimicrobial carriers able to deliver these products to target cells. They are able to kill some pathogenic bacteria such as E. coli or P. aeruginosa [176], [177]. Recently, they have also been confirmed to be efficient tools for fighting intracellular bacterial pathogens such as S. aureus. Those infections are difficult to treat because several classes of antibiotics are unable to reach the pathogen, and higher concentrations and a longer therapy time are needed. In these assays, OMVs from Cystobacter velatus and Sorangineae species exhibit low toxicity in various cell lines and primary immune cells, and are able to take them up into infected cells [177], [178]. From these results it is possible to envision the exploitation of OMVs from myxobacteria as novel therapeutic delivery systems to combat bacterial infections. The reports on M. xanthus OMV proteomics show that they are enriched (along with toxic proteins and antibiotics) in hypothetical proteins of unknown function which remain to be studied in depth [173], [176], [179], [180]. Moreover, a recent proteomic study of the OMV cargo from several M. xanthus strains seems to indicate that genetically similar strains of myxobacteria have diverse OMV proteomes [51], [181], giving rise to new research challenges.
Finally, it has been shown that myxobacterial predatory activity varies depending on the prey [77], [78], [182]. Moreover, Sutton et al. [183], using genome-wide association studies with 29 myxobacterial strains, identified different numbers of predation genes depending on the prey organisms. They suggested that the broad prey range of myxobacteria seems to be a consequence of the accumulation of arrangements of prey-specific predation genes, rather than possession of a set of general antibiotic genes. These results clearly indicate that the awakening of cryptic genes should be assayed in the presence of pathogens against which we are looking for a drug, since they may reinforce the positive results of the strategy of testing other bacteria as prey and assaying new products against resistant bacteria.
5. Summary and outlook
Development of AR by many pathogens is one of the main challenges for researchers in order to avoid a new era in which the number of deaths from infectious diseases that were thought to be under control increases to a level that represents an unacceptable health and economic cost. Finding new alternatives to kill these pathogens requires a focus on various different strategies, and one of these promising strategies is learning about the behaviors of bacterial predators and prey in nature. Understanding the strategies developed by predatory bacteria during evolution is helping researchers to emulate and use them in different antimicrobial approaches. Research on the use of predators as living antibiotics is yielding promising results, and although more research is required to demonstrate their efficacy in vivo, they represent a serious alternative to be considered. Currently, finding structural genes and regulatory elements involved in predation and the induction of silenced and cryptic genes or BGCs during these dynamic predatory processes is not only starting to help in the discovery of new antibiotics with novel mechanisms of action and new classes of bioactive natural products, but also facilitating the establishment of the best laboratory conditions for optimal scaled-up industrial production of metabolites. Finally, from the studies of the antagonistic predator–prey relationships, researchers are learning about the attack strategies used by the hunters, which are opening innovative research lines such as the use of OMVs as antimicrobial carriers. The mechanisms developed by the prey to defend itself are also proving useful in finding out about new determinants of AR. All of the studies reviewed here about bacterial predators are not only opening new doors for the discovery of new antibiotics, but also harnessing the enormous potential of bacterial predators as whole cell therapeutic agents. Taken together, these studies will help to prepare us for a near future when many antibiotics fail to treat MDRB. However, they require more support from government initiatives, along with changes in regulation to pave the way for valuable, efficacious, highly targeted, pathogen-specific antimicrobial therapies.
Declaration of Competing Interest
None.
CRediT authorship contribution statement
Juana Pérez: Writing - original draft, Conceptualization. Francisco Javier Contreras-Moreno: Writing - review & editing. Francisco Javier Marcos-Torres: Writing - review & editing. Aurelio Moraleda-Muñoz: Writing - review & editing, Funding acquisition. José Muñoz-Dorado: Writing - original draft, Funding acquisition, Conceptualization.
Acknowledgments
This work has been supported by the Spanish Government, grant BFU2016-75425-P to Aurelio Moraleda-Muñoz (70% funded by FEDER). Funder has no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. We thank Lola Contreras-Moreno for technical assistance with Fig. 1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.09.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fleming A.G. Responsibilities and opportunities of the private practitioner in preventive medicine. Can Med Assoc J. 1929;20:1–13. [PMC free article] [PubMed] [Google Scholar]
- 2.Domagk G. Ein beitrag zur chemotherapie der bakteriellen infektionen. Deut Med Wochenschr. 1935;61:250–253. [Google Scholar]
- 3.Schatz A., Bugie E., Waksman S.A. Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Proc Soc Exp Biol Med. 1944;55:66–69. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 5.Lewis K. The science of antibiotic discovery. Cell. 2020;181:29–45. doi: 10.1016/j.cell.2020.02.056. [DOI] [PubMed] [Google Scholar]
- 6.Blair J.A., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J.V. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 7.Crofts T.S., Gasparrini A.J., Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nature. 2017;15:422–434. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen H.K., Donato J., Wang H.H., Karen A., Cloud-Hansen K.A. Call of the wild: antibiotic resistance genes in natural environments. Nature Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 9.Bush K., Courvalin P., Dantas G., Davies J., Eisenstein B. Tackling antibiotic resistance. Nat Rev Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Costa V.M., King C.E., Kalan L., Morar M., Sung W.W.L. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 11.Van Boeckel T.P., Brower C., Gilbert M., Grenfell B.T., Levin S.A. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline, including tuberculosis. WHO/EMP/ IAU/2017.12. 2017; WHO: Geneva.
- 13.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 14.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D. Bad bugs, no drugs: no ESKAPE! an update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 15.Berendonk T.U., Manaia C.M., Merlin C., Fatta-Kassinos D., Cytryn E. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 16.Hernando-Amado S., Coque T.M., Baquero F., Martínez J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat Microbiol. 2019;4:1432–1442. doi: 10.1038/s41564-019-0503-9. [DOI] [PubMed] [Google Scholar]
- 17.Gudda F.O., Waigi M.W., Odinga E.S., Yang B., Carter L., Gao Y. Antibiotic-contaminated wastewater irrigated vegetables pose resistance selection risks to the gut microbiome. Environ Pollut. 2020;114752 doi: 10.1016/j.envpol.2020.114752. [DOI] [PubMed] [Google Scholar]
- 18.Pazda M., Kumirska J., Stepnowski P., Mulkiewicz E. Antibiotic resistance genes identified in wastewater treatment plant systems – a review. Sci Total Environ. 2019;697 doi: 10.1016/j.scitotenv.2019.134023. [DOI] [PubMed] [Google Scholar]
- 19.Nies D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 20.Blanco P., Hernando-Amado S., Reales-Calderón J., Corona F., Lira F. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms. 2016;4:14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L.G., Xia Y., Zhang T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017;11:651–662. doi: 10.1038/ismej.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ledingham K., Hinchliffe S., Jackson M., Thomas F., Tomson G. WHO Regional Office for Europe; Copenhagen, Denmark: 2019. Antibiotic resistance: using a cultural contexts of health approach to address a global health challenge. [Google Scholar]
- 23.O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. The Review on Antimicrobial Resistance. Wellcome Trust and the UK Department of Health. London, UK; 2014.
- 24.O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. The Review on antimicrobial resistance. Wellcome Trust and the UK Department of Health. London, UK; 2016.
- 25.Cooper M., Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 26.Silver L.L. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright G.D. Solving the antibiotic crisis. ACS Infect Dis. 2015;1:80–84. doi: 10.1021/id500052s. [DOI] [PubMed] [Google Scholar]
- 28.Reardon S. Bacterial arms race revs up. Nature. 2015;521:402–403. doi: 10.1038/521402a. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh C., Sarkar P., Issa R., Haldar J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol. 2019;27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Madhusoodanan J. Probing predatory bacteria as an antibacterial remedy. Proc Natl Acad Sci USA. 2019;116:22887–22890. doi: 10.1073/pnas.1917513116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C.H., Hsieh Y.H., Powers Z.M., Kao C.Y. Defeating antibiotic-resistant bacteria: exploring alternative therapies for a post-antibiotic era. Int J Mol. 2020;21:1061. doi: 10.3390/ijms21031061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurkevitch E., Davidov Y. Phylogenetic diversity and evolution of predatory prokaryotes. In: Jurkevitch E., editor. Predatory Prokaryotes-Biology, Ecology and Evolution. Springer Verlag; Heidelberg, Germany: 2007. pp. 11–56. [Google Scholar]
- 33.Chauhan A., Cherrier J., Williams H.N. Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad Sci USA. 2009;106:4301–4306. doi: 10.1073/pnas.0809671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H., Athar R., Zheng G., Williams H.N. Prey bacteria shape the community structure of their predators. ISME J. 2011;5:1314–1322. doi: 10.1038/ismej.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin J.N., Byrnes J.E.K., Cardinale B.J. Effects of predator richness on prey suppression: a metaanalysis. Ecology. 2013;94:2180–2187. doi: 10.1890/13-0179.1. [DOI] [PubMed] [Google Scholar]
- 36.Johnke J., Cohen Y., de Leeuw M., Kushmaro A., Jurkevitch E. Multiple micropredators controlling bacterial communities in the environment. Curr Opin Biotechnol. 2014;27:185–190. doi: 10.1016/j.copbio.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Kandel P.P., Pasternak Z., van Rijn J., Nahum O., Jurkevitch E. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol Ecol. 2014;89:149–161. doi: 10.1111/1574-6941.12342. [DOI] [PubMed] [Google Scholar]
- 38.Johnke J., Fraune S., Bosch T.C.G., Hentschel U., Schulenburg H. Bdellovibrio and like organisms are predictors of microbiome diversity in distinct host groups. Microb Ecol. 2019;79:252–257. doi: 10.1007/s00248-019-01395-7. [DOI] [PubMed] [Google Scholar]
- 39.Ezzedine J.A., Jacas L., Desdevises Y., Jacquet S. Bdellovibrio and like organisms in lake Geneva: an unseen elephant in the room? Front Microbiol. 2020;11:98. doi: 10.3389/fmicb.2020.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matin A., Rittenberg S.C. Kinetics of deoxyribonucleic acid destruction and synthesis during growth of Bdellovibrio bacteriovorus strain 109D on Pseudomonas putida and Escherichia coli. J Bacteriol. 1972;111:664–673. doi: 10.1128/jb.111.3.664-673.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosson R.A., Rittenberg S.C. Regulated breakdown of Escherichia coli deoxyribonucleic acid during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J Bacteriol. 1979;140:620–633. doi: 10.1128/jb.140.2.620-633.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monnappa A.K., Dwidar M., Mitchell R.J. Application of bacterial predation to mitigate recombinant bacterial populations and their DNA. Siol Biol Biochem. 2013;57:427–435. [Google Scholar]
- 43.Bratanis E., Andersson T., Lood R., Bukowska-Faniband E. Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Front Microbiol. 2020;11:662. doi: 10.3389/fmicb.2020.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez J., Moraleda-Muñoz A., Marcos-Torres F.J., Muñoz-Dorado J. Bacterial predation:75 years and counting! Environ Microbiol. 2016;18:766–779. doi: 10.1111/1462-2920.13171. [DOI] [PubMed] [Google Scholar]
- 45.Marshall R.C., Whitworth D.E. Is “wolf-pack” predation by antimicrobial bacteria cooperative? Cell behaviour and predatory mechanisms indicate profound selfishness, even when working alongside kin. BioEssays. 2019;41 doi: 10.1002/bies.201800247. [DOI] [PubMed] [Google Scholar]
- 46.Kumbhar C., Watve M. Why antibiotics: a comparative evaluation of different hypotheses for the natural role of antibiotics and an evolutionary synthesis. Nat Sci. 2013;5:26–40. [Google Scholar]
- 47.Kumbhar C., Mudliar P., Bhatia L., Kshirsagar A., Watve M. Widespread predatory abilities in the genus Streptomyces. Arch Microbiol. 2014;196:235–248. doi: 10.1007/s00203-014-0961-7. [DOI] [PubMed] [Google Scholar]
- 48.Berleman J.E., Kirby J.R. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao P., Dey A., Vassallo C.N., Wall D. How myxobacteria cooperate. J Mol Biol. 2015;427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keane R., Berleman J. The predatory life cycle of Myxococcus xanthus. Microbiology. 2016;162:1–11. doi: 10.1099/mic.0.000208. [DOI] [PubMed] [Google Scholar]
- 51.Furness E., Whitworth D.E., Zwarycz A. Predatory interactions between myxobacteria and their prey. In: Jurkevitch E., Mitchell R.J., editors. The ecology of predation at the microscale. Springer Nature Switzerland AG; Switzerland: 2020. pp. 1–36. [Google Scholar]
- 52.Thiery S., Kaimer C. The predation strategy of Myxococcus xanthus. Front Microbiol. 2020;11:2. doi: 10.3389/fmicb.2020.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sockett R.E. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol. 2009;63:523–539. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 54.Rotem O., Pasternak Z., Jurkevitch E. Bdellovibrio and like organisms. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes-deltaproteobacteria and epsilonproteobacteria. Springer Verlag; Berlin, Heidelberg, Germany: 2014. pp. 1–17. [Google Scholar]
- 55.Davidov Y., Huchon D., Koval S.F., Jurkevitch E. A new alpha proteobacterial clade of Bdellovibrio like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 2006;8:2179–2188. doi: 10.1111/j.1462-2920.2006.01101.x. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z., Kadouri D.E., Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soo R.M., Woodcroft B.J., Park D.H., Tyson G.W., Hugenholtz P. Back from the dead: the curious tale of the predatory cyanobacterium Vampirovibrio chlorellavorus. PeerJ. 2015;3 doi: 10.7717/peerj.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pasternak Z., Njagi M., Shani Y., Chanyi R., Rotem O. In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J. 2014;8:625–635. doi: 10.1038/ismej.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rendulic S., Jagtap P., Rosinus A., Eppinger M., Baar C. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 60.Tudor J., McCann M. Genomic analysis and molecular biology of predatory prokaryotes. In: Jurkevitch E., editor. Predatory prokaryotes-biology, ecology and evolution. Springer Verlag; Heidelberg, Germany: 2007. pp. 153–189. [Google Scholar]
- 61.Lambert C., Chang C.Y., Capeness M.J., Sockett R.E. The first bite-profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sester A., Korp J., Nett M. Secondary metabolism of predatory bacteria. In: Jurkevitch E., Mitchell R.J., editors. The ecology of predation at the microscale. Springer Nature Switzerland AG; Switzerland: 2020. pp. 127–154. [Google Scholar]
- 63.Liu G., Chater K.F., Chandra G., Niu G., Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev. 2013;77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landwehr W., Wolf C., Wink C. Actinobacteria and myxobacteria-two of the most important bacterial resources for novel antibiotics. In: Stadler M., Dersch P., editors. How to overcome the antibiotic crisis. Springer Switzerland; Switzerland: 2016. pp. 273–302. [DOI] [PubMed] [Google Scholar]
- 65.Kealey C., Creaven C.A., Murphy C.D., Brady C.B. New approaches to antibiotic discovery. Biotechnol Let. 2017;39:805–817. doi: 10.1007/s10529-017-2311-8. [DOI] [PubMed] [Google Scholar]
- 66.Reichenbach H. The ecology of the myxobacteria. Environ Microbiol. 1999;1:15–21. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 67.Rosenberg E., Varon M. Antibiotics and lytic enzymes. In: Rosenberg E., editor. Myxobacteria: development and cell interactions. Springer Verlag; New York: 1984. pp. 109–125. [Google Scholar]
- 68.Rashidan K.K., Bird D.F. Role of predatory bacteria in the termination of a cyanobacterial bloom. Microbiol Ecol. 2001;41:97–105. doi: 10.1007/s002480000074. [DOI] [PubMed] [Google Scholar]
- 69.Bull C.T., Shetty K.G., Subbarao K.V. Interactions between myxobacteria, plant pathogenic fungi, and biocontrol agents. Plant Dis. 2002;86:889–896. doi: 10.1094/PDIS.2002.86.8.889. [DOI] [PubMed] [Google Scholar]
- 70.Pham V.D., Shebelut C.W., Diodati M.E., Bull C.T., Singer M. Mutations affecting predation ability of the soil bacterium Myxococcus xanthus. Microbiology. 2005;151:1865–1874. doi: 10.1099/mic.0.27824-0. [DOI] [PubMed] [Google Scholar]
- 71.Berleman J.E., Chumley T., Cheung P., Kirby J.R. Rippling is a predatory behavior in Myxococcus xanthus. J Bacteriol. 2006;188:5888–5895. doi: 10.1128/JB.00559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hillesland K.L., Lenski R.E., Velicer G.J. Ecological variables affecting predatory success in Myxococcus xanthus. Microb Ecol. 2007;53:571–578. doi: 10.1007/s00248-006-9111-3. [DOI] [PubMed] [Google Scholar]
- 73.Morgan A.D., Maclean R.C., Hillesland K.L., Velicer G.J. Comparative analysis of Myxococcus predation on soil bacteria. Appl Environ Microbiol. 2010;76:6920–6927. doi: 10.1128/AEM.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mendes-Soares H., Velicer G.J. Decomposing predation: testing for parameters that correlate with predatory performance by a social bacterium. Microb Ecol. 2013;65:415–423. doi: 10.1007/s00248-012-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia R., Müller R. The family Myxococcaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The prokaryotes- Deltaproteobacteria and Epsilonproteobacteria. Springer Verlag; Berlin, Heidelberg: 2014. pp. 191–212. [Google Scholar]
- 76.Müller S., Strack S.N., Hoefler B.C., Straight P.D., Kearns D.B. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol. 2014;80:5603–5610. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pérez J., Jiménez-Zurdo J.I., Martínez-Abarca F., Millán V., Shimkets L.J. Rhizobial galactoglucan determines the predator pattern of Myxococcus xanthus and protects Sinorhizobium meliloti from predation. Environ Microbiol. 2014;16:2341–2350. doi: 10.1111/1462-2920.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Livingstone P.G., Morphew R.M., Whitworth D.E. Myxobacteria are able to prey broadly upon clinically-relevant pathogens, exhibiting a prey range which cannot be explained by phylogeny. Front Microbiol. 2017;8:1593. doi: 10.3389/fmicb.2017.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wenzel S.C., Müller R. The biosynthetic potential of myxobacteria and their impact in drug discovery. Curr Opin Drug Discov Develop. 2009;12:220–230. [PubMed] [Google Scholar]
- 80.Herrmann J., Fayad A.A., Müller R. Natural products from myxobacteria: novel metabolites and bioactivities. Nat Prod Rep. 2017;34:135–160. doi: 10.1039/c6np00106h. [DOI] [PubMed] [Google Scholar]
- 81.Hoffmann T., Krug D., Bozkurt N., Duddela S., Jansen R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat Commun. 2018;9:1–10. doi: 10.1038/s41467-018-03184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bader C.D., Panter F., Müller R. In depth natural product discovery – myxobacterial strains that provided multiple secondary metabolites. Biotechnol Adv. 2020;39 doi: 10.1016/j.biotechadv.2019.107480. [DOI] [PubMed] [Google Scholar]
- 83.Sockett R.E., Lambert C. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat Rev Microbiol. 2004;2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 84.Chatterjee A. Bdellovibrio bacteriovorus: life cycle and potential as a predatory renaissance. Adv Biotech. 2009;VIII:27–29. [Google Scholar]
- 85.Dwidar M., Monnappa A.K., Mitchell R.J. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012;45:71–78. doi: 10.5483/BMBRep.2012.45.2.71. [DOI] [PubMed] [Google Scholar]
- 86.Kadouri D.E., To K., Shanks R.M.Q., Doi Y. Predatory bacteria: a potential ally against multidrug-resistant Gram-negative pathogens. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allen H.K., Trachsel J., Looft T., Casey T.A. Finding alternatives to antibiotics. Ann N Y Acad Sci. 2014;1323:91–100. doi: 10.1111/nyas.12468. [DOI] [PubMed] [Google Scholar]
- 88.Negus D., Moore C., Baker M., Raghunathan D., Tyson J. Predator versus pathogen: how does predatory Bdellovibrio bacteriovorus interface with the challenges of killing Gram-negative pathogens in a host setting? Annu Rev Microbiol. 2017;71:441–457. doi: 10.1146/annurev-micro-090816-093618. [DOI] [PubMed] [Google Scholar]
- 89.Shatzkes K., Connell N.D., Kadouri D.E. Predatory bacteria: a new therapeutic approach for a post-antibiotic era. Future Microbiol. 2017;12:469–472. doi: 10.2217/fmb-2017-0021. [DOI] [PubMed] [Google Scholar]
- 90.Fratamico P.M., Cooke P.H. Isolation of bdellovibrios that prey on Escherichia coli O157:H7 and Salmonella species and application for removal of prey from stainless steel surfaces. J Food Safety. 1996;16:161–173. [Google Scholar]
- 91.Markelova N.Y. Predacious bacteria, Bdellovibrio with potential for biocontrol. Int J Hyg Environ Health. 2010;213:428–431. doi: 10.1016/j.ijheh.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Dashiff A., Junka R.A., Libera M., Kadouri D.E. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol. 2011;110 doi: 10.1111/j.1365-2672.2010.04900.x. 431e44. [DOI] [PubMed] [Google Scholar]
- 93.Otto S., Bruni E.P., Harms H., Wick L.Y. Catch me if you can: dispersal and foraging of Bdellovibrio bacteriovorus 109J along mycelia. ISME J. 2017;11:386–393. doi: 10.1038/ismej.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Atterbury R.J., Hobley L., Till R., Lambert C., Capeness M.J. Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol. 2011;77:5794–5803. doi: 10.1128/AEM.00426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Findlay J.S., Flick-Smith H.C., Keyser E., Cooper I.A., Williamson E.D. Predatory bacteria can protect SKH-1 mice from a lethal plague challenge. Sci Rep. 2019;9:7225. doi: 10.1038/s41598-019-43467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shatzkes K., Singleton E., Tang C., Zuena M., Shukla S. Predatory bacteria attenuate Klebsiella pneumoniae burden in rat lungs. mBio. 2016;7:e01847–e1916. doi: 10.1128/mBio.01847-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baker M., Negus D., Raghunathan D., Radford P., Moore C. Measuring and modelling the response of Klebsiella pneumoniae KPC prey to Bdellovibrio bacteriovorus predation, in human serum and defined buffer. Sci Rep. 2017;7:8329. doi: 10.1038/s41598-017-08060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Im H., Son S., Mitchell R.J., Ghim C.M. Serum albumin and osmolality inhibit Bdellovibrio bacteriovorus predation in human serum. Sci Rep. 2017;7:5896. doi: 10.1038/s41598-017-06272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iebba V., Totino V., Santangelo F., Gagliardi A., Ciotoli L. Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus cystic fibrosis isolates. Front Microbiol. 2014;5:280. doi: 10.3389/fmicb.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chanyi R.M., Koval S.F., Brooke J.S. Stenotrophomonas maltophilia biofilm reduction by Bdellovibrio exovorus. Environ Microbiol Rep. 2016;8:343–351. doi: 10.1111/1758-2229.12384. [DOI] [PubMed] [Google Scholar]
- 101.Willis A.R., Moore C., Mazon-Moya M., Krokowski S., Lambert C. Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr Biol. 2016;26:3343–3351. doi: 10.1016/j.cub.2016.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shatzkes K., Tang C., Singleton E., Shukla S., Zuena M. Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci Rep. 2017;7:43483. doi: 10.1038/srep43483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iebba V., Santangelo F., Totino V., Nicoletti M., Gagliardi A. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0061608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koval S.F., Bayer M.E. Bacterial capsules: no barrier against bdellovibrio. Microbiology. 1997;143:749–753. doi: 10.1099/00221287-143-3-749. [DOI] [PubMed] [Google Scholar]
- 105.Kadouri D., O'Toole G.A. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71 doi: 10.1128/AEM.71.7.4044-4051.2005. 4044e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nuñez M.E., Martin M.O., Chan P.H., Spain E.M. Predation, death, and survival in a biofilm: Bdellovibrio investigated by atomic force microscopy. Colloids Surf B. 2005;42:263–271. doi: 10.1016/j.colsurfb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 107.Kadouri D., Venzon N.C., O’Toole G.A. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol. 2007;73:605–614. doi: 10.1128/AEM.01893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Im H., Dwidar M., Mitchell R.J. Bdellovibrio bacteriovorus HD100, a predator of Gram-negative bacteria, benefits energetically from Staphylococcus aureus biofilms without predation. ISME J. 2018;12:2090–2095. doi: 10.1038/s41396-018-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Monnappa A.K., Dwidar M., Seo J.K., Hur J.H., Mitchell R.J. Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Sci Rep. 2014;4:3811. doi: 10.1038/srep03811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun Y., Ye J., Hou Y., Chen H., Cao J. Predation efficacy of Bdellovibrio bacteriovorus on multidrug-resistant clinical pathogens and their corresponding biofilms. Jpn J Infect Dis. 2017;70:485–489. doi: 10.7883/yoken.JJID.2016.405. [DOI] [PubMed] [Google Scholar]
- 111.Monnappa A.K., Bari W., Choi S.Y., Mitchell R.J. Investigating the responses of human epithelial cells to predatory bacteria. Sci Rep. 2016;6:33485. doi: 10.1038/srep33485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shanks R.M.Q., Davra V.R., Romanowski E.G., Brothers K.M., Stella N.A. An eye to a kill: Using predatory bacteria to control Gram-Negative pathogens associated with ocular infections. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0066723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gupta S., Tang C., Tran M., Kadouri D.E. Effect of predatory bacteria on human cell lines. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0161242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Romanowski EG, Stella NA, Brothers KM, Yates KA, Funderburgh ML, et al. Predatory bacteria are nontoxic to the rabbit ocular surface. Sci Rep 2016;6:30987. [DOI] [PMC free article] [PubMed]
- 115.Shatzkes K., Chae R., Tang C., Ramirez G.C., Mukherjee S. Examining the safety of respiratory and intravenous inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a mouse model. Sci Rep. 2015;5:12899. doi: 10.1038/srep12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shatzkes K., Singleton E., Tang C., Zuena M., Shukla S. Examining the efficacy of intravenous administration of predatory bacteria in rats. Sci Rep. 2017;7:1864. doi: 10.1038/s41598-017-02041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schwudke D., Linscheid M., Strauch E., Appel B., Zahringer U. The obligate predatory Bdellovibrio bacteriovorus possesses a neutral lipid A containing alpha-D-Mannoses that replace phosphate residues: similarities and differences between the lipid As and the lipopolysaccharides of the wild type strain B. bacteriovorus HD100 and its host-independent derivative HI100. J Biol Chem. 2003;278:27502–27512. doi: 10.1074/jbc.M303012200. [DOI] [PubMed] [Google Scholar]
- 118.Raghunathan D., Radford P.M., Gell C., Negus D., Moore C. Engulfment, persistence and fate of Bdellovibrio bacteriovorus predators inside human phagocytic cells informs their future therapeutic potential. Sci Rep. 2019;9:4293. doi: 10.1038/s41598-019-40223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shanks R.M.Q., Kadouri D.E. Predatory prokaryotes wage war against eye infections. Future Microbiol. 2014;9:429–432. doi: 10.2217/fmb.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Russo R., Kolesnikova I., Kim T., Gupta S., Pericleous A. Susceptibility of virulent Yersinia pestis bacteria to predator bacteria in the lungs of mice. Microorganisms. 2018;7:2. doi: 10.3390/microorganisms7010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Van Essche M., Quirynen M., Sliepen I., Van Eldere J., Teughels W. Bdellovibrio bacteriovorus attacks Aggregatibacter actinomycetemcomitans. J Dent Res. 2009;88:182–186. doi: 10.1177/0022034508329693. [DOI] [PubMed] [Google Scholar]
- 122.Dashiff A., Kadouri D.E. Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol. 2011;26:19–34. doi: 10.1111/j.2041-1014.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- 123.Kadouri D.E., Tran A. Measurement of predation and biofilm formation under different ambient oxygen conditions using a simple gas bag based system. Appl Environ Microbiol. 2013;79:5264–5271. doi: 10.1128/AEM.01193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Patini R., Cattani P., Marchetti S., Isola G., Quaranta G. Evaluation of predation capability of periodontopathogens bacteria by Bdellovibrio bacteriovorus HD100. An in vitro study. Materials. 2019;12:2008. doi: 10.3390/ma12122008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shemesh Y., Jurkevitch E. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ Microbiol. 2004;6:12–18. doi: 10.1046/j.1462-2920.2003.00530.x. [DOI] [PubMed] [Google Scholar]
- 126.Im H., Choi S.Y., Son S., Mitchell R.J. Combined application of bacterial predation and violacein to kill polymicrobial pathogenic communities. Sci Rep. 2017;7:14415. doi: 10.1038/s41598-017-14567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marine E., Milner D.S., Lambert C., Sockett R.E., Pos K.M. A novel method to determine antibiotic sensitivity in Bdellovibrio bacteriovorus reveals a DHFR dependent natural trimethoprim resistance. Sci Rep. 2020;10:5315. doi: 10.1038/s41598-020-62014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hobley L., Summers J.K., Till R., Milner D.S., Atterbury R.J. Dual predation by bacteriophage and Bdellovibrio can eradicate E. coli prey in situations where single predation cannot. J Bacteriol. 2020;202 doi: 10.1128/JB.00629-19. e00629-e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gupta S., Lemenze A., Donnelly R.J., Connell N.D., Kadouri D.E. Keeping it together: absence of genetic variation and DNA incorporation by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus during predation. Res Microbiol. 2018;169:237–243. doi: 10.1016/j.resmic.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 130.Herencias C., Salgado-Briegas S., Prieto M.A. Emerging horizons for industrial applications of predatory bacteria. In: Jurkevitch E., Mitchel R.J., editors. The ecology of predation at the microscale. Springer Nature Switzerland AG; Switzerland: 2020. pp. 189–194. [Google Scholar]
- 131.Jurkevitch E. The ecology of Bdellovibrio and like organisms in wastewater treatment plants. In: Jurkevitch E., Mitchel R.J., editors. The ecology of predation at the microscale. Springer Nature Switzerland AG; Switzerland: 2020. pp. 37–64. [Google Scholar]
- 132.Najnine F., Cao Q., Zhao Y., Cai J. Antibacterial activities of Bdellovibrio and like organisms in aquaculture. In: Jurkevitch E., Mitchel R.J., editors. The ecology of predation at the microscale. Springer Nature Switzerland AG; Switzerland: 2020. pp. 89–124. [Google Scholar]
- 133.Li Z., Ye X., Chen P., Ji K., Zhou J. Antifungal potential of Corallococcus sp. strain EGB against plant pathogenic fungi. Biol Control. 2017;110:10–17. [Google Scholar]
- 134.Li Z., Ye X., Liu M., Xia C., Zhang L. A novel outer membrane β-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J. 2019;13:2223–2235. doi: 10.1038/s41396-019-0424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ye X., Li Z., Luo X., Wang W., Li Y. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome. 2020;8:49. doi: 10.1186/s40168-020-00824-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Song C., Kumar A., Saleh M. Bioinformatic comparison of bacterial secretomes. Genom Proteom Bioinf. 2009;7:37–46. doi: 10.1016/S1672-0229(08)60031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tyson J., Sockett R.E. Nature knows best: employing whole microbial strategies to tackle antibiotic resistant pathogens. Environ Microbiol Rep. 2017;9:47–49. doi: 10.1111/1758-2229.12518. [DOI] [PubMed] [Google Scholar]
- 138.Lewis K. Antibiotics: Recover the lost art of drug discovery. Nature. 2012;485:439–440. doi: 10.1038/485439a. [DOI] [PubMed] [Google Scholar]
- 139.Johnston C.W., Magarvey N.A. Untwisting the antibiotic’ome. Nat Chem Biol. 2015;11:177–178. doi: 10.1038/nchembio.1757. [DOI] [PubMed] [Google Scholar]
- 140.Moloney M.G. Natural products as a source for novel antibiotics. Trends Pharmacol Sci. 2016;37:689–701. doi: 10.1016/j.tips.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 141.Mantravadi P.K., Kalesh K.A., Dobson R.C.J., Hudson A.O., Parthasarathy A. The quest for novel antimicrobial compounds: emerging trends in research, development, and technologies. Antibiotics (Basel) 2019;8:8. doi: 10.3390/antibiotics8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bode H.B., Bethe B., Hofs R., Zeeck A. Big effects from small changes: possible ways to explore Nature's chemical diversity. Chem Bio Chem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 143.Zarins-Tutt J.S., Barberi T.T., Gao H., Mearns-Spragg A., Zhang L. Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat Prod Rep. 2016;33:54. doi: 10.1039/c5np00111k. [DOI] [PubMed] [Google Scholar]
- 144.Antoraz S., Santamaría R., Díaz M., Sanz D., Rodríguez H. Toward a new focus in antibiotic and drug discovery from the Streptomyces arsenal. Front Microbiol. 2015;6:461. doi: 10.3389/fmicb.2015.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muñoz-Dorado J, Marcos-Torres FJ, García-Bravo E, Moraleda-Muñoz A, Pérez J. Myxobacteria: moving, killing, feeding, and surviving together Front Microbiol 2016;7:781. [DOI] [PMC free article] [PubMed]
- 146.Goldman B.S., Nierman W.C., Kaiser D., Slater S.C., Durkin A.S. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pérez J., Castañeda-García A., Jenke-Kodama H., Müller R., Muñoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci USA. 2008;105:15950–15955. doi: 10.1073/pnas.0806851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Muñoz-Dorado J, Moraleda-Muñoz A, Marcos-Torres FJ, ContrerasMoreno FJ, Martin-Cuadrado AB, et al. Transcriptome dynamics of the Myxococcus xanthus multicellular developmental program. eLife 2019;8:e5037. [DOI] [PMC free article] [PubMed]
- 149.Korp J., Vela-Gurovic M.S., Nett M. Antibiotics from predatory bacteria. Beilstein J Org Chem. 2016;12:594–607. doi: 10.3762/bjoc.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Weber T., Blin K., Duddela S., Krug D., Kim H.U. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gerth K., Jansen R., Reifenstahl G., Hofle G., Irschik H. The myxalamids, new antibiotics from Myxococcus xanthus (Myxobacterales). I. Production, physico-chemical and biological properties, and mechanism of action. J Antibiot. 1983;36:1150–1156. doi: 10.7164/antibiotics.36.1150. [DOI] [PubMed] [Google Scholar]
- 152.Irschik H, Trowitzsch-Kienast W, Gerth K, Höfle G, Reichenbach H. Saframycin Mx1, a new natural saframycin isolated from a myxobacterium. J Antibiot 1988;41:993–998. [DOI] [PubMed]
- 153.Trowitzsch-Kienast W., Gerth K., Wray V., Reichenbach H., Höfle G. Antibiotika aus gleitenden bakterien, LV-myxochromid A: ein hochungesättigtes lipopeptidlacton aus Myxococcus virescens. Liebigs Ann Chem. 1993;12:1233–1237. [Google Scholar]
- 154.Bois-Choussy M., Cristau P., Zhu J. Total synthesis of an atropdiastereomer of RP-66453 and determination of its absolute configuration. Angew Chem Int Ed. 2003;42:4238–4241. doi: 10.1002/anie.200351996. [DOI] [PubMed] [Google Scholar]
- 155.Krug D., Zurek G., Revermann O., Vos M., Velicer G.J. Discovering the hidden secondary metabolome of Myxococcus xanthus: a study of intraspecific diversity. Appl Environ Microbiol. 2008;74:3058–3068. doi: 10.1128/AEM.02863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Weissman K.J., Müller R. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep. 2010;27:1276–1295. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 157.Cortina N.S., Revermann O., Krug D., Müller R. Identification and characterization of the althiomycin biosynthetic gene cluster in Myxococcus xanthus DK897. ChemBioChem. 2011;12:1411–1416. doi: 10.1002/cbic.201100154. [DOI] [PubMed] [Google Scholar]
- 158.Cortina N.S., Krug D., Plaza A., Revermann O., Müller R. Myxoprincomide, a natural product from Myxococcus xanthus discovered by a comprehensive analysis of the secondary metabolome. Angew Chem Int Ed. 2012;51:811–816. doi: 10.1002/anie.201106305. [DOI] [PubMed] [Google Scholar]
- 159.Hug J.J., Müller R. Reference module in chemistry, molecular science and chemical engineering. Elsevier; 2020. Host development for heterologous expression and biosynthetic studies of myxobacterial natural products. [Google Scholar]
- 160.Findlay B.L. The chemical ecology of predatory soil bacteria. ACS Chem Biol. 2016;11:1502–1510. doi: 10.1021/acschembio.6b00176. [DOI] [PubMed] [Google Scholar]
- 161.Robertsen H.L., Musiol-Kroll E.M. Actinomycete-derived polyketides as a source of antibiotics and lead structures for the development of new antimicrobial drugs. Antibiotics. 2019;8:157. doi: 10.3390/antibiotics8040157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bentley S.D., Chater K.F., Cerdeño-Tárraga A.M., Challis G.L., Thomson N.R. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;9:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 163.Ikeda H., Ishikawa J., Hanamoto A., Shinose M., Kikuchi H. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 164.Traxler M.F., Kolter R. Natural products in soil microbe interactions and evolution. Nat Prod Rep. 2015;32:956. doi: 10.1039/c5np00013k. [DOI] [PubMed] [Google Scholar]
- 165.Xiao Y., Wei X., Ebright R., Wall D. Antibiotic production by myxobacteria plays a role in predation. J Bacteriol. 2011;193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.DePas W.H., Syed A.K., Sifuentes M., Lee J.S., Warshaw D. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus. Appl Environ Microbiol. 2014;80:7079–7087. doi: 10.1128/AEM.02464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang C., Liu X., Zhang P., Wang Y., Li Z. Bacillus licheniformis escapes from Myxococcus xanthus predation by deactivating myxovirescin a through enzymatic glucosylation. Environ Microbiol. 2019;21:4755–4772. doi: 10.1111/1462-2920.14817. [DOI] [PubMed] [Google Scholar]
- 168.Pérez J., Muñoz-Dorado J., Braña A.F., Shimkets L.J., Sevillano L. Myxococcus xanthus inducesactinorhodin overproduction and aerial mycelium formation by Streptomycescoelicolor. Microb Biotechnol. 2011;4:175–183. doi: 10.1111/j.1751-7915.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Müller S., Strack S.N., Ryan S.E., Kearns D.B., Kirby J.R. Predation by Myxococcus xanthus induces Bacillus subtilis to form spore-filled megastructures. Appl Environ Microbiol. 2015;81:203–210. doi: 10.1128/AEM.02448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Contreras-Moreno F.J., Muñoz-Dorado J., García-Tomsig N.I., Martínez-Navajas G., Pérez J. Copper and melanin play a role in Myxococcus xanthus predation on Sinorhizobium meliloti. Front Microbiol. 2020;11:94. doi: 10.3389/fmicb.2020.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Livingstone P.G., Millard A.D., Swain M.T., Whitworth D.E. Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lee N., Kim W., Chung J., Lee Y., Cho S. Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME J. 2020;14:1111–1124. doi: 10.1038/s41396-020-0594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Berleman J.E., Allen S., Danielewicz M.A., Remis J.P., Gorur A. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol. 2014;5:474. doi: 10.3389/fmicb.2014.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Remis J.P., Wei D., Gorur A., Zemla M., Haraga J. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol. 2014;16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gnopo Y.M.D., Watkins H.C., Stevenson T.C., DeLisa M.P., Putnam D. Designer outer membrane vesicles as immunomodulatory systems-reprogramming bacteria for vaccine delivery. Adv Drug Deliv Rev. 2017;114:132–142. doi: 10.1016/j.addr.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 176.Evans A.G.L., Davey H.M., Cookson A., Currinn H., Cooke-Fox G. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 177.Schulz E., Goes A., Garcia R., Panter F., Koch M. Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. J Control Release. 2018;290:46–55. doi: 10.1016/j.jconrel.2018.09.030. [DOI] [PubMed] [Google Scholar]
- 178.Goes A., Lapuhs P., Kuhn T., Schulz E., Richter R. Myxobacteria-derived outer membrane vesicles: potential applicability against intracellular infections. Cells. 2020;9:194. doi: 10.3390/cells9010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kahnt J., Aguiluz K., Koch J., Treuner-Lange A., Konovalova A. Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membranevesicles. J Proteome Res. 2010;9:5197–5208. doi: 10.1021/pr1004983. [DOI] [PubMed] [Google Scholar]
- 180.Whitworth D.E., Slade S.E., Mironas A. Composition of distinct sub- proteomes in Myxococcus xanthus: metabolic cost and amino acid availability. Amino Acids. 2015;47:2521–2531. doi: 10.1007/s00726-015-2042-x. [DOI] [PubMed] [Google Scholar]
- 181.Zwarycz A.S., Livingstone P.G., Whitworth D.E. Within-species variation in OMV cargo proteins: the Myxococcus xanthus OMV pan-proteome. Mol Omics. 2020 doi: 10.1039/d0mo00027. [DOI] [PubMed] [Google Scholar]
- 182.Livingstone PG, Morphew RM, Whitworth DE. Genome sequencing and pan-genome analysis of 23 Corallococcus spp. strains reveal unexpected diversity, with particular plasticity of predatory gene sets. Front Microbiol 2018;9:3187. [DOI] [PMC free article] [PubMed]
- 183.Sutton D., Livingstone P.G., Furness E., Swain M.T., Whitworth D.E. Genome-wide identification of myxobacterial predation genes and demonstration of formaldehyde secretion as a potentially predation-resistant trait of Pseudomonas aeruginosa. Front Microbiol. 2019;10:2650. doi: 10.3389/fmicb.2019.02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.