Abstract
Soybean mosaic virus (SMV), a member of the genus Potyvirus, is a prevalent and devastating viral pathogen in soybean-growing regions worldwide. Potyvirus-encoded P3 protein is reported to participate in virus replication, movements, and pathogenesis. This study provides evidence that the soybean (Glycine max) endo-1,3-beta-glucanase protein (designated as GmGLU) interacts with SMV-P3 by using a yeast two-hybrid system to screen a soybean cDNA library. A bimolecular fluorescence complementation assay further confirmed the interaction, which occurred on the cytomembrane in Nicotiana benthamiana cells. Subcellular localization experiment indicated that GmGLU localized in cytomembrane and could co-localized at PD with PD marker. The transient expression of GmGLU promoted the coupling of Turnip mosaic virus replication and cell-to-cell movement in N. benthamiana. Meanwhile, qRT-PCR experiment demonstrated that the expression of GmGLU which involved in callose regulation increased under SMV infection. Under SMV infection, callose deposition at PD was observed obviously by staining with aniline blue, which raise a physical barrier restricting cell-to-cell movement of SMV. When overexpression the GmGLU into the leaves under SMV infection, the callose induced by SMV was degraded. Coexpression the GmGLU and SMV in soybean leaves, callose was not found, whereas a large amount of callose deposition on soybean leaves which were only under SMV infection. The results show that GmGLU can degrade the callose induced by SMV infection and indicate that GmGLU may be an essential host factor involvement in potyvirus infection.
Keywords: Soybean mosaic virus, P3, GmGLU, interaction, callose
Highlights
-
-
GmGLU interacted with SMV-P3 in a screening of soybean cDNAs using mY2HS.
-
-
The interaction between GmGLU and SMV-P3 was confirmed using BiFC.
-
-
The subcellular location of GmGLU was on the cytomembrane and form the dots at plasmodesmata sites.
-
-
GmGLU was up-expressed in response to SMV infection.
-
-
GmGLU overexpression of soybean decreases callose deposition.
-
-
GmGLU overexpression promotes viral cell-to-cell movement and replication.
Introduction
Soybean mosaic virus (SMV), a member of the family Potyviridae, is the causative agent of one of the most well-known viral diseases affecting soybean (Kim et al., 2016). Specifically, the virions are transmitted by seeds and aphids, reducing the yield and quality of soybeans (Shukla et al., 1994; Yang and Gai, 2011). The SMV genome is a single positive-stranded RNA, about 9600 nucleotides long, containing an open reading frame (ORF) and 11 different multifunctional mature proteins (Urcuqui-Inchima et al., 2001; Chung et al., 2008; Wen and Hajimorad, 2010).
P3, a 43 kDa non-structural protein, is the most variable potyviral proteins (Eiamtanasate et al., 2007). Many previous studies have shown that P3 participates in virus replication, movement, and pathogenesis (Johansen et al., 2001; Jenner et al., 2003; Suehiro et al., 2004). Some studies further revealed that the P3 cistron may also function as a virulence determinant for the SMV elicitors of Rsv1-mediated resistance (Hajimorad et al., 2006, 2008, 2011; Chowda-Reddy et al., 2010; Wen et al., 2011, 2012; Wang et al., 2014). Previous studies have revealed that the Tobacco etch virus P3 protein is localized in the ER membrane and forms punctate inclusions in association with the Golgi apparatus (Cui et al., 2010). Moreover, the P3 punctate structure was found to traffic along the actin filaments and co-localize with the 6 kDa peptide-containing replication vesicles (Cui et al., 2010). The potyviral P3 cistron encodes two viral proteins, P3N-PIPO and P3, which have two distinct functions: P3N-PIPO is a dedicated MP (Wei et al., 2010) and P3 is an essential component of the viral replication complexes (Cui et al., 2017). Furthermore, P3N-PIPO interacts with P3 via the shared N-terminal domain to recruit viral replication vesicles for cell-to-cell movement (Chai et al., 2020). Therefore, we speculate that P3 may act as a bridge between virus replication and movement.
Previous studies have shown that P3 targets the host elongation factor eEF1A, inducing the associated unfolded protein response, which in turn facilitates SMV replication (Luan et al., 2016). It has been reported that P3 interacts with the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), thus contributing to the development of viral infection symptoms in host plants (Lin et al., 2011). Furthermore, previous reports have studied the interaction of soybean proteins with P3, including soybean actin-depolymerizing factor 2 (Lu et al., 2015), hypersensitive response-like lesion-inducing protein (Luan et al., 2019), endoplasmic reticulum homologous protein (GmRHP) (Cui et al., 2018), which might play important role in SMV infection. Taken together, these studies suggest that studying the function of P3 and its interactors can help us better understand viral infection.
Endo-1,3-β-glucanases (GLU) are grouped in the PR-2 family of PR proteins (Balasubramanian et al., 2012; Piršelová and Matušíková, 2013), which catalyze the hydrolysis of 1,3-β-glucan linkages and play important roles in plant defense and development. For instance, the previously reported that abscisic acid inhibits the transcription of 1,3-β-glucanase and thus remarkably strengthens the ability of plants to resist viruses (Mauch-Mani and Mauch, 2005; Fernández-Crespo et al., 2017). Callose, a 1,3-β-glucan which is a common substrate of GLU, is reversibly and transiently deposited in plant cell walls as part of the hypersensitive response to fungi, viruses, and abiotic stresses (Stone and Clarke, 1992; Kauss, 1996). Callose deposition occurs in the wall surrounding plasmodesmata [the intercellular connections between plant cells that allow cell-to-cell transport of sugars, amino acids, inorganic ions, proteins, and nucleic acids (reviewed by Lucas et al., 1993)] at both ends of the channel, compressing the plasma membrane inward, thus creating a narrowed neck region (Radford et al., 1998), which inhibits the transport of macromolecules through plasmodesma (PD). GLU as hydrolytic enzymes which specifically degrade callose, change the dynamic balance between callose synthesis and degradation, while promoting cell-to-cell viral movement and spread. Thus, callose is thought to act as a physical barrier limiting or preventing the spread of viruses in the host (Allison and Shalla, 1974; Iglesias and Meins, 2000). Previous study has shown that increased expression of GLU I in virus-infected cells can increase the size of local lesions and in the absence of the hypersensitive response, can promote virus spread (Gregor et al., 2001). As opposed to GLU overexpression, the deficiency in GLU results in reduced susceptibility to viral infection due to net increase in PD callose levels (Zavaliev et al., 2011).
In this study, we demonstrated that the soybean factor endo-1,3-β-glucanase (designated as GmGLU) interacts with SMV P3 in a mY2H analysis and BiFC. GmGLU transformed and transiently expressed into Nicotiana benthamiana leaf epidermal cells by agroinfiltration, formed punctate inclusions on the cytomembrane, and co-located with a PD marker at the PD in N. benthamiana. The expression of GmGLU in soybean leaves inoculated with SMV was higher than that in leaves mock-inoculated with PBS at 48 and 72 h post-infiltration. Furthermore, we demonstrated that overexpression of soybean GLU can decrease callose deposition at the PD which was induced by SMV. We also observed that the viral spread speed and genomic RNA accumulation significantly increased when coexpression GmGLU in N. benthamiana leaves. Our data support that GmGLU plays an important role in viral infection.
Materials and Methods
Plants, Viral Strains, and Plasmids
Seeds of Glycine max ‘Nannong 1138-2’ were obtained from the Soybean Improvement Center of Nanjing Agricultural University (Nanjing, China). Leaves of 2-weeks-old ‘Nannong 1138-2’ plants were inoculated via a point inoculation approach or direct rubbing by a brush or toothpick with SMV, isolate 6067-1 (strain SC15) (GenBank Accession: JF833015.1) in controlled environment chambers at 22°C with a 16-h light/8-h dark photoperiod. The N. benthamiana seedlings were grown in growth chambers under a 16-h light/8-h dark photoperiod at 22 to 24°C. Escherichia coli strain DH5a, Saccharomyces cerevisiae strain NMY51 (Dualsystems Biotech, Zurich, Switzerland), yeast expression vectors (pPR3-N, and pBT3-STE) (Dualsystems Biotech), and the pPR3-gateway vector (modified from pPR3-N) were used in this study. All vectors were constructed using Gateway technology. Agrobacterium tumefaciens strain EHA105 and the fluorescence expression vectors pEarleyGate104, pEarleyGate201-YC, pEarleyGate202-YN and the plant expression vector pEarleyGate201 were stored at the Jiangsu Province Academy of Agricultural Sciences (Nanjing, China).
Gene Cloning and Plasmid Construction
Gateway technology (Invitrogen, Burlington, ON, Canada) was used to generate all plasmid clones used in this work. Gene sequences were amplified by PCR using PrimeSTAR HS DNA Polymerase (Takara, Dalian, China). The resulting DNA fragments were purified and transferred by recombination into the entry vector pDONR221 (Invitrogen) using BP clonase II (Invitrogen) following the supplier’s recommendations. Insertions in the resulting pDONR clones were verified by DNA sequencing. The destination vectors, yeast expression vector (pPR3-gateway) or fluorescence expression vectors (pEarleyGate104, pEarleyGate201-YC, and pEarleyGate202-YN), or plant expression vector (pEarleyGate201) were produced using LR clonase II (Invitrogen) according to the conditions and procedures recommended by the supplier. The primer sequences used in this study are listed in Table 1.
TABLE 1.
Primer sequences used for cloning GLU genes.
| Name | Sequence (5′→ 3′)a |
| SMV-P3-F | ATTAACAAGGCCATTACGGCCGGGGAAGTGCAACAAAGGATGA |
| SMV-P3-R | AACTGATTGGCCGAGGCGGCCCCCTGTGCGGAGACATCTTCT GA |
| GmGLU-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGATTTTCTCAA GAGGCAAC |
| GmGLU-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCATTGAAACTGAG TTGGTATTTAGGT |
aThe restriction enzyme sequences or gateway sequences are underlined.
mY2H Analysis
The mY2H tests were performed using the Matchmaker DUAL membrane system (Clontech, Palo Alto, CA, United States) according to the manufacturer’s protocol. The DUAL membrane system was developed to identify and characterize protein–protein interactions between integral membrane proteins, membrane-associated proteins, and soluble proteins in their natural condition. The fusion vector pPR3-gateway-GmGLU was constructed by inserting the GmGLU gene into the pPR3-gateway vector. pPR3-gateway-GmGLU and pBT3-STE-P3 (constructed previously) were co-transformed into yeast cells using the small-scale lithium acetate transformation method. The yeast cells were then grown on SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp medium for 3–7 days at 30°C. Colonies growing on SD/-Ade/-His/-Leu/-Trp medium were transferred to SD/-Ade/-His/-Leu/-Trp/X-α-Gal solid medium and grown for 3–5 days at 30°C. Blue colonies were considered as positive clones. Yeast cells co-transformed with empty pBT3-STE-P3+pPR3-gateway, pBT3-STE+pPR3-gateway-GmGLU, and pBT3-STE+pPR3-gateway were used as a negative control, and those co-transformed with pBT3-STE-P3N-PIPO+pPR3-gateway-GmGOS1 were used as a positive control (Song et al., 2016). The experiments were repeated at least three times.
Agroinfiltration in N. benthamiana
All experiments were performed using N. benthamiana, an experimental host of SMV. For the BiFC assay, the A. tumefaciens strain EHA105 was transformed with pEarleyGate202-YN-GmGLU and pEarleyGate201-YC-P3 via cryoapplication. For the subcellular localization assay or overexpression, A. tumefaciens strain EHA105 was transformed with pEarleyGate104-GmGLU or pEarleyGate201-GmGLU, then cultured overnight with appropriate antibiotics at 28°C (Sparkes et al., 2006). For intercellular movement analysis, the infectious clone TuMV-GFP was described in Cui et al. (2017). The Agrobacterium cells were diluted to an optical density (OD600) of 0.5–0.6 for BiFC experiments (Tian et al., 2011) and 0.1–0.2 for subcellular expression and 0.001 for intercellular movement analyses. The diluted A. tumefaciens was injected into N. benthamiana and fluorescence was observed by confocal microscopy. Each BiFC experiment, subcellular localization assay and intercellular movement analysis was repeated at least three times, and each treatment included at least three independent N. benthamiana plants.
RNA Extraction and RT-PCR
RNA was isolated from 50 to 100 mg of frozen soybean leaf tissue using the total RNA isolation kit according to the manufacturer’s instructions (TIANGEN, Nanjing, China). The quality and quantity of RNA were measured with a nucleic acid and protein detection instrument. Reverse transcription (RT) and first-strand cDNA synthesis were carried out using Fast-RT SuperMix (TIANGEN, Nanjing, China). The primer pair SMV-CP-F (5′-ATGGTTGAGAGGGAAAGATCTATCTC-3′) and SMV-CP-R (5′-AGAGCTGCAGCCTTCATCTTG-3′) was used to detect soybean genomic RNA. An SMV coat protein gene DNA fragment at the expected size of 500 bp was amplified as follows: initial denaturation at 94°C for 2 min followed by 30 cycles of denaturation at 94°C for 0.5 min, annealing at 55°C for 0.5 min and elongation at 72°C for 1 min. The final extension step was at 72°C for 10 min.
Quantitative Reverse Transcription PCR
Quantitative reverse transcription PCR was carried out in a LightCycler 480 (Roche, Penzberg, Germany) using SYBR® Green I (Takara, Dalian, China). The primer pair GmGLU-F (5′-TATTTAGGTGATTTGTCAGGCC-3′) and GmGLU-R (5′-CCGCAATTTGATTAATCATGCC-3′) was used to detect soybean genomic RNA, and the primer pair GmTubulin-F (5′-CGAGTTCACAGAGGCAGAG-3′) and GmTubulin-R (5′-CACTTACGCATCACATAGCA-3′) was used to detect the reference gene Tubulin in G. max, which was used for normalization. The primer pair TuCP-F (5′-GGCACTCAAGAAAGGCAAGG-3′) and TuCP-R (5′-CTCCGTCAGTTCGTAATCAGC-3′) was used for detection of TuMV viral genomic RNA, and the primers NbActin-F (5′-GGGATGTGAAGGAGAAGTTGGC-3′) and NbActin-R (5′-ATCAGCAATGCCCGGGAACA-3′) for the reference gene Actin in N. benthamiana were used for normalization. The final reaction mixtures contained 250 nM of each primer in a volume of 20 μL. All genes were amplified under the same reaction conditions: 30 s at 95°C, followed by 40 cycles of 10 s at 95°C, 30 s at 53°C, and 34 s at 72°C. The specificity of amplification was verified using a melting curve analysis (60–95°C). qRT-PCRs with no template control were performed under the same conditions. All reactions were performed in triplicate, and the mean value was calculated. Cycle threshold (CT) values were determined using LightCycler 480 Real-Time PCR software (Roche, Penzberg, Germany) and were used for gene expression analysis. Bar charts and error lines were created using OriginLab OriginPro 8.5.
Aniline Blue Staining
Callose was stained by a modification of the aniline blue fluorochrome method (Conrath et al., 1998; Li et al., 2012). In brief, sections of about 1 cm × 1 cm were cut at different time points after inoculation from unifoliate leaves, which do not have large veins. Chlorophyll was cleared by heating the samples at 55°C in lactophenol (water: glycerol: phenol: lactic acid, 1:2:2:1 by volume) diluted with two volumes of ethanol for 30 min. The cleared leaf samples were stained in the dark with 0.1% aniline blue (Macklin, China) in 0.15M K2HPO4 buffer, pH 9.0, for 30–60 min. The leaves were observed under a confocal microscope. The experiment was repeated at least three times, and each treatment included at least three independent ‘Nannong 1138-2’ plants.
Confocal Microscopy
Epidermal cells of N. benthamiana were examined with Ultra View VOX inverted confocal microscope. For confocal microscopy, CFP was excited at 458 nm, and the emitted light was captured at 440 to 470 nm; YFP was excited at 514 nm, and the emitted light was captured at 525 to 650 nm; GFP was excited at 488 nm and the emitted light was captured at 505 to 555 nm. Post-acquisition images were processed by Adobe Photoshop software.
Results
Interaction Between GmGLU and SMV-P3 in Y2HS
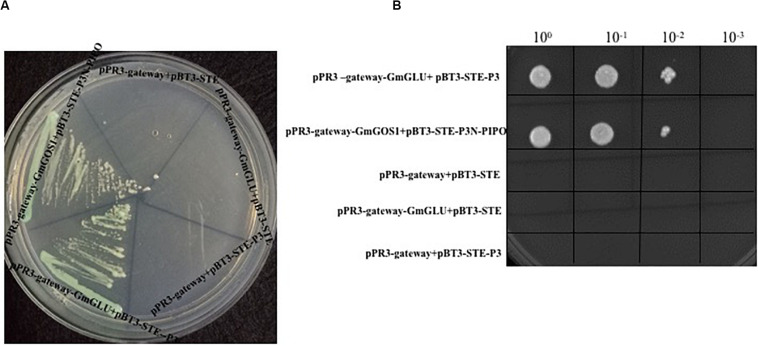
In a mY2HS analysis using the SMV-P3 protein as bait and a soybean cDNA library as prey, one candidate gene was identified and shared a 94.65% nucleotide sequence identity with the G. max endo-1,3-β-glucanase (AY461847.1). Thus, this isolated soybean factor was named GmGLU. The gene encoding SMV-P3 was inserted into pBT3-STE and the complete cDNA of soybean GmGLU (Accession Number: AY461847.1) was inserted into pPR3-gateway. The clones harboring pPR3-gateway-GmGLU+pBT3-STE-P3 grew well on SD/-Ade/-His/-Leu/-Trp/X-α-Gal medium (Figure 1A). Negative control P3N-PIPO cannot interact with GmGLU (Supplementary Figure S1). The positive control harboring pPR3-gateway-GmGOS1+pBT3-STE-P3N-PIPO also grew well on the selective medium, while the negative controls harboring pPR3-gateway-GmGLU+pBT3-STE\pPR3-gateway+pBT3-STE-P3 and pPR3-gateway+pBT3-STE did not grow on the selective medium. Different dilutions of the cells harboring pPR3-gateway-GmGLU+pBT3-STE-P3 grew as well as the positive control on SD/-Ade/-His/-Leu/-Trp medium (Figure 1B).
FIGURE 1.
Yeast two-hybrid assay to determine interaction between SMV P3 and GmGLU in Saccharomyces cerevisiae NMY51 cells using plasmids pPR3-gateway-GmGOS1 and pBT3-STE-P3N-PIPO (as positive control), pBT3-STE-P3+pPR3-gateway, pBT3-STE+pPR3-gateway-GmGLU, pPR3-gateway+pBT3-STE (as negative control), and pBT3-STE-P3+pPR3-gateway-GmGLU for transformation of yeast cells followed on SD/-Ade/-His/-Leu/-Trp medium containing X-α-Gal (A). Dilutions of pPR3-gateway-GmGLU+pBT3-STE-P3, positive control and negative control on SD/-Ade/-His/-Leu/-Trp medium (B).
Confirmation of Interaction by BiFC
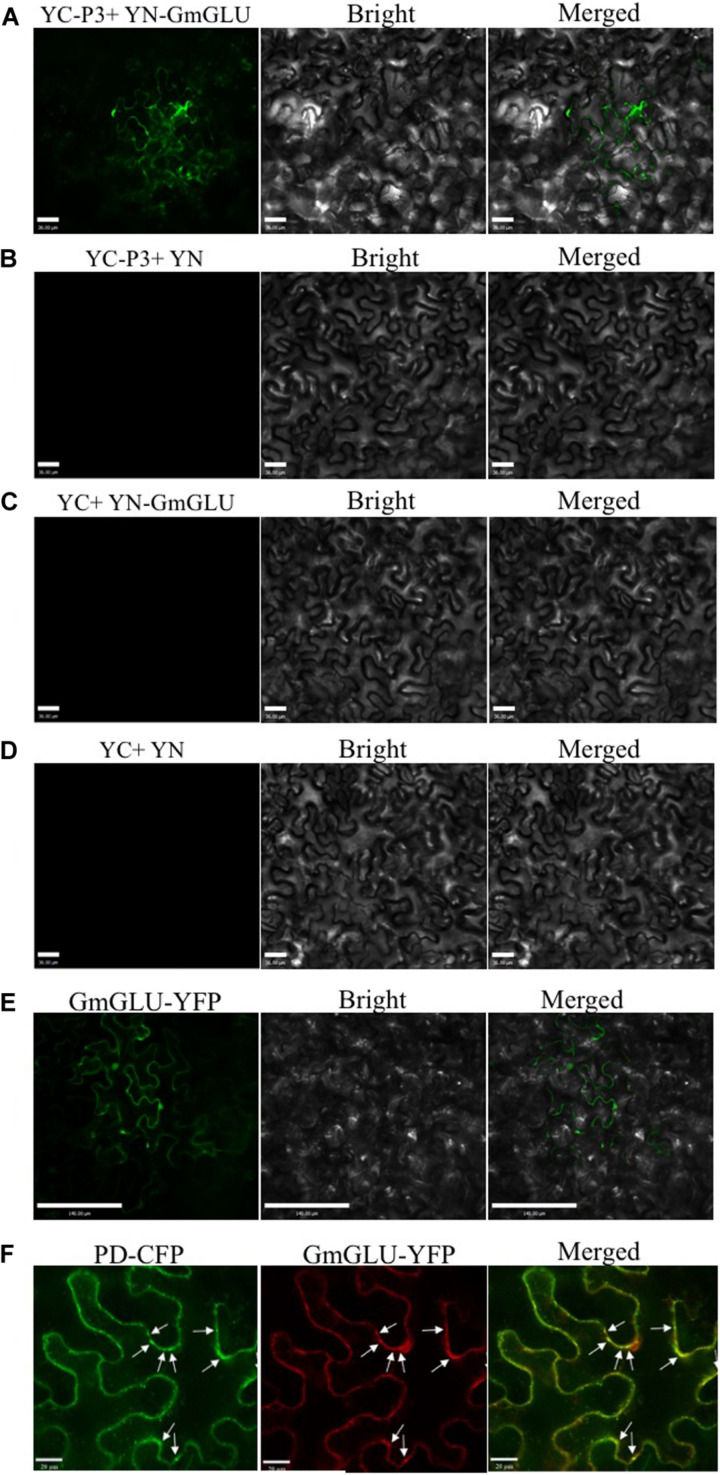
To further confirm the interaction between SMV-P3 and GmGLU in vivo, we recombined GmGLU into pEarlygate202-YN to create a fusion construct containing the N-terminal fragment of yellow fluorescent protein (YFP) and recombined SMV-P3 into pEarlygate201-YC to create a fusion construct containing the C-terminal fragment of YFP. These two vectors were co-transformed into N. benthamiana via Agrobacterium-mediated transformation, allowing the transient expression of fusion proteins. The green fluorescence indicative of the P3 and GmGLU combination was observed on the cytomembrane at 48 hpi (Figure 2A). No BiFC fluorescence was detected in the negative control samples expressing combinations with non-hybrid YC or/and YN (Figures 2B–D). These results confirmed that GmGLU could interact with SMV-P3 in vivo.
FIGURE 2.
Bimolecular fluorescence complementation assays demonstrating SMV P3 and GmGLU interaction in cytomembrane and epidermal cells of N. benthamiana. (A) SMV P3 and GmGLU were fused to N- and C-terminal of yellow fluorescent protein (YFP) respectively. YFP fluorescence in N. benthamiana leaves agroinfiltrated with pEarleyGate202-YN-GmGLU+pEarleyGate201-YC-P3. (B–D) Negative controls in the BiFC assay of GmGLU and SMV P3 interaction. Bar indicates 36 μm. (E) Fluorescence of YFP fused to GmGLU protein of host in N. benthamiana leaves under spectral confocal laser microscope. Bar indicates 140 μm. (F) Co-expression of plasmodesmata (PD) marker fused with cyan fluorescent protein (CFP) and GmGLU protein fused with YFP. GmGLU protein accumulated in dots on cytomembrane (PD marker sites). Bar indicates 20 μm.
Subcellular Localization of GmGLU in N. benthamiana Cells
To explore the possible functions of GmGLU, we analyzed its subcellular localization in N. benthamiana leaf epidermal cells. The gene was fused with YFP to form pEarleyGate104-GmGLU and agroinfiltrated into N. benthamiana. Using fluorescence microscopy at 48 hpi, the GmGLU protein fluoresced on the cytomembrane and formed small dots (Figure 2E). To investigate that where is small dots, the PD marker fused with cyan fluorescent protein (Wei et al., 2010; Park et al., 2017) and GmGLU protein fused with YFP coexpress in N. benthamiana leaf epidermal cells. The punctate inclusions on the cytomembrane from GmGLU were observed to overlap with PD marker on the points (Figure 2F). These results indicated that GmGLU was located at the PD site in leaf cells of N. benthamiana.
SMV Infection Induced the Overexpression of GmGLU
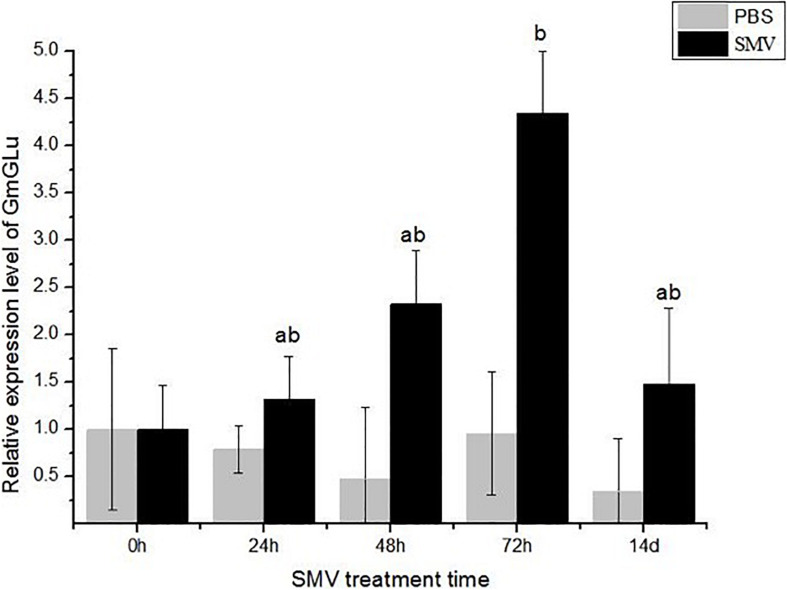
To investigate whether GmGLU is involved in response to SMV infection, we inoculated SMV onto the soybean variety ‘Nannong 1138-2’ and then detected the expression level of GmGLU at time-course after infection. The expression level of GmGLU in soybean leaves inoculated with SMV were significantly higher than those in leaves mock-inoculated with PBS at 24, 48, 72 hpi and 14 dpi (Figure 3). Additionally, a significant difference (P < 0.01) was found between the leaves treatment by SMV and PBS at 72 hpi (Figure 3). These findings confirmed that the infection of SMV induced the overexpression of GmGLU in soybean leaves.
FIGURE 3.
qRT-PCR analysis of GmGLU expression levels. Y-axes show relative expression levels of genes in SMV-infected and mock (PBS)-infected samples at different time points. Each error bar represents mean ± SD. Alphabetic symbols represent statistical significance: ab0.01 < P ≤ 0.05; bP ≤ 0.01.
SMV Infection Induces Callose Deposition
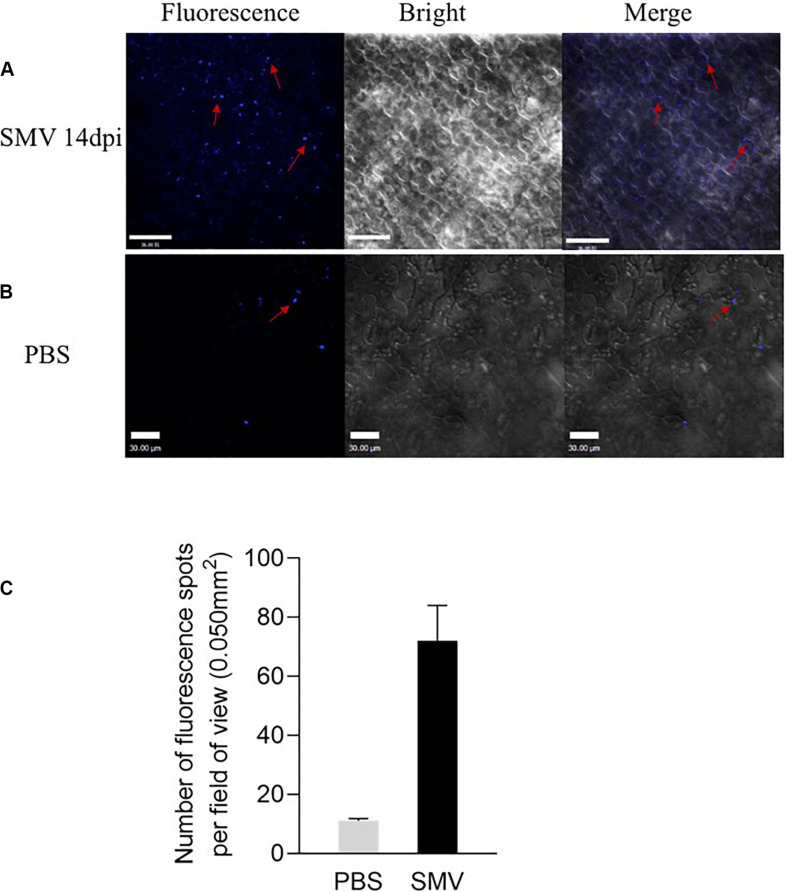
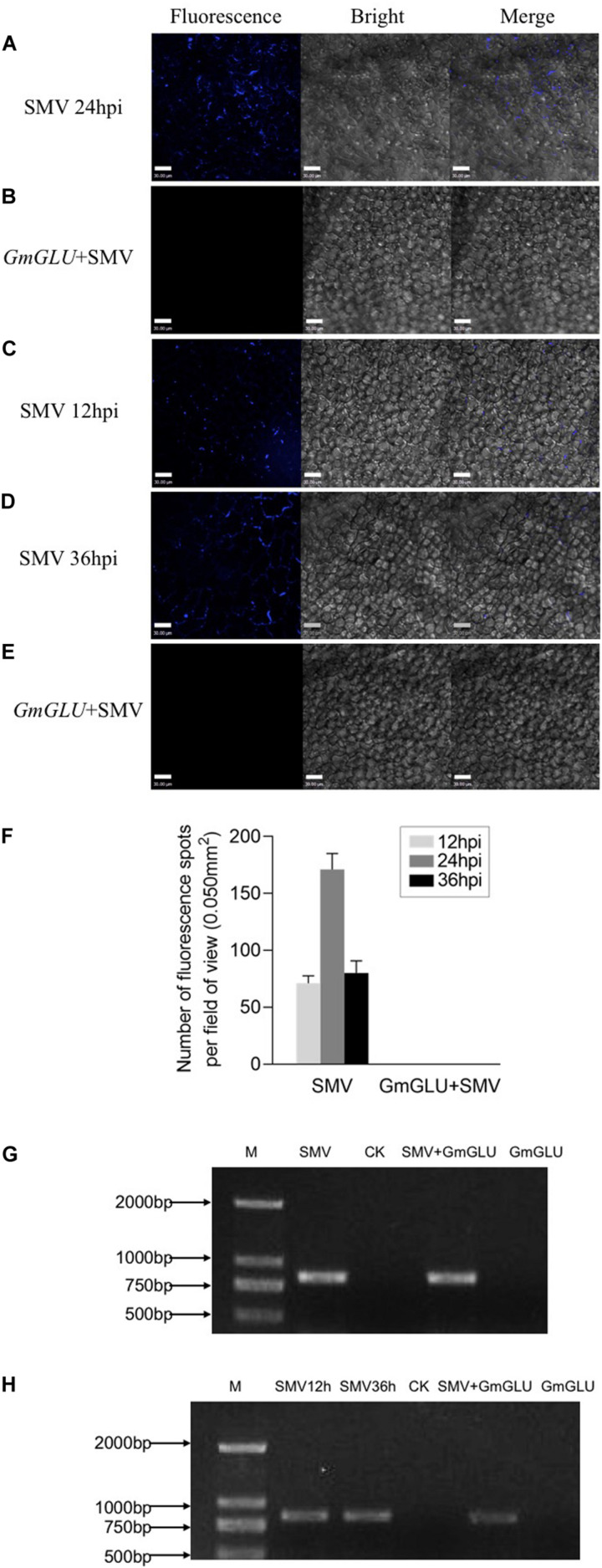
Callose can modulate the PD permeability, and the deposition of callose is one of the critical defense responses of host plants against viral infection. In order to detect the effect of SMV infection on callose deposition, we inoculated SMV into the soybean variety ‘Nannong 1138-2’ and then detected the deposition of callose 14 days after infection. PBS treatment was used as a negative control. Many points with blue fluorescence, unevenly distributed on the cell membrane, were observed by confocal microscopy (Figures 4A,C). The control group had a few points of callose deposition, caused by friction (Figures 4B,C). The accumulation of callose was quantified with the software ImageJ and was shown in Figure 4C. These results suggested that SMV infection can significantly induce callose deposition.
FIGURE 4.
Fluorescence spots at the inoculation sites detected from soybean cv. Nannong 1138-2 leaves inoculated with SMV strain SC-15. (A) Lots of Fluorescence spots were detected as uneven distribution on the cell membrane 14 days post-inoculation. (B) A few of callose deposition, caused by friction was observed as negative control. Bar indicates 30 μm. (C) Number of fluorescence spots per field of view (0.050 mm2), as indicated callose deposition in systemically infected leaves at 14 dpi. Three plants were examined in each treatment, two small areas of each plant were inoculated, and five fields of view are randomly observed in each area. Error bars represent the standard deviation from 30 fields of view and data were from three independent experiments.
GmGLU Decreases Callose Deposition at the PD
To further examine whether the soybean GmGLU plays a role in callose deposition at the PD, we inoculated SMV by toothpick on the leaves of soybean, where have been agroinfiltrated with GmGLU 24 h in advance. The leaves without agroinfiltration of GmGLU were used as control. After 24 h, we observed there was no callose deposition on soybean leaves with GmGLU overexpression (Figures 5B,F), whereas large callose depositions were found on soybean leaves which were only infected by SMV (Figures 5A,F). We therefore speculated that GmGLU inhibited the deposition of callose.
FIGURE 5.
GmGLU decreases callose deposition induced by SMV infection. GmGLU in the soybean leaves was overexpressed prior to the inoculation with SMV. (A) Confocal microscopic observation. There was a large amount of callose deposition on soybean leaves which were only infected by SMV. (B) There was no callose deposition on soybean leaves with overexpression GmGLU. We observed callose deposition through overexpressing GmGLU into soybean leaves which inoculated by SMV. (C,D) There were many the fluorescence spots formed by callose at 12 h and 36 post-SMV inoculation. (E) The fluorescence formed by callose at the inoculation point were disappeared following the transfection of GmGLU. Bar indicates 30 μm. (F) Number of fluorescence spots per field of view (0.050 mm2), as indicated callose deposition in primarily infected leaves. Three plants were examined in each treatment, two small areas of each plant were inoculated, and five fields of view are randomly observed in each area. Error bars represent the standard deviation from 30 fields of view and data were from three independent experiments. (G,H) Detection of viral CP-RNA by RT-PCR analysis in the upper leaves. The viral CP-RNA was detected in the SMV-infected soybean cv. Nannong 1138-2 (G: lane 1, lane 3; H: lane 1, lane 2, lane 4). The viral CP-RNA was not detected from the negative control and GLU-treatment (G: lane 2, lane 4; H: lane 3, lane 5). Independent experiments were performed three times.
To further confirm this hypothesis, we observed callose deposition through overexpressing GmGLU into SMV-infected leaves. At 12 h post-SMV inoculation, a few fluorescence spots resulting from callose formation were found by staining with aniline blue (Figures 5C,F). We then transfected GmGLU into the SMV-infected leaves. At 36 h post-SMV inoculation, we found that the fluorescence formed by callose at the inoculation point disappeared following the transfection of GmGLU (Figures 5E,F), while there was considerable callose deposition on soybean leaves which were only infected with SMV (Figures 5D,F). No fluorescent spots produced by callose deposition was detected on leaves only inoculated with GmGLU and healthy plant leaves (Supplementary Figures S2A,B). This result suggests that GmGLU could degrade callose. Further, the viral RNA on the leaves of the inoculation was detected by RT-PCR analysis to prove that the plants were indeed infected by SMV (Figures 5G,H). These results indicated that the overexpression of soybean GmGLU decreases callose deposition induced by SMV infection at the PD.
Overexpression of GmGLU Promotes Viral Proliferation
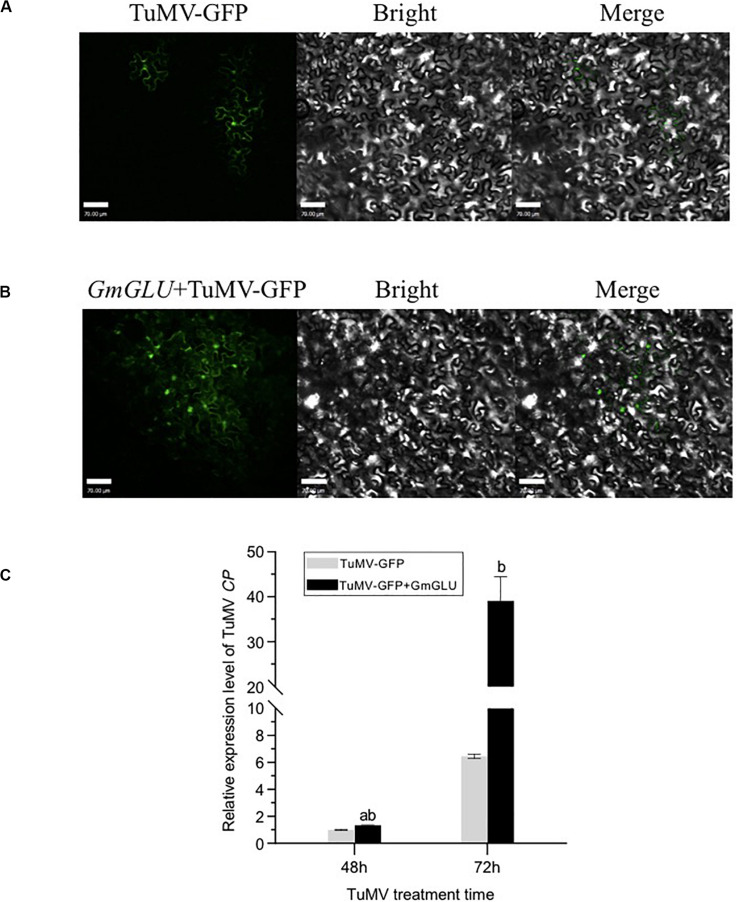
Systemic viral spread was monitored with a recombinant form of TuMV-GFP (Cui et al., 2017). The infectious clone TuMV-GFP and pEarleyGate201-GmGLU were agroinfiltrated into N. benthamiana leaf cells, and the spread of fluorescence due to TuMV-GFP was monitored 3.5 days later. As Figure 6B shows, leaves co-infected with TuMV-GFP/GmGLU-201 showed a cluster of virus-infected cells, while sporadic luminescence occurred at the site of the initial infection in N. benthamiana leaves agroinfiltrated with the control TuMV-GFP (Figure 6A). The virus spread significantly increased of the GLU-treated plants compared with the control (Figure 6B). These results suggested that the movement of viruses is facilitated in the plant with GmGLU overexpression.
FIGURE 6.
Expression of GmGLU promote the viral proliferation. (A,B) Confocal microscopic observation of viral intercellular movement of TuMV-GFP, TuMV-GFP/GmGLU-201. Images were taken 3.5 days post-agroinfiltration. Independent experiments were performed three times. Typical imaging was shown. Bar indicates 70 μm. (C) qRT-PCR detection of the viral accumulation level in N. benthamiana leaves transfected with TuMV-GFP, TuMV-GFP/GmGLU-201. Leaves were collected at 48 h post-transfection. Viral RNA was quantified by qRT-PCR analysis of the CP RNA level using the N. benthmiana ACTIN gene transcripts as the internal control. TuMV-GFP genomic RNA level was normalized to 1. Statistical differences from three biological replicates, determined by unpaired two-tailed Student’s test, are indicated by ab and b as follows: ab0.001 < P ≤ 0.01; bP ≤ 0.001. Each error bar represents mean ± SD.
To test if GmGLU overexpression affects virus accumulation, we carried out qRT-PCR assay. N. benthamiana leaves were inoculated with the infectious clone TuMV-GFP and GmGLU-201, qRT-PCR was performed to quantify TuMV genomic RNA 48 and 72 h post-transfection. We found that GmGLU overexpression significantly promotes TuMV genomic RNA accumulation compared with control leaves which were only agroinfiltrated by TuMV-GFP (Figure 6C). Taken together, these results indicated that GmGLU overexpression promotes viral cell-to-cell movement and accumulation.
Discussion
Soybean mosaic virus is composed of a single-stranded, positive-sense RNA genome and coat protein (CP) that protects nucleic acid. Since viruses lack their own reproduction system, they achieve systemic infection by interacting with host proteins by taking advantage of the host’s cellular mechanisms for replication and intracellular/intercellular movement (Gergerich and Dolja, 2006). If the virus loses the ability to move, infection will be limited to an initial infection site or infected organ (Hong and Ju, 2017).
Previous reports indicated that the successful infection of plants by most viruses depends on cell-to-cell movement via PD and rapid long distance movement through the sieve tubes of the phloem (Lucas, 1995; Van Bel, 2003; Lucas and Lee, 2004; Oparka, 2004). For plant viruses, the ability of crossing cell walls through PD into adjacent healthy cells is necessary to establish systemic infection (Carrington et al., 1996; Seron and Haenni, 1996). However, virions are too large to move through the PD, and thus transport through PD is usually limited to small molecules less than 1 kDa in size (Lucas, 1995). To overcome this limitation, most viruses encode MPs, which interact with host proteins to increase the SEL of the PD and help virions pass through PD into adjacent cells (Lucas, 1995; Iglesias and Meins, 2000). Our results suggest that GLU interacts with P3 to reduce the SEL, which can play a role in potyviral intercellular spread in infected plants.
Callose is a linear 1,3-β-glucan playing an important role on the plant allergy defense response (Piršelová and Matušíková, 2013). The callose content at PD determines the aperture of the PD channel and thus controls cell-to-cell transport of macromolecules (Bucher et al., 2001; Levy and Epel, 2009; Zavaliev et al., 2011). Once the accumulated amount of callose is down-regulated, it will cause PD to relax and allow more macromolecules trafficking (Beffa et al., 1996; Iglesias and Meins, 2000). The net callose level at PD is determined by the balance between the activity of callose synthase and 1,3-β-glucanase, because loss of either enzyme activity affects callose accumulation and transport through PD (Guseman et al., 2010; Vaten et al., 2011). The accumulation of callose in susceptible hosts is very low or similar to uninfected plants (Krasavina et al., 2002; Zavaliev et al., 2011). In this study, we explored the mechanism by which GLU degrades callose by targeting P3, and our results provide evidence for the regulatory effect of GmGLU on callose under SMV infection.
In this study, GmGLU interacted with SMV-P3 in a screening of soybean cDNAs using mY2HS. The expression of GmGLU in soybean leaves inoculated with SMV was increased. GLU is a PR protein, located on the cytomembrane and forms the dots at plasmodesmata sites, playing important roles in plant defense and development (Leubner-Metzger and Meins, 1999; Wan et al., 2011). GLU are cell wall enzymes that catalyze the callose and enhances viral transmission by degrading callose on PD (Deom et al., 1992; Zavaliev et al., 2011). The role of the GLU in virus spread has been described in TMV (Bucher et al., 2001; Gregor et al., 2001). Infection of tobacco with TMV led to a 20-fold increase in class I GLU levels (Vogeli-Lange et al., 1988; Epel, 2009). Furthermore, by combining SMV infection with aniline blue staining, we demonstrate that callose deposition induced by SMV infection was decreased when soybean GLU is overexpressed. The viral spread speed and genomic RNA accumulation significantly increased under overexpression of GmGLU. This conclusion is consistent with the research of Bucher et al. (2001). It is widely believed that the deposition of callose is a critical defense response of host plants against viral infection (Li et al., 2012). These data suggest that GmGLU regulate the callose deposition at the PD to allow viral intercellular trafficking in viral infection by interacting with SMV-P3.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at NCBI (AY461847.1).
Author Contributions
XYC conceived the project. XYC and XC supervised the work. FS and YW performed most of the experiments with assistance from FZ, XY, HC, and XHC. YW, FS, and XYC wrote the manuscript with contributions from all the authors. All authors analyzed the data.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Aiming Wang (Agriculture and Agri-Food Canada) for providing the pPR3-N and pBT3-STE vectors and Dr. Yuhai Cui (Agriculture and Agri-Food Canada) for providing pEarleyGate202-YN and pEarleyGate201-YC vectors.
Abbreviations
- BiFC
bimolecular fluorescence complementation
- ER
endoplasmic reticulum
- GmGLU
soybean (Glycine max) endo-1,3-beta-glucanase
- MP
movement protein
- mY2HS
membrane yeast two-hybrid system
- PBS
phosphatic buffer solution
- PD
plasmodesmata
- PR
pathogenesis-related
- PVX
Potato virus X
- PVY
Potato virus Y
- SEL
size exclusion limit
- SMV
Soybean mosaic virus
- TMV
Tobacco mosaic virus
- TuMV
Turnip mosaic virus.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31101411), National Key Research and Development Project (2018YFE0112200 and 2016YFE0203800), the Key R&D Project of Jiangsu Province (BE2019376), and Jiangsu Agricultural Industry Technology System [JATS(2019)399]. The research was carried out in the joint lab of China-AAFC for legume research in JAAS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.536771/full#supplementary-material
Yeast two-hybrid assay used to determine that SMV P3N-PIPO and GmGLU cannot interact in Saccharomyces cerevisiae NMY51 cells using plasmids pPR3-gateway-GmGLU+pBT3-STE-P3 (1) and pPR3-gateway-GmGOS1+pBT3-STE-P3N-PIPO (2) as positive control, pPR3-gateway+pBT3-STE-P3 (4) and pPR3-gateway+pBT3-STE-P3N-PIPO (5) and pPR3-gateway-GmGLU+pBT3-STE (6) and pPR3-gateway+pBT3-STE (7) as negative control, pPR3-gateway-GmGLU+pBT3-STE-P3N-PIPO (3) for transformation of yeast cells followed on SD/-Ade/-His/-Leu/-Trp medium containing X-α-Gal. In addition, negative controls 4–7 and pPR3-gateway-GmGLU+pBT3-STE-P3N-PIPO could not grow on the selective medium.
No fluorescent spots produced by callose deposition were detected on leaves only inoculated with GmGLU and healthy plant leaves. (A) No callose deposits can be detected on leaves overexpressing GmGLU. (B) No callose deposits can be detected on healthy plant leaves. Bar indicates 30 μm.
References
- Allison A. V., Shalla T. A. (1974). The ultrastructure of local lesions induced by potato virus X: a sequence of cytological events in the course of infection. Phytopathology. 64 784–793. 10.1094/Phyto-64-784 [DOI] [Google Scholar]
- Balasubramanian V., Vashisht D., Cletus J., Sakthivel N. (2012). Plant β-1,3-glucanases: their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 34 1983–1990. 10.1007/s10529-012-1012-6 [DOI] [PubMed] [Google Scholar]
- Beffa R. S., Hofer R. M., Thomas M., Meins F., Jr. (1996). Decreased susceptibility to viral disease of beta-1,3-glucanase-deficient plants generated by antisense transformation. Plant Cell 8 1001–1011. 10.2307/3870211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher G. L., Tarina C., Heinlein M., Serio F. D., Meins F., Jr., Iglesias V. A. (2001). Local expression of β-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28 361–369. 10.1046/j.1365-313x.2001.01181.x [DOI] [PubMed] [Google Scholar]
- Carrington J. C., Kasschau K. D., Mahajan S. K., Schaad M. C. (1996). Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8 1669–1681. 10.1105/tpc.8.10.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai M. Z., Wu X. Y., Liu J. H., Fang Y., Luan Y. M., Cui X. Y., et al. (2020). P3N-PIPO interacts with P3 via the shared N-terminal domain to recruit viral replication vesicles for cell-to-cell movement. J. Virol. 94:e01898-19. 10.1128/JVI.01898-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowda-Reddy R. V., Sun H. Y., Chen H. Y., Poysa V., Ling H., Gijzen M., et al. (2010). Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype Soybean. Mol. Plant Microbe Interact. 24 37–43. 10.1094/MPMI-07-10-0158 [DOI] [PubMed] [Google Scholar]
- Chung B. Y., Miller W. A., Atkins J. F., Firth A. E. (2008). An over lapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U.S.A. 105 5897–5902. 10.1073/pnas.0800468105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U., Klessig D. F., Bachmair A. (1998). Tobacco plants perturbed in the ubiquitin-dependent protein degradation system accumulate callose, salicylic acid, and pathogenesis-related protein. Plant Cell Rep. 17 876–880. 10.1007/s002990050501 [DOI] [PubMed] [Google Scholar]
- Cui X. Y., Lu L., Wang Y., Yuan X. X., Chen X. (2018). The interaction of Soybean reticulon homology domain protein(GmRHP) with Soybean mosaic virus encoded P3 contributes to the viral infection. Biochem. Biophys. Res. Commun. 495 2105–2110. 10.1016/j.bbrc.2017.12.043 [DOI] [PubMed] [Google Scholar]
- Cui X. Y., Wei T. Y., Chowda-Reddy R. N., Sun G. Y., Wang A. M. (2010). The Tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffics along actin microfilaments. Virology 397 56–63. 10.1016/j.virol.2009.11.015 [DOI] [PubMed] [Google Scholar]
- Cui X. Y., Yaghmaiean H., Wu G. W., Wu X. Y., Chen X., Thornc G., et al. (2017). The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex. Virology 510 147–155. 10.1016/j.virol.2017.07.016 [DOI] [PubMed] [Google Scholar]
- Deom C. M., Lapidot M., Beachy R. N. (1992). Plant virus movement proteins. Cell 69 221–224. 10.1016/0092-8674(92)90403-Y [DOI] [PubMed] [Google Scholar]
- Eiamtanasate S., Juricek M., Yap Y. K. (2007). C-terminal hydrophobic region leads PRSV P3 protein to endoplasmic reticulum. Virus Genes 35 611–617. 10.1007/s11262-007-0140-x [DOI] [PubMed] [Google Scholar]
- Epel B. L. (2009). Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host β-1,3 glucanases. Semin. Cell Dev. Biol. 20 1074–1081. 10.1016/j.semcdb.2009.05.010 [DOI] [PubMed] [Google Scholar]
- Fernández-Crespo E., Navarro J., Serra-Soriano M., Finiti I., Garciìa-Agustiìn P., Pallás V., et al. (2017). Hexanoic acid treatment prevents systemic MNSV movement in cucumis melo plants by priming callose deposition correlating SA and OPDA accumulation. Front. Plant Sci. 8:1793. 10.3389/fpls.2017.01793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergerich R., Dolja V. (2006). Introduction to plant viruses, the invisible foe. Plant Health Instr. 10.1094/PHI-I-2006-0414-01 [DOI] [Google Scholar]
- Gregor L. B., Corina T., Manfred H., Di Serio F., Meins F., Iglesias V. A. (2001). Local expression of enzymatically active class I β-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28 361–369. 10.1046/j.1365-313X.2001.01181.x [DOI] [PubMed] [Google Scholar]
- Guseman J. M., Lee J. S., Bogenschutz N. L., Peterson K. M., Virata R. E., Xie B., et al. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137 1731–1741. 10.1242/dev.049197 [DOI] [PubMed] [Google Scholar]
- Hajimorad M. R., Eggenberger A. L., Hill J. H. (2006). Strain-specific P3 of Soybean mosaic virus elicits Rsv1-mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1-genotype Soybean. Virology 345 156–166. 10.1016/j.virol.2005.09.055 [DOI] [PubMed] [Google Scholar]
- Hajimorad M. R., Eggenberger A. L., Hill J. H. (2008). Adaptation of Soybean mosaic virus avirulent chimeras containing P3 sequences from virulent strains to Rsv1-genotype soybeans is mediated by mutations in HC-Pro. Mol. Plant Microbe Interact. 21 937–946. 10.1094/mpmi-21-7-0937 [DOI] [PubMed] [Google Scholar]
- Hajimorad M. R., Wen R. H., Eggenberger A. L., Hill J. H., Saghai Maroof M. A. (2011). Experimental adaptation of an RNA virus mimics natural evolution. J. Virol. 85 2557–2564. 10.1128/JVI.01935-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. S., Ju H. J. (2017). The plant cellular systems for plant virus movement. Plant Pathol. 33 213–228. 10.5423/PPJ.RW.09.2016.0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias V. A., Meins M. F., Jr. (2000). Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 21 157–166. 10.1046/j.1365-313x.2000.00658.x [DOI] [PubMed] [Google Scholar]
- Jenner C. E., Wang X., Tomimura K., Ohshima K., Ponz F., Walsh J. A. (2003). The dual role of the Potyvirus P3 protein of Turnip mosaic virus as a symptom and avirulence determinant in Brassicas. Mol. Plant Microbe Interact. 16 777–784. 10.1094/MPMI.2003.16.9.777 [DOI] [PubMed] [Google Scholar]
- Johansen I. E., Lund O. S., Hjulsager C. K., Laursen J. (2001). Recessive resistance in Pisum sativum and Potyvirus pathotype resolved in a gene-forcitron correspondence between host and virus. J. Virol. 75 6609–6614. 10.1128/JVI.75.14.6609-6614.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H. (1996). “Callose synthesis,” in Membranes: Specialized Functions in Plants, ed. Smallwood M. (Oxford: BIOS Press; ), 77–92. [Google Scholar]
- Kim H. J., Kim M. J., Pak J. H., Im H. H., Lee D. H., Kim K. H., et al. (2016). RNAi-mediated Soybean mosaic virus (SMV) resistance of a Korean Soybean cultivar. Plant Biotechnol. Rep. 10 257–267. 10.1007/s11816-016-0402-y [DOI] [Google Scholar]
- Krasavina M. S., Malyshenko S. I., Raldugina G. N., Burmistrova N. A., Nosov A. V. (2002). Can salicylic acid affect the intercellular transport of the Tobacco mosaic virus by changing plasmodesmal permeability? Russ. J. Plant Physiol. 49 71–77. 10.1023/a:1013760227650 [DOI] [Google Scholar]
- Leubner-Metzger G., Meins F. (1999). “Functions and regulation of plant β-1,3-glucanases (PR-2),” in Pathogenesis-Related Proteins in Plants, ed. Datta S. K. (Boca Raton, FL: CRC Press; ), 49–76. [Google Scholar]
- Levy A., Epel B. L. (2009). “Cytology of the (1-3)-beta-glucan (callose) in plasmodesmata and sieve plate pores,” in Chemistry, Biochemistry and Biology of (1-3)-beta-Glucans and Related Polysaccharides, ed. Bacic A. (London: Academic Press; ), 439–463. 10.1016/b978-0-12-373971-1.00013-3 [DOI] [Google Scholar]
- Li W. L., Zhao Y. S., Liu C. J., Yao G. B., Wu S. S., Hou C. Y., et al. (2012). Callose deposition at plasmodesmata is a critical factor in restricting the cell-to-cell movement of Soybean mosaic virus. Plant Cell Rep. 31 905–916. 10.1007/s00299-011-1211-y [DOI] [PubMed] [Google Scholar]
- Lin L., Luo Z., Yan F., Lu Y., Zheng H., Chen J. (2011). Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes. 43 90–92. 10.1007/s11262-011-0596-6 [DOI] [PubMed] [Google Scholar]
- Lu L., Wu G. W., Xu X. M., Luan H. X., Zhi H. J., Cui J., et al. (2015). Soybean actin-depolymerizing factor 2 interacts with Soybean mosaic virus-encoded P3 protein. Virus Genes 50 333–339. 10.1007/s11262-014-1150-0 [DOI] [PubMed] [Google Scholar]
- Luan H. X., Liao W. L., Niu H. P., Cui X. Y., Chen X., Zhi H. J. (2019). Comprehensive analysis of Soybean mosaic virus P3 protein interactors and hypersensitive response-like lesion-inducing protein function. Int. J. Mol. Sci. 20:3388. 10.3390/ijms20143388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H. X., Shine M. B., Cui X. Y., Chen X., Ma N., Kachroo P., et al. (2016). The Potyviral P3 protein targets eukaryotic elongation factor 1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 172 231–234. 10.1104/pp.16.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W. J. (1995). Plasmodesmata-intercellular channels for macromolecular transport in plants. Curr. Opin. Cell Biol. 7 673–680. 10.1016/0955-0674(95)80109-X [DOI] [PubMed] [Google Scholar]
- Lucas W. J., Ding B., Van der Schoot C. (1993). Plasmodesmata and the supracellular nature of plants. New Phytol. 125 435–476. 10.1111/j.1469-8137.1993.tb03897.x [DOI] [PubMed] [Google Scholar]
- Lucas W. J., Lee J. Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5 712–726. 10.1038/nrm1470 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B., Mauch F. (2005). The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 8 409–414. 10.1016/j.pbi.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Oparka K. J. (2004). Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 9 33–41. 10.1016/j.tplants.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Park S. H., Li F. F., Renaud J., Shen W. T., Li Y. Z., Guo L. H., et al. (2017). NbEXPA1, an α-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. Plant J. 92 846–861. 10.1111/tpj.13723 [DOI] [PubMed] [Google Scholar]
- Piršelová B., Matušíková I. (2013). Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol. Plant 35 635–644. 10.1007/s11738-012-1103-y [DOI] [Google Scholar]
- Radford J. E., Vesk M., Overall R. L. (1998). Callose deposition at plasmodesmata. Protoplasma 201 30–37. 10.1007/bf01280708 [DOI] [Google Scholar]
- Seron K., Haenni A. L. (1996). Vascular movement of plant viruses. Mol. Plant Microbe Interact. 9 435–442. 10.1094/MPMI-9-0435 [DOI] [PubMed] [Google Scholar]
- Shukla D. D., Ward C. W., Bunt A. A. (1994). The Potyviridae. Wallilngford: C.A.B. International. [Google Scholar]
- Song P. W., Chen X., Wu B. Y., Gao L., Zhi H. J., Cui X. Y. (2016). Identification for soybean host factors interacting with P3N-PIPO protein of Soybean mosaic virus. Acta Physiol. Plant 38:131 10.1007/s11738-016-2126-6 [DOI] [Google Scholar]
- Sparkes A., Kearns R. J., Hawes A. C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generat ion of stably transformed plants. Nat. Protoc. 1 2019–2025. 10.1038/nprot.2006.286 [DOI] [PubMed] [Google Scholar]
- Stone B. A., Clarke A. C. (1992). Chemistry and Biology of 1,3-β-Glucans. Bundoora, VI: La Trobe University Press. [Google Scholar]
- Suehiro N., Natsuaki T., Watanabe T., Okuda S. (2004). An important determinant of the ability of Turnip mosaic virus to infect Brassica spp. and/or Raphanus sativus is in its P3 protein. J. Gen. Virol. 85 2087–2098. 10.1099/vir.0.79825-0 [DOI] [PubMed] [Google Scholar]
- Tian G., Lu Q., Zhang L., Kohalmi S. E., Cui Y. H. (2011). Detection of protein interactions in plant using a gateway compatible bimolecular fluorescence complementation (BiFC) system. J. Vis. Exp. 55 e3473. 10.3791/3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urcuqui-Inchima S., Haenni A. L., Bernardi F. (2001). Potyvirus proteins: a wealth of functions. Virus Res. 74 157–175. 10.1016/S0168-1702(01)00220-9 [DOI] [PubMed] [Google Scholar]
- Van Bel A. J. E. (2003). The phloem, a miracle of ingenuity. Plant Cell Environ. 26 125–149. 10.1046/j.1365-3040.2003.00963.x [DOI] [Google Scholar]
- Vaten A., Dettmer J., Wu S., Stierhof Y. D., Miyashima S., Yadav S. R., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21 1144–1155. 10.1016/j.devcel.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Vogeli-Lange R., Hansen-Gehri A., Boller T., Meins F., Jr. (1988). Induction of the defense-related glucanohydrolases, β-1,3-glucanase and chitinase, by Tobacco mosaic virus infection of tobacco leaves. Plant Sci. 54 171–176. 10.1016/0168-9452(88)90110-0 [DOI] [Google Scholar]
- Wan L. L., Zha W. J., Cheng X. Y., Liu C., Lv L., Liu C. X., et al. (2011). A rice-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233 309–323. 10.1007/s00425-010-1301-z [DOI] [PubMed] [Google Scholar]
- Wang Y., Khatabi B., Hajimorad M. R. (2014). Amino acid substitution in P3 of Soybean mosaic virus to convert avirulence to virulence on Rsv4-genotype soybean is influenced by the genetic composition of P3. Mol. Plant Pathol. 16 301–307. 10.1111/mpp.12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T. Y., Zhang C. W., Hong J., Xiong R. Y., Kasschau K. D., Zhou X. P., et al. (2010). Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 6:e1000962. 10.1371/journal.ppat.1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen R. H., Hajimorad M. R. (2010). Mutational analysis of the putative pipo of Soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology 400 1–7. 10.1016/j.virol.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Wen R. H., Khatabi B., Ashfield T., Saghai Maroof M. A., Hajimorad M. R. (2012). The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant Microbe Interact. 26 203–215. 10.1094/MPMI-06-12-0156-R [DOI] [PubMed] [Google Scholar]
- Wen R. H., Saghai Maroof M. A., Hajimorad M. R. (2011). Amino acid changes in P3, and not the overlapping pipo-encoded protein, determine virulence of Soybean mosaic virus on functionally immune Rsv1-genotype soybean. Mol. Plant Pathol. 12 799–807. 10.1111/j.1364-3703.2011.00714.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. H., Gai J. Y. (2011). Identification, inheritance and gene mapping of resistance to a virulent Soybean mosaic virus strain SC15 in soybean. Plant Breed. 130 128–132. 10.1111/j.1439-0523.2010.01797.x [DOI] [Google Scholar]
- Zavaliev R., Ueki S., Epel B. L., Citovsky V. (2011). Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248 117–130. 10.1007/s00709-010-0247-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Yeast two-hybrid assay used to determine that SMV P3N-PIPO and GmGLU cannot interact in Saccharomyces cerevisiae NMY51 cells using plasmids pPR3-gateway-GmGLU+pBT3-STE-P3 (1) and pPR3-gateway-GmGOS1+pBT3-STE-P3N-PIPO (2) as positive control, pPR3-gateway+pBT3-STE-P3 (4) and pPR3-gateway+pBT3-STE-P3N-PIPO (5) and pPR3-gateway-GmGLU+pBT3-STE (6) and pPR3-gateway+pBT3-STE (7) as negative control, pPR3-gateway-GmGLU+pBT3-STE-P3N-PIPO (3) for transformation of yeast cells followed on SD/-Ade/-His/-Leu/-Trp medium containing X-α-Gal. In addition, negative controls 4–7 and pPR3-gateway-GmGLU+pBT3-STE-P3N-PIPO could not grow on the selective medium.
No fluorescent spots produced by callose deposition were detected on leaves only inoculated with GmGLU and healthy plant leaves. (A) No callose deposits can be detected on leaves overexpressing GmGLU. (B) No callose deposits can be detected on healthy plant leaves. Bar indicates 30 μm.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at NCBI (AY461847.1).