Abstract
The worldwide outbreak of SARS-CoV-2, severe acute respiratory syndrome coronavirus 2 as a novel human coronavirus, was the worrying news at the beginning of 2020. Since its emergence complicated more than 870,000 individuals and led to more than 43,000 deaths worldwide. Considering to the potential threat of a pandemic and transmission severity of it, there is an urgent need to evaluate and realize this new virus’s structure and behavior and the immunopathology of this disease to find potential therapeutic protocols and to design and develop effective vaccines. This disease is able to agitate the response of the immune system in the infected patients, so ARDS, as a common consequence of immunopathological events for infections with Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV, and SARS-CoV-2, could be the main reason for death. Here, we summarized the immune response and immune evasion characteristics in SARS-CoV, MERS-CoV, and SARS-CoV-2 and therapeutic and prophylactic strategies with a focus on vaccine development and its challenges.
Keywords: SARS-CoV-2, Vaccine, Immune response
1. Introduction
The pandemic occurrences of the SARS-CoV infection were observed in November 2002 in Guangdong, Southern China, that infected over 8422 humans and caused 916 deaths worldwide during 2002–2003 [1], [2]. The MERS-CoV was firstly reported in Saudi Arabia in the year of 2012 and led to 2494 laboratory-confirmed infected cases, 858 mortalities, and 38 deaths since September 2012 [3], [4], [5], [6], [7]. Coronavirus disease 2019 (COVID-19) as a novel coronavirus, SARS-CoV-2 that is first identified in a seafood market in Wuhan, Hubei Province, China, have been recent outbreaks in China and many other countries around the world [2], [8]. Since February 23, 2020, approximately 76,936 cases have been reported in the Chinese Mainland and also 1,875 cases outside of the Mainland [9]. Moreover, over 9,700 cases were confirmed in China, and 106 cases were identified in 19 other countries on 30 January 2020 [9], [10]. According to the last WHO reports, on 4 August 2020, there have been 18,142,718 confirmed cases of COVID-19, and 691,013 deaths, globally. Also, approximately 88 574 cases have been reported in China that accompanied with 4678 deaths [11].
CoVs virulent is mainly related to some defined virulence genes that are able to antagonize the responses of the cellular innate immune system in most cases. During virus infection, an immune response against the virus is triggered by host factors [12]. Nonetheless, it is valuable to consider that immunopathogenesis as a process of disease development is related to severe (out of control) immune response, which can lead to lung damage, malfunction, and probably the reduced capacity. Thus, viral interactions with the innate immune system play a primary role in displaying of infection outcome [13]. In other words, it can be referred that the insufficient or malfunction of the immune system could potentially lead to higher viral replication and consequence tissue damages.
Some structural and non-structural proteins have been involved in human CoV pathogenesis and could be considered for vaccine production from highly attenuated strains of common human CoVs [14], [15], [16], [17], [18], [19]. Effective therapeutic and protective strategies like vaccine designing that can be promptly applied against new emergent strains is a research priority, as these viruses spread rapidly out of heterogeneous and zoonotic [20]. Virus vaccines must have the following characteristics to be utilized in human cases: immunogenicity, safety, broad-spectrum stability, and suitability for long-lasting immunity induction. To achieve these factors, comprehensive knowledge of the humoral and cellular immune responses and their defensive roles, virus molecular structure, genetic configuration, antigens, biology, and pathogenesis could be helpful in vaccine investigations. Knowledge about the details and complexity of different serotypes related to CoV is an important step for designing vaccines or a human neutralizing monoclonal antibody, which is recognized as a promising therapeutic method against the infection linkedCoV or possible prophylactic agents [21], [22], [23].
Along with MERS-CoV, the SARS-CoV spike (S) glycoprotein is known as the main target for defensive immunity in vivo condition; however, SARS-CoV has different immunological effects than other beta coronaviruses with limited antiviral cross-reactivity by antibodies. Indeed, the diversity of important immunogenic factors of the S protein should be addressed for the vaccine development to supply broad protection [24]. Actually, among all viral proteins, S and nucleocapsid (N) attract the most interest among scientists for vaccine production against MERS-CoV. In contrast, other functional proteins like envelope (E) protein and a non-structural protein (NSP)16 are chosen as potential immunogens in vaccine design [25], [26], [27].
Approximately 89% of the SARS-CoV-2 nucleotide sequence is approximately similar to that of SARS-like coronaviruses. Due to this, the early advancement of the potential SARS-CoV-2 vaccine is planned to manufacture based on those advanced earlier for SARS-CoV [28].
Researchers are working seriously to find possible cures for saving human lives and producing vaccines to prevent possible future infections. In this review, we summarized the immunopathology of COVID-19, SARS-CoV, MERS-CoVs, and vaccine development strategies, progresses, and challenges for this emerging virus.
2. Coronaviruses
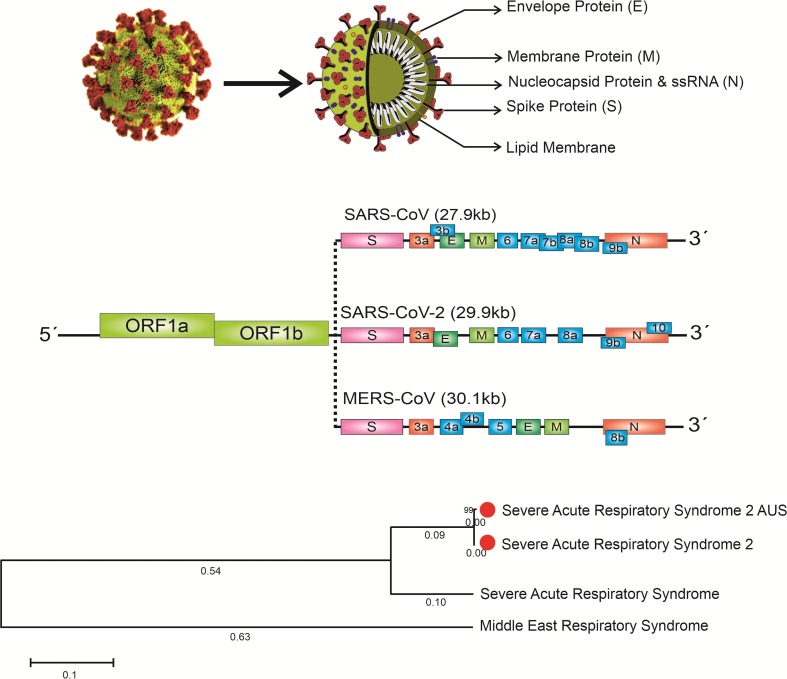
Coronaviruses, as known as zoonotic pathogens, belong to the Coronaviridae family of the order Nidovirales. The Coronavirus genome consists of a single‐stranded positive‐sense RNA (+ssRNA) (~30 kb) that is linked with a 3′poly-A tail (crown-shape peplomers with 80–160 nM in size) and a structure of 5′‐terminal cap [29]. Coronaviruses have the largest genomes (26.4e31.7 kb) among all known RNA viruses. SARS-CoV as a single strand virus covered by a lipid membrane ((M), E, and S glycoproteins) [30] appears to mediate viral entry to the host target cells, and this entrance could be facilitated via ACE2 as the functional receptor [31].
The S protein is an essential stimulator for the induction of neutralizing antibodies. Moreover, the receptor-binding domain (RBD) in the S1 subunit of S protein is composed of multiple conformational structures of neutralizing epitopes that are significant markers for vaccine development (Fig. 1 ).
Fig. 1.
The genomic structure and phylogenetic tree of coronaviruses and coronavirus Spike Glycoprotein.
It is proposed that recombinant proteins that contain RBD directly or vectors encoding the RBD sequence may be used as an ideal safe vaccine for the prevention of SARS-CoV infection. Therefore, the RBD fragment in the middle part of the S1 subunit is considered as a useful vaccine against the SARS-CoV challenge [32].
MERS-CoV is responsible for MERS infection, which belongs to the genus ß-coronavirus, a positive sense, single-stranded RNA ß-coronavirus. Since this virus is basically originated from bats as a zoonotic disease, it has been suggested that bats are the most natural reservoir of MERS-CoV [33]. MERS-CoV genome consists of at least 10 ORFs, encoding for 4 structural proteins like SARS-CoV, including S, E, M, five accessory proteins listed as ORF3, ORF4a, ORF4b, ORF5, and ORF8b, N proteins and 16 NSP (NSP1-NSP16) [34]. SARS-CoV and newly appeared COVID-19 are the members of the same beta coronavirus subgroup, they only have 70% similarity at the genome level, and interestingly the novel group has been found to be genetically different from SARS-CoV [35]. This isolated novel ß-coronavirus, like other typical coronaviruses, has different ORFs (at least ten). ORF1a/b, as the first one, can be recognized about 75% of viral RNA, which is used for the production of two large poly-proteins. The complex of viral replicase transcriptase is formed by pp1a and pp1ab poly-proteins in MERS-CoV and SARS-CoV due to processing into 16nsp1-nsp16 [36].
The nsp proteins mediate the rearrangement of membranes originated from the rough endoplasmic reticulum, resulting in vesicles with double-membrane formation where viral replication and transcription occur [37], [38]. While SARS-CoV-2 binds to the ACE2 receptor, cellular receptor dipeptidyl peptidase 4 (DPP4 also called CD26) is the target of MERS-S via the RBD in the N-terminal surface subunit (S1). The molecular interaction of MERS-CoV with hCD26 has been delineated. The S1 domain is responsible for the detection of hCD26, situated in a C-terminal 240-residue RBD with a core and an external subdomain [39]. It was reported that the binding of the surface S1 unit facilitates the attachment of the virus to the surface of target host cells. Additionally, S protein cleavage at the S1/S2 and the S2 site is linked to priming processes through cellular proteases, which entails virus fusion and cellular membranes, driven via the S2 subunit. Engagement of ACE2 by SARS-S as the entry receptor [40] leads to the cellular serine protease TMPRSS2 employment for priming of S protein [41], [42].
The functional characterization and defined antigenicity of S protein make this protein an important target for vaccine development [43]. The 3D structure of the RBD region in the S protein regulates the formation of the van der Waals forces [24]. It is reported that Lys 31 residue can be critically recognized on the human ACE2 receptor by 394 Glu residue in the RBD domain of SARS-CoV-2 [44]. Briefly, after receptor attachment, the S protein undergoes a conformation change, which facilitates the fusion of viral envelope with the cell membrane via the endosomal pathway, resulting in the RNA release of SARS-CoV-2 into the target cells. Genome RNA is initially translated into the polyproteins of viral replicase (pp1a and 1ab), then can be cleaved into small fragments by viral proteinases. Discontinuous transcription of polymerase activity could lead to the production of different subgenomic mRNAs that can be translated later to the viral proteins. Finally, viral proteins and RNAs assembling into virions in the Golgi and ER organelles stimulates the transportation of vesicles to release [45].
3. Immune response
3.1. Innate immunity
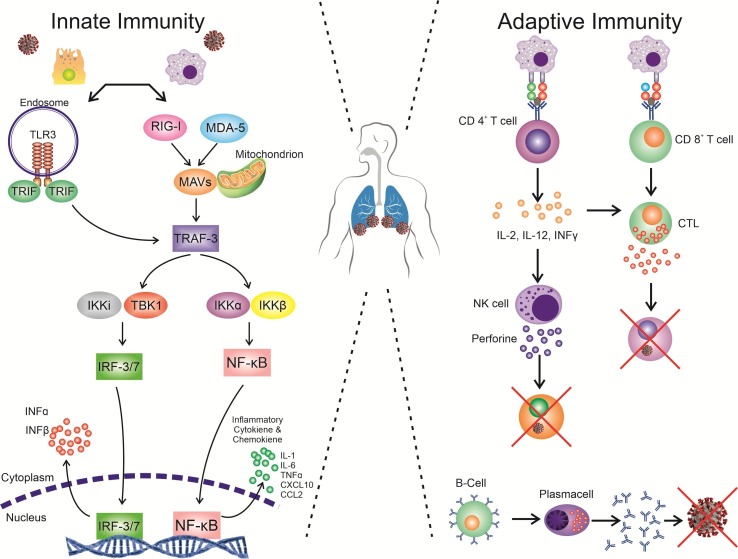
The launch response of the immune system to the invading of a microorganism such as a virus is directly related to the host sensing of the target organism and its linked constituents like uncapped viral RNA or the cellular stress response and consequent biological changes or damages due to infection [46]. This response could be primarily conducted by germline-encoded pattern recognition receptors (PRRs) such as Toll-Like Receptors (TLRs) or Retinoic acid-inducible gene I (RIG-I)-like from the components receptors (RLRs) that enable to detect PAMPs (Pathogen Associated Molecular Patterns) originated of a virus or its replication intermediates, promoting the initial antiviral signaling cascades in response to the infection (Fig. 2 ) [47]. RLRs has known RNA sensors that are localized in the cytosol include three main members: laboratory of genetics and physiology 2 (LGP2), RIG-I, and melanoma differentiation-associated protein 5 (MDA5). RLRs are the essential sensors for virus infections that mediate effectively the transcriptional induction of various genes associated with the antiviral response of the host and type I interferons [48]. More interestingly, these sensors not only able to recognize viral RNAs but also the mislocalized or misprocessed cellular RNAs (unusual forms). Such unusual cellular RNAs processing or localization could potentially indicate the infection or possible occurrence of sterile inflammatory pathologies; therefore, the activation of RLRs is crucial in immunopathology concept [49]. MDA5, and RIG-I, as Interferon Stimulated Genes (ISGs), are mainly transcribed during the infection of the cells by SARS-CoV under in vitro condition. It is reported that Murine coronavirus (MHV) could be detected by MDA5 in microglial cells and brain macrophages, and in oligodendrocyte cells, by MDA5 and RIG-I. Though it has not been identified yet whether SARS-CoV infection can be recognized by MHV or RLRs, SARS-CoV is more likely to have similar intermediates for the replication process, as putative ligands for RLRs sensors; therefore, SARS-CoV is able to be potentially recognized by the same protocoles [47].
Fig. 2.
Innate and adaptive immunity in SARS-COV2 infection. Immune response primarily conducted by PRRs like TLRs or RLRs that enable the detection of PAMPs originated from a virus or its replication intermediates, promoting the initial antiviral signaling cascades in response to the infection, including inflammatory cytokine production. The presentation of antigen can consequently induce humoral and cellular immunity in the human body, which are enhanced by B and T cells (virus-specific cells).
Toll-like receptors (TLRs) as one of the PRRs, could be expressed by several types of immune cells. [50]. TLRs can be expressed either extracellular such as TLR1, TLR2, TLR4, TLR5, and TLR6 on the surface of the cells or intracellular like TLR3, TLR7, TLR8, and TLR9 in the cytosolic compartment, where they detect the genetic material of microorganisms [51].
Viruses are mainly characterized based on the complexity of their genomes and also divided according to their replication mechanism (Baltimore classification). The nucleic acid sequence of viral genomes could have consisted of either RNA or DNA, single-stranded or double-stranded, continuous or segmented, and negative or positive in polarity [52], [53].
Though there is no proof for the direct implication of TLR in SARS-CoV detection, it was reported that TLR4 could potentially detect the viral related glycoproteins of RSV presented mainly on the lung epithelium’s surface cells as an essential cofactor for the entrance of respiratory viruses. It has also been identified that TLR4 plays as a protective factor against the activity of MHV-1 in an animal investigation model for SARS- respiratory disease. Further studies also reported that by the injection of MA15-SARS-CoV to a mouse model and human dendritic cells, the transcriptional level of TLRs increased. Additionally, TLR3 activation plays a protective effect in the infected mouse model by SARS-CoV.
RNA viruses can be internalized into the cells via different mechanisms, including binding to the surface receptors, such as ACE2 for SARS-CoV or CEACAM1 for MHV and plasma fusion. This internalization process could expose the viral genomic (RNA) to the dsRNA sensing system (MDA5, TLR3, and RIGI) in the cells. These proteins could induce IFNb and the production of IFNβ protein through initiating of the IRF-3 cascade signaling. IFNβ protein can bind INFAR1 (IFNα/β receptors) on the surface of the surrounding or same cells, activating the signaling pathway of Stat1 that is associated with the activation of several anti-viral genes with ISRE promoter. This could raise a question that how coronaviruses, either human(OC43, NL63, SARS-CoV, and 229E) or animal (MHV, IBV, and TGEV), interact with the IFN system (type I) [54].
Antiviral cytokine IFN not only has a vital role in controlling many viral infections but also they could promote the process of viral clearance through programing of the adaptive immune response. However, the aberrant response of IFN, cytokine, and ISGs was recognized in those patients suffering from an advanced level of SARS disease compared to the healthy group with innate immune regulated SARS disease [54], [55].
Several reports indicate that IFN (Type I) could inhibit the growth of SARS-CoV in the cell culture and also the viral replication process in different animal models such as a mouse or cynomolgus macaques. Further investigations confirm that the SARS-CoV-infected animal model with a deficiency in IFN receptors such as Type I or Type III show more advanced stages in the replication process of viruses, particularly in the lungs. The expression of IFN receptor (Type III) was identified 24 h before the transcripts of Type I IFN in 2B4 cells, showing a delay in Type I IFN signaling and proving the protective function of Type III IFN in response to SARS-CoV infection [56]. Overall, studies in animal models of SARS-CoV-1 and MERS-CoV infection demonstrated that failure to induce IFN-I response in early stages correlates with the severity of the disease. Of note, these models may be indicated that timing is critical, as IFN-I response is protective in the beginning stage of the disease but is pathologic in the late stage [57].
Although SARS-CoV could effectively infect alveolar epithelial or airway cells, the infection of different hematopoietic cells like dendritic cells (DCs) and monocyte-macrophages is failed. The infection of DCs with SARS-CoV could also induce the lower levels of antiviral cytokines (IFN-αβ) expressions and control the pro-inflammatory cytokines and chemokines up-regulations such as TNF, IL-6, CCL2, CCL3, CCL5, and CXCL10 [58]. In the same way, macrophages infected with SARS-CoV indicate a considerable delay but a significant rise in pro-inflammatory cytokines levels, particularly IFN [59]. Also, airway epithelial cells that are infected by SARS-CoV represent a large production of CCL2, CCL3, CCL5, and CXCL10 [56]. It is thought that delayed but higher amounts of chemokines and cytokines productions could lead to the deregulation or malfunction of the innate immune response to the infection. It was found that the serum levels of chemokines (IL-8, CXCL10, CCL2, and CXCL9) and pro-inflammatory cytokines (IL-12, IFN-γ, IL-6, TGFβ, and IL-1) were increased in the patients with an advanced level of SARS infection compared to the patients with an uncomplicated level of SARS infection [60].
However, infected patients with SARS in advanced levels had a meager amount of anti-inflammatory cytokines such as IL-10. Despite the increased level of chemokines and pro-inflammatory cytokines in SARS-infected patients, the levels of ISGs such as CCL-2 and CXCL10 and IFNs like IFN-γ and IFN-α were enhanced in the infected patients compared to healthy control or the infected patients at a moderate level. These data indicate the possible role of ISGs and IFNs in the immunopathogenesis of infected patients with SARS. Therefore, it seems that dysregulation and exaggeration of chemokine or cytokine in response to SARS-CoV in macrophages, AECs, and DCs could play a considerable function in the pathogenesis of SARS [60].
MERS-CoV, similar to SARS-CoV, could infect airway epithelial cells, inducing the responses of pro-inflammatory cytokines like IL-1β, IL-6, IL-8, and IFNs significantly but delayed. Though MERS-CoV is able to be replicated in either naïve or activated DCs and monocyte-macrophages, activated T cells can only support the MERS-CoV replication [53]. This is a significant contrast that SARS-CoV could abortively infect T cells, monocyte-macrophages. The infection of THP-1 cells, dendritic cells, and DCs by MERS-CoV induce the higher production levels of chemokines and pro-inflammatory cytokines but in a delayed manner. However, IFN-α/β induction through DCs and monocyte-macrophages was not a significant except for the pDCs, which produce a remarkable amount of IFNs during the infection of MERS-CoV. It was documented that the serum levels of chemokines such as CCL5, IL-8, and CXCL-10 were increased considerably in individuals infected with an advanced level of MERS in comparison with those who are suffering from moderate disease.
The higher amount of these factors in the serum of MERS-infected patients is correlated with increased numbers of monocyte and neutrophil in the peripheral blood cells and lungs, showing the possible function of these cells in the pathology of the lungs [60].
The kappa light-chain-enhancer of activated B cells (NF-κB) as a well-known transcriptional nuclear factor is the most critical regulator for the induction of different pro-inflammatory cytokines such as IL-6 and IL-8, the early IFN-β expression upon viral infection and innate immunity system. More interestingly, it has been proved that the induction of IL-8, TNF-α, and IL-6 could have occurred in the cells with spike protein overexpression (a protein of SARS-CoV) through the NF-κB pathway [61], [62].
Neutrophils, as well-known immune cells, are presented in several types of lung diseases related to ARDS (acute respiratory distress syndrome) and probably could contribute to the acute injury of lungs. Neutrophils have been poorly investigated in aspects of their response to viral infection, particularly viral disease in the respiratory system. To be able to reach the target site (the potential pathogenic site), neutrophils breakdown the collagen in pulmonary tissue (extracellular matrix (ECM)) by the production of MMP9 proteins. The production of TNF in the surrounding cells could increase the release of MMP9, leading to the absorption of more numbers of neutrophils towards the inflammation or infection. Subsequently, neutrophils start phagocytosis and degranulation of neutrophil myeloperoxidase, elastase, neutrophil extracellular traps [63], and reactive oxygen species (ROS), which contribute to the pathogenic microorganism clearance.
Neutrophils as a first line of immune defense in the beginning of a viral infection, increase in the number of these kinds of immune cells as well as their raised lifespans, composed the important phenomenon in the patients with COVID-19. Despite their protective effect against viral infections, neutrophils could injure in host tissues infected with a virus. Activation and degranulation of these cells in the microenvironment of infected cells can damage the host tissue and worsen the disease outcome. The neutrophils, via the induction of pro-inflammatory cytokines, as well as ROS production, could worsen the disease manifestation [64]. It is suggested that severe host response in the infected patients with COVID-19 is due to improper activation of leukocytes such as neutrophils in peripheral blood. These results indicate that when the normal signals to moderate inflammation process are lost, specifically under cytokine storm conditions or a signaling interruption between neutrophils and macrophages, can cause progressive and consequently uncontrollable inflammation [65].
Netosis is known as specific cell death of neutrophils whereby NETS (decondensed chromatin fibers, granule components, and histones) are deployed in order to kill and immobilize pathogens while accelerating its death process. Though its main aim is to restrict the pathogenic spread, some viruses or bacteria could lead to excessive formation of NETs [66]. In other words, severe production of NET can induce a cascade reaction in an inflammatory procedure that is related to the promotion of metastasis in cancer disease, micro thrombosis facilitation, and damaging of surrounding tissues, resulting in permanent destroy of organs such as renal, pulmonary, or probably cardiovascular systems [65].
The lung infection by HPAI, MERS-CoV, influenza A virus, and SARS-CoV is reported to cause a severe acute lung injury such as ARDS that is characterized by considerable deterioration to the alveolar epithelium and neutrophils infiltration [67]. A study conducted by Chen and et al. showed that 38% of the patients infected by COVID-19 had higher levels of neutrophils [68]. The critical question is that neutrophilia is driven whether by virus-induced cytopathy or viral infection response.
ARDS, as a common consequence of immunopathological events for infections with MERS-CoV, SARS-CoV, and SARS-CoV-2, could be the main reason for death. In a study was represented that 6 out of 41 cases that infected by SARS-CoV-2 died because of ARDS [69]. It has been reported that one of the critical detrimental mechanisms of ARDS is the uncontrollable inflammatory response that leads to the release of a huge concentration of chemokines and pro-inflammatory cytokines by the immune system in SARS-CoV infection. Similar to individuals infected by SARS-CoV, patients with an advanced level of MERS-CoV infection reveal increased amounts of pro-inflammatory cytokines and chemokines in serum samples compared to the patient with a moderate level of infection. The cytokine storm can induce a detrimental attack on the immune system, leading to ARDS or organ failures that can cause death or severe infection condition in the patients [70].
Histological tests of lungs from individuals infected by SARS indicated a remarkable cellular infiltrate in the alveoli and interstitium, which includes macrophages and neutrophils as predominant cells. These data are associated with the increment of monocyte and neutrophil numbers and decrement of CD8+ and CD4+ T cells in the patients infected with fatal SARS.
In addition to the immunohistochemical test indicated that MERS-CoV could mainly infect alveolar and airways epithelial cells, macrophages, and endothelial cells. The severity of lung damage is related to uncontrolled levels of macrophages and neutrophils infiltration into the lungs and higher counts of these cells in the sample of peripheral blood in the MERS-infected patients [60].
3.2. Adaptive immunity
Antigenic peptides are predominately expressed by the MHC (major histocompatibility complex) in humans and can be detected by CTLs (virus-specific cytotoxic T lymphocytes). Therefore, recognition of the fundamental mechanism of the SARS-CoV-2 antigen presentation could significantly contribute to comprehend the COVID-19 pathogenesis. Because there have not been any reports regarding this issue yet, some information, according to the previous studies on MERS-CoV and SARS-CoV were described in this part. The SARS-CoV antigen presentation is mainly dependent on MHC I and partially MHC II. The presentation of antigen can consequently induce humoral and cellular immunity in the human body, which are enhanced by B and T cells (virus-specific cells) (Fig. 2) [70], [71].
3.2.1. Humoral immunity
Similar to other viral infections, the antibody profile has a common production pattern of IgG and IgM antibodies in the response of the SARS-CoV infection. The IgM antibodies specified for SARS can be detected up to 12 weeks, though the IgG antibody could be identified after a long time-frame, indicating that IgG antibody play probably the protective function and IgG antibody is mainly either S-specific and N specific [71].
The infection of the SARS-CoV induced the process of seroconversion at the beginning of 4 days after the onset of the disease and was recognized in most patients for 14 days. It was reported that long-lasting neutralizing and some specific IgG antibodies were even detectable after 2 years since the infection time. The seroconversion process is observed for the infection of MERS-CoV at the end of the second or third weeks of the onset. It has been studied that the response of weak and delayed antibody is related to the severe disease condition in both kinds of coronavirus infection. Moreover, all serum samples from infected patients could neutralize the detrimental function of the SARS-CoV-2 in vitro condition, showing the possible successful increment of humoral responses. However, the humoral responses of some specific antibodies are whether associated with the severity of the disease is not clear yet [72].
Although the response of successful neutralizing is indicated in most cases, higher titers are also linked to more intense clinical symptoms, proposing that the response of a robust antibody could not be sufficient alone to prevent severe diseases. In the previous epidemic of SARS-CoV-1, it was indicated that neutralizing titers were considerably lower in the recovered patients than decreased patients. This has brought a challenge that the responses off antibodies to such viruses could potentially lead to pulmonary pathology though the induction of antibody-dependent enhancement (ADE) as an interesting phenomenon that concerns many researchers in this field.
This phenomenon can be seen when virus-specific IgG antibodies (non-neutralizing) contribute to the entrance of virus particles to several cells such as monocytes or macrophages via expressed Fc-receptor (FcR), inducing the activation of inflammatory procedure in these cells. Research on the infected rhesus macaques with SARS-CoV-1 showed that anti-S IgG is contributed in increasing the number of macrophages and monocytes in the lung and severe acute lung injury (ALI) [57]. Jaume et al. and Yip et al. also reported that though anti-S antibodies inhibit the entrance of viruses into permissive cells, they increase the infection by attaching to the IgG Fc receptor-II positive (FcγRII + ) cells, such as macrophages or B cells [73]. Furthermore, ADE is recognized in the isolation of monoclonal antibodies from a patient infected with MERS-CoV. ADE of disease is also a common concern for the development of vaccines and antibody therapies because the mechanisms that motivate antibody protection against any virus have a theoretical potential to amplify the infection or elicit harmful immunopathology. This phenomenon occurs in MERS, SARS-CoV-1, Zika, HIV, and dengue virus infection and vaccination [74]. Vaccine-specific variations in ADE could occur for many reasons, including modifications in vaccine protein glycosylation, vaccine adjuvant, and previous exposure to other CoV strains. Multiplex approaches developed for influenza can be rapidly adapted to this use, particularly to evaluate the balance between vaccine-induced protection from infection against the increased risk of severe disease with the following infection despite vaccination [75]. However, there is not available data that supports ADE contributed by sera in animal models such as vaccinated rats with SARS-CoV-2 RBD and immunized macaques with the SARS-CoV-2 vaccine [76], [77].
SARS-CoV-2, as a known mucosal targeted virus, is expected to produce secretory IgA (sIgA) and also promote the mucosal immunity strongly. It is valuable to refer that the function of sIgA in the infection of COVID-19 has studied limitedly, though SARS-CoV-2 could enter into the body via respiratory mucosa, and sIgA plays a crucial role in mucosal defenses. Moreover, different reports regarding COVID-19 infection have indicated that the presence of IgA antibody in infected patients with SARS-CoV-2 and vaccinated with anti-SARS (administered either sub-lingually or intranasally. These reports show the importance of studying the role of sIgA secretions in infected patients with COVID-19 and recognition of its antiviral function in respiratory tract mucosa, inflammation, and immune response [78].
Regarding the function of systemic and mucosal IgA in the infection of COVID-19, IgA induction by using lactoferrin to promote the signaling pathway of canonical TGF-β, or retinoic acid to increase the responses of lactoferrin-induced IgA could be a potential and novel method for COVID-19 therapy. Yu et al. reported, however, the enhanced response of IgA was detected in severe COVID-19, which may show its damaging effects. It is proposed that COVID-19 is possible a part of an IgA-mediated disease (associated with IgA vasculitis and deposition), which explains the related organ injuries in COVID-19 infection such as kidney injury or acute pulmonary embolism [79].
3.2.2. Cellular immunity
The migrated cells (DCs) from the lungs to the T-cell area can activate naïve CD8+ T cells over the viral infection, resulting in the differentiation and proliferation of them into CTLs [80]. CD8+ T and CD4+ T lymphocytes (mainly CD8+ T lymphocytes) employ different cell-associated mediators such as FasL, perforin, or granzyme to promote the apoptosis process in target cells. Because the induction of cytolysis needs to the engagement of antigen receptors in T lymphocytes by viral complexes of peptide/MHC molecules, the apoptosis mediated by T –cells mainly restricts the infected cells. As a significant exception, the mediated cytolysis by T lymphocytes plays a negligible function in the development of injured tissue generated by the adaptive immune system upon viral infection. However, the inflammatory mediators (soluble) derived from T cells such as MIP-1a TNF and IFNγ are able to deteriorate even uninfected cells and also increase the infiltration of injury-promoting innate immune cells [71].
CD4+ T cells are able to contribute to the production of antibodies, induce the activity of cytotoxic T lymphocyte (CTL) in CD8+ T cells, and act as functional memory cells. T helper cells have essential roles in the regulation of B cell proliferation, differentiation and switching of immunoglobulin class. In particularly Tfh cells also contribute to the somatic mutation of B cells in the germinal centers. [81]. ICOS and CD40 ligand (CD40L) are abundantly expressed in Tfh cells, which its ligation with ICOSL and CD40 is fundamental for the B cell's immune responses [82].
Cytotoxic CD4+ T cells (ThCTL) are mainly determined by their cytotoxic function and/or phenotype. ThCTL can also be detected in the peripheral blood mononuclear cells (PBMCs) (human species) in severe viral infections such as HCMV (human cytomegalovirus), HIV-1 (human immunodeficiency virus 1), and hepatitis viruses. A study that was conducted by Brown et al. indicated that, in vitro generated CD4+ cells could get cytolytic characteristic and have protective effect on influenza virus infection in combination with B cell help. In another study they indicated that, functional ThCTL could be found as a large effector population in lung (besides PBMC) as well following influenza infection in mouse model and play the cytotoxic role in the infection loci. It was reported that, effector profiles of CD4+ cells is distinct in the lung than draining lymph node (DLN) and that IFN-γ production via CD4+ cells at the site of infection could play a protective role. Moreover, CD4+ cells using perforin-mediated cytotoxicity that get in the lung, but not the DLN, are able to augment recovery from lethal influenza virus infection [83], [84], [85]. More studies are in need to evaluate these cells in COVID-19.
The most current reports reveal that the numbers of CD8+ T and CD4+ cells are considerably reduced in the peripheral blood of the patients infected with SARS-CoV-2; however, their over-activation is a proof for high positive fractions of HLA-DR (as described CD4+ 3.47% and CD38+ (CD8 39.4%). Likewise, the response of SARS-CoV-infected patients at the acute phase is related to the severe reduction of CD4+ T and CD8+ T cells. In 12 patients recovering from mild COVID-19, robust T cell responses specific for viral N, M, and S proteins were detected by IFN-γ ELISPOT. In the absence of antigen, CD8+ T and CD4+ T memory cells can even persist for several years in recovered patients with the SARS-CoV infection and capable of continuing the proliferation of T cells, IFN-g production, and DTH response.
A study reported that the infection of SARS-CoV peptide is able to be recognized in response to specific memory T cells in 14 out of 23 recovered patients with SARS infection. It has also been reported that specific CD8+ T cells can indicate a similar influence on MERS-CoV removal in mice. These investigations could probably provide useful data about rational vaccines designing for SARS-CoV-2 infection [15]. The rise of CD8+ T cells in MERS-CoV-infected patients at the early phase could be associated with the severity of the disease, and T helper cells (Th1 type) are recognized at the convalescent stage. The cells of airway memory CD4+ T are specified for conserved epitope play a protective role against lethal function in animal models and can also observe cross-reaction between the MERS-CoV and the SARS-CoV.
The strong response of T cells associated remarkably with higher amounts of neutralizing antibody; however, more Th2 cytokines in serum such as IL-4, IL-5, and IL-10 were identified in the fatal individuals. As neutrophils have a detrimental effect on all infections, the protective or possible destructive roles of Th17 against the infection of human coronavirus remains unclear [72].
4. Immune evasion by SARS-CoV and MERS-CoV
MERS-CoV and SARS-CoV can utilize different strategies to escape from the immune system to stay alive in host cells. Over the replication cycle of SARS-CoV, the dsRNA intermediates segregation of viruses in DMVs (Double Membrane Vesicles) is able to cover viral PAMPS from PRRs recognition in the cytosol. It has not been clarified whether the products of viral dsRNA or ssRNA degradation could be sequestered in DMVs or detected by PRRs. Due to the lack of a cap (50 nucleotides) in viral mRNAs, the eukaryotic mRNAs can be distinguished, and many viruses such as SARS-CoV have a sophisticated mechanism for mimicking the host capping system [47]. Although IFN-I has a significant protective effect on the infection of the SARS-CoV and MERS-CoV, the IFN-I pathway is controlled in the infected animal model. It is indicated that accessory protein 4a belongs to MERS-CoV is able to inhibit IFN induction at the MDA5 activation level via interaction directly with ds RNA. Moreover, ORF5, ORF4b, ORF4a, and membrane proteins of this virus could block IRF3 (nuclear transport of IFN regulatory factor 3) and subsequent IFN β promoter activation. Coronavirus can also possibly affect the presentation of antigen. As an example, it was shown that the expression of genes is linked to the presentation of antigens is remarkably down-regulated after MERS-CoV infection [70].
Reportedly, SARS-CoV, through encoding nsp3-deubiquitinase [18], nsp3-macrodomain, nsp1, ORF6, ORF9b, and ORF3b, could subvert the antiviral reaction by antagonizing of ISG and IFN responses. While nsp1 blocks the IFN responses through inhibiting STAT1 phosphorylation, nsp3 could impair the responses of IFN by undetected mechanisms. In addition, some structural proteins, such as nucleocapsid and membrane proteins, could dampen the signaling process of IFN [60]. While M protein is able to suppress the production of type I IFN by controlling the formation of the TRAF3-containing complex, the suppression mechanism of N protein is unknown [86]. Either nsp7 or nsp15 originated from SARS-CoV were considered as IFN antagonists; however, the functional mechanism has not been recognized yet. Of note, nsp15, as a known inhibitor for MAVS-induced apoptosis, could also act via an IFN-independent mechanism [87]. Furthermore, acute SARS-CoV papain-like proteases (PLP) and human coronavirus (HCoV) NL63 could antagonize the signaling of the innate immune system contributed by stimulator of interferon genes (STING) such as MITA, ERIS, or MYPS. The membrane-anchored PLP domain expression in SARS-CoV (PLpro-TM) or human HCoV-NL63 (PLP2-TM) blocks the activation of IRF-3 dependent promoters and STING-mediated of IRF-3 nuclear translocation [88].
The presence of immunosenescence in older individuals impacts slightly on the innate immunity system, but significantly on the responses of T cell-dependent adaptive immunity. More evidence shows that the level of proinflammatory cytokines is considerably increased in elderly mice, and alveolar macrophages are sensitive to IFN-γ activation. [78]. In other words, immunosenescence and probably comorbid disorders are able to induce viral cytokine storm in older individuals, leading to the failure of life-threatening respiratory and multisystemic involvement. Therefore individual medicine can be developed in older people according to their personal medical history [89].
5. Vaccine development and its challenges
5.1. Inactivated whole virus
Inactivated Whole Virus or Whole killed Virus vaccine (WKV) is remarkable because of its safety, cost-effectiveness, easily prepared, and not involving laborious genetic manipulation [90]. Several inactivated vaccines have been produced for both SARS-CoV and MERS-CoV virions and tested in several animal models. U.V and formaldehyde were utilized to completely inactivate the whole virus as a novel vaccine that could significantly increase antibodies concentrations against the viral proteins of SARS-CoV [91]. Furthermore, cellular immunity activated has been shown by stimulation of IFNγ and interleukin-4 production, and also inhibition of SARS-CoV replication in the respiratory tract of mice vaccine by SARS-CoV (Utah) plus Al(OH)3 as an adjuvant [92].
Qu et al. have shown that formaldehyde-inactivated-SARS-CoV strain GZ50 prompts neutralizing antibodies up to 1:640 when administered intranasally. Specific IgA was detected in the tracheal lung fluid of immunized mice when used either alone or with cholera toxin B (CTB) or polyethylene glycol (PEG) as an adjuvant [93]. In another study, formaldehyde-inactivated-SARS-CoV strain NS-1 represented an effective response, both humoral and mucosal immunity against SARS-CoV infection in monkeys [94]. The improved humoral immune response due to the application of the inactivated virus with adjuvants, either MF59 or alum associated with the stimulation of the CD4+ T cells, has been observed even after gene-based vaccination [95]. In other two studies conducted by Roberts and See, a whole-killed (deactivated by β-propiolactone) SARS-CoV vaccine or β-propiolactone (BPL) inactivated, and a mixture of two adenovirus-based vectors, one expressing the N and other expressing the S protein (named Ad S/N vaccine), were examined in BALB/c mice, golden Syrian hamsters, and 129S6/SvEv mice, respectively. The results of the first study showed the efficiency of the WKV associated with the higher neutralizing antibody titers compared to that of the Ad S/N vaccine. Nonetheless, intranasally administration of S/N limited noticeably the SARS-CoV replication in the lungs [96], [97]. Similar results were reported for the neutralizing antibodies production with various protection levels using inactivated SARS-CoV virion vaccines with or without adjuvant in different animal models [63], [96], [98], [99], [100], [101], [102], [103], [104], [105]. Moreover, SARS-CoV vaccine studies in mice lead to the observation of Th2-type immune responses, and also eosinophilia. It was indicated that oligomers of the SARS-CoV S protein act as an immunogen and cause eosinophilia in animals [98], [99]. These observed side effects are significant and should be accurately evaluated before using this type of vaccine in humans. Also, if genetically attenuated viruses were utilized in the starting point of virus inactivation process, the vaccine-induced eosinophilia would be probably abated.
The application results of the inactivated SARS-CoV vaccine to 36 human cases revealed its high level of safety, well tolerating, and the ability to induce neutralizing antibodies. Nonetheless, the responses of SARS-CoV-specific IFN-γ-secreting T-cells have been identified in the mice vaccinated by a mixture of WKV and Ad S/N vaccine. Therefore, it has appeared that WKV vaccines are reliably safe and able to induce either SARS-CoV neutralizing antibodies or T lymphocytes, though the mechanism of action is still unknown [106].
Chemical-based inactivated MERS-CoV vaccine where the viruses are deactivated by formaldehyde, could trigger neutralizing antibodies in mice without the activation of any T-cell responses, as reported by Deng et al. [107]. However, introducing of adjuvant (alum and CpG ODN) to these vaccines could improve the protective immunity in the mice received human dipeptidyl peptidase4, hDPP4 [107]. Wirblich et al. designed an inactivated whole-vaccines by mixture vectors encode a fusion protein such as the S1 domain of MERS-CoV linked to the C-terminus of G protein (RABV, rabies virus).
In MERS-CoV combined RABV, the S1 domain was combined with the RABV particles (BNSP333-S1). The responses of neutralizing antibodies were recognized after applying of the vaccines in mice with considerable safety [108]. Despite the advantages of IWV-based vaccine responses, such as neutralizing antibodies induction and reduction of the viral load in hDPP4 transgenic mice, unwanted responses like hypersensitivity lung immunopathologic reactions were observed [109], [110]. Besides the investigation of inactivated vaccines in animals, phase I of human trials was performed with or without adjuvants. The vaccine caused strong responses against the RBD, and the RBD-specific antibodies in the antisera could efficiently block receptor binding and virus entry when they combined with adjuvants like Freund’s complete or incomplete. In spite of their high level of efficacy, they led to eosinophilia and other related symptoms of immunopathological disorders [101]. Therefore, it is necessary to develop novel vaccines with a high level of both antibodies and T-cells response; however, limited eosinophilic reactions. Although to the complex compositions of whole-cell antigens, the evaluation and quality control of WKV is complicated, several institutions started WKV development by different strains with confident low or no pathogenicity (Table 1 ) [111].
Table 1.
Potential preclinical vaccine candidates against SARS-CoV, MERS-CoV, and SARS-CoV2.
| Vaccine type | Antigen target | Administration route | Reference |
|---|---|---|---|
| Potential vaccine candidates against SARS-CoV | |||
| Inactivated Whole Virus | SARS-CoV (UV-V)/ TLR | SC, IP, IN | [63] |
| SARS-CoV- BPL | IM/IN | [91] | |
| SARS-CoV (Utah)/S protein + Al(OH)3 | IN | [92] | |
| SARS-CoV | IM | [94] | |
| SARS-CoV | IN or IM | [97] | |
| SARS-CoV | IN | [98] | |
| SARS-CoV | IM/IN | [99] | |
| SARS-CoV + Alum | IN | [104] | |
| SARS-CoV (Advax delta inulin adjuvant) | IM | [105] | |
| SARS-CoV- BPL-MF59 | IN | [162] | |
| SARS-CoV | IP | [208] | |
| SARS- CoV /NF-κB inhibitors | IP | [134] | |
| SARS-CoV (MA15) | IN | [209] | |
| Live -attenuated vaccine | SARS-CoV ΔNSP16 | IN | [19] |
| SARS-CoV (MAwt)/ MA-ExoN | IN | [128] | |
| SARS-CoV-ΔE | IN | [132] | |
| TCID-SARS-CoV | IN, IP | [133] | |
| SARS- CoV /NF-κB inhibitors | IN | [134] | |
| DNA vaccine | N protein | IP | [193] |
| S, M, N, or E protein | IM | [192] | |
| S protein | IM | [191] | |
| S protein | IM | [191] | |
| Recombinant vaccine | RBD protein | – | [178] |
| Subunit vaccines | RBD-Fc protein | ID, IM | [101] |
| S318-510 protein | SC | [154] | |
| S (14-762) protein | SC | [155] | |
| S ectodomain | IN | [157] | |
| trimeric Spike protein | IP, SC | [158] | |
| RBD-CHO protein | SC | [159] | |
| S1-fold on and S2 domain protein | SC, IM | [160] | |
| M protein | ID | [166] | |
| N protein | SC | [167] | |
| N protein | IP | [168] | |
| S2 protein | SC | [210] | |
| SARS-CoVΔNSP16/ExoN | IN | [211] | |
| trimeric Spike protein | IP, SC | [212] | |
| Potential vaccine candidates against MERS-CoV | |||
| Inactivated whole virus | S protein+ (alum + CpG) | IM | [107] |
| Chimeric RABV/S1 protein | IM | [108] | |
| MERS-CoV | IM | [109] | |
| MERS-CoV | IN | [213] | |
| Live -attenuated vaccine | MERS-CoV mutant | ND | [125] |
| rMERS-CoV-RFP, or rSARS-CoV/cDNA | In vitro | [126] | |
| Truncated, soluble variant of S protein | IP | [214] | |
| MERSS-CoVΔNSP16 (mutant) | IN | [215] | |
| N protein | IP | [216] | |
| DNA vaccine | DNA encoding S protein | IM followed by EP | [194] |
| DNA encoding S1 protein | IN | [196] | |
| Full-length (pS) or S1-subunit (pS1) | IM | [189] | |
| Recombinant vaccine | RBD protein | IM or SC | [143] |
| S protein | IM and IN | [180] | |
| RBD-Fc protein | IM | [177] | |
| RBD | IM | [39] | |
| Recombinant adenovirus-based vaccine | S protein | IN | [182] |
| Recombinant vaccine | N-terminal domains (NTD) of S protein | IM | [140] |
| RBD protein | IN | [144] | |
| RBD protein | IM | [191] | |
| Combination vaccines(protein and DNA) | S glycoprotein and subunits | IM | [139] |
| Subunit vaccines | EMC/S protein | IM | [107] |
| RBD protein | SC | [136] | |
| trimer S protein | IM | [137] | |
| S1 protein | SC | [141] | |
| S1 protein | IM | [142] | |
| S1 protein (S1-Fc variants) | ND | [146] | |
| RBD-Fc | SC | [148] | |
| RBD Trimer | IM | [217] | |
| RBD-Fc | SC | [195] | |
| RBD- Fc, EMC/2012 + Addavax | IM | [218] | |
| IM (Intramuscular), IN (Intranasal), IP (Intraperitoneal), SC (Subcutaneous), EP (Electroporation). | |||
Although the Th2 immune responses, which induce production of IL-4, may be sufficient for protection against viral infections, defense requires Th1 responses, including cytotoxic T cells and Th1 cytokine production such as IFN-γ. In order to this purpose achieve, using some adjuvants such as Polyinosinic-polycytidylic acid (poly-IC), which is a synthetic dsRNA, has been suggested. The adjuvant provides activating the immune responses of host defense, both innate and adaptive immunity, especially when combined with viral antigens. In a study, it also showed, poly- IC and Polyriboinosinic-polyribocytidylic acid (poly- ICLC) could enhance the production of IFN-α, -β, and IFN -γ, which lead to inhibition of CoV replication [112], [113], [114].
TLRs, especially TLR3, are the dominant elements as immunomodulators that have been used in vaccine designing, enable to response against different viruses. In fact, TLR signaling pathways is a key for differentiating of CD4+ T cells into Th1 cells, which induce IFN-γ production, and lead to a change of Ab class from IgM to IgG2 [115].
On the other hand, the combination of TLR3 with poly-IC, which has a wide range of target cells, produce good immune responses against viral infection [116]. In a study by Barnard et al., the combination of IFN-α and Ampligen® (poly I: poly C12U) were evaluated against SARS-CoV, strain Urbani in BALB/c mice. The results demonstrated that the compounds not only induce IFN production but also protect against death in lethal models of disease as well as suppress the virus replication in the lung tissues and improve the pathogenesis effects of the virus [117], [118], [119].
In another study which conducted by Zhao et al., intranasal pre-treatment of Poly I-C (a TLR3 agonist) in C57BL/6 mice infected with SARS-CoV (MA15) leads to expression of IFN-β, IFN-γ, IL-1, TNF genes, virus loads reduction, and pathological effects in mice lungs [116].
Although the high stability of WKV has been proved during several studies, its efficacy is low and needs reminders of the immune system by repeated immunization [120]. In a study performed by Gao et al., PiCoVacc, as an inactivated SARS-CoV-2 vaccine, evaluated in mice, rats, and macaques at different doses of vaccine during three times immunization. The study results showed partial or complete protection in macaques without any notable pathology and progress of infection [121].
Several randomized, double-blinded clinical trial studies conducted by investigaters against COVID-19 show low or mild efficacy with adverse reactions after each dose vaccination. Additional results about safety and also immunogenicity of the WKV-COVID-19 vaccine will be provided via the following phase3 clinical trial [122].
5.2. Live attenuated vaccines
Though live attenuated vaccines application has been limited due to the risk of reversion to a virulent type, they are mainly known as strong immunogenic, so a single administration in the absence of any adjuvant can provoke sufficient and effective protective immunity. Furthermore, they can stimulate strong cellular and antibody responses and often induce immunity persisting for several decades, by only one dose administration [123]. Live-attenuated vaccines either viruses with reduced fidelity capacity (mutated in nsp14), partial viruses (viruses eliminated in the envelope section (E) of protein), or the attenuated vector of viruses (AAV adenovirus, measles virus, parainfluenza, VSV, poxvirus, and rabies virus) are nontoxic and can trigger antibody and T-cell responses against SARS and MERS coronaviruses [124].
Based on the investigation, a MERS-CoV live attenuated vaccine in response to was designed by eliminating of the E gene from the MERS-CoV genome as a recombinant form (rMERS-CoV) by Almazan et al. This engineered virus could not cause infection and replicate in one cycle. Biosafety problems related to the risk of virulence reversion could occur by vaccines based on the live attenuated viruses, while rMERS-CoV lacking E gene is propagation defective and prevents directed reverse to virulence, allowing safer alternative approaches [125].
Awareness of MERS-CoV molecular clone and recombinant viruses presenting special genes such as red fluorescent protein (RFP), provides procedures for developing antiviral agents and designing of LAV. In a study conducted by Scobey et al., the transfected cDNA (full-length) recovered some recombinant viruses (rMERS-CoV) for inserting into the component clones. Furthermore, the accessory protein ORF5 was deleted and replaced with tomato red fluorescent protein (rMERS-RFP). These recombinants (rMERS-CoV-without ORF5, MERS-CoV-RFP, and rMERS-CoV) were replicated in the high titers, while MERS lacking ORF3–5 indicated decreased titer compared to rMERS-CoV [126]. A live-attenuated measles virus (MV) vaccine that encodes the MERS-CoV spike glycoprotein (MERS-S) and nucleocapsid protein (MERS-N) was characterized in a study by Bodmer et al. The results showed that replication-competent MV-MERS-S(H) vaccine could trigger strong neutralizing antibody titers and cellular immunity (IFN-γ and TNF-α) in adult mice by a fraction of MERS CoV-specific CD8+ T cells and MV-specific CD4+ T cells. Furthermore, the recombinant vaccine of MV expressing MERS-CoV-N induces N-specific T cell responses in vaccinated animals.
In general, live-attenuated vaccines can be recognized by the innate immune system like dendritic cells expressing pattern-recognition receptors, including the TLRs. However, many viruses can induce different signaling pathways of receptors and activation of immune cells [127].
Replication fidelity as a dominant virulence factor in coronaviruses is about 20-fold greater than other RNA viruses, which is associated with a 3′→5′ exonuclease (ExoN) activity. Considering this issue, a live- attenuated vaccine against SARS-CoV, according to mouse-adapted SARS-CoV (MAwt) was investigated and found that MA-ExoN as a stable mutator phenotype could profoundly reduce fidelity after regular administration and attenuate pathogenesis in young, old and immunocompromised BALB/c mice. The obtained results indicated that high amounts of neutralizing antibodies are generated by the vaccinated mice with MA-ExoN. Also, stable weakening of CoVs and, possibly, other RNA viruses can be performed by ExoN inactivation [128], [129].
Another effective attenuated vaccine based on the deletion of the full-length E gene of SARS-CoV (SARS-CoV-ΔE) or the carboxy-terminal region of nsp1 protein was developed in the cell culture or in vivo in BALB/c mice. The attenuation of these recombinant viruses by attenuating of E and nsp1 genes mutations showed protective effects via induction of both T cells and antibody responses in the vaccinated animal model against the lethal parental virus [130].
Ribose 2′-O-methylation of viral as a molecular pattern of mRNA, which is associated with the viral nonstructural protein nsp16, disrupts the induction of type I interferon production. On the other hand, type I interferon induction dependent on the cytoplasmic RNA sensor Mda5 (melanoma differentiation-associated protein 5) in viruses with deficient in 2′-O-methyltransferase. Therefore, the study by Züst et al. showed that mutants lacking 2′-O-methyltransferase activity-induced large production of type I interferon. The attenuation mixture of wild-type and mutant NSP16 (CoV 2 = O MTase mutation) was examined in C57BL/6 mice, which indicated good efficiency in old mice and present rationale reasons for producing live attenuated coronavirus vaccines [131].
The efficiency of another live-attenuated SARS-CoV vaccine was evaluated in the golden Syrian hamster using a recombinant SARS-CoV without the E gene (rSARS-CoV-E). The high titers of serum-neutralizing antibodies were discovered. This antibody prevents the replication process of heterologous SARS-CoV and homologous (SARS-CoV Urbani) in the respiratory tract of animal models [132]. Also, a high neutralizing antibody titer was obtained by rVSV-S vaccination in a study done by Vogel et al. [133]. A live-attenuated vaccine comprising a recombinant SARS coronavirus without the E gene was also investigated, and it was found that the rSARS-CoV-E vaccine is immunogenic and effective in hamsters as an animal model, despite being attenuated in the replication process in the respiratory tract [132], [134].
The IWV and LAV are the oldest vaccines developed with new technology and could be potentially known as the first vaccine for SARS-CoV-2 in clinical trial investigations. Different research departments in China have isolated the various strains of SARS-CoV-2 and begun to manufacture these kinds of vaccines. A new deoptimized live-attenuated vaccine against SARS-CoV-2 has been designed in India (Codagenix/Serum Institute) by the sequence of the viral genome as named “rationally designed.” It is in the pre-clinical stage, and the anticipated time is in summer 2020 (Table 2 ) [111]. Along with, recombinant SARS-CoV-2 vaccine that incorporates the adenovirus type 5 vector (Ad5-nCoV), designed by CanSino Biological Incorporation, Beijing Institute of Biotechnology, Canadian Center for Vaccinology comes into Phase I, Phase II, and Phase I/II respectively [Id. NCT04313127, Id.NCT04341389, Id.NCT04398147].
Table 2.
Potential clinical trial vaccine candidates against SARS-CoV, MERS-CoV, and SARS-CoV2.
| vaccine type | Antigen target | Administration route | The clinical trial dose regimen | Phase | Reference |
|---|---|---|---|---|---|
| Potential vaccine candidates (clinical trial) cf against SARS-CoV | |||||
| DNA vaccine | S protein | IM | 3 dose vaccination | Phase I | [184] |
| S protein | EP | Three-injection vaccination regimen (0.67, 2, and 3 mg DNA/dose) followed by electroporation | Phase I | Id.NCT02670187 | |
| S protein | IM | 5 × 10^9 and 2.5 × 10^9 vp ChAdOx1 | Phase I | Id.NCT03399578 | |
| Potential vaccine candidates (clinical trial) against MERS | |||||
| DNA vaccine | S protein | IM | 0·67 mg, 2 mg, or 6 mg GLS-5300 intramuscular injection at baseline, week 4, and week 12 followed immediately by co-localized intramuscular electroporation | Phase I | [191] |
| Potential vaccine candidates (clinical trial) against SARS-CoV2 | |||||
| Inactivated whole virus | SARS-Alum | IM | 2 doses: 0, 14 days | Phase I/II |
NCT04383574 NCT04342583 |
| SARS | IM | 2 doses (0,14 or 0,21 days) | Phase I/II | ChiCTR2000031809 | |
| SARS | IM | 2 doses (0,14 or 0,21 days) | Phase I/II | ChiCTR2000032459 | |
| SARS | IM | 2 doses (0, 28 days) | Phase I/II | NCT04412538 | |
| SARS-CoV-2 (PiCoVacc mixed with alum adjuvants) | IM | various doses | Pre-clinical | [121] | |
| DNA vaccine | S protein | ID followed by EP | Two ID injections of 1.0 mg (total 2.0 mg per dosing visit) | Phase I | [200] |
| mRNA vaccine | S protein | IM | 0.5 ml [mL] of mRNA-1273 on Days 1 and 29 in the deltoid muscle and will be followed through 12 months post-second vaccination (Day 394) | Phase I | [174] |
| Recombinant SARS-CoV-2 (Adenovirus Type 5 Vector) | Adenovirus Type 5 Vector | IM | Dose-escalating phase I clinical trial in healthy 18 to 60 years of age (5E10, 1E11, 1.5 E11 VP Ad5-nCoV at 18 to 60 years old) on Days 1 in the deltoid muscle. 1 × 10^11vp, 5 × 10^10vp, and placebo of Ad5-nCoV administered through 1.0 mL intramuscular injection in the deltoid muscle on Day 0. A total of 96 healthy adult volunteers will be vaccinated in phase I stepwise according to the dose-escalation design from the younger adults (18 to <55) to the older adults (65 to <85). There are 2 dosage levels used in this phase: 5E10vp and 10E10vp, and 2 dose schedules. |
*Phase I **Phase II ***Phase I/II |
Id. CT04313127 Id. NCT04341389 Id.CT04398147 |
*CanSino Biological Incorporation, **Beijing Institute of Biotechnology, **Canadian Center for Vaccinology.
5.3. Subunit vaccines
To develop both MERS and SARS vaccines, the most focus of the subunit vaccine was on the recombinant S protein (RBD) synthesized in heterologous expression systems. The subunit vaccines can be designed by full-length S protein and its subunits (S1, S2), N-terminal domain (NTD) with or without adjuvants to improve their immunogenicity [135]. In fact, the RBD in the S1 subunit plays a critical role in the binding process of the virus to host cellular receptors [136]. High-titer neutralizing antibodies can be induced by the subunit vaccine based on both RBD and non-RBD S protein (full-length) like the S2 subunit. As an example, a recombinant S protein trimer of MERS-CoV (MERS S-2P) can fuse to the DPP4 receptor, and RBD, S2-specific neutralizing antibodies produced. However, neutralizing antibodies induction was observed when this protein was utilized against divergent pseudotyped MERS-CoV in mice [137], [138], [139], [140], [141], [142]. Contrary, partial effectiveness in the protection of immunized macaques and reduction of pneumonia and viral load as indicated by the RBD of S protein vaccination in Lan et al. study [143]. It was also reported that various fragments of the RBD, including 358–588 [144], 367–588 [145], 377–588, and 367–606 [39] could be used to vaccine development. The fragment of RBD containing residues 377–588 of MERS-CoV is a critical neutralizing domain and induces the highest humoral responses and neutralizing antibodies in immunized animals, especially when it fused with Fc of human IgG (S377-588-Fc) [144]. In another study, S377-588-Fc of MERS-CoV with MF59 as an adjuvant evaluated in BALB/c mice at different doses. The results showed strong humoral and cellular immune responses in the immunized mice at the lowest doses [136]. Similar results indicated that MERS-CoVneutralization could rise by the immune response against the 358-to-588, 1-to-357, and S1-Fc 1-to-747variants [146]. It was proved that the fragment-comprising residues 377–588 of MERS CoV could sufficiently protect hDPP4-Tg and Adenovirus (Ad) Ad/hDPP4-transduced mice against MERS-CoV without inducing of immunological toxicity or eosinophilic immune development [140], [144], [146], [147], [148]. Also, the residues of 367–588 or 367–606 in the MERS-CoV S1 subunit contain RBD [145], [149], [39]. In another investigation, it was found that intranasal administration of the RBD domain of the S protein in MERS-CoV can lead to more powerful response of local mucosal immune system in the lung tissue in comparison with the subcutaneous immunization [150].
Similar to MERS-CoV, several non-neutralizing immunodominant domains are presented in the S protein of SARS-CoV (full–length) and are responsible for attaching the receptor to cellular ACE2, that may assist to the immunogenicity of main neutralizing domains or trigger detrimental immune responses [151], [152], [153], [154]. In addition to being immunogenic, these subunit vaccines indicate a high safety level and represent inconsiderable side effects. In several studies on SARS–CoV challenges, the role of the S protein in the induction of serum-neutralizing antibodies and the production of protective immune responses by S protein in the animal models (mice, monkey) were confirmed [154], [155], [156], [157], [158], [159], [160]. Also, it was observed that CD4+ and the responses of CD8+ T-cells could be triggered by SARS–CoV S protein [161], [162]. Other subunit vaccines that are able to induce protective responses in mice and rabbits have been designed based on SARS-CoV RBD. It was discovered in another report that a chimeric protein(RBD linked to RBD-Fc) could provoke neutralizing antibodies in the different immunized animals(mice and rabbits), which could last for months and protect most of the vaccinated animal models from SARS-CoV infection [163]. Alongside RBD, several studies on the neutralizing activity of S1 protein or non-neutralizing antibody responses of S2 protein have confirmed the defensive efficiency of S protein fragments (S1 and S2) against SARS-CoV infection [155], [160], [164]. It has also been found that other subunit vaccines produced based on non-S structural proteins such as M proteins can lead to the neutralizing activity in the animal model [165].
Moreover, the identified protective response of immunodominant epitopes of M proteins (M1-31 and M132-161) that were obtained from the serum of SARS recovered patients in the immunized mouse and rabbit have been shown to cause immunogenicity and can trigger IgG specific antibodies in rabbits [166]. Various investigations showed that the CoV N protein could be considered as another antigen candidate in SARS-CoV vaccine production [167], [168]. Unlike M protein, the interaction of antibodies with CoV N proteins could provoke the activity of no virus-neutralizing under in vitro condition [154]. However, in in-vivo conditions, the induction of cell-mediated immunity by the protein can cause the protection to be activated [106]. The defensive mechanism of N- and M-based SARS subunit vaccines is still unidentified.
SARS-CoV-2 S protein subunits such asS1 subunit that consist of two domains including the C terminal domain (CTD) linked RBD domain and NTD, and the S2 subunit consists of the membrane fusion peptide (FP), a membrane-proximal external region (MPER), and a transmembrane domain (TM) [169]. Several institutes have recently developed many subunit vaccines against SARS-CoV-2. All of these vaccines are in the pre-clinical stage. Drosophila S2 insect cell expression system VLPs (Virus-like particles) is a developed strategy that has been currently used by ExpreS2ion. Virus-like particles or multiple recombinant structural proteins have enclosed a cell membrane lipid envelope or nanostructures enclosing the capsid proteins within itself. Also, it can fold the antigenic proteins of several microorganisms, such as bacteria, fungi, insects, mammalian cell lines, and even in transgenic plants. VLPs can act as non-replicating vectors for subunit and live-attenuated vaccine production in the absence of its genome [170].
Other subunit candidate vaccines based on S protein have been developed by WRAIR/USAMRIID, University of EpiVax, Georgia, and Queensland/GS, which are in pre-clinical stages. Adjuvanted S protein trimer introduced by Clover Biopharmaceuticals/Glaxo and Sanofi Pasteur/Glaxo Smith Kline institutes. Also, Ii-Key peptide and plant-based subunit vaccines are produced by iBio/CC-Pharming and Generex/EpiVax, respectively. In addition, full-length S-trimer/nanoparticle plus Matrix M by NovaVax and S1 or RBD protein by Bio/CC-Pharming were listed in the WHO list March 2020 (Table 1) [171], [172].
5.4. Recombinant vaccines
For an extended period, recombinant vaccines were widely produced in yeast or mammalian cells, which both are expensive expression platforms. The main disadvantages of such systems include high costs for preparation culture media and the risk of contamination by human pathogens. Additionally, the production of recombinant vaccines in bacterial systems was not successful due to improper folding of eukaryotic peptides and the occurrence of inclusion bodies in bacterial hosts [173]. Most of the recombinant vaccines are designed based on highly purified recombinant proteins or subunits of pathogens (Table 1).
5.4.1. Recombinant vector vaccines (RVVs)
Recombinant vector vaccines (RVVs) are produced based on bacterial or live viral vectors that are designed to represent various exogenous antigens [174]. RVVs appear to be the most promising types of live vaccines for the development of a transmissible vaccine platform [175].
Overall, gene-based vaccines are able to stimulate the responses of both cellular and humoral immune systems strongly, and viral vectors could potentially be an effective approach for antigen-encoding genes delivery and antigen presentation. To be applied as an effective vaccine carrier, the provided viral vector should be firstly safe and then present pathogen-specific antigens efficiently to the immune system [176]. RVVs appears to remain effective, even when genetically are not in a stable form and prone to reversion to insert free vector. These types of vaccines seem to be the most promising transmissible vaccine platform [175].
It has been shown that although recombinant RBD vaccines were formulated with Freund's adjuvant and Sigma adjuvant system® (monophosphoryl-lipid A and trehalose dicorynomycolate adjuvant) to elicit neutralizing antibodies with strong protective immunity in the vaccinated animals, they significantly show reduced or eliminated antibody-dependent immune enhancement (ADE) and other related detrimental inflammatory or immune responses [177]. It has been also demonstrated that a recombinant SARS-CoV RBD protein fused with human Fc produces highly strong immune responses, leading to the completely protected vaccinated mice against SARS-CoV infection [163]. Considering the robust capacity of RBD for the induction of neutralizing antibody, the recombinant proteins linked RBD can be utilized for the development of SARS-CoV vaccines [178]. Another study showed that long-term protective immunity in animal models could induce highly robust neutralizing antibodies after the administration of SARS-CoV RBD proteins [179].
What makes S protein of MERS-CoV as a key target for vaccine development is the immunogenic property that can induce neutralizing antibodies. For this reason, a chimeric virus was constructed based on the vesicular stomatitis virus (VSV) through the replacement of the G gene by the MERS-CoV S gene (VSVΔG-MERS) to develop a vaccine against MERS-CoV disease [180]. Previous studies on MERS have revealed that subcutaneous (s.c.) administration of the recombinant protein (RBD-Fc) can promote the potent ability of the vaccine to induce systemic neutralizing antibody responses in the vaccinated mice. Furthermore, intranasal (i.n.) use of MERS-CoV RBD-Fc is able to induce the responses of the humoral immune system compared to the one provoked by s.c. Uses, including the release of neutralizing antibodies, but more strong systemic responses of cellular and local mucosal immune responses in mouse lungs [150]. Other vaccines have been developed against the MERS-CoV infection highlights the function of RBD on the spike protein of MERS-CoV and the protective role of the recombinant NTD (rNTD) of spike proteins as an effective vaccine formulation in female BALB/c mice [38]. Studies on the development of rRBD vaccination against MERS-CoV infection have shown that while the RBD mainly induces most of the responses of the host immune system, this type of vaccine-induced only partial protection against the infection in both mice and non-human primates [143], [181].
It has been demonstrated systemic responses of lung resident memory T-cells, secretory IgA, and IgG could be induced by the recombinant adenovirus-based vaccine, which expresses MERS-CoV S protein when utilized intranasally into BALB/c mice, providing a stable neutralizing immunity to MERS spike pseudotyped virus; therefore, it is suggested that this vaccine has probably protective role against MERS-CoV infection [182]. The responses of the immune system to these provided vaccines were assessed in BALB/c mice, and obtained results indicated that RV based vaccines are able to induce earlier antibody responses and higher amounts of cellular immunity compared to the GEM particle vector [40]. Based on an animal study, the induction of neutralizing antibodies and the consequence of protection from the viral of SARS particles in mice emerged by the recombinant vaccine [183]. As discussed, another vaccine that is developed for clinical trial phase I is a DNA-based vaccine against SARS-COV [184]. Besides the vaccines undergoing phase I clinical trials, one of the notable COVID-19 vaccine candidates is a recombinant formulated vaccine in oral tablet form, which is used the genome sequence of SARS-CoV-2 and can induce strong systemic and mucosal immunity (Table 1) [185].
A clinical trial study conducted by Zhu et al. on recombinant adenovirus type-5 (Ad5) vectored vaccine against COVID-19 expressing the S glycoprotein of SARS-CoV-2 virus e is tolerable and also immunogenic in healthy adults 28 days after vaccination. The peaked of specific humoral responses against SARS-CoV-2 was shown at day 28 post-vaccination in healthy adult persons, and rapid, specific T-cell responses were peaked from day 14 post-vaccination suggesting the potential investigation of Ad5 vectored COVID-19 vaccine that makes it a strong warranty for further investigation. Additional suggestions on the safety and also immunogenicity of the Ad5 vectored COVID-19 vaccine will be provided via an ongoing phase 2 trial in China (NCT04341389).
Along with, recombinant SARS-CoV-2 vaccine that incorporates the adenovirus type 5 vector (Ad5-nCoV), designed by CanSino Biological Incorporation, Beijing Institute of Biotechnology, Canadian Center for Vaccinology comes into Phase I, Phase II, and Phase I/II respectively [Id. NCT04313127, Id.NCT04341389, Id.NCT04398147] [186], [187].
5.5. DNA vaccines
DNA vaccines represent two significant benefits compared to protein-based vaccines: (i) the easy DNA manipulation and (ii) low cost of production [188]. In addition, in contrast to the other protein-based subunit vaccines, DNA vaccines can lead to the Th1-biased immune response [189].
The success in the outcome of vaccination is linked to both antibody and T-cell-mediated immunity, and generally, among different vaccines, only live-recombinant vaccines can effectively induce cellular immunity. It has been demonstrated that DNA vaccines encode proteins from pathogens induce both humoral and cellular immune responses. What makes DNA vaccine to induce a cellular immune response is the mimicking of the live viruses effects, which produce antigenic proteins and efficiently represented by MHC class I; therefore, inducing the responses of CD8+ T-cells [190].
Several successful DNA vaccines have been developed for the functional proteins of SARS-CoV, such as the one designed for S, and also those identified for M and N proteins [191]. A DNA vaccine that is provided based on M protein expression has been proved that is able to provoke neutralizing antibody and the activity of cytotoxic T-lymphocytes in mice [192]. The N-protein peptide N220 of the SARS-CoV synthesized via bioinformatics assessment supplies essential information for designing of therapeutic vaccines against SARS-COV infection [193].
The responses of humoral and cellular immune systems can be induced by the application of DNA vaccines in response to MERS-CoV and SARS-CoV infection [194]. It has been reported that DNA encodes the whole S protein to provoke neutralizing antibodies and then robust the immunity mediated via T cell in animal models such as mice, macaques, and camels. Mitigated distinguishing clinical signs and symptoms of MERS-CoV infection in macaques, including pneumonia, were observed in immunized macaques [192]. MERS-CoV-specific antibody and the response of T-cells can be initiated by DNA vaccines as effective vaccine candidates in non-human primates [194]. Vaccines that can express the MERS-CoV RBD, induced robust neutralizing antibodies in mice as well as in the response of T-cells in non-human primates [195], [143].
The results of a recent study indicate that DNA vaccines that express the full-length subunit of S1 against MERS-CoV could significantly represent higher amounts of S1-specific antibodies (Abs). It is suggested that while plasmids expressing full S protein induce Th2 response and are associated with minimized risk of the immunopathologies, DNA vaccine expressing S1 subunit can be considered as a potential vaccine candidate. It needs to note that full-length DNA vaccines against MERS-CoV lead to considerable titers of IgG2a and IgG2b antibodies (Th1-skewed response) along with very subtle S1-specific CD8+ IFN-γ response. This result suggests that raised amounts of all IgG isotypes in a balanced Th1/Th2 response is generated by pS1-immunization with markedly increased CD8+ IFN-γ response compared to the pS group [189]. Another study supports the use of the plasmid encoding S1 protein for the development of DNA vaccines against MERS-CoV S1 protein. The obtained results confirmed the strong protective humoral and cellular immune responses of DNA based vaccines in mice and also antigen-specific CD4+ and CD8+ T cells secreting IFN-c and other generated cytokines provoked by gene-based vaccines [196].
The previous reports on MERS have revealed that although subcutaneous vaccination with RBD of MERS-CoV S protein-induced systemic humoral immune responses, much stronger local mucosal immune responses, those induced by intranasal vaccination are strong and more robust [150]. Recently animal studies in rhesus macaques, mice, and came demonstrated immunization with a MERS-CoV vaccination, results in the activation of T-cell responses and increased production of Th1 and Th2 cytokines [194].
The combination vaccines (protein and DNA), encodings protein, which is in the preclinical phase, and DNA-based vaccines with moderate and mild symptoms in Phase I and II clinical trials are two other vaccines against MERS-CoV [139], [189].
In a recent animal study on DNA vaccine, which encodes more than 700 residues of the S1 protein, the defined vaccine induces strong protective responses of the antigen-specific humoral and cellular immune responses in mice [196]. Based on the fact that SARS-CoV-2 shares approximately high nucleotide similarly (89%) to SARS-like coronaviruses, the strategies for designing of SARS-CoV-2 vaccines are built based on the previous advances for SARS. Furthermore, genomic analyses indicated that either SARS-CoV or SARS-CoV-2 exhibits genomic similarities in the receptor-binding domain that can directly bind to the human receptor ACE2, suggesting its essential implications for the designing of novel vaccines [197].
A DNA vaccine candidate targeting MERS have advanced into clinical trials [Id.NCT02670187]. Another vaccine candidate against MERS has been recently processed to phase I/II clinical trial as a DNA vaccine [198]. Furthermore, an efficient vaccine candidate named MERS001 for MERS-CoV with ChAdOx1 that encodes the S protein of the virus is developed at phase I clinical trial [Id. NCT03399578].
To date, some companies experience the designing of DNA-based vaccines that encode S protein against the COVID-19 challenge since March 13, 2020, according to the WHO report, including Inovio Pharmaceuticals (USA), Sanofi Pasteur/BARDA (France/USA), Sciences/Evvivax (USA/Italy), Takis Biotech/Applied DNA [199].
Clover Biopharmaceuticals could produce a protein trimer vaccine for SARS-CoV-2 (S-Trimer)
by Trimer-Tag© technology application via a rapid expression system (mammalian cell-culture), and they will examine the preclinical safety soon [111].
Induction of anti-S antibodies in an animal model like mice and following SARS virus-specific neutralizing antibody and the responses of T-cells in the first phase of the human trial by using DNA vaccine candidates is well documented [184]. To date, all vaccine candidates against the SARS-COV2 have recently moved into clinical development. There are two vaccine candidates, the first one that is entered to Phase I clinical trials is a DNA plasmid encoding S protein (INO-4800) delivered by electroporation [200], while another one is a recombinant vaccine that is produced by using of an adenovirus vector 5 (Ad5- nCoV) that encodes the S protein with full length, which is undergoing phase I trials since 12th April 2020 [197].
Results from phase I from the first DNA vaccine candidate against MERS coronavirus to enter clinical trials, dose-ranging study of GLS-5300, reported from 75 adults aged 18–50 years, received 0·67 mg, 2 mg, or 6 mg GLS-5300 intramuscular injection at baseline, week 4, and week 12 followed immediately by co-localized intramuscular electroporation (Table 2) [201].
5.6. mRNA vaccines
RNA-based technologies for the generation of vaccines have attracted tremendous attention as a result of its safety and long-lasting immune response in human and animal models during two past decades [202]. Based on an NIH report, a new mRNA vaccine named mRNA-1273 has been recently produced by Moderna in cooperation with the National Institute of Allergy and Infectious Diseases (NIAID), which is one of two candidate vaccines entered into Phase 1 human clinical trial and tested on 45 enrolled healthy adult volunteers. mRNA-1273 encodes Spike (S) protein in a prefusion stabilized form to mimic the natural course of viral infection. Moderna is a well-known biotechnology company with previous experience in mRNAs-based vaccines designing encoding antigenic proteins full-length S, S1, or S2 for SARS-CoV and MERS-CoV. Lipid nanoparticle (LNP)-encapsulated mRNA cocktail encoding VLP is another mRNA-based vaccine against COVID-19, which is introduced by Lin Jinzhong and his colleagues. They optimized and formulated an mRNA cocktail containing three genes with the ability to generate VLPs with structural and morphological features similar to native nCoV-2019 antigens (Table 1, Table 2) [203]. Of more than 100 candidate vaccines developing worldwide, Moderna's mRNA COVID-19 vaccine and CanSino's non-replicating adenovirus type-5 (Ad5), which is a vectored COVID-19 vaccine, are both entered in to phase 1 clinical trial zhu [204].
5.7. Virus-like particle vaccines
VLP vaccines are engineered products formed by viral proteins expression in various protein expression systems with the capacity of proteins self-assembly, which broadly served against highly virulent emerging viruses, including coronaviruses [205]. Medicago is an example of these companies which synthesized the SARS-CoV-2 VLP vaccine via its proprietary plant-based system. The company announced that the SARS-CoV-2 VLP vaccine would undergo human trials by July or August 2020 (Table 1) [206].
6. Future perspectives
The disruption of the host immune system by the COVID-19 is the main characteristic of an outbreak of SARS-CoV-2 diseases. The innate and adaptive immune response is considered as a central factor of antiviral defense. However, in patients infected with SARS-CoV-2, the virus could paralysis the immune protection by evasion of the immune response. Therefore, to achieve more information about the COVID-19 and immune response interaction, further investigation is required.
There is limited information about SARS-CoV-2 infection and more questions than responses for the lately recognized virus, such as the structural features, etiology, or related epidemiology, pathogenesis, and immunopathogenesis, etc. Access to necessary information on highly pathogenic SARS-CoV2, like identifying of mediators that manage the immune response, the involved pathways, and the mechanism regulate virus behavior, will help the design and development of successful antiviral therapeutics and vaccine candidates. To stop disease progression and the immune response promotion, it is necessary to formulate an effective vaccine.
High mutation rates in SAR-CoV-2 are the critical subject in vaccine design, specifically when it can affect the binding affinity of the virus. A SARS-Coronavirus -2 variant carrying the spike protein amino acid mutation D614G has grown into the most dominant form in the global pandemic. Active tracking of variant frequencies shown a recurrent pattern of G614 increase at several geographic levels: regional, national and community. The shift arisen even in local epidemics where the original D614 form was well recognized prior to institution of the G614 variant. The consistency of this pattern was highly important, suggesting that the G614 variant may have a fitness benefit. Studies have shown that in infected individuals, G614 is related with lower RT-PCR cycle thresholds, indicative of higher upper respiratory tract viral loads, but not with increased disease severity [207]. Even though difficult infectiousness of the G614 variant may completely reason for its persistence and rapid spread, other aspects should also be measured. These include epidemiological influences because viral spread also is determined by on whom it infects, and epidemiological influences can also cause changes in genotype frequency to mimic evolutionary forces. In all possibility, an arrangement of evolutionary selection for G614 and the founder’s special effects of being presented into highly mobile and connected populations may have together contributed, in part, to its rise. The G-clade mutations in the 5 UTR or in the RdRP protein might also have effects [208].
In addition, there could be immunological consequences resulting from the G614 change in Spike. The G614 variant is sensitive to neutralization by polyclonal convalescent serum, which is boosting in terms of immune involvements, but it will be vital to determine whether the D614 and G614 forms of SARS-CoV-2 are differentially vaccine-elicited antibodies or sensitive to neutralization by antibodies produced in response to infection with either type of the virus [209].
Despite fast developing in vaccines, it will probably come too late to have an impact on the first wave of a potential pandemic. Still, critical lessons can be attained for the development of potential vaccines against rapidly emerging viruses. Notably, if the virus to be maintained within their host communities, SARS-CoV-2 vaccines will be critical to reducing morbidity and mortality. Moreover, as SARS-CoV-2 targets the mucosa of the respiratory tract, developing a vaccine that boosts the robust immunity through the in route as an efficient strategy to stop COVID-19 infection. The immunogenicity of vaccines is determined via numerous factors; the route of administration is one of the important factors. Therefore, the administration of vaccines in an optimal route and the appropriate adjuvant to ensure an optimal immune response could play a significant role in the development and successful application of SARS-CoV-2 vaccines.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Schwartz D.A., Graham A.L.J.V. Potential maternal and infant outcomes from (Wuhan) coronavirus 2019-nCoV infecting pregnant women: lessons from SARS, MERS, and other human coronavirus infections. Viruses. 2020;12(2):194. doi: 10.3390/v12020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J.Y. Lee, et al., The clinical and virological features of the first imported case causing MERS-CoV outbreak in South Korea, 2015, BMC Infectious Diseases 17(1) (2017) 498. [DOI] [PMC free article] [PubMed]
- 4.Peeri N.C., The SARS, et al. MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020;49(3):717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W.-j., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui D.S.J.C.i.c.m. Epidemic and emerging coronaviruses (severe acute respiratory syndrome and middle east respiratory syndrome) Clin. Chest Med. 2017;38(1):71–86. doi: 10.1016/j.ccm.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W.-H., et al. The SARS-CoV-2 vaccine pipeline: An overview. Curr. Trop. Med. Rep. 2020;(7):61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jernigan D.B. Update: public health response to the coronavirus disease 2019 outbreak—United States, February 24, 2020. MMWR. Morbidity and mortality weekly report. CDC. 2020;69(8):216–219. doi: 10.15585/mmwr.mm6908e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giwa A., Desai A., Jagoda A. Novel coronavirus COVID-19: an overview for emergency clinicians. Emerg. Med. Pract. 2020;22(2):1–21. [PubMed] [Google Scholar]
- 11.WHO, 2020.
- 12.Fehr A.R., et al. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7(6) doi: 10.1128/mBio.01721-16. e01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C., et al. ACS Publications; 2020. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angelini M.M., et al. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4(4) doi: 10.1128/mBio.00524-13. e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minskaia E., et al. Discovery of an RNA virus 3′→ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U S A. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shokri S., et al. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J. Cell Physiol. 2019;234(3):2143–2151. doi: 10.1002/jcp.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan K., et al. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faure E., et al. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menachery V.D., et al. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J. Virol. 2014;88(8):4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prompetchara E., Ketloy C., Palaga T.J.A.P.J.A.I. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 21.Pascal K.E., et al. Pre-and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. U S A. 2015;112(28):8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muth D., et al. Infectious Middle East respiratory syndrome coronavirus excretion and serotype variability based on live virus isolates from patients in Saudi Arabia. J. Clin. Microbiol. 2015;53(9):2951–2955. doi: 10.1128/JCM.01368-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corman V.M., et al. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88(19):11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z., et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. U S A. 2007;104(29):12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almazán F., et al. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Res. 2014;189:262–270. doi: 10.1016/j.virusres.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sui J., et al. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J. Virol. 2014;88(23):13769–13780. doi: 10.1128/JVI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffers S.A., et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U S A. 2004;101(44):15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu F., et al. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. BioRvix. 2020 [Google Scholar]
- 29.Wang Y., et al. A recombinant infectious bronchitis virus from a chicken with a spike gene closely related to that of a turkey coronavirus. Arch. Virol. 2020;165(3):703–707. doi: 10.1007/s00705-019-04488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navas-Martín S., Weiss S.R. Coronavirus replication and pathogenesis: implications for the recent outbreak of severe acute respiratory syndrome (SARS), and the challenge for vaccine development. J. Neurovirology. 2004;10(2):75–85. doi: 10.1080/13550280490280292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong S.K., et al. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du L., et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Memish Z.A., et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19(11):1819. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voss M.W., et al. Bridging animal and human models of exercise-induced brain plasticity. Trends Cognitive Sci. 2013;17(10):525–544. doi: 10.1016/j.tics.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gralinski L.E., Menachery V.D. Return of the Coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fehr A.R., Perlman S. Coronaviruses. Springer; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knoops K., et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6(9) doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu G., et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling Y., et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glowacka I., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shulla A., et al. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85(2):873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E., et al. Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine. 2014;32(45):5975–5982. doi: 10.1016/j.vaccine.2014.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shereen M.A., et al. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braciale T.J., Hahn Y.S. Immunity to viruses. Immunol. Rev. 2013;255:1. doi: 10.1111/imr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virology. 2012;2(3):264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoneyama M., et al. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020:1–15. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satoh T., Akira S. Toll-like receptor signaling and its inducible proteins. Myeloid Cells in Health and Disease: A Synthesis. 2017:447–453. [Google Scholar]
- 51.Vijay K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int. Immunopharmacol. 2018;59:391–412. doi: 10.1016/j.intimp.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novoa R.R., et al. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell. 2005;97(2):147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lau S.K., et al. Delayed induction of proinflammatory cytokines and suppression of innate antiviral response by the novel Middle East respiratory syndrome coronavirus: implications for pathogenesis and treatment. J. Gen. Virol. 2013;94(12):2679–2690. doi: 10.1099/vir.0.055533-0. [DOI] [PubMed] [Google Scholar]
- 54.Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133(1):101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao J., et al. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5(10) doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikawa T., et al. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vabret N., et al. Immunology of COVID-19: current state of the science. Immunity. 2020 doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Law H.K., et al. Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunology. 2009;10(1):35. doi: 10.1186/1471-2172-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dosch S.F., Mahajan S.D., Collins A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-κB pathway in human monocyte macrophages in vitro. Virus Res. 2009;142(1–2):19–27. doi: 10.1016/j.virusres.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Channappanavar R., Perlman S. Seminars in Immunopathology. Springer; 2017. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Al von Messling V., Griffin D.E. How respiratory viruses overcome mucosal defenses and exploit the unique environment of the respiratory tract. Curr. Opin. Virol. 2012;2(3):221. doi: 10.1016/j.coviro.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iwata-Yoshikawa N., et al. Effects of Toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-inactivated severe acute respiratory syndrome-related coronavirus vaccine. J. Virol. 2014;88(15):8597–8614. doi: 10.1128/JVI.00983-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hemmat N., et al. Neutrophils, crucial, or harmful immune cells involved in coronavirus infection: a bioinformatics study. Front. Genet. 2020;11:641. doi: 10.3389/fgene.2020.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barnes B.J., et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong J.J.M., et al. Insights into the immuno-pathogenesis of acute respiratory distress syndrome. Ann. Translational Med. 2019;7(19) doi: 10.21037/atm.2019.09.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Camp J.V., Jonsson C.B. A role for neutrophils in viral respiratory disease. Front. Immunol. 2017;8:550. doi: 10.3389/fimmu.2017.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X., et al. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorski S.A., Hufford M.M., Braciale T.J. Recent insights into pulmonary repair following virus-induced inflammation of the respiratory tract. Curr. Opin. Virology. 2012;2(3):233–241. doi: 10.1016/j.coviro.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 73.Yip M.S., et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virology J. 2014;11(1):1–11. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arvin A.M., et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020:1–14. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 75.Wang J., Zand M.S. The potential for antibody-dependent enhancement of SARS-CoV-2 infection: Translational implications for vaccine development. J. Clin. Transl. Sci. 2020:1–11. [Google Scholar]
- 76.Quinlan B.D., et al. The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. IMMUNITY-D-20-00389. 2020 [Google Scholar]
- 77.Bao L., et al. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020 [Google Scholar]
- 78.García L.F. Immune response, inflammation, and the clinical spectrum of COVID-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu H.-Q., et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur. Respir. J. 2020 doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen X., et al. Host immune response to influenza A virus infection. Front. Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nurieva R.I., Chung Y. Understanding the development and function of T follicular helper cells. Cell. Mol. Immunol. 2010;7(3):190–197. doi: 10.1038/cmi.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Q., et al. Molecular basis of the differentiation and function of virus specific follicular helper CD4+ T cells. Front. Immunol. 2019;10:249. doi: 10.3389/fimmu.2019.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marshall N.B., Swain S.L. Cytotoxic CD4 T cells in antiviral immunity. Biomed Res. Int. 2011;2011 doi: 10.1155/2011/954602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown D.M., et al. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J. Virol. 2012;86(12):6792–6803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown D.M., et al. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 2006;177(5):2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 86.Siu K.-L., et al. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;11(2):141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Wit E., et al. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun L., et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perrotta F., et al. COVID-19 and the elderly: insights into pathogenesis and clinical decision-making. Aging Clin. Exp. Res. 2020:1–10. doi: 10.1007/s40520-020-01631-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.C.Y. Yong, et al., Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus, 10 (2019) 1781. [DOI] [PMC free article] [PubMed]
- 91.A. Roberts, et al., Animal models and vaccines for SARS-CoV infection. 133(1) (2008) 20–32. [DOI] [PMC free article] [PubMed]
- 92.Spruth M., et al. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. 2006;24(5):652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.D. Qu, et al., Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. 23(7) (2005) 924–931. [DOI] [PMC free article] [PubMed]
- 94.J. Zhou, et al., Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. 23(24) (2005) 3202–3209. [DOI] [PMC free article] [PubMed]
- 95.W.-p. Kong, et al., Modulation of the immune response to the severe acute respiratory syndrome spike glycoprotein by gene-based and inactivated virus immunization. 79(22) (2005) 13915–13923. [DOI] [PMC free article] [PubMed]
- 96.A. Roberts, et al., Immunogenicity and protective efficacy in mice and hamsters of a β-propiolactone inactivated whole virus SARS-CoV vaccine, 23(5) (2010) 509–519. [DOI] [PMC free article] [PubMed]
- 97.R.H. See, et al., Comparative evaluation of two severe acute respiratory syndrome (SARS) vaccine candidates in mice challenged with SARS coronavirus, 87(3) (2006) 641–650. [DOI] [PubMed]
- 98.M. Bolles, et al., A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge 85(23) (2011) 12201–12215. [DOI] [PMC free article] [PubMed]
- 99.C.-T. Tseng, et al., Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus, 7(4), 2012. [DOI] [PMC free article] [PubMed]
- 100.S. Xiong, et al., Immunogenicity of SARS inactivated vaccine in BALB/c mice. 95(2) (2004) 139–143. [DOI] [PMC free article] [PubMed]
- 101.Y., He, et al., Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry, 325(2) (2004) 445–452. [DOI] [PMC free article] [PubMed]
- 102.K.G. Lokugamage, et al., Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. 26(6) (2008) 797–808. [DOI] [PMC free article] [PubMed]
- 103.M.E. Darnell, et al., Severe acute respiratory syndrome coronavirus infection in vaccinated ferrets, 196(9) (2007) 1329–1338. [DOI] [PMC free article] [PubMed]
- 104.R.H. See, et al., Severe acute respiratory syndrome vaccine efficacy in ferrets: whole killed virus and adenovirus-vectored vaccines. 89(9) (2008) 2136–2146. [DOI] [PubMed]
- 105.Y. Honda-Okubo, et al., Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. 89(6) (2015) 2995–3007. [DOI] [PMC free article] [PubMed]
- 106.Roper, R.L. and K.E.J.E.r.o.v. Rehm, SARS vaccines: where are we? 2009. 8(7): p. 887-898. [DOI] [PMC free article] [PubMed]
- 107.Y. Deng, et al., Enhanced protection in mice induced by immunization with inactivated whole viruses compare to spike protein of middle east respiratory syndrome coronavirus. 7(1) (2018) 1–10. [DOI] [PMC free article] [PubMed]
- 108.Wirblich C., et al. One-health: a safe, efficient, dual-use vaccine for humans and animals against Middle East respiratory syndrome coronavirus and rabies virus. J. Virol. 2017;91(2):e02040–16. doi: 10.1128/JVI.02040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Agrawal S., et al. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J. Virol. 2015;89(7):3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin J., et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir. Ther. 2007;12(7):1107–1113. [PubMed] [Google Scholar]
- 111.Zhang J., et al. Progress and prospects on vaccine development against SARS-CoV-2. Vaccines. 2020;8(2):153. doi: 10.3390/vaccines8020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martins K.A., Bavari S., Salazar A.M. Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev. Vaccines. 2015;14(3):447–459. doi: 10.1586/14760584.2015.966085. [DOI] [PubMed] [Google Scholar]
- 113.Chattopadhyay S., Sen G.C. dsRNA-activation of TLR3 and RLR signaling: gene induction-dependent and independent effects. J. Interferon Cytokine Res. 2014;34(6):427–436. doi: 10.1089/jir.2014.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buniello A., et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Totura A.L., et al. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. MBio. 2015;6(3) doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhao J., et al. Intranasal treatment with poly (I· C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012;86(21):11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barnard D.L., Kumaki Y. Recent developments in anti-severe acute respiratory syndrome coronavirus chemotherapy. Future Virology. 2011;6(5):615–631. doi: 10.2217/fvl.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.P. Mosaddeghi, et al., Therapeutic approaches for COVID-19 based on the dynamics of interferon-mediated immune responses, 2020.
- 119.Barnard D.L., et al. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antiviral Chem. Chemother. 2006;17(5):275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 120.Calina D., et al. Towards effective COVID-19 vaccines: Updates, perspectives and challenges. Int. J. Mol. Med. 2020;46(1):3–16. doi: 10.3892/ijmm.2020.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao Q., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Organization W.H. DRAFT landscape of COVID-19 candidate vaccines. World. 2020 [Google Scholar]
- 123.S.A. Plotkin, W.A. Orenstein, O. PA., Vaccines. 5th edn ed. Vol. 5th edn. 2008, Philadelphia: Saunders/Elsevier.
- 124.Perlman S., Vijay R.J.I.J.o.I.D. Middle East respiratory syndrome vaccines. Int. J. Infect. Dis. 2016;47:23–28. doi: 10.1016/j.ijid.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Almazan F., et al. Engineering a replication-competent, propagationdefective middle east respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4:e00650–e713. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.T. Scobey, et al., Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. 110(40) (2013) 16157–16162. [DOI] [PMC free article] [PubMed]
- 127.Pulendran B., Ahmed R.J.N.i. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12(6):509. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Graham R.L., et al. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18(12):1820. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Subbarao K., et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jimenez-Guardeno J.M., et al. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015;11(10):1005215. doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Züst R., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lamirande E.W., et al. A live attenuated severe acute respiratory syndrome coronavirus is immunogenic and efficacious in golden Syrian hamsters. J. Virol. 2008;82(15):7721–7724. doi: 10.1128/JVI.00304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vogel L.N., et al. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV) Vaccine. 2007;25(12):2173–2179. doi: 10.1016/j.vaccine.2006.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.DeDiego M.L., et al. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.!!! INVALID CITATION !!! (41, 53, 96-105).
- 136.Tang J., et al. Optimization of antigen dose for a receptor-binding domain-based subunit vaccine against MERS coronavirus. Hum. Vaccin. Immunother. 2015;11(5):1244–1250. doi: 10.1080/21645515.2015.1021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pallesen J., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U S A. 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li K., et al. Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase. J. Infect. Dis. 2016;213(5):712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang L., et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat. Commun. 2015;6(1) doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiaming L., et al. The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine. 2017;35(1):10–18. doi: 10.1016/j.vaccine.2016.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Y., et al. Receptor-binding domain of MERS-CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS-CoV infection. Hum. Vaccin. Immunother. 2017;13(7):1615–1624. doi: 10.1080/21645515.2017.1296994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Adney D.R., et al. Efficacy of an adjuvanted Middle East respiratory syndrome coronavirus spike protein vaccine in dromedary camels and alpacas. Viruses. 2019;11(3):212. doi: 10.3390/v11030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lan J., et al. Recombinant receptor binding domain protein induces partial protective immunity in rhesus macaques against Middle East respiratory syndrome coronavirus challenge. EBioMedicine. 2015;2(10):1438–1446. doi: 10.1016/j.ebiom.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ma C., et al. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments—the importance of immunofocusing in subunit vaccine design. Vaccine. 2014;32(46):6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen Y., et al. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87(19):10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mou H., et al. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013;87(16):9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jiang L., et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci. Transl. Med. 2014;6(234):234ra59. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 148.Du L., et al. Identification of receptor-binding domain in S protein of the novel human coronavirus MERS-CoV as an essential target for vaccine development. J. Virol. 2013 doi: 10.1128/JVI.01048-13. 01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Du L., et al. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016;7(1):1–9. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ma C., et al. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: Implication for designing novel mucosal MERS vaccines. Vaccine. 2014;32(18):2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weingartl H., et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.See R.H., et al. Rapid response research-SARS coronavirus vaccines and application of processes to other emerging infectious diseases. Curr. Immunol. Rev. 2005;1(2):185–200. [Google Scholar]
- 153.Berger A., et al. Severe acute respiratory syndrome (SARS)—paradigm of an emerging viral infection. J. Clin. Virol. 2004;29(1):13–22. doi: 10.1016/j.jcv.2003.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Du L., et al. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393(1):144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bisht H., et al. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. U S A. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bukreyev A., et al. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hu M.C., et al. Intranasal protollin-formulated recombinant SARS S-protein elicits respiratory and serum neutralizing antibodies and protection in mice. Vaccine. 2007;25(34):6334–6340. doi: 10.1016/j.vaccine.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kam Y.W., et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcγRII-dependent entry into B cells in vitro. Vaccine. 2007;25(4):729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Du L., et al. A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral Immunol. 2010;23(2):211–219. doi: 10.1089/vim.2009.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li J., et al. Immunogenicity and protection efficacy of monomeric and trimeric recombinant SARS coronavirus spike protein subunit vaccine candidates. Viral Immunol. 2013;26(2):126–132. doi: 10.1089/vim.2012.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huang J., et al. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine. 2007;25(39–40):6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Stadler K., et al. SARS vaccine protective in mice. Emerg. Infect. Dis. 2005;11(8):1312–1314. doi: 10.3201/eid1108.041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.He Y., et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guo Y., et al. Elicitation of immunity in mice after immunization with the S2 subunit of the severe acute respiratory syndrome coronavirus. DNA Cell Biol. 2005;24(8):510–515. doi: 10.1089/dna.2005.24.510. [DOI] [PubMed] [Google Scholar]
- 165.Yang Z.-y., et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.He Y., et al. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2005;43(8):3718–3726. doi: 10.1128/JCM.43.8.3718-3726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu S.-J., et al. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24(16):3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zheng N., et al. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine. 2009;27(36):5001–5007. doi: 10.1016/j.vaccine.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Syomin B., Ilyin Y.J.M.B. Virus-like particles as an instrument of vaccine production. Mol. Biol. 2019;53(3):323–334. doi: 10.1134/S0026893319030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mukherjee R.J.J.o.B. Global efforts on vaccines for COVID-19: Since, sooner or later, we all will catch the coronavirus. J. Biosci. 2020;45(1):68. doi: 10.1007/s12038-020-00040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kahandal S.S. Recent developments on COVID-19 and associated human coronavirus disease therapeutic agents, and vaccines. ACS Cent. Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Franklin S.E., Mayfield S.P. Prospects for molecular farming in the green alga Chlamydomonas reinhardtii. Curr. Opin. Plant Biol. 2004;7(2):159–165. doi: 10.1016/j.pbi.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 174.Bull J.J., Smithson M.W., Nuismer S.L. Transmissible viral vaccines. Trends Microbiol. 2018;26(1):6–15. doi: 10.1016/j.tim.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Murphy A.A., Redwood A.J., Jarvis M.A. Self-disseminating vaccines for emerging infectious diseases. Expert Rev. Vaccines. 2016;15(1):31–39. doi: 10.1586/14760584.2016.1106942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Souza A., et al. Recombinant viruses as vaccines against viral diseases. Braz. J. Med. Biol. Res. 2005;38(4):509–522. doi: 10.1590/s0100-879x2005000400004. [DOI] [PubMed] [Google Scholar]
- 177.Du L., et al. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25(15):2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jiang S., He Y., Liu S. SARS vaccine development. Emerg. Infect. Dis. 2005;11(7):1016. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Song Z., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu R., et al. A recombinant VSV-vectored MERS-CoV vaccine induces neutralizing antibody and T cell responses in rhesus monkeys after single dose immunization. Antiviral Res. 2018;150:30–38. doi: 10.1016/j.antiviral.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lan J., et al. Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the Middle East respiratory coronavirus (MERS-CoV) receptor-binding domain as an antigen. PLoS ONE. 2014;9(11) doi: 10.1371/journal.pone.0112602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim M.H., Kim H.J., Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length Spike protein of Middle East respiratory syndrome coronavirus. PLoS ONE. 2019;14(7) doi: 10.1371/journal.pone.0220196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen W.-H., et al. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Tropical Med. Rep. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Martin J.E., et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a Phase I clinical trial. Vaccine. 2008;26(50):6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Vaxart. PipelineReview.com. Vaxart Announces Initiation of Coronavirus Vaccine Program. 2020 [cited 2020 January 31]; Available from: https:// investors.vaxart.com/news-releases/news-release-details/vaxart-announcesinitiation-coronavirus-vaccine-program; Accessed on: 09.02.2020.
- 186.https://clinicaltrials.gov/ct2/show/NCT04341389. A Phase II Clinical Trial to Evaluate the Recombinant Vaccine for COVID-19 (Adenovirus Vector) (CTII-nCoV). 2020.
- 187.https://www.trialsitenews.com/cansino-biologics-ad5-ncov-the-first-covid-19-vaccine-to-phase-ii-clinical-trials/. CanSino Biologics’ Ad5-nCoV the First COVID-19 Vaccine to Phase II Clinical Trials, 2020.
- 188.Leitner W.W., Ying H., Restifo N.P. DNA and RNA-based vaccines: principles, progress and prospects. Vaccine. 1999;18(9–10):765–777. doi: 10.1016/s0264-410x(99)00271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Al-Amri S.S., et al. Immunogenicity of candidate MERS-CoV DNA vaccines based on the spike protein. Sci. Rep. 2017;7:44875. doi: 10.1038/srep44875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gurunathan S., et al. DNA vaccines: a key for inducing long-term cellular immunity. Curr. Opin. Immunol. 2000;12(4):442–447. doi: 10.1016/s0952-7915(00)00118-7. [DOI] [PubMed] [Google Scholar]
- 191.Roper R.L., Rehm K.E. SARS vaccines: where are we? Expert Rev. Vaccines. 2009;8(7):887–898. doi: 10.1586/erv.09.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Okada M., et al. Development of vaccines and passive immunotherapy against SARS corona virus using SCID-PBL/hu mouse models. Vaccine. 2007;25(16):3038–3040. doi: 10.1016/j.vaccine.2007.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cheung Y.-K., et al. Induction of T-cell response by a DNA vaccine encoding a novel HLA-A* 0201 severe acute respiratory syndrome coronavirus epitope. Vaccine. 2007;25(32):6070–6077. doi: 10.1016/j.vaccine.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Muthumani K., et al. A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015;7(301) doi: 10.1126/scitranslmed.aac7462. p. 301ra132-301ra132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang N., et al. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chi H., et al. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine. 2017;35(16):2069–2075. doi: 10.1016/j.vaccine.2017.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim E., et al. Microneedle array delivered recombinant coronavirus vaccines: Immunogenicity and rapid translational development. EBioMedicine. 2020 doi: 10.1016/j.ebiom.2020.102743. 102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yong C.Y., et al. Recent advances in the vaccine development against middle east respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect. Dis. Therapy. 2020:1–20. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Le T.T., et al. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 201.Modjarrad K., et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19(9):1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.C. Biopharmaceuticals, Clover initiates development of recombinant subunit-trimer vaccine for wuhan coronavirus (2019-ncov), 2020.
- 203.fudan.edu.cn. Towards an effective mRNA vaccine against 2019-nCoV. 2020 [cited 2020 7 MAR]; Available from: https://www.fudan.edu.cn/en/2020/0307/c344a104281/page.htm.
- 204.Zhu F.-C., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. The Lancet. 2020 doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Josefsberg J.O., Buckland B. Vaccine process technology. Biotechnol. Bioeng. 2012;109(6):1443–1460. doi: 10.1002/bit.24493. [DOI] [PubMed] [Google Scholar]
- 206.businesswire. Medicago Announces Production of a Viable Vaccine Candidate for COVID-19. 2020 [cited 2020 MAR 12]; Available from: https://www.businesswire.com/news/home/20200312005345/en/.
- 207.Korber B., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4) doi: 10.1016/j.cell.2020.06.043. pp. 812–827. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gudbjartsson D.F., et al. Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2027653. [DOI] [PubMed] [Google Scholar]
- 209.Korber B., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Guo J.-P., et al. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology. 2004;324(2):251–256. doi: 10.1016/j.virol.2004.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Menachery V.D., et al. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 2018;92(17):e00710–18. doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jaume M., et al. SARS CoV subunit vaccine: antibodymediated neutralisation and enhancement. Mediccine. 2012;18(2):31–36. [PubMed] [Google Scholar]
- 213.Tao X., et al. Characterization and demonstration of the value of a lethal mouse model of Middle East respiratory syndrome coronavirus infection and disease. J. Virol. 2016;90(1):57–67. doi: 10.1128/JVI.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Malczyk A.H., et al. A highly immunogenic and protective Middle East respiratory syndrome coronavirus vaccine based on a recombinant measles virus vaccine platform. J. Virol. 2015;89(22):11654–11667. doi: 10.1128/JVI.01815-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Menachery V.D., et al. Middle east respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere. 2017;2(6) doi: 10.1128/mSphere.00346-17. e00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bodmer B.S., et al. Live-attenuated bivalent measles virus-derived vaccines targeting Middle East respiratory syndrome coronavirus induce robust and multifunctional T cell responses against both viruses in an appropriate mouse model. Virology. 2018;521:99–107. doi: 10.1016/j.virol.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tai W., et al. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016;499:375–382. doi: 10.1016/j.virol.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nyon M.P., et al. Engineering a stable CHO cell line for the expression of a MERS-coronavirus vaccine antigen. Vaccine. 2018;36(14):1853–1862. doi: 10.1016/j.vaccine.2018.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]