Abstract
Atopic asthma is a chronic lung disease of lower airways caused mainly due to action of T-helper (Th) 2 type cytokines, eosinophilic inflammation, mucus hypersecretion and airway remodelling. Interleukin (IL)-33 increases type 2 immunity polarization in airway playing critical role in eosinophilic asthma. On the other hand, NLRP3 inflammasome activation results in the release of caspase-1 (Casp-1) which, in its turn, promotes IL-33 inactivation. Recent studies have shown associations between NLRP3 variants and inflammatory diseases. However, no study with genes in NLRP3 inflammassome route has been conducted so far with asthma and atopy in any population to date. Blood samples were collected from 1246 asthmatic and non-asthmatic children. Associations were tested for single nucleotide polymorphism (SNP)s in NLRP3 and CASP1 with asthma and markers of atopy and in cultures stimulated with Blomia tropicalis (Bt) mite crude extract. The T allele of rs4925648 (NLRP3) was associated with increased asthma risk (OR 1.50, P = 0.005). In addition, the T allele of rs12130711 polymorphism, whithin the same gene, acted as a protector factor for asthma (OR 0.78, P = 0.038). On the other hand, the C allele of rs4378247 NLRP3 variant was associated with lower levels of IL-13 production when peripheral blood cells were stimulated with Bt (OR 0.39, P = 4E-04). In addition, the greater the number of risk alleles in IL33/NLRP3/CASP1 route the greater was the risk for asthma. The T allele of rs7925706 CASP1 variant was also associated with increased risk for asthma (OR 1.47, P = 0.008). In addition, this same allele increased the eosinophil counts in blood (mm3) in asthmatic individuals compared with non-asthmatic (P = 0.0004). These results suggest that NLRP3 and CASP1 polymorphisms may be associated with susceptibility for asthma and markers of atopy in our population.
Keywords: Asthma, Allergy, Inflammasome, Variants, NLRP3, CASP1
1. Introduction
Asthma is a disease that affect the life of people, being more prevalent in childhood [1]. Blomia tropicalis (Bt) mite rapidly settles in homes of climates countries such as Brazil [2], being the main asthma-induced allergen in the tropics [3], [4], [5], [6]. Atopic asthma is characterized by inflammation of airways, with predominance of Th2-type cytokines, as IL-13, and eosinophilic inflammation [7]. These cytokine may induce airway hyperreactivity via direct effects on several cells that form and make up the airway [8].
IL-33 has the ability to polarize the antigen-driven atopic asthma response [9]. This cytokine is commonly expressed in epithelial cells and acts as one of the signals in response to damage [10], [11]. IL-33 release activates several innate and adaptative immune cells through its membrane receptor (ST2L) and induces Th2-type cytokines, polarizing the inflammation for a type 2 immune response [7], [12].
Interestingly, inflammasomes are a variety of intracellular of multiprotein complexes that can be activated by several stimuli resulting in pathogens invasion through the airway epithelium constantly exposed, playing an important role in both innate and adaptive immune responses. Among the inflammasomes AIM2, IPAF, NLRP1 and NLRP3, this last one has prominent role in various inflammatory diseases [13], [14], [15], [16]. In this context, the airway invasion by potentially antigens becomes an enabling environment for activation of the NLRP3 complex (NLRP3, ASC and Pro-caspase-1), that culminates in the release of Casp-1. Casp-1, in its turn, promotes inactivation of IL-33 [15], [17], [18].
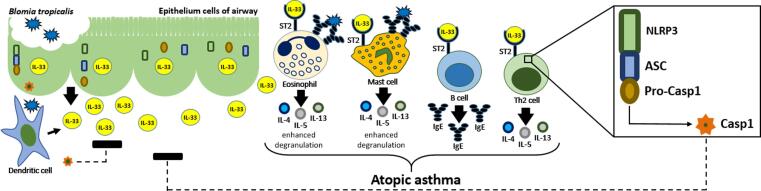
One recent study showed that activation of NLRP3 inflamassome and release of caspase-1 reduced the allergic airway inflammation induced by mite and orchestrated by IL-33 [19]. Fig. 1 shows the immunopathogenesis of Bt-induced atopic asthma and key role of IL-33/NLRP3 inflamassome route activation.
Fig. 1.
Immunopathogenesis of Bt-induced atopic asthma in the IL-33/NLRP3/CASP1 route. The contact of Bt with cells of the aerial epithelium results in damage and consequent release of IL-33. This, in turn, favors the Th2 response profile by activating several innate and adaptive immune cells, with high production of proinflammatory cytokines, also increasing degranulation and specific IgE production. On the other hand, activation of the intracellular NLRP3 receptor, present in the cytoplasm of the cells involved in this response, results in NLRP3 inflammasome formation and Casp-1 release. Casp-1 acts by inhibiting the IL-33 action and, consequently, attenuates the inflammation caused by this cytokine in airway.
The immunopathogenesis of asthma is multi-faceted, involving genetic regulation of gene related to cytokines, transcription factors as well as epigenetic regulators [20], [21]. Moreover, it is known that genetic variations can be differentially distributed according to ethical and geographical origins [22]. Thus, genetic variants in NLRP3/CASP1 genes may affect the susceptibility to asthma and atopy in certain populations [23].
Recent genetic studies have shown polymorphisms in NLRP3 associated with autoimmune and inflammatory diseases [24], such as type 1 diabetes and inflammatory bowel disease [25], [26], primary gout [27], type-2 diabetes mellitus and insulin resistance [28], [29] and ankylosing spondylitis [30]. However, associations between variants in NLRP3 have never been explored before in the context of allergic diseases [24].
Thus, considering the key role of such genes on the inactivation of IL-33 and decreased Th2 immune response, our goal was to verify if genetic variations on NLRP3 and CASP1 genes affect asthma symptoms and allergy, which from the best of our knowledge, was never reported before.
2. Materials and methods
2.1. Study populations and ethical approvals
This is a population-based study where the data presented herein were obtained from 1246 individuals from 5 to 11 years old enrolled in SCAALA Program, in Salvador city, northeastern Brazil [7], [31], [32], [33], [34], [35], as previously described. The complete information of study design can be found elsewhere (Barreto at al 2006). In SCAALA-Salvador cohort, asthma was defined according to the International Study of Allergy and Asthma in Childhood (ISAAC) and only children without any other respiratory and/or chronic condition were assessed. In addition to that, since some consanguineous relatives were included in the original cohort and those individuals genetically identified as relatives (degree of kinship), they were also excluded of this analysis. Also, we excluded individuals with genotyping call rate bellow than 10%. Thus, from originally 1445 individuals from SCAALA-Salvador cohort, a total of 1246 individuals with complete dataset were included in this work. The free and informed consent form as well as the ISAAC phase II questionnaire, was signed and answered by the legal guardian of each children. The Ethics Committee of the Collective Health Institute approved this study (register 003-05/CEP-ISC).
2.2. Asthma definition
We considered asthmatic children as those with information of diagnostic of asthma or wheezing in the past 12 months and at least one of the following clinic conditions: sleep interrupted by the wheezing; wheezing >4 times; wheezing during physical activity. All the other children belonged to the non-asthmatics group.
2.3. Obtention of blood sample and markers of atopy
Heparinized blood was colleted and the serum frozen until the moment of use. The measurement of anti-Bt IgE was performed according to the manufacturer's instructions (Phadia Diagnostics AB, Uppsala, Sweden). Skin prick test (SPT) using extract of Bt was applied on forearm of each subject [7] as described previously. In addition, we have used IL-13 as a marker of Th2 immune response. To this end, whole blood cultures were performed and IL-13 accumulation in culture upon Bt (40 μg/ml) extract stimulation was measured in supernatants [7] as previously described.
2.4. IL-13 levels in whole blood cultures
The concentration of IL-13 in supernatants in pg/mL was performed by sandwich ELISA, following all manufacturer's instructions (BD PharMingen, San Diego, CA, USA). Regarding IL-13 levels, children who presented variation in concentration between the lowest and highest points of the standard curve (62.5/4000) we considered as responders for IL-13 [7], as previously described.
2.5. DNA extraction and genotyping analysis
White blood cells from each children were used for DNA extration, according to the Flexigene@DNA Kit (Qiagen, Hilden, Germany) protocol. For genotyping, we used a BeadChip panel from Illumina with 2.5 Million of variants [7], as described previously. After genotyping, we proceeded with extraction of all SNPs from the region of the two genes of interest, NLRP3 and CASP1. For this, we delimit the start and end positions of both genes as show in assembly GRCh37.p13 as follows: in chromosome 1, of 1,247,579,247 to 247,612,410 on NLRP3 and in chromosome 11, of 104,896,235 to 104,905,884 on CASP1 (ncbi.nlm.nih.gov). After this stage, we obtained a total of 66 SNPs in the NLRP3 gene and 8 SNPs in the CASP1 gene. We then proceed with the application of the following filters for quality control: genotyping error rate ≤ 10%; Hardy-Weinberg equilibrium p < 0.05 and minor allele frequency (MAF) p < 1% [36]. After applying those filters, we obtained 56 markers in NLRP3 and 6 in CASP1, which were included in the analysis. Of these, 17 were statistically significant for NLRP3 and 2 for CASP1. We did not have any significant associations for ASC (data not shown).
2.6. Functional in silico analysis
General information regarding each variant such as position, function and chromosome were accessed in (ncbi.nlm.nih.gov) and 1000 genomes (internationalgenome.org). In addition, we also used the RegulomeDB (http://www.regulomedb.org) which attributes a score of 1 to 6 for each SNP and the lower the score the greater the probability of a certain SNP being involved in regulatory processes. The scores of 1a to 1f, 2a to 2c and 3a to 3b are likely to affect binding. The 4 to 6 scores have less binding evidence [37] and 7 score has no available information. In rSNPBase (http://rSNP.psych.ac.cn/) we accessed information about regulation in terms of proximal, distal and post-transcriptional regulation of the SNPs included in this study.
2.7. Statistical analysis
Analysis of association using logistic regressions by means of multivariated models adjusted by sex, age, helminth infection and individual ancestry (PC1 and PC2 – which classify subjects according to their ethnic characteristics [38], was performed between variants in NLRP3 and CASP1 and asthma, severity of asthma symptoms, positivity to SPT and specific IgE, using PLINK 1.9 software. We used the additive model for all the genetic analyses presented herein. The additive model very well represents the dominant model, and we do not see much advantage in making the recessive model in our population due to the low number of individuals having the rare allele in homozygosis in certain analysis [39], thus we decided to present results using the additive model while both, the recessive and dominant models, are presented in the supplementary material (Tables 1 and 2, supplementary material). In addition, power analyzes were conducted using PGA v 2.0 software [40] considering a Relative Risk (RR) of 1.5, which is consistent with an expected moderate effect size for SNPs associated with complex outcomes, and different genetic models (additive, recessive, and dominant) (see Supplementary Table 3). To create LD plot we use Haploview software. GraphPad 6 software was used for statistical analysis with eosinophil number using Mann–Whitney test. For haplotype analysis we used SNPStats platform (https://www.snpstats.net/start.htm) [41]. For genetic risk score analysis, we used alleles of variants in IL-33/inflammasome NLRP3 route that presented risk for asthma in our population. The A, T and T alleles of rs12551256 (IL33), rs4925648 (NLRP3) and rs7925706 (CASP1), respectively. We assigned a score from 0 to 4 for each individual according to the number of risk alleles and proceeded to logistic regression of each group with risk alleles compared to individuals without any risk alleles. P < 0.05 was considered as significant.
3. Results
The descriptive analysis of 942 controls and 274 asthmatics enrolled in the present study is shown in Table 1. Statistically significant differences were observed for all analyzed phenotypes, with p-value < 0.001. The asthmatic group was younger and had more males. Also, they had high levels of specific IgE for Bt and were more reactive to SPT to Bt and upon Bt stimulation of peripheral blood cells, also produced more IL-13 than non-asthmatics (Table 1).
Table 1.
Population of study and demographic characteristics.
| Children group (1246) |
|||||
|---|---|---|---|---|---|
| Variables | Control | % | Asthma | % | P-value |
| 942* | 75.90% | 274* | 22% | ||
| Age | |||||
| ≤5 | 314 | 33.30% | 132 | 48.20% | <0.001 |
| 6–7 | 337 | 35.80% | 88 | 32.10% | |
| ≥8 | 291 | 30.90% | 54 | 19.70% | |
| Sex | |||||
| Male | 506 | 53.50% | 151 | 55.10% | <0.001 |
| Female | 436 | 46.10% | 123 | 44.90% | |
| Specific IgE for | |||||
| Bt | 288 | 30.60% | 128 | 46.70% | <0.001 |
| SPT for | |||||
| Bt | 192 | 20.40% | 76 | 27.70% | <0.001 |
| Cytokine production by Bt stimulated peripheral blood cells | |||||
| IL-13 | 53 | 5.60% | 18 | 6.60% | <0.001 |
30 subjects presented data miss. For this analysis was used chi-squared test.
Table 2 presents characteristics of all polymorphisms in NLRP3 and CASP1, explored in our study. All SNPs had MAF > 1% and the majority, intron variant. In the same Table 2, the attributable RegulomeDB score is also presented where 2.86% of the studied SNPs were classified as 1f score; 8,57% as 3a; 65,71% as having 4 to 6 and 20% with no information. From now on, only variants with significant associations with the phenotypes explored herein, will be presented bellow.
Table 2.
Characterization of all polymorphisms in NLRP3 and CASP1, included in analysis of study.
| Gene | CHR | SNP | A1 | A2 | MAF | FUNCTION | RegulomeDB |
|---|---|---|---|---|---|---|---|
| NLRP3 | 1 | rs56383829 | T | C | 0.49 | intron variant | 5 |
| NLRP3 | 1 | rs3806265 | C | T | 0.47 | intron variant | 5 |
| NLRP3 | 1 | rs10159239 | G | A | 0.39 | intron variant | 5 |
| NLRP3 | 1 | rs12137901 | C | T | 0.30 | intron variant, upstream variant 2 KB | 4 |
| NLRP3 | 1 | rs12130711 | T | C | 0.28 | intron variant | 1f |
| NLRP3 | 1 | rs4925659 | A | G | 0.28 | intron variant | 4 |
| NLRP3 | 1 | rs10925023 | T | G | 0.28 | intron variant | 7 |
| NLRP3 | 1 | rs200927356 | G | A | 0.27 | intron variant | 5 |
| NLRP3 | 1 | rs72553860 | G | A | 0.26 | intron variant, upstream variant 2 KB | 4 |
| NLRP3 | 1 | rs4925650 | A | G | 0.26 | intron variant | 7 |
| NLRP3 | 1 | rs4378247 | C | T | 0.24 | intron variant | 3a |
| NLRP3 | 1 | rs12565738 | T | C | 0.24 | intron variant | 5 |
| NLRP3 | 1 | rs4925654 | A | G | 0.21 | intron variant | 6 |
| NLRP3 | 1 | rs12728998 | T | C | 0.18 | intron variant | 7 |
| NLRP3 | 1 | rs7525979 | T | C | 0.15 | synonymous codon | 5 |
| NLRP3 | 1 | rs79796552 | G | A | 0.14 | intron variant | 6 |
| NLRP3 | 1 | rs4925648 | T | C | 0.13 | intron variant, upstream variant 2 KB | 7 |
| NLRP3 | 1 | rs10925022 | G | A | 0.12 | intron variant | 5 |
| NLRP3 | 1 | rs36021952 | A | G | 0.11 | intron variant | 3a |
| NLRP3 | 1 | rs72771992 | G | T | 0.11 | utr variant 5 prime | 7 |
| NLRP3 | 1 | rs74154644 | T | C | 0.11 | intron variant | 5 |
| NLRP3 | 1 | rs45624634 | A | C | 0.11 | intron variant | 4 |
| NLRP3 | 1 | rs4925651 | T | G | 0.10 | intron variant | 5 |
| NLRP3 | 1 | rs4925543 | A | G | 0.10 | synonymous codon | 5 |
| NLRP3 | 1 | rs7524914 | C | T | 0.09 | intron variant | 6 |
| NLRP3 | 1 | rs73136263 | T | G | 0.03 | utr variant 5 prime | 7 |
| NLRP3 | 1 | rs55914518 | A | G | 0.02 | intron variant | 6 |
| NLRP3 | 1 | rs35433972 | T | C | 0.02 | intron variant | 5 |
| NLRP3 | 1 | rs116096527 | A | G | 0.01 | intron variant | 5 |
| CASP1 | 11 | rs79180143 | A | C | 0.28 | intron variant, utr variant 3 prime | 7 |
| CASP1 | 11 | rs531542 | A | G | 0.14 | intron variant | 3a |
| CASP1 | 11 | rs580253 | A | G | 0.14 | intron variant, synonymous codon | 7 |
| CASP1 | 11 | rs7925706 | T | C | 0.13 | intron variant, utr variant 5 prime | 4 |
| CASP1 | 11 | rs556205 | G | T | 0.08 | intron variant | 6 |
| CASP1 | 11 | rs75775137 | G | A | 0.06 | intron variant | 5 |
CHR, chromosome; SNP, single nucleotide polymorphism; A1, minor allele; A2, ancestral allele; MAF, minor allele frequency; utr, untranslated region.
Polymorphic alleles of variants in NLRP3 and CASP1 showed significant associations with asthma, asthma severity, IL-13 levels, specific IgE and SPT for Bt (see Table 3). The majority of NLRP3 and CASP1 SNPs were associated with protection rather than with risk for asthma and its severity, as well as, atopy markers. The T, A, G, T and T alleles of rs12130711, rs36021952, rs72771992, rs56383829 and rs74154644, respectively, were negatively associated with asthma (OR 0.78, 95% CI: 0.62–0.98, P = 0.038), (OR 0.69, 95% CI: 0.49–0.98, P = 0.038), (OR 0.67, 95% CI: 0.48–0.94, P = 0.019), (OR 0.82, 95% CI: 0.67–0.99, P = 0.046) and (OR 0.69, 95% CI: 0.49–0.98, P = 0.038). The A and T alleles of rs4925654 and rs12728998 were associated with mild asthma symptoms (OR 0.62, 95% CI: 0.44–0.88, P = 0.009) and (OR 0.68, 95% CI: 0.48–0.98, P = 0.042), respectively. Also the C, T, A and T alleles of rs4378247, rs12565738, rs45624634, and rs7525979, respectively, were negatively associated with IL-13 production (OR 0.39, 95% CI: 0.22–0.67, P = 0.0004), (OR 0.24, 95% CI: 0.09–0.65, P = 0.004), (OR 0.41, 95% CI: 0.24–0.70, P = 0.001) and (OR 0.38, 95% CI: 0.19–0.75, P = 0.006) when peripheral blood cells were stimulated with Bt. In addition, the A allele of rs79180143 was associated with a lower SPT reactivity (OR 0.79, 95% CI: 0.63–0.99, P = 0.046) against the Bt crude extract. On the other hand, the C, T, G, A and T alleles of polymorphisms rs12137901, rs4925648, rs72553860, rs55914518 and rs7925706, respectively, were positively associated with asthma (OR 1.28, 95% CI: 1.04–1.57, P = 0.016), (OR 1.50, 95% CI: 1.14–1.98, P = 0.005), (OR 1.38, 95% CI: 1.11–1.72, P = 0.004), (OR 1.85, 95% CI: 1.04–3.29, P = 0.034) and (OR 1.47, 95% CI: 1.11–1.96, P = 0.008). Also, both A alleles of rs116096527 and rs4925659 were positively associated with sIgE levels (OR 2.25, 95% CI: 1.13–4.48, P = 0.043) and SPT positivity (OR 1.30, 95% CI: 1.05–1.61, P = 0.021), both against Bt whole extract.
Table 3.
Significant associations between variants on NLRP3 and CASP1 with asthma, severity asthma symptoms, IL-13 production, anti-Bt IgE and SPT for Bt.
| Gene | SNP | Model | Geno | Aff | Unaff | ORa | CI 95% | P-value* |
|---|---|---|---|---|---|---|---|---|
| Asthma | ||||||||
| NLRP3 | rs12137901 | Additive | C/C | 33 | 79 | 1.28 | 1.04–1.57 | 0.016 |
| C/T | 111 | 359 | ||||||
| T/T | 116 | 457 | ||||||
| NLRP3 | rs4925648 | Additive | T/T | 11 | 11 | 1.5 | 1.14–1.98 | 0.005 |
| T/C | 63 | 183 | ||||||
| C/C | 186 | 696 | ||||||
| NLRP3 | rs72553860 | Additive | G/G | 28 | 51 | 1.38 | 1.11–1.72 | 0.004 |
| G/A | 104 | 335 | ||||||
| A/A | 129 | 509 | ||||||
| NLRP3 | rs55914518 | Additive | A/A | 1 | 0 | 1.85 | 1.04–3.29 | 0.034 |
| A/G | 17 | 35 | ||||||
| G/G | 243 | 860 | ||||||
| NLRP3 | rs12130711 | Additive | T/T | 12 | 75 | 0.78 | 0.62–0.98 | 0.038 |
| T/C | 101 | 369 | ||||||
| C/C | 148 | 451 | ||||||
| NLRP3 | rs36021952 | Additive | A/A | 2 | 12 | 0.69 | 0.49–0.98 | 0.038 |
| A/G | 42 | 190 | ||||||
| G/G | 216 | 688 | ||||||
| NLRP3 | rs72771992 | Additive | G/G | 3 | 18 | 0.67 | 0.48–0.94 | 0.019 |
| G/T | 37 | 174 | ||||||
| T/T | 219 | 697 | ||||||
| NLRP3 | rs56383829 | Additive | T/T | 52 | 236 | 0.82 | 0.67–0.99 | 0.046 |
| T/C | 134 | 436 | ||||||
| C/C | 75 | 223 | ||||||
| NLRP3 | rs74154644 | Additive | T/T | 2 | 12 | 0.69 | 0.49–0.98 | 0.038 |
| T/C | 42 | 190 | ||||||
| C/C | 217 | 692 | ||||||
| CASP1 | rs7925706 | Additive | T/T | 8 | 9 | 1.47 | 1.11–1.96 | 0.008 |
| T/C | 65 | 185 | ||||||
| C/C | 177 | 675 | ||||||
| Severity asthma symptoms | ||||||||
| NLRP3 | rs4925654 | Additive | A/A | 3 | 45 | 0.62 | 0.44–0.88 | 0.009 |
| A/G | 37 | 357 | ||||||
| G/G | 101 | 613 | ||||||
| NLRP3 | rs12728998 | Additive | T/T | 2 | 34 | 0.68 | 0.48–0.98 | 0.042 |
| T/C | 35 | 321 | ||||||
| C/C | 104 | 660 | ||||||
| IL-13 production in Blomia tropicalis-stimulated blood cell cultures | ||||||||
| NLRP3 | rs4378247 | Additive | C/C | 0 | 59 | 0.39 | 0.22–0.67 | 0.0004 |
| C/T | 15 | 394 | ||||||
| T/T | 50 | 570 | ||||||
| NLRP3 | rs12565738 | Additive | T/T | 1 | 67 | 0.41 | 0.24–0.70 | 0.001 |
| T/C | 14 | 385 | ||||||
| C/C | 53 | 588 | ||||||
| NLRP3 | rs45624634 | Additive | A/A | 0 | 10 | 0.24 | 0.09–0.65 | 0.004 |
| A/C | 4 | 209 | ||||||
| C/C | 64 | 821 | ||||||
| NLRP3 | rs7525979 | Additive | T/T | 0 | 33 | 0.38 | 0.19–0.75 | 0.006 |
| T/C | 9 | 268 | ||||||
| C/C | 59 | 739 | ||||||
| Anti-Blomia tropicalis IgE | ||||||||
| NLRP3 | rs116096527 | Additive | A/A | 0 | 2 | 2.25 | 1.13–4.48 | 0.043 |
| A/G | 17 | 10 | ||||||
| G/G | 383 | 756 | ||||||
| SPT for Blomia tropicalis | ||||||||
| NLRP3 | rs4925659 | Additive | A/A | 25 | 69 | 1.3 | 1.05–1.61 | 0.021 |
| A/G | 117 | 334 | ||||||
| G/G | 118 | 495 | ||||||
| CASP1 | rs79180143 | Additive | A/A | 21 | 74 | 0.79 | 0.63–0.99 | 0.046 |
| A/C | 81 | 355 | ||||||
| C/C | 154 | 446 | ||||||
Geno, genotype; Aff, affected; Unaff, no affected; OR, odds ratio; CI, confidence interval.
Adjusted by gender, age, helminth infection, ancestry markers.
P-value corrected.
Table 4 presents polymorphisms in NLRP3 and CASP1 and data on the impact of such SNPs in gene expression based in experimental studies, according to rSNPBase. Regarding variants in NLRP3, the SNPs rs12137901, rs4925659, rs72553860, rs4925648, rs36021952, rs72771992, rs74154644 were related with proximal regulation only, while the rs56383829 was related with distal regulation. Regarding variants in CASP1, the SNPs rs79180143 and rs7925706 were related with proximal regulation and with RNA binding protein-mediated regulation. We performed haplotypes analysis in NLRP3 gene. The following haplotypes for SNPs rs72553860, rs12137901, rs36021952 and rs74154644 have shown significant associations with asthma (H1-GCGC; OR 1.43, 95% CI: 1.09–1.87, P = 0.011); (H2-GCC; OR 1.39, 95% CI: 1.09–1.77, P = 0.008); (H3-GCG; OR 1.38, 95% CI: 1.08–1.76, P = 0.009); (H4-GC; OR 1.37, 95% CI: 1.10–1.70, P = 0.005) (see Table 5). On the other hand, the following SNPs rs45624634, rs12565738, rs7525979 and rs4378247 were associated with IL-13 production in Bt-stimulated blood cell cultures in haplotype analysis (H1-CTC; OR 0.53, 95% CI: 0.29–0.98, P = 0.042); (H2-CCT; OR 0.41, 95% CI: 0.21–0.81, P = 0.011); (H3-ATC; OR 0.26, 95% CI: 0.09–0.71, P = 0.009); (H4-CCTT; OR 0.13, 95% CI: 0.03–0.56, P = 0.006); (H5-ATCT; OR 0.11, 95% CI: 0.01–0.80, P = 0.030) (see Table 5).
Table 4.
Possible mechanisms where by SNPs on NLRP3 and CASP1 may affect these genes expression, according to rSNPBase.
| Gene | CHR | SNP | PR | DR | RNA BPMR |
|---|---|---|---|---|---|
| NLRP3 | 1 | rs12137901 | Yes | No | No |
| NLRP3 | 1 | rs4925659 | Yes | No | No |
| NLRP3 | 1 | rs72553860 | Yes | No | No |
| NLRP3 | 1 | rs4925648 | Yes | No | No |
| NLRP3 | 1 | rs36021952 | Yes | No | No |
| NLRP3 | 1 | rs72771992 | Yes | No | No |
| NLRP3 | 1 | rs74154644 | Yes | No | No |
| NLRP3 | 1 | rs56383829 | No | Yes | No |
| CASP1 | 11 | rs79180143 | Yes | No | Yes |
| CASP1 | 11 | rs7925706 | Yes | No | Yes |
CHR, chromosome; SNP, single nucleotide polymorphism; PR, Proximal regulation; DR, Distal regulation; BPMR, binding protein mediated regulation.
Table 5.
Haplotype analysis between rs72553860, rs12137901, rs36021952 and rs74154644 on asthma risk and between rs45624634, rs12565738, rs7525979 and rs4378247 in decreasing IL-13 production Bt-stimulated, both in NLRP3 gene.
| Frequencies |
||||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype | rs72553860 | rs12137901 | rs36021952 | rs74154644 | Case | Control | Odds Ratioa/95% Confidence interval | P-value |
| Reference | A | T | G | C | 0.57 | 0.55 | 1 | – |
| Haplotype association with risk for asthma | ||||||||
| 1 asthma | G | C | G | C | 0.19 | 0.25 | 1.43 (1.09–1.87) | 0.011 |
| 2 asthma | G | C | – | C | 0.23 | 0.21 | 1.39 (1.09–1.77) | 0.008 |
| 3 asthma | G | C | G | – | 0.23 | 0.22 | 1.38 (1.08–1.76) | 0.009 |
| 4 asthma | G | C | – | – | 0.26 | 0.24 | 1.37 (1.10–1.70) | 0.005 |
| Happlotype | rs45624634 | rs12565738 | rs7525979 | rs4378247 | Case | Control | Odds Ratioa/95% Confidence interval | P-value |
| Reference | C | C | C | T | 0.48 | 0.47 | 1 | – |
| Haplotype association with IL-13 | ||||||||
| 1 IL-13 | C | T | C | – | 0.12 | 0.13 | 0.53 (0.29–0.98) | 0.042 |
| 2 IL-13 | C | C | T | – | 0.12 | 0.13 | 0.41 (0.21–0.81) | 0.011 |
| 3 IL-13 | A | T | C | – | 0.08 | 0.09 | 0.26 (0.09–0.71) | 0.009 |
| 4 IL-13 | C | C | T | T | 0.09 | 0.1 | 0.13 (0.03–0.56) | 0.006 |
| 5 IL-13 | A | T | C | T | 0.06 | 0.07 | 0.11 (0.01–0.80) | 0.030 |
Adjusted for confounding variables.
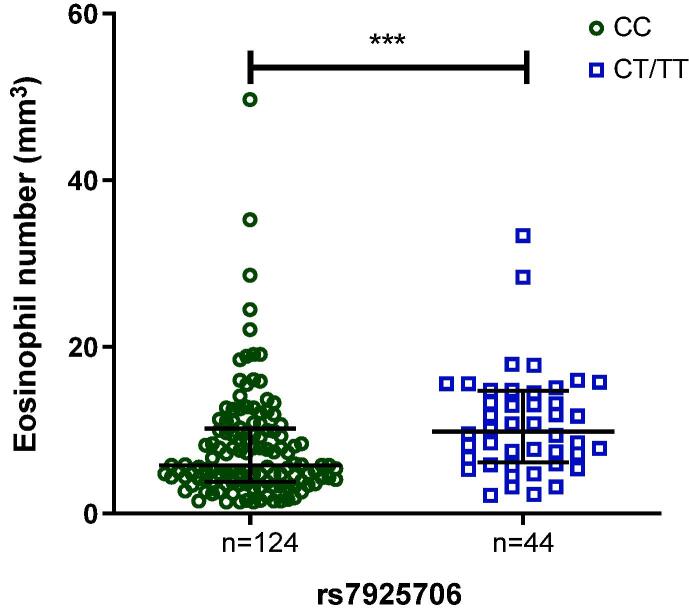
In a previous study with this same population, the G allele of rs12551256 (IL33) was associated with protection to asthma [7]. Therefore, the most common allele for this particular SNP (the A allele) in our population was associated to asthma risk (OR 1.41, 95% CI: 1.06–1.88, P = 0.017). In order to perform a risk score analysis [42], we analyzed the A allele of rs12551256 (IL33) along with the T allele of rs4925648 (NLRP3) and the allele and T allele of rs7925706 (CASP1) to check if increasing the number of risk allele, the greater is the risk for asthma (see Table 6). And indeed, we were able to confirm that hypothesis where the greater the number of risk alleles in those genes all together, the greater is the risk for asthma in our population. So as can be seen in Table 6, subjects with one risk allele had more than the double of risk for asthma. In the individuals with two or three risk alleles, the risk for asthma is considerable increased. The maximum risk was found in individuals with four risk alleles which represented a risk five times greater for asthma in relation to the reference group (which subjects have no risk alleles). In addition, when we looked for the effect of variants in both NLRP3 and CASP1 on eosinophils in blood from asthmatics patients, only rs7925706 variant from CASP1 was able to modulate eosinophil counts in blood from asthmatics subjects (Fig. 3).
Table 6.
Genetic risk score with the A allele of rs12551256 (IL33) + T allele of rs4925648 (NLRP3) + T allele of rs7925706 (CASP1) both association with risk for asthma.
| Asthma |
||||
|---|---|---|---|---|
| Risk allele | Controls | Cases | Odds Ratioa/95% Confidence interval | P-value |
| 0 | 55 (17.6%) | 6 (8%) | 1.00 | – |
| 1 | 257 (82.4%) | 69 (92%) | 2.47 (1.01–6.04) | 0.029 |
| 2 | 368 (87%) | 113 (95%) | 3.00 (1.24–7.24) | 0.006 |
| 3 | 150 (73.2%) | 50 (89.3%) | 3.15 (1.27–7.86) | 0.006 |
| 4 | 32 (36.8%) | 16 (72.7%) | 5.77 (1.89–17.60) | 9E-04 |
Adjusted for confounding variables.
Fig. 3.
Eosinophil number per mm3 in blood of asthmatic subjects according to genotype of CASP1 SNP rs7925706. Bars represent the median of eosinophils number according with each group genotyp, represented by its holders. CC: median = 5.800 and CT/TT: median = 9.850. The analyis of the data was by Manny-Whitney test. ***P < 0.001.
4. Discussion
NLRP3 receptor is one of the proteins that make up the multiprotein complex that forms the NLRP3 inflammasome, that in their state of activation, results in activation of Casp-1. This protein complex also acts as a key role in both innate and adaptive immunities. In addition, in Th2 cells from mice, NLRP3 inflammasome positively regulates Th2 programming, promoting Th2 differentiation as a transcription factor [14], [43].
A model of mice-induced pulmonary inflammation in NLRP3 and CASP1-deficient showed an increased airway inflammation, with influx of eosinophils, Th2 cytokines profile and IL-33, thus, demonstrating that the Th2 response generated by IL-33 was regulated by Casp-1. Casp-1 activation via NLRP3 inflammasome leading to, downregulation of IL-33 and, thus, Th2 response [19].
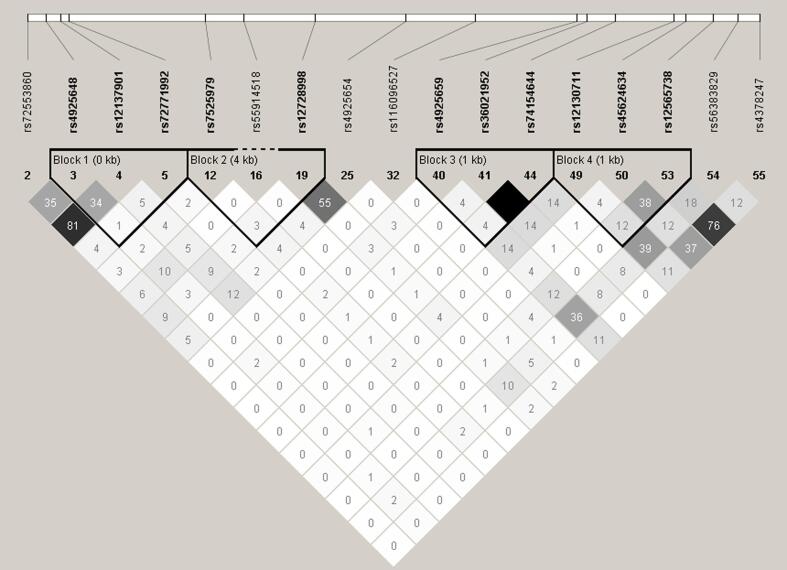
In this present study, we reported several polymorphisms in NLRP3 and CASP1 never described before to be associated to asthma and Bt-induced allergy. Regarding variants in NLRP3, the G, C and T alleles of rs72553860, rs12137901, and rs4925648, respectively, and the T allele of CASP1 SNP rs7925706 were associated with susceptibility to asthma. The first two SNPs (rs72553860 and rs12137901) are in high LD (r2 = 0.81) (Fig. 2). The CASP1 polymorphism, rs7925706 (T allele), besides of being associated with asthma was also associated with an increased number of eosinophils (mm3) in blood among asthmatic subjects (see Fig. 3).
Fig. 2.
LD plot of significance analysis of 17 SNPs on NLRP3, analyzed in our population. Top bar indicates the physical location of each variant. The colors of each square vary according to the degree of LD, being the total equilibrium in color white with value of r2 equal to 0 and total unbalance in color black with value of r2 equal to one. The different shades of gray show an intermediate imbalance with a value of r2 > 0 and < 0.8. This analysis was perfomed with Haploview software.
In the same direction, the A and A alleles of rs4925659 and rs116096527 implied in increased Bt-specific IgE and SPT reactivity. Thus, we hypothesize that a greater release of IL-33 in response to Bt in the airways may occur, contributing to Th2 inflammation.
NLRP3 acts as a transcription factor in T helper cells, polarizing them to a Th2 profile [14]. Thus, NLRP3 inflammasome and Casp-1 may be activated upon activation of IL-33/ST2 pathway. Although we cannot confirm that based on our findings, our hypothesis is that SNPs in NLRP3/CASP1 pathway may lead to a low binding of transcription factor inducing a lower expression of the NLRP3 and CASP1. Decreased activation such genes, results in lower inactivation of IL-33 and thus larger Th2-inflammation with increased release of markers of allergy such as IgE and IL-13 studied herein and increasing the risk for asthma.
Recent studies have shown that IL-33 cytokine is the key element that links early events after lung epithelial cells contact with the allergen and Th2 response, resulting on asthma [9], [44]. An experimental mice model receiving CD4+ effector Th2 cells, exposed to Bt antigens have developed lung eosinophilia, hyperplasia of smooth muscle cells and goblet cell, as well as overproduction of IgE and mucus production through IL-13 [6].
On the other hand, regarding variants in NLRP3, the A, T, T, G and T alleles of rs36021952, rs74154644, rs12130711, rs72771992 and rs56383829 respectively, were protector factors for asthma in our population. The first and the second SNPs (rs36021952 and rs74154644) are in total LD (r2 = 1) (see Fig. 2). Both the A and T alleles of NLRP3 polymorphisms rs4925654 and rs12728998 respectively, were associated with decreasing severity of asthma symptoms. The C, T and A alleles of NLRP3 SNPs rs4378247, rs12565738, rs45624634 and rs7525979 respectively, were negatively associated with IL-13 production in peripheral blood cells when stimulated with Bt and whereas the first two SNPs (rs4378247 and rs12565738) are in moderate LD (r2 = 0.76) (see Fig. 2). Carriers of A allele of CASP1 variant rs79180143 were less likely to have a positivity SPT reaction for Bt.
Interestingly, in the present study most of the observed associations between analyzed SNPs and the investigated outcomes were related to protection to Bt-induced allergy/asthma. In other words, the most common alleles for these SNPs in our population were risk alleles. These results are in some way in agreement with epidemiological findings that demonstrate high prevalence of asthma and other allergic conditions in our population [34], [45], [46]. However, these are complex diseases and likely influenced by dozens or hundreds of genes [47], [48]. Thus, these results need to be interpreted with caution, so that they do not lead to mistaken conclusions regarding the genetic predisposition of our population to these conditions.
For the NLRP3 gene, the nine SNPs originally identified as associated with asthma symptoms were reduced to four independent regions after a clumping analysis, two of which were positively associated and the other two negatively associated with asthma outcome (see Supplementary Table 4). This suggests that different variants within this gene may have antagonistic effects in relation to the risk of asthma, which is a matter for future investigation.
We also conducted a haplotype analysis in the NLRP3 gene for asthma and IL-13 production. We showed that haplotypes with two, three or four alleles increased risk of asthma (see Table 5). On the other hand, we also have shown a high protection for IL-13 production upon Bt stimulation in addition of three or four alleles in another haplotype described here (see Table 5). No significant result was obtained after the haplotypes analysis in CASP1 (data not shown). In addition, we performed a polygenic risk score with A allele of IL33 rs12551256 variant previously described [7], as well as T and T alleles of NLRP3 rs4925648 and CASP1 rs7925706 polymorphisms, respectively, both risk alleles described here for asthma, and again, the greater the number of the risk alleles in IL33/NLRP3/CASP1 route, the greater the risk for asthma (see Table 6). No previous work was found in literature taking into account variants occurring together in such genes for asthma or allergy.
5. Conclusion
In summary, our findings demonstrate associations between polymorphisms in NLRP3 inflammassome pathway with asthma and allergy and that variants in IL33/NLRP3/CASP1 route, together, increase asthma risk. We also have shown the effect of CASP1 variant rs7925706 (T allele) in eosinophil counts in blood among asthmatic individuals. Our data reinforce a critical role of NLRP3 and CASP1 on asthma and allergic inflammation. Additional studies are need to better understand the role and the impact of such polymorphisms on asthma and allergy as means of potential targets for therapeutical intervetions.
CRediT authorship contribution statement
Gerson A. Queiroz: Conceptualization, Methodology, Validation, Formal analysis, Writing - original draft. Raimon R. Silva: Validation, Writing - original draft. Anaque O. Pires: Validation, Writing - original draft. Ryan S. Costa: Writing - review & editing. Neuza M. Alcântara-Neves: Resources, Writing - review & editing. Thiago M. Silva: Formal analysis, Validation. Mauricio L. Barreto: Supervision, Resources, Writing - review & editing. Sergio C. Oliveira: Writing - review & editing. Camila A. Figueirêdo: Supervision, Resources, Writing - review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We thank the SCAALA (Social Change of Asthma and Allergy in Latin America) cohort for making this study possible.
Ethics
The Ethics Committee of Federal University of Bahia approved this study (register 003-05/CEP-ISC).
Financial support
Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001, National Research Council, Brazil (CNPq) and Foundation for Research Support of the State of Bahia (FAPESB). This study was also funded by Wellcome Trust, United Kingdom (Grant no. 072405/Z/03/Z).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cytox.2020.100032.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sole D., Aranda C.S., Wandalsen G.F. Asthma: epidemiology of disease control in Latin America – short review. Asthma Res. Pract. 2017;3:4. doi: 10.1186/s40733-017-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guilleminault L., Viala-Gastan C. Blomia tropicalis: a house dust mite in the tropics. Rev. Mal. Respir. 2017 doi: 10.1016/j.rmr.2016.10.877. [DOI] [PubMed] [Google Scholar]
- 3.Arruda L.K., Rizzo M.C., Chapman M.D., Fernandez-Caldas E., Baggio D., Platts-Mills T.A. Exposure and sensitization to dust mite allergens among asthmatic children in Sao Paulo, Brazil. Clin. Exp. Allergy. 1991;21(4):433–439. doi: 10.1111/j.1365-2222.1991.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 4.Serravalle KaM M., Jr. Ácaros na poeira domiciliar na cidade de Salvador-Bahia. Rev. Bras. Alerg. Imunopatol. 1999;22:19–24. [Google Scholar]
- 5.Sopelete M.C., Silva D.A., Arruda L.K., Chapman M.D., Taketomi E.A. Dermatophagoides farinae (Der f 1) and Dermatophagoides pteronyssinus (Der p 1) allergen exposure among subjects living in Uberlandia, Brazil. Int. Arch. Allergy Immunol. 2000;122(4):257–263. doi: 10.1159/000024407. [DOI] [PubMed] [Google Scholar]
- 6.Chua Y.L., Liong K.H., Huang C.H., Wong H.S., Zhou Q., Ler S.S. Blomia tropicalis-specific TCR transgenic Th2 cells induce inducible BALT and severe asthma in mice by an IL-4/IL-13-dependent mechanism. J. Immunol. 2016;197(10):3771–3781. doi: 10.4049/jimmunol.1502676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Queiroz G.A., Costa R.S., Alcantara-Neves N.M., de Oliveira Nunes, Costa G., Barreto M.L., Carneiro V.L. IL33 and IL1RL1 variants are associated with asthma and atopy in a Brazilian population. Int. J. Immunogenet. 2017;44(2):51–61. doi: 10.1111/iji.12306. [DOI] [PubMed] [Google Scholar]
- 8.Gour N., Wills-Karp M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 2015;75(1):68–78. doi: 10.1016/j.cyto.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sjoberg L.C., Nilsson A.Z., Lei Y., Gregory J.A., Adner M., Nilsson G.P. Interleukin 33 exacerbates antigen driven airway hyperresponsiveness, inflammation and remodeling in a mouse model of asthma. Sci. Rep. 2017;7(1):4219. doi: 10.1038/s41598-017-03674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prefontaine D., Lajoie-Kadoch S., Foley S., Audusseau S., Olivenstein R., Halayko A.J. Increased expression of IL-33 in severe asthma: evidence of expression by airway smooth muscle cells. J. Immunol. 2009;183(8):5094–5103. doi: 10.4049/jimmunol.0802387. [DOI] [PubMed] [Google Scholar]
- 11.Prefontaine D., Nadigel J., Chouiali F., Audusseau S., Semlali A., Chakir J. Increased IL-33 expression by epithelial cells in bronchial asthma. J. Allergy Clin. Immunol. 2010;125(3):752–754. doi: 10.1016/j.jaci.2009.12.935. [DOI] [PubMed] [Google Scholar]
- 12.Kim H.Y., Chang Y.J., Subramanian S., Lee H.H., Albacker L.A., Matangkasombut P. Innate lymphoid cells responding to IL-33 mediate airway hyperreactivity independently of adaptive immunity. J. Allergy Clin. Immunol. 2012;129(1) doi: 10.1016/j.jaci.2011.10.036. pp. 216–27 e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cassel S.L., Joly S., Sutterwala F.S. The NLRP3 inflammasome: a sensor of immune danger signals. Semin. Immunol. 2009;21(4):194–198. doi: 10.1016/j.smim.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruchard M., Rebe C., Derangere V., Togbe D., Ryffel B., Boidot R. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat. Immunol. 2015;16(8):859–870. doi: 10.1038/ni.3202. [DOI] [PubMed] [Google Scholar]
- 15.Eltom S., Belvisi M.G., Stevenson C.S., Maher S.A., Dubuis E., Fitzgerald K.A. Role of the inflammasome-caspase1/11-IL-1/18 axis in cigarette smoke driven airway inflammation: an insight into the pathogenesis of COPD. PLoS ONE. 2014;9(11):e112829. doi: 10.1371/journal.pone.0112829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santana P.T., Martel J., Lai H.C., Perfettini J.L., Kanellopoulos J.M., Young J.D. Is the inflammasome relevant for epithelial cell function? Microbes Infect. 2016;18(2):93–101. doi: 10.1016/j.micinf.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Cayrol C., Girard J.P. The IL-1-like cytokine IL-33 is inactivated after maturation by caspase-1. Proc. Natl. Acad. Sci. U.S.A. 2009;106(22):9021–9026. doi: 10.1073/pnas.0812690106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao W., Hu Z. The enigmatic processing and secretion of interleukin-33. Cell. Mol. Immunol. 2010;7(4):260–262. doi: 10.1038/cmi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madouri F., Guillou N., Fauconnier L., Marchiol T., Rouxel N., Chenuet P. Caspase-1 activation by NLRP3 inflammasome dampens IL-33-dependent house dust mite-induced allergic lung inflammation. J. Mol. Cell. Biol. 2015;7(4):351–365. doi: 10.1093/jmcb/mjv012. [DOI] [PubMed] [Google Scholar]
- 20.Tumes D.J., Papadopoulos M., Endo Y., Onodera A., Hirahara K., Nakayama T. Epigenetic regulation of T-helper cell differentiation, memory, and plasticity in allergic asthma. Immunol. Rev. 2017;278(1):8–19. doi: 10.1111/imr.12560. [DOI] [PubMed] [Google Scholar]
- 21.Russell R.J., Brightling C. Pathogenesis of asthma: implications for precision medicine. Clin. Sci. (Lond). 2017;131(14):1723–1735. doi: 10.1042/CS20160253. [DOI] [PubMed] [Google Scholar]
- 22.Tan M.S., Yu J.T., Jiang T., Zhu X.C., Wang H.F., Zhang W. NLRP3 polymorphisms are associated with late-onset Alzheimer's disease in Han Chinese. J. Neuroimmunol. 2013;265(1–2):91–95. doi: 10.1016/j.jneuroim.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Lamkanfi M., Dixit V.M. Inflammasomes and their roles in health and disease. Annu. Rev. Cell Dev. Biol. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- 24.Hitomi Y., Ebisawa M., Tomikawa M., Imai T., Komata T., Hirota T. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J. Allergy Clin. Immunol. 2009;124(4) doi: 10.1016/j.jaci.2009.07.044. pp. 779–85 e6. [DOI] [PubMed] [Google Scholar]
- 25.Miao Z., Li C., Chen Y., Zhao S., Wang Y., Wang Z. Dietary and lifestyle changes associated with high prevalence of hyperuricemia and gout in the Shandong coastal cities of Eastern China. J. Rheumatol. 2008;35(9):1859–1864. [PubMed] [Google Scholar]
- 26.Martinon F. Mechanisms of uric acid crystal-mediated autoinflammation. Immunol. Rev. 2010;233(1):218–232. doi: 10.1111/j.0105-2896.2009.00860.x. [DOI] [PubMed] [Google Scholar]
- 27.Meng D.M., Zhou Y.J., Wang L., Ren W., Cui L.L., Han L. Polymorphisms in the NLRP3 gene and risk of primary gouty arthritis. Mol. Med. Rep. 2013;7(6):1761–1766. doi: 10.3892/mmr.2013.1429. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y., Zhang D., Zhang L., Fu M., Zeng Y., Russell R. Variants of NLRP3 gene are associated with insulin resistance in Chinese Han population with type-2 diabetes. Gene. 2013;530(1):151–154. doi: 10.1016/j.gene.2013.07.082. [DOI] [PubMed] [Google Scholar]
- 29.Wang S., Fang F., Jin W.B., Wang X., Zheng X.S. Investigation into the association between NLRP3 gene polymorphisms and susceptibility to type 2 diabetes mellitus. Genet. Mol. Res. 2015;14(4):17447–17452. doi: 10.4238/2015.December.21.15. [DOI] [PubMed] [Google Scholar]
- 30.Zhao S., Chen H., Wu G., Zhao C. The association of NLRP3 and TNFRSF1A polymorphisms with risk of ankylosing spondylitis and treatment efficacy of etanercept. J. Clin. Lab. Anal. 2017 doi: 10.1002/jcla.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Figueiredo C.A., Barreto M.L., Rodrigues L.C., Cooper P.J., Silva N.B., Amorim L.D. Chronic intestinal helminth infections are associated with immune hyporesponsiveness and induction of a regulatory network. Infect. Immun. 2010;78(7):3160–3167. doi: 10.1128/IAI.01228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figueiredo C.A., Alcantara-Neves N.M., Amorim L.D., Silva N.B., de Carvalho L.C.P., Cooper P.J. Evidence for a modulatory effect of IL-10 on both Th1 and Th2 cytokine production: the role of the environment. Clin. Immunol. 2011;139(1):57–64. doi: 10.1016/j.clim.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barreto M.L., Cunha S.S., Alcantara-Neves N., Carvalho L.P., Cruz A.A., Stein R.T. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study) BMC Pulm Med. 2006;6:15. doi: 10.1186/1471-2466-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcantara-Neves N.M., de S.G.B.G., Veiga R.V., Figueiredo C.A., Fiaccone R.L., da Conceicao J.S. Effects of helminth co-infections on atopy, asthma and cytokine production in children living in a poor urban area in Latin America. BMC Res. Notes. 2014;7:817. doi: 10.1186/1756-0500-7-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lima L.C., Queiroz G.A., Costa R.D.S., Alcantara-Neves N.M., Marques C.R., Costa G.N.O. Genetic variants in RORA are associated with asthma and allergy markers in an admixed population. Cytokine. 2019;113:177–184. doi: 10.1016/j.cyto.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34(6):591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pires A.O., Queiroz G.A., de Jesus Silva M, da Silva R.R., da Silva H.B.F., Carneiro N.V.Q. Polymorphisms in the DAD1 and OXA1L genes are associated with asthma and atopy in a South American population. Mol. Immunol. 2018;101:294–302. doi: 10.1016/j.molimm.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Lima-Costa M.F., Rodrigues L.C., Barreto M.L., Gouveia M., Horta B.L., Mambrini J. Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative) Sci. Rep. 2015;5 doi: 10.1038/srep09812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lettre G., Lange C., Hirschhorn J.N. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet. Epidemiol. 2007;31(4):358–362. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- 40.Menashe I., Rosenberg P.S., Chen B.E. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sole X., Guino E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 42.Conran C.A., Na R., Chen H., Jiang D., Lin X., Zheng S.L. Population-standardized genetic risk score: the SNP-based method of choice for inherited risk assessment of prostate cancer. Asian J. Androl. 2016;18(4):520–524. doi: 10.4103/1008-682X.179527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C.S., Shin D.M., Jo E.K. The role of NLR-related protein 3 inflammasome in host defense and inflammatory diseases. Int Neurourol J. 2012;16(1):2–12. doi: 10.5213/inj.2012.16.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston L.K., Bryce P.J. Understanding interleukin 33 and its roles in eosinophil development. Front Med. (Lausanne) 2017;4:51. doi: 10.3389/fmed.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigues L.C., Newcombe P.J., Cunha S.S., Alcantara-Neves N.M., Genser B., Cruz A.A. Early infection with Trichuris trichiura and allergen skin test reactivity in later childhood. Clin. Exp. Allergy. 2008;38(11):1769–1777. doi: 10.1111/j.1365-2222.2008.03027.x. [DOI] [PubMed] [Google Scholar]
- 46.Figueiredo C.A., Amorim L.D., Alcantara-Neves N.M., Matos S.M.A., Cooper P.J., Rodrigues L.C. Environmental conditions, immunologic phenotypes, atopy, and asthma: new evidence of how the hygiene hypothesis operates in Latin America. J. Allergy Clin. Immunol. 2013;131(4) doi: 10.1016/j.jaci.2013.01.016. pp. 1064–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morales E., Duffy D. Genetics and gene-environment interactions in childhood and adult onset asthma. Front. Pediatr. 2019;7:499. doi: 10.3389/fped.2019.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mammen J.R., Arcoleo K. Understanding the genetics of asthma and implications for clinical practice. J Am Assoc Nurse Pract. 2019;31(7):384–387. doi: 10.1097/JXX.0000000000000246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.