SUMMARY
There is an unmet clinical need for improved tissue and liquid biopsy tools for cancer detection. We investigated the proteomic profile of extracellular vesicles and particles (EVPs) in 426 human samples from tissue explants (TEs), plasma, and other bodily fluids. Among traditional exosome markers, CD9, HSPA8, ALIX, and HSP90AB1 represent pan-EVP markers, while ACTB, MSN, and RAP1B are novel pan-EVP markers. To confirm that EVPs are ideal diagnostic tools, we analyzed proteomes of TE- (n = 151) and plasma-derived (n = 120) EVPs. Comparison of TE EVPs identified proteins (e.g., VCAN, TNC, and THBS2) that distinguish tumors from normal tissues with 90% sensitivity/94% specificity. Machine-learning classification of plasma-derived EVP cargo, including immunoglobulins, revealed 95%sensitivity/90% specificity in detecting cancer. Finally, we defined a panel of tumor-type-specific EVP proteins in TEs and plasma, which can classify tumors of unknown primary origin. Thus, EVP proteins can serve as reliable biomarkers for cancer detection and determining cancer type.
In Brief
A comprehensive proteomic analysis of extracellular vesicles and particles (EVPs) from 426 human samples identifies pan-EVP markers, biomarkers for EVP isolation, for cancer detection and determining cancer type.
Graphical Abstract
INTRODUCTION
Pathologists routinely employ tissue biopsies, when accessible, to diagnose cancer, cancer spread, and measure treatment response. Meanwhile, liquid biopsies are minimally invasive, can be obtained serially, and may detect cancer at an earlier, more curable stage. As expectations for liquid biopsy technologies for early cancer detection grow, exosomes may provide a valuable resource.
Exosomes are 30–150 nm nanovesicles of endosomal origin, enriched in nucleic acids, lipids, and proteins (O’Driscoll, 2015; Thakur et al., 2014) that mediate intercellular communication in normal physiology and pathology (Johnstone et al., 1987; Maas et al., 2017; Skog et al., 2008; Yáñez-Mó et al., 2015). Previously, we reported the prognostic and functional importance of tumor-derived exosome proteins in tumor progression, immune regulation, and metastasis (Costa-Silva et al., 2015; Hoshino et al., 2015; Peinado et al., 2012; Rodrigues et al., 2019). Moreover, we deconvoluted the heterogeneity of extracellular nanoparticles, defining three distinct subpopulations, small exosomes (Exo-S), large exosomes (Exo-L), and exomeres (Zhang and Lyden, 2019) that we collectively refer to as extracellular vesicles and particles (EVPs).
Mounting evidence suggests that EVPs could be used for early cancer detection, prognosis, and to guide therapy (Chen et al., 2017). EVPs are actively released into the peripheral circulation at concentrations of >109 vesicles/mL, providing ample material for downstream analyses (Colombo et al., 2014). Mass spectrometry-based proteomic profiling is emerging as a strategy to gain insight into the biology and clinical potential of circulating EVPs (Choi et al., 2015). Despite the public availability of several EVP protein databases, (e.g., Vesiclepedia, EVpedia, ExoCarta) (Kalra et al., 2012; Kim et al., 2015; Mathivanan and Simpson, 2009), much remains unknown about EVP proteomes, including: (1) markers for reliable isolation of EVPs in humans, regardless of tissue source; (2) markers to distinguish cancer versus non-cancer; and (3) markers unique to specific primary tumors (e.g., lung, pancreas, breast, etc.). To address this gap in knowledge, we sought to define EVP protein signatures that distinguish cancer patients from healthy individuals.
To identify universal EVP markers and improve the isolation of human EVPs, we analyzed 497 human and murine samples by proteomic profiling. Among conventional exosome markers, heat shock cognate 71 kDa protein (HSPA8), heat shock protein HSP 90-beta (HSP90AB1), CD9, and programmed cell death 6-interacting protein (ALIX) were the most prominent markers found in human-derived EVPs isolated from cells, tissues, and most biofluids. We identified 13 additional proteins shared by >50% of human samples, thus drastically expanding the panel of human EVP markers.
By examining EVP proteomes of paired tumor and adjacent tissue from viable surgical specimens of pancreatic and lung cancer patients, we identified cancer-specific EVP protein signatures. Moreover, by comparing matched tissue explant (TE)- and plasma-derived EVPs, we found tumor-associated EVP proteins unique to the plasma of cancer patients and determined that EVP plasma proteins were derived from the tumor microenvironment, distant organs, and the immune system. Next, we analyzed the tissue and plasma EVP proteomes of stage I–IV cancers from several pediatric and adult cancers, and compared them to non-tumor tissues and healthy control (HC) plasma. Random forest classification of EVP proteomes revealed cancer detection specificities and sensitivities of 90% and 94% for tissues, and 95% and 90% for plasma, respectively. Importantly, plasma-derived EVP cargo could distinguish among cancer types in patients. These data suggest that tumor-associated EVP proteins can serve as biomarkers for early-stage cancer detection and classify uncertain primary tumor types.
RESULTS
Proteomic Characterization of Human EVPs
We used sequential ultracentrifugation (SUC) to isolate EVPs from 497 normal and cancer-associated human and murine-derived samples, including cell lines, tissues, plasma, and other bodily fluids (Figure 1A; Table S1). All EVP samples isolated by SUC represent a heterogeneous population categorized into three prominent sub-populations that include exomeres (non-vesicular particles <50 nm) and two exosome subpopulations (exo-S 50–70 nm; exo-L 90–120 nm) (Zhang et al., 2018) (Figure 1B). Heterogeneous EVP populations were characterized in terms of size range (30–150 nm) and morphology via nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM), respectively (Figures 1B, S1A, and S1B). We constructed a database of EVP proteomes from 426 human samples, which included resected normal and malignant tissues (n = 131), blood plasma (n = 120), cell lines (n = 115), blood serum (n = 7), bone marrow (n = 20), lymphatic fluid (n = 13), and bile duct fluid specimens (n = 20) from 152 control and 274 cancer samples (Figure 1A). The cancer patient-resected tissue and plasma samples analyzed included both adult cancers (pancreatic, lung, breast, and colorectal carcinomas and melanoma) and pediatric cancers (neuroblastoma and osteosarcoma). The average number of unique proteins detected in the EVPs was 862 (25% to 75% percentile, 310 to 1,282 proteins), with the lowest diversity in plasma and serum (an average of 265 and 273 proteins in human plasma and serum, respectively, and 210 proteins in murine plasma) and the highest numbers of proteins in TE EVPs (average of 1,482 and 1,523 proteins in human and murine TEs, respectively) (Figure S1C). Although for some specific exosomal proteins, concentrations increased with cancer stage (Peinado et al., 2012), we did not observe differences between non-tumor and tumor samples in the number of distinct EVP proteins detected for most sources except cell lines (Figure S1D).
Figure 1. Proteomic Characterization of EVPs Obtained from 497 Samples from Seven Different Sources.
(A) EVPs from 426 human and 71 murine samples were analyzed by liquid chromatography tandem-mass spectrometry (LC-MS/MS).
(B) Centrifugation protocol and workflow for EVP enrichment (left, *sucrose cushion was applied to PaCa samples), representative nanoparticle tracking analysis (middle), and transmission electron microscopy imaging (right) of EVPs from human control (HC) plasma. Scale bar represents 200 nm.
(C) Pearson correlation of EVP protein expression among samples types. Larger and darker circles depict a higher correlation between samples.
(D) Positivity for 11 conventional exosomal protein markers across different tissue types. The frequency (%) of samples expressing the specified protein is noted in each box. Darker red depicts higher frequency.
(E) The frequency (%) of samples from each source positive for the 13 newly defined EVP markers. Proteins found in >50% of all human samples were identified. Annotation as in (D).
(F) GO analysis for the 13 common EVP proteins listed in (E).
(G) Western blot of EVPs isolated from human cell lines, PaCa TEs, as well as HC and PaCa plasma for conventional and newly identified EVP markers.
(H) ExoView analyses performed on EVPs isolated from HC plasma for conventional and newly identified EVP markers. Error bars represent the standard error of the mean.
To evaluate the overall correlation between EVP proteomes derived from different sources, we performed a Pearson correlation analysis comparing specimen types (plasma versus TEs) and species (human versus murine) for all tumor and non-tumor EVP samples. The sample source was the strongest determinant of EVP protein signatures (Figure 1C). EVP proteins from human plasma overlapped best with human serum-derived EVPs (r2 = 0.92), followed by human bone marrow (r2 = 0.65) and lymphatic fluid EVPs (r2 = 0.64), and correlated least with human cell line (r2 = 0.15) and TE-derived EVPs (r2 = 0.24), suggesting the complexity of plasma and lymph EVP proteomes may drive the divergence from tissue EVP proteomes (Figure 1C). In terms of inter-species differences, the proteomes of human and murine cell line- and TE-derived EVPs were similar (r2 = 0.85 and 0.78, respectively), whereas the proteomes of plasma-derived EVPs largely differed between mice and humans (r2 = 0.52) (Figure 1C). These observations held true whether tumor samples or non-tumor samples were analyzed together or separately (Figures 1C and S2A). These data suggest that, in general, EVP profiles differ significantly depending on the tissue source and species, and murine plasma-derived EVP proteomes cannot be used to guide liquid biopsy studies in patients.
Unbiased EVP Proteome Analysis Identifies 13 Common EVP Biomarkers in Humans
To identify ubiquitous pan-EVP markers for improved isolation from various human and murine sources, we investigated the frequency of specific proteins found in EVPs from different sources. Traditional exosomal markers (e.g., tetraspanins, heat shock proteins) were investigated first, and of 11 conventional exosomal markers examined (Thery et al., 2006), HSPA8 was the only protein found in >50% of EVP samples from all sources (Figure 1D). Remarkably, although CD63 was present in 89% of the murine cell line-derived EVP samples examined, it was detected less frequently in tissue-derived EVPs and rarely, if ever, in EVPs isolated from biofluids of either human or mouse origin (Figure 1D). Among the human cell line-derived EVP proteins, all of the established exosome markers, except CD63, were present in ≥77% of 115 human cell line-derived samples (Figure 1D), supporting the idea that SUC specifically enriches preparations in exosomes. Importantly, interrogation of extracellular nanoparticle sub-population proteomics data revealed that of the traditional exosome markers, CD9, TSG101, and CD81 were detected in Exo-S, Exo-L, as well as exomeres (Figure S2B). For mouse cell line-derived EVPs, all 11 markers were highly represented (≥86%) (Figure 1D). However, for human plasma or serum, CD9 and HSPA8 were the only proteins found at ≥ 50% frequency (Figure 1D), suggesting that pan-exosome markers currently used to verify exosomal origin in cell culture do not translate directly to patient-derived biofluids. These findings were similar regardless of whether we analyzed tumor- or non-tumor-derived specimens (Figure S2C) and highlight the need to identify novel pan-EVP markers in human specimens.
Therefore, to identify proteins found at high frequency in all human-derived EVPs, irrespective of source, we searched for proteins that met a threshold of ≥ 50% representation across specimens. Of 11,000 human EVP proteins, only 13 matched this criterion (Figures 1E and S2D). Gene Ontology (GO) analysis demonstrated that the vast majority of these proteins, including alpha-2-macroglobulin (A2M), β2-microglobulin (B2M) (Zagorac et al., 2012), stomatin (STOM) (Mairhofer et al., 2002; Snyers et al., 1999), filamin A (FLNA), fibronectin 1 (FN1), gelsolin (GSN), hemoglobin subunit beta (HBB), galectin-3-binding protein (LGALS3BP), ras-related protein 1b (RAP1B), beta-actin (ACTB), and joining chain of multimeric IgA and IgM (JCHAIN) are proteins trafficked through endosomes and likely markers of endocytosis/exocytosis (Figure 1F; Table S2). Of these 13 molecules, ACTB, moesin (MSN) (Muriel et al., 2016), and RAP1B (Pizon et al., 1994) represent pan-exosome/exomere markers that can be identified in human Exo-S/Exo-L as well as exomeres, whereas STOM is a specific exosome marker found only in Exo-S/Exo-L that can thus distinguish exosomes from exomeres (Figure S2B). We next validated several of these EVP markers in cell lines, patient TE, and plasma by immunoblotting (Figure 1G). Moreover, we used a second anti-body-based assay, the ExoView platform (Nanoview Biotech, Inc.), and detected B2M and MSN on the surface of plasma-derived EVPs from three independent donors (Figure 1H); thus, these proteins could be employed to improve affinity-based protocols for EVP isolation from human plasma/biofluids/tissues. These EVP proteins may represent bona fide exosomal markers. Because these markers were present at a similar frequency regardless of whether the samples were of tumor or non-tumor origin, they could be used to improve exosome isolation from any and all human samples.
Identification of Tissue-Specific Tumor-Derived EVP Proteins in Patients
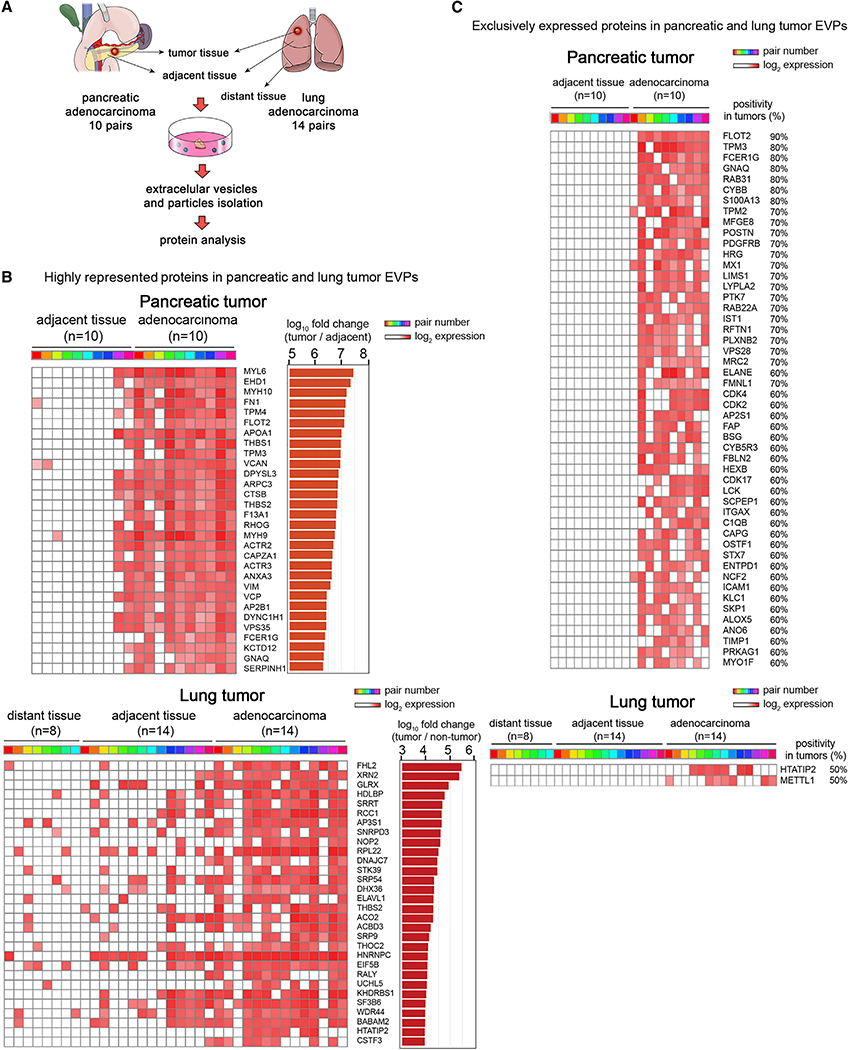
To identify EVP proteins that could be used as diagnostic biomarkers for cancer patients, we first sought to identify shared and unique tumor-specific EVP proteins by performing a pairwise comparison between tumor tissue (TT) EVP proteomes, as tumor EVP-enriched sources, and matched non-tumor adjacent tissue (AT) EVP proteomes. TT and AT were resected from 10 patients with pancreatic adenocarcinoma (PaCa) and 14 patients with lung adenocarcinoma (LuCa), and EVPs were isolated for pairwise comparison (Figure 2A). In addition, we obtained eight non-tumor distant tissues (DTs) resected from LuCa patients, because non-malignant tissues collected distally from tumor sites are less likely to be affected by tumor-secreted factors. EVPs isolated from these DTs were included as a third group in the comparison (Figure 2A).
Figure 2. PaCa- and LuCa-Specific EVP Protein Cargo in Surgically Removed TEs.
(A) Diagram of TE culture method for EVPs isolation from paired TT and AT and matched DT (for LuCa).
(B) Top 30 proteins highly represented in PaCa TT compared to AT (top), and LuCa TT compared to AT and DT (bottom). The heatmap is based on proteins found >50% of TT at levels >10-fold higher in TT than AT (FDR <0.05). Colored boxes identify sample pairs.
(C) Top 50 proteins never found in AT but found in >50% of PaCa (top), and the two proteins found in 50% of LuCa TT and never found in AT or DT (bottom).
Distinct EVP proteins with potential biomarker value and biological relevance in PaCa and LuCa were identified by analyzing EVP proteins most enriched in TT as compared to AT and DT. We searched for EVP proteins present in ≥50% of the samples, and, of those, we selected the ones showing a 10-fold or larger increase compared to AT or AT/DT with a false discovery rate (FDR) of <0.05. Based on these criteria, 356 and 123 EVP proteins were identified as TT-enriched proteins in PaCa and LuCa, respectively (top 30 proteins in Figure 2B; complete list in Tables S3 and S4). Of the >600 EVP proteins highly expressed in both PaCa and LuCa TT, we identified 11 shared EVP proteins: versican (VCAN), cathepsin B (CTSB), thrombospondin 2 (THBS2), septin 9 (SEPTIN9), basigin (BSG), fibulin 2 (FBLN2), four and a half LIM domains 2 (FHL2), inosine triphosphatase (ITPA), galectin-9 (LGALS9), splicing factor 3b subunit 3 (SF3B3), and calcium/calmodulin dependent serine protein kinase (CASK) (Tables S3 and S4). Classification of the pathways related to the enriched proteins from PaCa TT-derived EVPs using GO Term Finder revealed that PaCa EVP-packaged proteins were involved in epithelial mesenchymal transition (EMT) (i.e., FN1, VCAN, tropomyosin alpha-4 chain [TPM4], dihydropyriminase-related protein 3 [DPYSL3], THBS2, thrombospondin 1 [THBS1], serpine H1 [SERPINH1], and vimentin [VIM]) and associated with cytoskeleton, filament assembly, and the extracellular matrix (ECM) (i.e., FN1, myosin-10 [MYH10], actin-related protein 2/3 complex subunit 3 [ARPC3], myosin-9 [MYH9], THBS1, THBS2, tropomyosin alpha-3 chain [TPM3], and TPM4) consistent with studies reporting changes in stiffness and ECM deposition in PaCa (Nielsen et al., 2016; Procacci et al., 2018) (Figure S3). For LuCa, Myc targets (small nuclear ribonucleoprotein Sm D3 [SNRPD3], AP-3 complex subunit sigma-1 [AP3S1], heterogeneous nuclear ribonucleoproteins C1/C2 [HNRNPC], and 60 ribosomal protein L22 [RPL22]) and RNA processing (5′−3′ exoribonuclease 2 [XRN2], tRNA (cytosine(72)-C(5))-methyltransferase, NSUN6 [NOP2], SNRPD3, cleavage stimulation factor subunit 3 [CSTF3], ATP-dependent DNA/RNA helicase DHX36 [DHX36], serrate RNA effector molecule homolog [SRRT], RNA-binding protein Raly [RALY], ELAV-like protein 1-A [ELAVL1], HNRNPC, RPL22, and THO complex subunit 2 [THOC2]) were highly represented in TT-derived EVPs (Figure S4). Additionally, Gene Set Enrichment Analysis (Subramanian et al., 2005) revealed that EMT, coagulation, and actin signaling pathways were highly enriched in PaCa, whereas cell cycle, metabolic, and RNA processing pathways were significantly enriched in LuCa (Figures S3 and S4). Although EMT was found to be highly represented in PaCa EVPs (p < 0.001), it was not significant in LuCa EVPs (p = 0.49). Similarly, RNA processing pathways were not enriched in PaCa EVPs (p = 0.77). Our finding that PaCa and LuCa TT EVP cargo is distinct and related to discreet cellular processes suggests that EVP protein packaging is heterogeneous across tumor types and reflects tumor biology.
In addition to examining EVP proteins overrepresented in TT, we also mined our dataset for EVP proteins exclusive to TT versus AT/DT and generated a list of proteins detected in ≥ 50% of either PaCa or LuCa TT samples but never found in AT or DT (Figure 2C). Although we identified over 50 proteins, including ECM-related and pro-inflammatory proteins (e.g., periostin [POSTN], S100A13), exclusive to PaCa TT-derived EVPs, we found only two proteins, HIV-1 Tat interactive protein 2 (HTATIP2) and methyltransferase like 1 (METTL1), that were unique for LuCa TT-derived EVPs (Figure 2C). Notably, among the top 30 EVP proteins enriched in PaCa TT (Figure 2B), four proteins (flotillin 2 [FLOT2], TPM3, Fc fragment of IgE receptor [FCER1G], and G protein subunit alpha Q [GNAQ]) overlapped with proteins found solely in tumor EVPs (Figure 2C). In LuCa, one protein identified in the TT versus AT/DT comparison, HTATIP2, overlapped with EVP proteins present exclusively in tumor EVPs (Figures 2B and 2C; Tables S3 and S4), further validating these proteins as having PaCa- and LuCa-specific biomarker potential. Collectively, these data suggest that cancer EVP proteins may reflect selective packaging and could discriminate among cancer types.
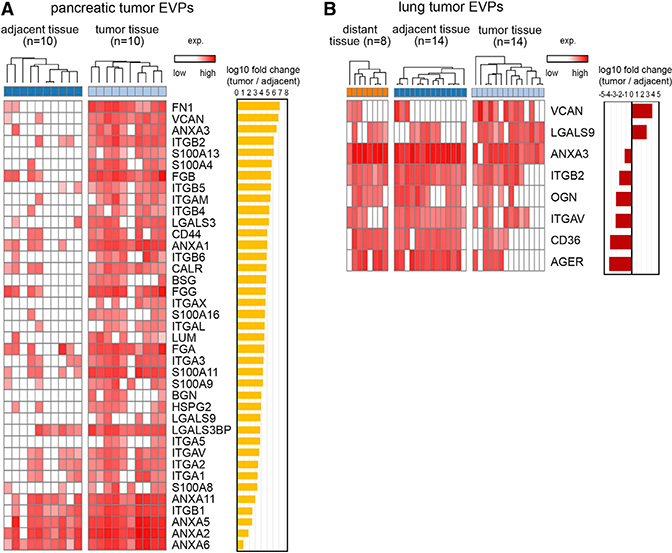
Specific DAMP Molecules Are Packaged in TT-Derived EVPs
Because tumor-derived exosomes interact with the immune system (Becker et al., 2016), we asked whether specific proteins involved in eliciting immune responses, such as damage-associated molecular pattern (DAMP) proteins, which have key roles in cancer development and tumor progression (Hernandez et al., 2016) (Table S5), are packaged in TT-derived EVPs. We found 39 EVP DAMPs (e.g., VCAN) that were highly enriched in PaCa TT-derived EVPs versus AT-derived EVPs (Figure 3A). Of these, six proteins present in TT-derived were never found in AT-derived EVPs: S100A13, BSG, LGALS9, biglycan (BGN), and integrins (ITGs) α5 and αX. Similar analyses revealed two abundantly expressed DAMP EVP proteins, VCAN and LGALS9 in LuCa (Figure 3B). These DAMPs are effective pro-inflammatory molecules (e.g., LGALS9, S100A13, and BGN) or receptors for pro-inflammatory cytokines (e.g., BSG and ITGs) (Hernandez et al., 2016). Notably, VCAN and LGALS9 were highly enriched in both PaCa and LuCa TT EVPs, suggesting that they represent EVP inflammatory response markers shared across cancers (Figures 3A and 3B). Interestingly, certain DAMPs, such as annexin A3 (ANXA3), and several ITGs (e.g., ITGB2 and ITGAV) were enriched in LuCa, but not PaCa AT/DT EVPs. This finding may reflect the presence of cancer-associated stroma in AT/DT (Figure 3B) and further emphasizes that the non-tumor-derived EVP proteome is as informative as the tumor-derived EVP proteome in identifying specific cancer types. Collectively, unique DAMPs present in cancer or non-cancer EVPs may help delineate the pro-tumoral versus immunogenic roles of DAMP molecules.
Figure 3. EVP DAMP Molecules Enriched in PaCa and LuCa.
(A and B) EVP proteins enriched in (A) PaCa TT and found in >50% of TT samples; and (B) LuCa TT or AT/DT, found in >50% of TT or AT/DT samples, respectively, with >10-fold difference and FDR <0.05. Paired t test was used to calculate FDR.
See also Table S5.
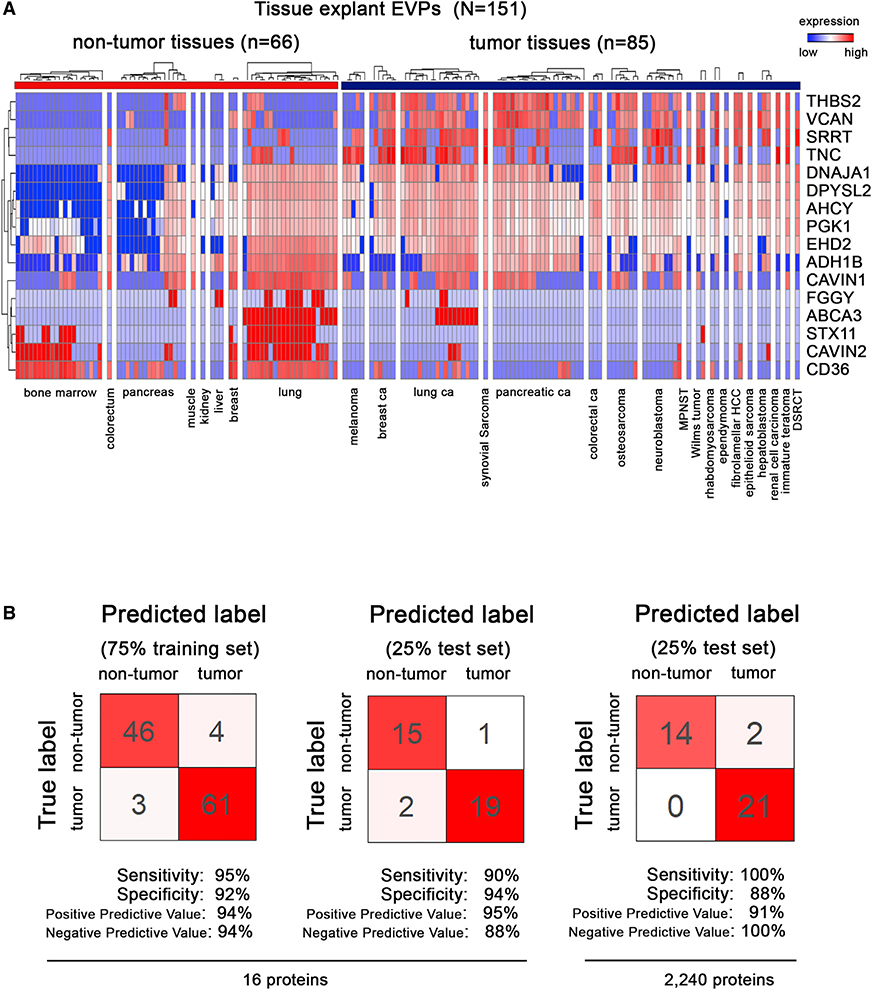
Analysis of Tissue-Derived EVP Proteins across Multiple Cancers Identifies Tumor-Associated EVP Signatures
Having identified TT-specific EVP proteins, we next set out to determine whether comparing TT-derived and non-TT-derived EVP proteomes could distinguish cancer from non-cancer. We analyzed 131 tissue explant- and 20 bone marrow-derived EVP samples. Eighty-five samples were isolated from TT, whereas 66 were classified as non-TT (Figure 4A). We employed random forest classification, which is robust to noise and overfitting, to identify a subset of EVP proteins that accurately discriminates between HC and patients with tumors. To train and subsequently test the model, samples were evenly partitioned based on sample type (i.e., control sample or tumor sample) and 75% of samples were used as a training set with the remaining 25% representing the independent test set. Based on 16 EVP proteins, applying 10-fold cross-validation to the training set yielded a sensitivity (true positive rate) of 95% and specificity (true negative rate) of 92% (Figure 4B). When applied to the independent test set samples, the model based on a subset of EVP proteins achieved 90% sensitivity and 94% specificity, whereas basing the model on all 2,240 proteins detected in TE EVPs achieved 100% sensitivity and 88% specificity. This result is likely driven in part by tissue-specific field effects, as sensitivity and specificity improve when focusing on specific tissue types. Analysis of larger sample sizes is required to further validate this model and inform on tissue-specific tumor-associated EVP signatures.
Figure 4. Identification of Tumor-Associated EVP Signatures in Surgically Removed Tissue Explants from Multiple Tumor Types.
(A) Proteins with the highest predictive values in classifying tumor and non-tumor samples by random forest algorithm.
(B) Classification error matrix using a random forest classifier of 75% training set and 25% test set, for the 16 proteins from (A) (left), and all 2,240 tissue explant EVP proteins (right). The number of samples identified is noted in each box.
See also Table S6.
Despite the inherent tissue-specific variation, we identified a combination of proteins most likely to distinguish cancer from non-cancer (Figure 4A; Table S6). Notably, THBS2 and VCAN, EVP proteins highly enriched in both PaCa and LuCa TT, were predictive in identifying cancer, suggesting that these proteins could be used as pan-cancer EVP markers. Moreover, specific EVP adhesion markers (e.g., CD36, tenascin C [TNC], THBS2, and VCAN) and metabolic enzymes (e.g., all-trans-retinol dehydrogenase [NAD(+)] ADH1B/alcohol dehydrogenase 1B [ADH1B], adenosylhomocysteinase [AHCY], and phosphoglycerate kinase 1 [PGK1]) may be pan-cancer markers (Table S6).
Because proteomic databases are periodically updated, and as proof of principle that our biomarker identification pipeline is largely independent of database changes, we reanalyzed the entire cell line, TE, and plasma datasets against the most recent iteration of the UniProt Complete HUMAN proteome (February, 2020: 74,788 sequences) (see STAR Methods). Using the updated dataset, we achieved 90% sensitivity/94% specificity in cancer detection (Figures 4A, 4B, S5A, and S5B).
Tumor, Peritumoral Microenvironment, and Distant Stroma EVP Proteins Contribute to Tumor-Associated EVP Signatures in Plasma
Plasma remains the most readily accessible source for liquid biopsies. Therefore, to understand the characteristics and composition of tumor-associated EVP proteins, we first sought to determine which of these proteins are present in the plasma of PaCa and LuCa patients. Then, we investigated whether these proteins originated from the TT, AT/DT, or elsewhere.
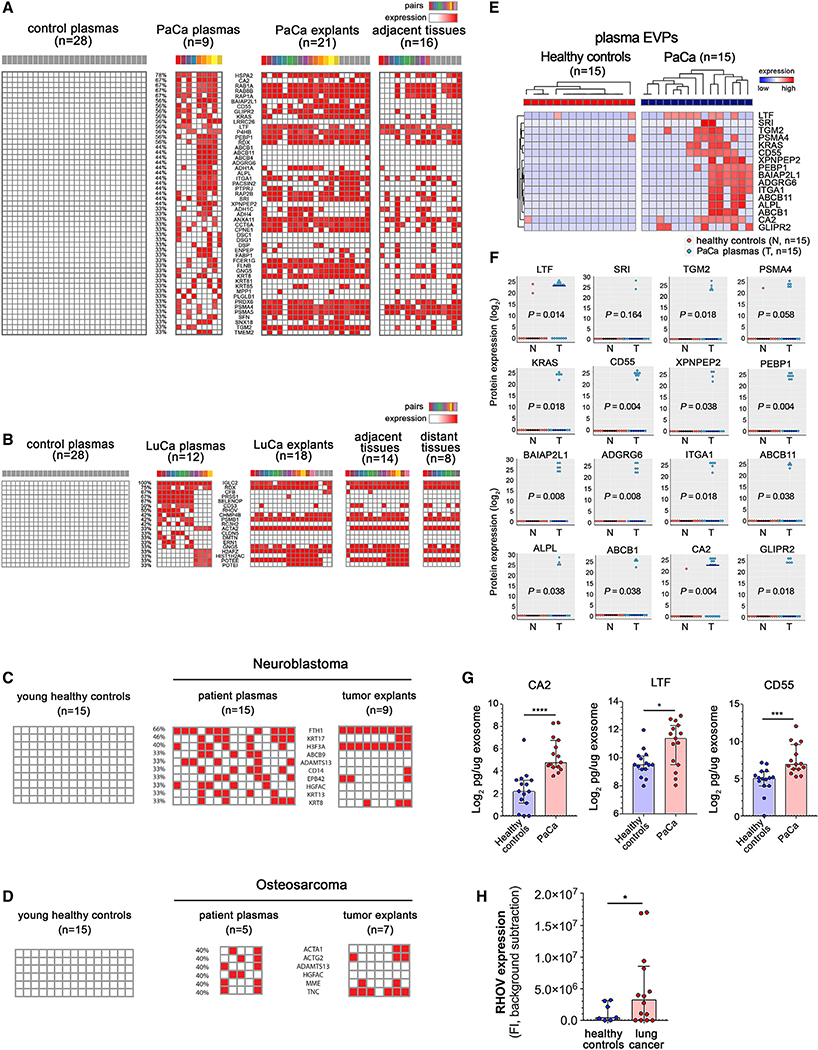
We analyzed the plasma EVP proteomes of 9 patients with PaCa (78% stage II and 22% stage III) and 12 patients with LuCa (50% stage I, 42% stage II, and 8% stage III), selecting EVP proteins found in >30% of patient plasma but never in the plasma of 28 healthy adult controls. We found 51 and 19 plasma-derived EVP proteins unique to PaCa and LuCa, respectively (Figures 5A and 5B). To identify the likely source of these EVPs, we compared these plasma-derived EVP proteins with TT, AT, and DT-derived EVP proteomes (Figures 5A and 5B). Interestingly, proteins such as brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAIAP2L1), alkaline phosphatase, tissue-nonspecific isozyme (ALPL), receptor-type tyrosine-protein phosphatase eta (PTPRJ), high-affinity immunoglobulin epsilon receptor subunit gamma (FCER1G), and cell surface hyaluronidase (TMEM2), were present in both plasma- and TT-derived PaCa EVPs, but were packaged at extremely low levels or were undetectable in all of the 16 AT-derived EVP samples, suggesting that these proteins most likely originate from pancreatic tumor cells (Figure 5A). KRAS, an oncoprotein that drives PaCa, was frequently packaged in TT EVPs (76%) and could be detected in plasma EVPs of patients with PaCa. To our surprise, many proteins, such as leucine-rich repeat-containing protein 26 (LRRC26), ATP-dependent translocase ABCB1 (ABCB1), bile salt export pump (ABCB11), adhesion G-protein coupled receptor G6 (ADGRG6), desmocollin-1 (DSC1), desmoglein-1 (DSG1), keratin, type II cuticular Hb1 (KRT81), and plasminogen-like protein B (PLGLB1), were absent or packaged at low levels in both TT- and AT-derived EVPs, but were found exclusively in PaCa patient plasma-derived EVPs, suggesting that these EVP proteins originate from distant organs (DOs), or immune cells. That these proteins were never found in plasma EVPs from HC reinforces the idea that cancer is a systemic disease that alters EVP cargo of DO and immune cells.
Figure 5. Identification and Validation of Tumor-Associated Protein Cargo in Tissue- and Plasma-Derived EVPs.
(A and B) Proteins exclusively found in >30% of (A) PaCa or (B) LuCa patient plasma-derived EVP samples but never found in HC plasma-derived EVP samples (left) were matched to explant-derived EVPs (PaCa TT and AT; LuCa TT, AT, and DT; right). The colored boxes identify sample pairs.
(C and D) EVP proteins found in >30% (C) of neuroblastoma or (D) osteosarcoma plasma-derived EVPs but never in plasma EVPs from HC (left), and their presence in tumor explant EVPs (right).
(E) Top 20 PaCa plasma markers validated by targeted MS in an independent cohort of PaCa and HC.
(F) Log2 protein expression of the data presented in (E). p values calculated by Wilcoxon rank-sum test.
(G) ELISA validation of CA2, LTF, and CD55 in plasma EVP from 15 PaCa and 15 HC. *p < 0.05, ***p = 0.0001, ****p < 0.0001. p values are calculated by Mann-Whitney U test. Error bars represent median with interquartile range.
(H) ELISA validation of RHOV in plasma EVP from 14 LuCa and 7 HC. *p < .05. p value calculated by t test. Error bars represent median with interquartile range.
For LuCa, we identified 19 plasma EVP proteins present in more than 30% of patients (Figure 5B). Unlike PaCa, all proteins detected in LuCa TT were also found in AT and most of DT. Proteins such as selenoprotein P (SELENOP), rho-related GTP-binding protein RhoV (RHOV), roquin-2 (RC3H2), claudin-5 (CLDN5), dematin (DMTN), and serine/threonine-protein kinase/endoribonuclease IRE1 (ERN1), were only detected in plasma, but not in TT, AT, or DT, supporting the systemic nature of cancer. For example, the liver-derived SELENOP, frequently found in plasma-derived EVPs from LuCa patients, was never detected in lung-derived EVPs, suggesting LuCa affects liver function.
To demonstrate that these observations were not restricted to LuCa and PaCa, or adult cancers in general, we examined TT- and plasma-derived EVPs isolated from advanced stage patients with two of the most frequent pediatric solid cancers: neuroblastoma and osteosarcoma (Figures 5C and 5D; Table S1). Pediatric cancers are fast-growing, overtaking the organ where they originate, therefore rendering AT harvesting very challenging. We analyzed TT-derived EVPs from 9 neuroblastoma and 7 osteosarcoma patients and plasma-derived EVPs from 15 neuroblastoma and 5 osteosarcoma patients (Figures 5C and 5D). Plasma-derived EVPs from 15 pediatric HC were also assessed (Table S1). We focused our analyses on EVP proteins detected in >33% of cancer patient plasma but never in any of the control subject plasma. In neuroblastoma, we found 10 plasma EVP proteins, ferritin heavy chain (FTH1), keratin, type I cytoskeletal 17 (KRT17), histone H3.3 (H3F3A), ATP-binding cassette sub-family B member 9 (ABCB9), a disintegrin and metalloproteinase with thrombospondin motifs 13 (ADAMTS13), CD14, erythrocyte membrane protein band 4.2 (EPB42), hepatocyte growth factor activator (HGFAC), keratin, type I cytoskeletal 13 (KRT13), and KRT8 (Figure 5C), related to cellular proliferation/cell cycle and differentiation. In osteosarcoma, we identified 6 plasma EVP proteins, actin, alpha skeletal muscle (ACTA1), actin, gamma-enteric smooth muscle (ACTG2), ADAMTS13, HGFAC, neprilysin (MME), and TNC, related to tissue morphogenesis (Figure 5D). Interestingly, EVP protein cargo reflected the cell of origin of each cancer (osteoblast versus neuroblast).
To validate the top PaCa plasma EVP proteins, we reanalyzed the samples based on the most recent protein database and confirmed the top EVP protein list (Figure S6A). We then selected the top 20 EVP proteins found exclusively in PaCa plasma but never in the plasma of 28 HC (see STAR Methods for full protein list). Next, we employed a targeted MS approach, parallel reaction monitoring (PRM), to quantify these 20 proteins in plasma-derived EVPs isolated from an independent cohort of 15 PaCa and 15 HC (Table S1). Eighty percent of the markers, such as carbonic anhydrase 2 (CA2), lactoferrin (LTF), BAIAP2L1, KRAS, phosphatidyletholamine-binding protein 1 (PEBP1), and CD55 were validated through this approach (Figures 5E and 5F). In addition, three of these PaCa-specific EVP proteins, CA2, LTF, and CD55, were validated by ELISA (Figure 5G). Collectively, our data demonstrate that 16/20 PaCa EVP proteins identified by unbiased EVP proteomics are present at higher levels in PaCa EVPs compared to HC. Wethen reanalyzed the LuCa dataset based on the most recent protein database and found RHOV to be consistently elevated in LuCa EVPs (Figure S6B). Using ELISA, we confirmed a significant increase in RHOV levels in 14 LuCa EVPs relative to 7 HC plasma EVPs (Figure 5H; Table S1).
Taken together, our data demonstrate that plasma-derived EVPs originate from various sources, and EVP proteomic analyses can identify cancer-type-specific plasma EVP protein profiles in resectable and advanced disease. By comparing plasma-derived and tissue-derived EVP proteins, we could distinguish between tumor-derived, adjacent tissue-derived and distant organ EVPs. Furthermore, plasma EVP protein signatures of cancer patients were distinct from those of control subjects and were cancer-type-specific, suggesting that EVP protein profiles could serve as a liquid biopsy tool to detect cancer and differentiate among cancer types.
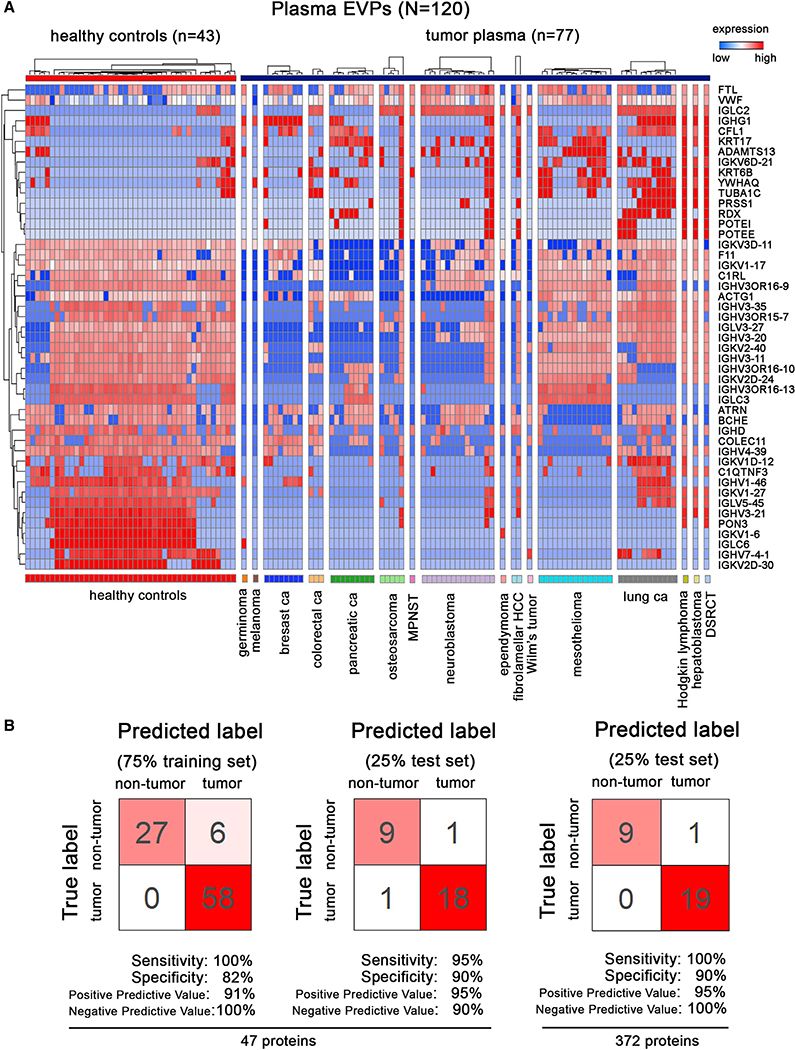
Analysis of Plasma-Derived EVP Proteins across Multiple Cancers Identifies Tumor-Associated EVP Signatures
Employing random forest classification, in the same manner described for tissue samples, we explored tumor-associated plasma EVP signatures. We analyzed 120 plasma-derived EVP proteomes from77 cancer patients with 16 different cancer types, including breast, lung, or pancreatic carcinoma, mesothelioma, and neuroblastoma, and 43 HC subjects (Figure 6A). Ten-fold cross-validation of the training set yielded 100% sensitivity and 82% specificity (Figure 6B). Our model achieved 95% sensitivity and 90% specificity, showing that a combination of different immunoglobulin-related proteins was most predictive for detecting cancer (Figures 6Aand6B; TableS7). When all 372 proteins detected in plasma EVPs were used to generate the model, 100% sensitivity and 92%specificitywere achieved (Figure 6B). Notably, predictive proteins that discriminate cancer versus non-cancer included not only plasma-derived EVP proteins present in cancer patients, but also those proteins found in normal plasma EVPs that are absent or present at low levels in cancer patient plasma EVPs, further supporting the notion that cancer versus non-cancer discrimination should also take into account those EVP proteins that are lost in cancer (Figure 6A). Furthermore, we also reanalyzed the plasma dataset based on the most recent protein database and confirmed the top protein list as well as the validation results with 100% sensitivity/80% specificity (Figures S6C and S6D). Taken together, our results suggest that plasma-derived EVP proteins could be useful as liquid biopsy tests for cancer detection.
Figure 6. Identification of Tumor-Associated EVP Signatures in Plasma From Patients with Various Cancers.
(A) EVP proteins with the highest predictive values in classifying tumor and non-tumor plasma samples by random forest algorithm.
(B) Classification error matrix using random forest classifier of 75% training set and 25% test set, for the 47 proteins in A (left), and all 372 plasma EVP proteins (right). The number of samples identified is noted in each box.
See also Table S7.
Patient Tumor Tissue-Derived EVP Proteomics Classify Cancer Types
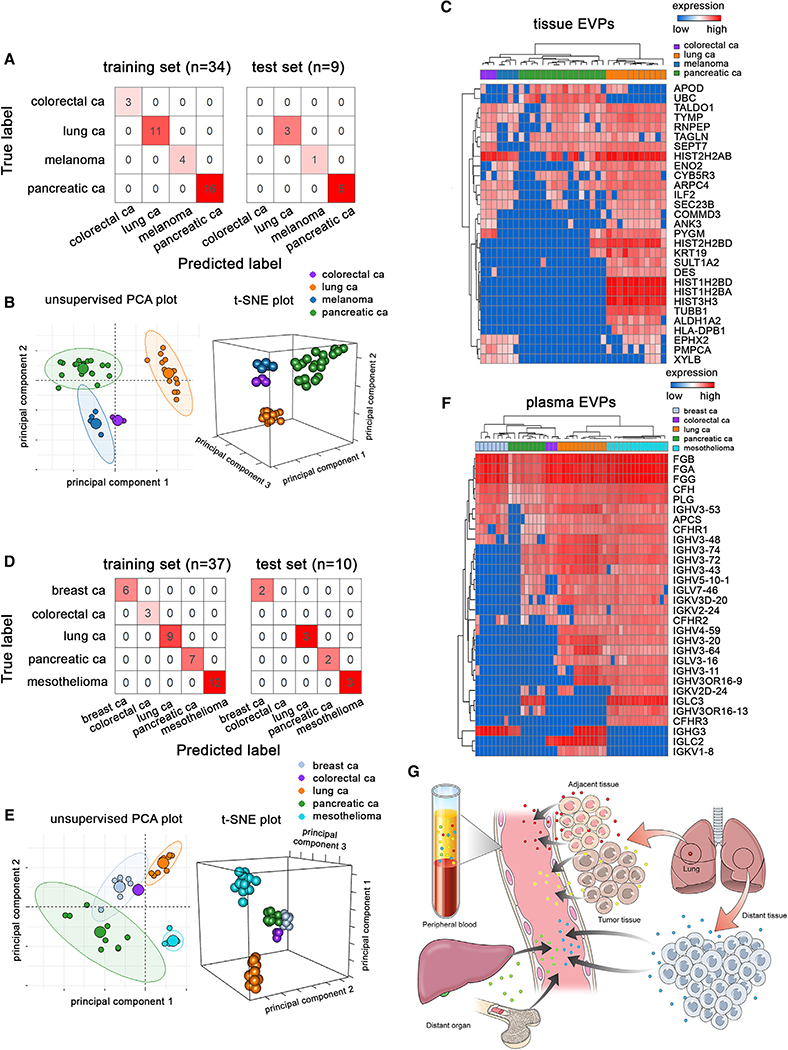
We next sought to determine if a patient’s EVP protein signature could be assigned to a particular cancer type. We analyzed EVP proteins derived from tissues obtained from the primary tumor or sentinel lymph nodes of patients with four different cancer types: melanoma, colorectal, pancreatic, and lung cancer (Figure 7A; Table S1). To identify protein signatures that can discriminate between the four tumor types, we employed random forest classification and t-Stochastic Neighbor Embedding (t-SNE) for visualization. We were able to correctly discriminate every tumor sample, as summarized in a confusion matrix (Figure 7A). Unsupervised principal component analysis (PCA) and supervised 3D t-SNE plot were used to visualize the differences among samples (Figure 7B). Feature selection by random forest identified 29 EVP proteins, some of which are related to immune function, as having the highest predictive value for distinguishing among the four cancers analyzed (Figure 7C). Based on these EVP proteins, samples clustered together according to the primary tumor type. Interestingly, based on the t-SNE visualization and random forest classifier results, tumor specificity of EVP signatures was independent of cancer staging and could distinguish between cancers even at early stages, especially in PaCa and LuCa (Figure 7B). Thus, EVP profiles of tissue biopsies (i.e., lymph nodes) could aid in classifying cancer types, supporting a diagnosis that can lead to a more specific treatment plan for patients with cancer of unknown primary tumor origin.
Figure 7. Tumor-Derived EVP Profiles Classify Primary Tumor of Origin.
(A) Classification error matrix for 75% training and 25% test sets from tissue explant-derived EVPs.
(B) Unsupervised 2D (left) and supervised 3D (right) t-SNE plots representing proteins in (C).
(C) Proteins with the highest predictive value in the random forest algorithm based on primary tumor tissue-derived EVPs. Primary tumor tissue (n = 38; colon [n = 3, stage 0 = 1, stage III = 2], lung [n = 14, stage I = 7, stage II = 5, stage III = 2], and pancreas [n = 21, stage I = 1, stage II = 15, stage III = 5]) or tumor-positive draining lymph nodes from melanoma patients (n = 5, stage III = 5) were analyzed.
(D) Classification error matrix for the 75% training and 25% test sets from plasma-derived EVPs.
(E) Unsupervised 2D (left) and supervised 3D (right) t-SNE plots representing proteins in (F).
(F) Proteins with the highest predictive value as determined by random forest algorithm based on plasma-derived EVP differences relative to primary tumor type. Samples included breast cancer (n = 8, stage I = 1, stage II = 2, stage IV = 5), colorectal cancer (n = 3, stage 0 = 1, stage III = 2), lung cancer (n = 12, stage I = 6, stage II = 5, stage III = 1), pancreatic cancer (n = 9, stage 2 = 7, stage 3 = 2), and mesothelioma (n = 15, stage I = 2, stage III = 1, stage IV = 1, not available [NA] = 11).
(G) Model illustrating sources of plasma EVPs reflecting a combination of TT-, AT/DT-, and DO-derived EVPs.
Plasma-Derived EVP-Based Liquid Biopsies Classify Cancer Types
Because tissue biopsies are not always available to confirm tumor type, we performed a similar analysis using plasma-derived EVP proteomes from patients with five different cancers, including breast, colorectal, lung, and pancreatic cancers as well as mesothelioma. Even though the majority of plasma-derived EVPs are of hematopoietic origin (Caby et al., 2005), feature selection of EVP proteins by random forest analysis revealed a strong association within the same tumor type, as demonstrated by the training versus test set classifier results, heatmap, and 3D t-SNE projection (Figures 7D–7F). Similar to our analysis of cancer versus non-cancer plasma, among the 30 EVP proteins that could distinguish between cancer types, immunoglobulins were the top family of proteins found at high frequency in most plasma-derived EVP samples, especially in mesothelioma and LuCa (Figure 7F). Importantly, we found that samples cluster based on primary tumor type regardless of cancer stage for all five cancer types analyzed. These findings constitute proof of principle that plasma-derived EVPs proteomes represent bona fide tumor-specific signatures capable of distinguishing cancer types, independent of their stage. Overall, tissue- and plasma-derived EVP proteomes can be beneficial in determining tumor type for a diagnosis in patients with cancer of unknown primary tumor origin.
DISCUSSION
Liquid biopsy tests show promise for early cancer detection, tumor classification, and monitoring treatment responses. The billions of EVPs circulating in bodily fluids could represent an essential component of the liquid biopsy test. Despite previous exosomal protein biomarker studies (Castillo et al., 2018; Gangoda et al., 2017; Hurwitz et al., 2016; Ji et al., 2013) a consensus on EVP markers is lacking due to limited EVP proteomic datasets from human samples and appropriate controls to guide data analysis and interpretation.
Here, we performed a large-scale, comprehensive analysis of EVP proteomes from 426 human cancer and non-cancer samples derived from various cells, tissues, and bodily fluids. Several standard exosome markers, including CD63, TSG101, flotillins, and ALIX, were not well represented in human plasma, suggesting a need for additional pan-exosome markers for EVP purification and detection. We identified markers for EVP isolation from liquid biopsies, such as MSN, FLNA, STOM, and the RAP1B. Importantly, TT, non-tumor tissue, and plasma EVPs are heterogeneous populations (Jeppesen et al., 2019; Zhang et al., 2019); therefore, future work will determine the contribution of exosomes and exomeres to proteomic signatures.
Our proof of principle analysis in patients identified proteins expressed at significantly higher levels or found exclusively in TT-derived EVPs, as compared to AT- and DT-derived EVPs. Proteins involved in EMT, coagulation, and actin signaling pathways were enriched in PaCa EVPs, whereas cell cycle, metabolic, and RNA processing pathways abounded in LuCa EVPs. Over 40 EMT-related proteins (e.g., ECM molecules, ITGs, and proteases) were uniquely packaged in PaCa EVPs and may reflect the degree of tumor stromal infiltration. Noticeably absent were the nuclear EMT proteins SNAIL, SLUG, ZEB, and TWIST, because transcription factors are rarely packaged into EVPs. Conversely, proteins associated with RNA processing, but not EMT-associated proteins, were present in LuCa but not PaCa EVPs, further illustrating the tumor specificity of EVP protein packaging. Interestingly, proteins involved in clotting/thrombosis, such as Factors II, III, and IX and THBS2 in PaCa and THBS2 in LuCa, were highly packaged in tumor EVPs, consistent with the life-threatening thrombosis observed in these patients.
Among the proteins highly enriched in PaCa and LuCa TT, we found 11 shared tumor-specific EVP proteins including ECM molecules (BSG and VCAN), FBLN2, and immunomodulators, such as LGALS9. In contrast, the vast majority of highly enriched TT EVP proteins were exclusive to each tumor type, highlighting cancer heterogeneity across tumor types at the EVP level. By expanding our analysis to 18 different cancer types compared to various control samples (e.g., AT/DT and breast reduction tissues), we identified 16 proteins that best defined both adult and pediatric cancer, many of which represent adhesion molecules (i.e., VCAN and THBS2) underscoring the importance of ECM in cancer. Interestingly, we found many DAMP molecules, which are essential to normal immune function and sterile inflammation associated with tissue repair, in both tumor and non-tumor EVPs (Wolchok et al., 2010). However, we also identified DAMP molecules specific to tumor EVPs, such as S100A4 and S100A13, BSG and LGALS9, which may induce immune suppression and tumor-promoting inflammation. These data are consistent with our previous findings that tumor EVPs transfer their cargo to recipient cells at distant sites, generating pro-inflammatory pre-metastatic niches that support future metastasis (Costa-Silva et al., 2015; Hoshino et al., 2015).
In LuCa, HTATIP2, which is secreted following HIV infection and associated with HIV-associated neurocognitive disorders, was specifically packaged in TT-derived EVPs. Because tumor EVPs disseminate systemically and disrupt the blood-brain barrier (Chen et al., 2016; Rodrigues et al., 2019), EVP HTATIP2 may contribute to the paraneoplastic syndrome described in LuCa patients. Furthermore, epigenetic changes drive cancer progression in LuCa, (Duruisseaux and Esteller, 2018); therefore, it was not surprising that EVP METTL1 was exclusively detected in LuCa TT. Thus, tumor-derived EVPs may drive epigenetic changes in the tumor microenvironment and distant organs.
EVPs reflect the systemic effects of cancer, the cancer-associated changes occurring not only in the developing primary tumor, but also the tumor microenvironment, distant organs (e.g., liver), and the immune system (Figure 7G). Thus far, we showed that cancer-associated circulating EVPs are derived from TT, the tumor microenvironment, distant organs and immune cells in cancer patients, and healthy control EVP signatures are as informative in identifying cancer as cancer-derived EVPs. Importantly, plasma-derived EVPs were replete with immunoglobulins, which was the most highly represented family of proteins distinguishing normal and cancer samples, as well as between cancer types. This finding is in accordance with studies demonstrating that tumor-infiltrating and systemic B cell responses are both predictive and indicative of responses to immunotherapy (Helmink et al., 2020). Interestingly, many of the plasma-derived EVP proteins specific for the organ where the cancer originated were shared between TT and AT/DT, suggesting that the tumor microenvironment is a major contributor to cancer-associated EVPs in plasma.
By examining cancer-associated plasma EVPs from a diversity of stage I to stage IV cancer patients, we could detect tumor-associated EVP protein signatures prior to the development of distant metastases, suggesting that plasma-circulating EVP proteins could be used as biomarkers for early cancer detection. Our proof of principle studies provide a rationale for a concerted effort to rigorously screen patients with genetic predispositions to cancer (germline BRCA1 and P53 mutations) or those with pro-inflammatory conditions (i.e., pancreatitis, ulcerative colitis, and Crohn’s disease) predisposing them to cancer development. Screening for PaCa in these individuals may lead to early diagnosis, prior to clinical manifestations, allowing for the administration of potentially curative radiation/surgical therapies. Examining specific tumor-associated EVP protein profiles in tissues and plasma should be part of the standard-of-care monitoring strategy.
Up to 5% of patients admitted at major cancer centers are diagnosed with “cancer of unknown primary origin,” and their treatment consists of a combination of several highly cytotoxic therapies (Stella et al., 2012; Varadhachary and Raber, 2014). We showed that different cancer types, including PaCa, LuCa, breast cancer, colorectal cancer, and mesothelioma, can be distinguished through specific combinations of EVP proteins, derived from either tumor tissues or plasma. These cancer-type-specific EVP protein signatures could be used as a liquid biopsy tool to help diagnose and guide treatments for these patients.
Taken together, our findings support the idea that tumor-associated EVP proteins could be used as biomarkers for early-stage cancer detection, treatment response, and potentially for diagnosing tumors of unknown primary origin. These findings could lead to the development of novel and improved methods for total or tumor-derived EVP isolation and implementing routine plasma EVP-based screening in the clinic.
Limitations of Study
This proof of principle study uses human tissue and plasma EVP proteomes to identify early cancer detection biomarkers and classify tumors of unknown primary origin. Although we employed widely used, standard EVP isolation methodologies, advanced technologies will be required to minimize contaminants, especially in the plasma, and to further validate key EVP proteins highlighted here. Moreover, dissecting the contribution of EVP subpopulations, such as Exo-L, Exo-S vesicles, and exomeres to these biomarker signatures may further strengthen diagnostic interpretations. Last but not least, because plasma circulating EVP proteomes reflect systemic host responses, and function rather than genotype, further studies on large patient cohorts will be required to directly compare their power, sensitivity, and specificity to Food and Drug Administration (FDA)-approved tests based on circulating DNA or plasma protein detection as standard, routine diagnostic tools for early cancer detection in the clinic.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, David Lyden (dcl2001@med.cornell.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The MS-based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride) and is available via ProteomeXchange with identifier PXD018301. The code supporting the current study has not been deposited in a public repository as it does not contain newly generated software or custom code, but is available from the corresponding author upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and cell culture
B16-F10, B16-F1, 4T1, 67NR, 168FARN, CT26, K7M2, Melan-A, LLC1 (LL/2), HIEC-6, NIH/3T3, H2373, H-MESO-1, human mesothelial cells LP-9, ORT and HCG27 (gifts from Dr. A. Shukla), NAMALWA, MKN45, C4–2B, WERE-Rb-1, Y79, DLD-1, HEK293, MDA-MB-231 series (parental, −1833, −4175 and −831, gifts from Dr. J. Massague; −4173 and −4180, gifts from Dr. A. Minn; 231BR, gift from Dr. P. Steeg) (Bos et al., 2009; Kang et al., 2003; Minn et al., 2005; Yoneda et al., 2001), SW620, HCT116 (Horizon Discovery), uveal melanoma (gift from Dr. V. Rajasekhar), 131/4–5B2 and 131/8–2L (gifts from Dr. R. Gladdy) (Cruz-Munoz et al., 2008), CCG9911 and CLS1 (gifts from Dr. A. Kentsis), MCF10A, MDA-MB-468, VCAP, HIEC, HT29, MiaPaca2, Kasumi, SNU1, SNU16, LNCaP, human rhabdomyosarcoma CT10 and RD (gifts from Dr. R Gladdy), human osteosarcoma Saos-2 and U2OS and human Ewing sarcoma SK-NP-DW (gifts from Dr. A Narendran), PaCa cell lines PANC-1, AsPC-1, Pan02 (purchased from the National Cancer Institute Tumor Repository) and NIH 3T3 cell were cultured in DMEM, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) and 10% FBS. Human melanoma cells (SK-Mel03, A375M and A375P were obtained from MSKCC), human prostatic carcinoma cell lines PC3 and DU145, as well as human PaCa cell lines BXPC-3, HPAF-II, human LuCa cell lines LLC, PC-9, H1650, H1975, H292, H358, H2228, A549, 1118A and ET2B (PC-9, ET2B and 1118A, gifts from Dr. P. Gao and J. Bromberg), human leukemia cell line Nalm6, K-562 (DSMZ) and NB-4 (DSMZ) cells and murine breast cancer cell line E0771 were cultured in RPMI, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) and 10% FBS. Human breast cancer cell line SK-BR-3 was cultured in McCoy’s 5a Medium Modified, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) and 10% FBS. WI-38 cells were cultured in MEM alpha, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) and 10% FBS. Human osteosarcoma cell line 143B, human Ewing sarcoma cell line SKES1, human neuroblastoma SK-N-BE(2) and IMR5 (gifts from Dr. A. Narendran) were cultured in RPMI, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), non-essential amino acids, sodium pyruvate, HEPES and 10% FBS. Unless stated otherwise, cell lines were obtained from American Type Culture Collection. Human and mouse cell lines were authenticated using STR profiling by commercial providers. Mycoplasma testing was performed prior to EVP isolation for all of the cell lines using the ATCC Mycoplasma testing kit. All cells were maintained in a humidified incubator with 5% CO2 at 37ºC and routinely tested and confirmed to be free of mycoplasma contamination. When collecting conditioned media for EVP isolation, FBS (GIBCO, Thermo Fisher Scientific) was first depleted of EVPs by ultracentrifugation at 100,000 xg for 70 min. Cells were cultured for 3–4 days and supernatant was collected before cells reached confluency.
Primary cell cultures
Primary HMEC strains were generated and maintained as described previously (Labarge et al., 2013). Human mammary epithelia were derived from discarded reduction mammoplasty tissue in accordance with applicable legal and ethical standards per the internal review board at City of Hope; IRB#15418. S1 and T4–2 cells (gift from Dr. M Bissell) (Weaver et al., 2002) were grown in H14 medium on collagen-coated tissue culture flasks. HepG (gift from Dr. R Schwartz) were cultured in collagen-coated plates in DMEM, supplemented with 10% FBS. Human mammary epithelial cells and fibroblast cell lines N253_LEP, N253_MEP, N255_MEP, N274_fibroblast and N274_MEP (gifts from Dr. M Bissell) were cultured in DMEM/F12, supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml) and 10% FBS.
Human specimens and processing
Fresh human tumor tissues were obtained from patients surgically treated at Memorial Sloan Kettering Cancer Center (MSKCC) (see Table S1 for age, gender and stage). All individuals provided informed consent for tissue donation according to protocols approved by the institutional review board of MSKCC (IRB 11–033A, 16–774, 16–1514 and 15–015, MSKCC; IRB 0604008488, WCM). The study is compliant with all relevant ethical regulations regarding research involving human participants. Weanalyzed all the available human specimens. No statistical method was used for sample size estimation. The study does not involve any clinical trials or randomization into experimental groups.
Tissue samples
Millimeter-sized fresh tumor and peritumoral adjacent tissue were harvested from patients with localized PaCa undergoing resection with curative intent (either pancreaticoduodenectomy or distal pancreatectomy) at MSKCC. The ages ranged from 3 to 88. The tissue was placed in ice-cold PBS within minutes of collection and submitted for downstream processing and analysis. The pancreatic tissue collection was conducted through the Precision Pathology Biobanking Center (PPBC), Department of Pathology, MSKCC. PPBC separated a biopsy of tumor tissue and procured a separate biopsy of peritumoral non-involved pancreas (AT) wherever there was a sufficient resection margin. Tissues were cut into small pieces and cultured for 24 hours in serum-free RPMI, supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). Conditioned media was processed for EVP isolation. LuCa, breast cancer, colorectal cancer, DSRCT, epithelioid sarcoma, fibrolamellar sarcoma, fibromeller HCC, hepatoblastoma, immature teratoma, renal cell carcinoma, melanoma, MPNST, neuroblastoma, osteosarcoma, rhabdomyosarcoma, synovial sarcoma and Wilms’ tumor were collected from patients undergoing resection at MSKCC and processed as described above.
Human plasma/serum samples
Plasma samples were collected from patients or healthy controls. Sample size varied from 0.4–6 mL of plasma. The ages ranged from 1 to 83. Blood samples collected in lavender-top EDTA tubes were kept at room temperature for 10 minutes followed by 10 minute centrifugation at 500 × g, 20 minute centrifugation at 3,000 × g, 20 minute centrifugation at 12,000 × g, and the supernatant was collected and stored at −80C for EVP isolation. Samples were thawed on ice and centrifuged at 12,000 × g for 20 min to remove large microvesicles. EVPs were collected by spinning at 100,000 × g for 70min. EVPs were washed in PBS and pelleted again by ultracentrifugation in a Beckman Coulter Optima XE or XPE ultracentrifuge. The final EVP pellet was resuspended in PBS, and protein concentration was measured by BCA (Pierce, Thermo Fisher Scientific). Five micrograms of EVP protein were used for mass spectrometry analysis. Serum samples were collected in serum collection tubes with spray-coated silica. Sample size varied from 0.4–6 mL of serum. All of the samples were then processed using the same protocol employed for plasma samples for EVP isolation and proteomics analysis.
Bone Marrow
Three mililiters of bone marrow plasma from healthy donors were purchased from HemaCare and stored at −80C for EVP isolation. The ages ranged from 28–69 years and 80% of subjects were male. The entire sample volume available was then processed using the same protocol employed for plasma samples for EVP isolation and proteomics analysis.
Human lymphatic fluid
A volume of 0.7–15 mL of lymphatic fluid was collected after radical lymphadenectomy from routinely used sucking drainage. To ensure that the sample of lymph fluid did not contain any surgical debris, only the fluid released between 24 and 48 hours was collected (the first 24 hour batch was discarded). Samples were centrifuged (500 xg, 10 minutes followed by 20 minute centrifugation at 3,000 xg), and the supernatant was collected and stored at −80C for EVP isolation. The entire sample volume available was then processed using the same protocol employed for plasma samples for EVP isolation and proteomics analysis. Age and gender information were not obtained for these samples.
Human bile duct fluid
With the approval of the MSKCC IRB 10–118, a bile bank was established in 2010 and prospectively maintained for patients undergoing resection of hepatopancreatobiliary cancer. Bile was collected for the bank by needle cannulation of the common bile duct at the time of operation. Patients had pathologically confirmed extra-hepatic cholangiocarcinoma when the bile was collected. The ages ranged from 56–92 years and 75% were male. Bile was snap-frozen in liquid nitrogen and stored at −80C until analysis. One milliliter of bile from each patient was used for EVP isolation and analysis. One milliliter of ice-cold PBS was added to each thawed bile fluid, and the mixture was homogenized by repeated pipetting followed by EVP isolation. The entire sample volume available was then processed using the same protocol employed for plasma samples for EVP isolation and proteomics analysis.
Mouse specimens and processing
All mouse experiments were performed in accordance with Institutional Animal Care and Use Committees (IACUC) and American Association for Laboratory Animal Science (AAALAS) guidelines (Weill Cornell Medicine animal protocol 0709–666A). Female 6–8 weeks old wild-type C57BL/6 and BALB/c mice, and immunocompromised NOD/SCID/γc−/− (NSG) and athymic nude mice (Foxn1nu), and PyMT (Tg(MMTV-PyVT)634Mul) mice were obtained from Jackson Laboratories. Pdx1Cre;Lsl-KrasG12D;Lsl-TP53R172H (KPC) mice were obtained from Dr. B. Stanger (Hingorani et al., 2005; Stanger et al., 2005). Animals were monitored for stress, illness or abnormal tissue growth, and euthanized if health deteriorated. Animals were not involved in any previous procedures nor received any drugs. Animals were provided water and chow ad libitum and maintained in a pathogen-free facility. To isolate EVPs from plasma of tumor-bearing mice, 1 × 106 melanoma, breast, pancreatic, or lung tumor cells were injected into nude mice. Mouse blood (250 μl) was drawn from the retro-orbital sinus through a capillary tube (Fisher Scientific) into a BD EDTA microtainer blood collection tube (Fisher Scientific) when tumor size was over 800 mm3. From non-tumor bearing mice, 0.25–1 mL of blood was drawn from the retro-orbital sinus through a capillary tube (Fisher Scientific) into a BD EDTA microtainer blood collection tube (Fisher Scientific). The plasma of mice within the same group was pooled for EVP isolation. Tumor and non-tumor tissues were cut into small pieces (around 1 mm3) and cultured for 24h in serum-free RPMI, supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). Conditioned media and plasma were processed for EVP isolation as described above.
METHOD DETAILS
EVP purification, characterization and analyses
EVPs were purified by sequential ultracentrifugation (Figure 1A). Cell contamination was removed from 3–4 day cell culture supernatant, bodily fluids or resected tissue culture supernatant by centrifugation at 500 × g for 10 min. To remove apoptotic bodies and large cell debris, the supernatants were then spun at 3,000 × g for 20 min, followed by centrifugation at 12,000 × g for 20 min to remove large microvesicles. Finally, EVPs were collected by ultracentrifugation in 4 or 31 mL ultracentrifugation tubes (#355645 and #355631, Beckman Coulter) at 100,000 × g for 70min. EVPs were washed in PBS and pelleted again by 100,000 × g ultracentrifugation in 50.4Ti or 70Ti fixed-angle rotors in a Beckman Coulter Optima XE or XPE ultracentrifuge. For PaCa samples, conditioned media was processed for EVP isolation with the final step using sucrose cushion to remove adipose and insoluble material contamination as previously described (Lamparski et al., 2002). One milliliter of sucrose density cushion, composed of 20mM Tris, 30% sucrose, deuterium oxide (D2O), pH 7.4, was overlayed with PBS-re-suspended EVP pellets in a ultracentrifuge tube and spun at 100,000 × g for 70 min. The final EVP pellet was resuspended in PBS, and protein concentration was measured by BCA (Pierce, Thermo Fisher Scientific). EVP size and particle number were analyzed using the LM10 or DS500 nanoparticle characterization system (NanoSight, Malvern Instruments) equipped with a violet laser (405 nm). Samples were subjected to mass spectrometry in triplicate for cell lines where amounts were sufficient, and the stages of obtaining proteomic raw data were blinded for the experimental group. No randomization or stratification of samples into groups was necessary, and thus none were performed.
Data-dependent analysis of EVP samples
Enriched EVP samples (typically 5ug - adjusted based on BCA measurements) were dried by vacuum centrifugation and re-dissolved in 30–50uL 8M Urea/50mM ammonium bicarbonate/10mm DTT. Following lysis and reduction, proteins were alkylated using 20 or 30mM iodoacetamide (Sigma). Proteins were digested with Endopeptidase Lys C (Wako) in < 4M urea followed by trypsination (Promega) in < 2M Urea. Peptides were desalted and concentrated using Empore C18-based solid phase extraction prior to analysis by high resolution/high mass accuracy reversed phase (C18) nano-LC-MS/MS. Typically, 30% of samples were injected. Peptides were separated on a C18 column (12 cm / 75 μm, 3 μm beads, Nikkyo Technologies) at 200 or 300 nl/min with a gradient increasing from 1% Buffer B/95% buffer A to 40% buffer B/60% Buffer A in typically 90 or 120 min (buffer A: 0.1% formic acid, buffer B: 0.1% formic acid in 80% acetonitrile). Mass spectrometers (Q-Exactive, Q-Exactive Plus, Q-Exactive-HF or Fusion Lumos, Thermo Scientific) were operated in data dependent (DDA) positive ion mode.
Proteomic database search
High resolution/high mass accuracy nano-LC-MS/MS data was processed using Proteome Discoverer 1.4.1.14 (Thermo-Scientific, 2012)/Mascot 2.5 (Perkins et al., 1999). Human data was queried against the UniProt’s Complete HUMAN proteome (February, 2020: 74,788 sequences). Mouse data was queried against UniProt’s Complete MOUSE proteome (March, 2020; 55,412 sequences) using the following parameters: Enzyme: Trypsin/P, maximum allowed missed cleavage sites: 2, monoisotopic precursor mass tolerance: 10 ppm, monoisotopic fragment mass tolerance: 0.02 Da, dynamic modifications: Oxidation (M), Acetyl (Protein N-term), static modification: Carbamidomethyl (C). Percolator was used to calculate peptide False Discovery Rates (FDR), which was calculated per file. A false discovery rate (FDR) of 1% was applied to each separate LC-MS/MS file. For EVP enriched samples that had been in contact with Fetal Bovine Serum (FBS, exemplified by samples that originated from cell culture) an FBS specific database was concatenated to the human and mouse databases when querying the data. For plasma and tissue samples, solely the sequences of porcine trypsin and Endopeptidase LysC were concatenated to the human and mouse databases.
Targeted MS analysis
Using the library search results, a set of target peptides from proteins of interest (CA2, CD55, GLIPR2, KRAS, P4HB, PEBP1, PSMA4, PACSIN2, TGM2, PTPRJ, ABCB1, XPNPEP2, ADGRG6, ABCB11, ITGA1, LTF, ALPL, SRI, LRRC26 and BAIAP2L1, see Table S8 for list of targeted peptides) was selected and a time-scheduled parallel reaction monitoring (PRM) method was designed. One microgram of EVP protein from each sample (15 healthy, 15 pancreatic cancers) was digested as described above then combined and fractionated by high-pH reverse phase spin columns (Pierce, cat# 84868) according to manufacturer specifications, yielding a total of 9 fractions. Each fraction was injected into the Orbitrap Fusion Lumos (Thermo Scientific) operating in data-dependent mode with quadrupole isolation and HCD fragmentation. MS1 resolution was set to 60k and MS2 resolution was set to 30k. Each fraction was injected twice, with the first injection scanning from 350–650 m/z and the second injection scanning from 640–1200 m/z. Separation was achieved using a 120mm*100μm pulled-emitter fused silica column packed with 3μm C18 (Nikkyo Technos) coupled to an Easy 1200 nLC HPLC system (Thermo Scientific). Solvent A was 0.1 formic acid in water and solvent B was 0.1% formic acid, 80% acetonitrile in water. Peptides were separated at 300nL/min across a linear gradient ranging from 2%–35% B over 70 minutes followed by a sharp increase to 90% B over 1 minute and 17 minutes washing at 90% B. Raw data was searched as described above, using solely the proteolytic enzymes as contaminants. Raw data from the targeted experiment was analyzed as described above, except for using a fixed PSM validation rather than an FDR-based correction.
Transmission electron microscopy (TEM)
For negative staining TEM analysis, 0.1 μg/μl of EVPs in PBS were placed on a formvar/carbon coated grid and allowed to settle for 1 min. The sample was blotted and negatively stained with 4 successive drops of 1.5% (aqu) uranyl acetate, blotting between each drop. Following the last drop of stain, the grid was blotted and air-dried. Grids were imaged with a JEOL JSM 1400 (JEOL, USA, Ltd, Peabody, MA) transmission electron microscope operating at 100Kv. Images were captured on a Veleta 2K × 2K CCD camera (Olympus-SIS, Munich, Germany).
Asymmetric-flow field-flow fractionation (AF4) fractionation
Exosome subpopulations (exomeres, < 50 nm with an average of 35 nm in diameter; Exo-S, 60–80 nm in diameter; Exo-L, 90–120 nm in diameter and small exosome vesicles) were separated using AF4 as previously described (Zhang et al., 2018; Zhang and Lyden, 2019). Briefly, samples were separated in a short channel (144mmlength, Wyatt Technology, Santa Barbara) with a 10 kDa molecular weight cutoff (MWCO) Regenerated Cellulose membrane (Millipore) on the accumulation bottom wall and a 490 μm spacer (channel thickness). The fractionation was operated by the Eclipse AF4 system (Wyatt Technology). Lastly, the system was eluted twice. Chemstation software (Agilent Technologies) with an integrated Eclipse module (Wyatt Technology) was used to operate the AF4 flow and Astra 6 (Wyatt Technology) was used for data acquisition and analysis. One hundred micrograms of protein per sample (at 1 μg/μl, i.e., 100 μl) isolated using the sequential ultracentrifugation method were spun at 12,000 × g for 5 min before loading onto the AF4 system (to remove aggregates) and then injected using the autosampler.
Wester Blot Analysis
EVPs were harvested in RIPA buffer (Sigma, 20–188) supplemented with a protease and phosphatase inhibitor cocktail (Thermofisher, 78440). 5 μg (cell lines and tissue samples) and 20 μg (plasma) of proteins were diluted with sample buffer, run on Novex 4%–20% Tris-glycine gels (Life Technologies, XP04122BOX) and transferred onto PVDF membranes (Thermofisher, 88520). Membranes were sequentially blocked with 1X TBS containing 5% BSA (w/v) and 0.1% Tween20 (v/v), incubated with antibodies against conventional exosome, and newly discovered EVP markers (see Key Resources Table) overnight at 4ºC, washed 3 times with 1X PBS containing 0.1% Tween20 (v/v), incubated with horseradish peroxidase-conjugated anti-mouse (Santa Cruz biotechnology, sc-516102) or anti-rabbit (Jackson Immunoresearch, 111–035-144) secondary antibodies and washed again to remove unbound antibody. Bound antibody complexes were detected with ECL (GE healthcare, RPN2209).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CD9 (clone MM2/57) | Millipore | Cat#CBL162; RRID: AB_2075914 |
| Mouse monoclonal anti-CD81 (clone B11) | Santa Cruz | Cat#cs166029; RRID: AB_2275892 |
| Mouse monoclonal anti-Tsg101 (clone c-2) | Santa Cruz | Cat#sc7965: RRID AB_671392 |
| Rabbit polyclonal anti-beta-actin | Cell Signaling | Cat#4967; RRID: AB_330288 |
| Mouse monoclonal anti-syntenin-1 | Santa Cruz | Cat#sc100336; RRID: AB_2183156 |
| Mouse monoclonal anti-Mac-2BP | Santa Cruz | Cat#sc374541; RRID: AB_10989981 |
| Mouse monoclonal anti-stomatin | Santa Cruz | Cat#sc376869 |
| Mouse monoclonal anti-beta-2-microglobulin (clone G-10) | Santa Cruz | Cat#sc46697; RRID: AB_626749 |
| Mouse monoclonal anti-moesin (clone E-10) | Santa Cruz | Cat#sc13122; RRID: AB_627962 |
| Mouse monoclonal anti-RhoV (clone F-2) | Santa Cruz | Cat#sc515072 |
| Mouse monoclonal anti-moesin (clone E-10) AF647 | Santa Cruz | Cat#sc13122; RRID: AB_627962 |
| Mouse beta-2-microglobulin (BBM.1) | Santa Cruz | Cat#sc13565; RRID: AB_626748 |
| Biological Samples | ||
| Human pancreatic cancer and adjacent normal pancreas tissue; matched blood | Memorial Sloan Kettering Cancer Center | See Table S1 for a list of patients included |
| Human lung cancer and adjacent or distal normal lung tissue; matched blood | Memorial Sloan Kettering Cancer Center | See Table S1 for a list of patients included |
| Human malignant resected tissues | Memorial Sloan Kettering Cancer Center | See Table S1 for a list of patients included |
| Healthy donor blood | Memorial Sloan Kettering Cancer Center | See Table S1 for a list of donors included |
| Human bodily fluids (bile, lymph) | Memorial Sloan Kettering Cancer Center | See Method Details |
| Human bone marrow | HemaCare | N/A |
| Patient-derived xenografts (PDX) | Dr. V. Rajasekhar | N/A |
| Critical Commercial Assays | ||
| Human DAF ELISA Kit (CD55) | Abcam | Cat#ab256405 |
| Human Carbonic Anhydrase 2 (CA2) ELISA Kit | Abcam | Cat#ab222881 |
| Human Lactoferrin ELISA Kit | Abcam | Cat#ab200015 |
| Deposited Data | ||
| Proteomic MS data | This paper | https://www.ebi.ac.uk/pride/archive/projects/PXD018301/ |
| Experimental Models: Cell Lines | ||
| Human: MDA-MB-231 | Dr. J. Massague; Minn et al., 2005 | N/A |
| Human: MDA-MB-1833 | Dr. J. Massague; Kang et al., 2003 | N/A |
| Human: MDA-MB-4175 | Dr. J. Massague; Minn et al., 2005 | N/A |
| Human: MDA-MB-831 | Dr. J. Massague; Bos et al., 2009 | N/A |
| Human: MDA-MB-4173 | Dr. A. Minn; Minn et al., 2005 | N/A |
| Human: MDA-MB-4180 | Dr. A. Minn; Minn et al., 2005 | N/A |
| Human: MDA-MB-231BR | Dr. P. Steeg; Yoneda et al., 2001 | N/A |
| Human: uveal melanoma | Dr. V. Rajasekhar | N/A |
| Human: 131/4-5B2 | Dr. R. Gladdy; Cruz-Munoz et al., 2008 | N/A |
| Human: 131/8-2L | Dr. R. Gladdy; Cruz-Munoz et al., 2008 | N/A |
| Human: CCG9911 | Dr. A. Kentis | N/A |
| Human: CLS1 | Dr. A. Kentis | N/A |
| Human: MCF10A | ATCC | CRL-10317 |
| Human: MDA-MB-468 | ATCC | HTB-132 |
| Human: VCAP | ATCC | CRL-2876 |
| Human: HT-29 | ATCC | HTB-38 |
| Human: MIA PaCa-2 | ATCC | CRM-CRL-1420 |
| Human: Kasumi-1 | ATCC | CRL-2724 |
| Human: SNU-1 | ATCC | CRL-5971 |
| Human: SNU-16 | ATCC | CRL-5974 |
| Human: LNCap | ATCC | CRL-1740 |
| Human: HCT116 | ATCC | CCL-247 |
| Human: SW620 | ATCC | CCL-227 |
| Human: Rhabdomyosarcoma | Dr. R. Gladdy | CT-10, RD |
| Human: Saos-2 | ATCC | HTB-85 |
| Human: U-2 OS | ATCC | HTB-96 |
| Human: SK-NP-DW | Dr. A. Narendran | N/A |
| Human: PANC-1 | ATCC | CRL-1469 |
| Human: AsPC-1 | ATCC | CRL-1682 |
| Human: melanoma | MSKCC | SK-Mel03 |
| Human: melanoma | MSKCC | A375M |
| Human: melanoma | MSKCC | A375P |
| Human: PC-3 | ATCC | CRL-1435 |
| Human: DU 145 | ATCC | HTB-81 |
| Human: BxPC-3 | ATCC | CRL-1687 |
| Human: HPAF-II | ATCC | CRL-1997 |
| Human: NCI-H1650 | ATCC | CRL-5883 |
| Human: NCI-H1975 | ATCC | CRL-5908 |
| Human: NCI-H292 | ATCC | CRL-1848 |
| Human: NCI-H358 | ATCC | CRL-5807 |
| Human: NCI-H2228 | ATCC | CRL-5935 |
| Human: A549 | ATCC | CRM-CCL-185 |
| Human: 1118A | Drs. P. Gao, J. Bromberg | N/A |
| Human: ET2B | Drs. P. Gao, J. Bromberg | N/A |
| Human: PC-9 | Drs. P. Gao, J. Bromberg | N/A |
| Human: Nalm6 | ATCC | CRL-3273 |
| Human: K-562 | DSMZ | ACC10 |
| Human: NB-4 | DSMZ | ACC207 |
| Human: SK-BR-3 | ATCC | HTB-30 |
| Human: mammary epithelial cells | City of Hope | N/A |
| Human: mammary epithelial and fibroblasts | Dr. M. Bissell | N/A |
| Human: 143B | ATCC | CRL-8308 |
| Human: SK-ES-1 | ATCC | HTB-86 |
| Human: SK-N-BE(2) | ATCC | CRL-2271 |
| Human: IMR5 | Dr. A. Narendran | N/A |
| Human: HepG2 | Dr. R. Schwartz | N/A |
| Human: S1 | Dr. M. Bissell; Weaver et al., 2002 | N/A |
| Human: T4-2 | Dr. M. Bissell; Weaver et al., 2002 | N/A |
| Human: MCF-7 | ATCC | HTB-22 |
| Human: WI38 | ATCC | CCL-75 |
| Human: HIEC-6 | ATCC | CRL-3266 |
| Human: NIH/3T3 | ATCC | CRL-1658 |
| Human: H2373 | NCI-H2373 | N/A |
| Human: H-MESO-1 | NCI-DCTD | N/A |
| Human: LP-9 | Dr. A. Shukla | CVCL_E109 |
| Human: ORT | Dr. A. Shukla | CVCL_N815 |
| Human: HGC27 | Dr. A. Shukla | CVCL_1279 |
| Human: NAMALWA | ATCC | CRL-1432 |
| Human: MKN45 | ACCEGEN | ABC-TC0687 |
| Human: C4-2B | ATCC | CRL-3315 |
| Human: WERI-Rb-1 | ATCC | HTB-169 |
| Human: Y79 | ATCC | HTB-18 |
| Human: DLD-1 | ATCC | CCL-221 |
| Human: HEK293 | ATCC | CRL-1573 |
| Mouse: B16-F0 | ATCC | CRL-6322 |
| Mouse: B16-F1 | ATCC | CRL-6323 |
| Mouse: B16-F10 | ATCC | CRL-6475 |
| Mouse: 4T1 | ATCC | CRL-2539 |
| Mouse: 67NR | Dr. J. Bromberg | N/A |
| Mouse: 168FARN | Barbara Ann Karmanos Cancer Center | N/A |
| Mouse: Pan02 | NCI Tumor Repository | N/A |
| Mouse: E0771 | Dr. J. Bromberg | N/A |
| Mouse: LLC1 (LL/2) | ATCC | CRL-1642 |
| Mouse: CT26 | ATCC | CRL-2683 |
| Mouse: K7M2 | ATCC | CRL-2836 |
| Mouse: Melan-A | Ximbio | CVCL_4624 |
| Experimental Models: Organisms/Strains | ||
| Mouse: wild type C57BL/6 | Jackson Laboratory | Cat#000664 |
| Mouse: PyMT (Tg(MMTV-PyVT)634Mul) | Jackson Laboratory | Cat#002374 |
| Mouse: Pdx1Cre;Lsl-KrasG12D;Lsl-TP53R172H (KPC) | Dr. B. Stanger; Hingorani et al., 2005 | N/A |
| Mouse: NOD/SCID/γc−/− (NSG) | Jackson Laboratory | Cat#005557 |
| Mouse: wild type BALB/c | Jackson Laboratory | Cat#000651 |
| Mouse: athymic nude (Foxn1nu) | Jackson Laboratory | Cat#002019 |
| Software and Algorithms | ||
| Proteome Discoverer 1.4.1.14 | Thermo Scientific | https://www.thermofisher.com/us/en/home.html |
| Mascot 2.5 | Matrix Science | http://www.matrixscience.com/ |
| R, v3.2.5 | The R Foundation | https://www.r-project.org |
| GSEA, MSigDB v5.1 | UC San Diego and Broad Institute | https://www.broadinstitute.org/gsea/msigdb/index.jsp |
Exoview
The ExoView (NanoView Biosciences) human tetraspanin kit was used to analyze the samples. Chips were prescanned for background signal followed by overnight incubation with plasma-derived EVPs in Incubation Solution (1:100 dilution; 5mL of plasma were spun for EVP isolation and the pellet was resuspended in 100 ul of PBS). Chips were then washed with Solution A, followed by antibody incubation in IF Blocking Solution (final concentration of 0.1 ug/ml). Chips were then washed again with Solution A followed by Solution B and then DI water before drying. Chips were then imaged with ExoView R100 reader using the ExoScan 2.5.5 acquisition software. The data were then analyzed using ExoViewer 2.5.0 with sizing thresholds set to 50 to 200nm diameter. The number of positive particles detected per ug of EVP protein for each fluorescence channel was calculated.
ELISA
Carbonic anhydrase 2 (CA2), lactoferrin (LTF) and CD55 were measured using commercially available ELISAs (abcam222881, 200015, 256405, respectively). Four micrograms of EVP protein were used for CA2 measurements and 0.5ug of EVP protein were used for LTF and CD55 assays. For the CD55 assay the EVPs were diluted in the kit’s cell extraction buffer. The protein amounts were calculated against the kit’s standards.
For markers where ELISA kits were not commercially available, an “in-house” indirect ELISA assay (exoELISA) was designed. Briefly, EVPs were resuspended in 0.2 M sodium bicarbonate buffer (pH 9.4) and immobilized on a 96-well plate (0.5–1 ug of EVPs per well) overnight at 4ºC. EVPs were washed 3 times with PBS, permeabilized with 0.2% Triton X-100 (in TBS) and then blocked with TNB buffer (TSA biotin system, Perkin Elmer), incubated with Rho V antibody (4 ug/mL, SantaCruz, sc-515072) or mouse IgG1 isotype control (4 ug/mL, MAB002, R&D) for 16h at 4ºC and then with a fluorescently labeled anti-mouse secondary antibody (10 ng/mL, ThermoFisher Scientific), both diluted in in TNB buffer. EVPs were washed 3 times for 5 minutes with PBS between each passage. Fluorescent intensity (FI) was measured using a SpectraMax® iD5 plate reader (Molecular Devices), and the FI of the isotype control wells was subtracted from the FI value of each matched sample before representation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Computational analyses
Software tools used for this study are available as open source R packages (https://www.r-project.org, v3.2.5; R Core Team, 2013). For key analyses these include: ‘limma’ for QC, analysis and exploration of proteomic expression data; ‘fgsea’ for gene set enrichment analysis and gene-gene correlations; ‘randomForest’, ‘PAM’ and ‘caret’ for training and plotting classification and regression models. Additional data exploration results were generated using custom functions in ‘skitools’ (https://github.com/mskilab/skitools).
Tandem MS data were queried against a database using Proteome Discoverer v1.4/MASCOT software. The relative abundance of a given protein was calculated from the average area of the three most intense peptide signals. For this software, this abundance measure ranges approximately 4 orders of magnitude, resulting in a lower signal range of 0.8–1.2 × 106 that can be integrated for proteins of low abundance. Proteins for which area intensities were below the minimum range or were not detected were assigned an area of zero. For the proteins that were identified by multiple UniProt ID, the probe (based on UniProt ID) values were collapsed at the protein level using the probe with the maximum intensity.
For EVP protein frequency analysis based on presence and absence of the proteins, protein abundance was not considered; proteins were classified as detected or not detected across all samples. For pairwise comparison of PaCa and LuCa, we considered proteins as tumor exclusive markers if they were detected in at least two of the TT samples and not detected in any of the AT/DT samples. The same criteria were applied for identifying exclusive markers across plasma samples. Marker selection and heatmap generation were conducted using the software GENE-E (https://www.broadinstitute.org/software/gene-e). Proteins were sorted by signal-to-noise statistic, (μA - μB)/(αA + αB) where μ and α represent the mean and standard deviation of proteomic expression, respectively, for each class (Golub et al., 1999). Next, the signal to noise marker selection tool from GENE-E was used to identify fraction-specific markers with 1,000 permutations. To identify enriched proteins, a fold change cut-off of ≥ 10 was applied to select tumor-specific markers (FDR < 0.05). This list was further filtered for those proteins detected in at least half of TT samples (i.e., at least 2 out of 4 samples). For plasma analysis in PaCa and LuCa samples, EVP proteins that were never found in healthy control plasma but found in at least two of the patient samples were chosen. For supervised random forest, we used the entire proteomic expression dataset.
For Gene Set Enrichment Analysis (GSEA), we used the entire proteomic expression dataset (Subramanian et al., 2005). Gene sets from Molecular signatures database (MSigDB, https://www.broadinstitute.org/gsea/msigdb/index.jsp) v5.1 were used for GSEA (H: 50 hallmark gene sets; CS:KEGG: 186 canonical pathways from Kyoto Encyclopedia of Genes and Genomes [KEGG] pathway database; C5: 825 gene sets based on Gene Ontology [GO] term) (Liberzon et al., 2011). The default parameters were used to identify significantly enriched gene sets.
Random Forest is a machine learning method that combines the output of an ensemble of regression trees to predict the value of a response variable. The use of this method reduces the risk of over-fitting and makes the method robust to outliers and noise in the input data. We used Recursive Feature Elimination (RFE) provided by the caret R package for feature selection using default options and determined the minimal number of top features with the best accuracy according to the variable importance measure. We divided the data into a training set and an independent test set. Heatmaps based on random forest algorithm were generated to find highest predictive values. To identify enriched proteins, a fold change cut-off of > 10 or < 1/10 for tissue explant dataset or > 4 or < 1/4 for plasma dataset was applied to select tumor- or non-tumor specific markers (FDR < 0.05). Next, the Random Forest algorithm (RFE algorithm) was applied to identify biomarker differentiating tumor from non-tumor samples. To visualize high-dimensional datasets, t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm was applied to generate t-SNE plots using the ‘Rtsne’ R package.
Statistical analysis
All statistical analyses were performed using the statistical software R. Statistical significance was calculated by two-tailed Student’s t test or Wilcoxon rank-sum test unless specified otherwise in the figure legend. Data are expressed as mean ± SEM. A p-value < 0.05 in biological experiments or FDR < 0.05 after multiple comparison correction in proteomics data analysis was considered statistically significant.
Supplementary Material
Figure S1. Characterization of EVPs from Murine and Human Cell Lines, Tissue Explants and Bodily Fluids, Related to Figure 1
(A) TEM images for EVPs isolated from a human cell line (Panc-1), human plasma (colon cancer), human explant (melanoma), human lymphatic fluid, human bile duct fluid, a mouse cell line (B16-F10) and mouse tissue explant (lung tissue).
(B) Nanosight profiles showing size distribution for EVPs isolated from a human cell line (HCT116), human plasma (melanoma), human explant (pancreatic cancer), human lymphatic fluid, human bile duct fluid, a mouse cell line (4T1), mouse plasma (B16-F10) and mouse tissue explant (lung tissue).
(C) Average number of EVP proteins detected by MS for each source: all samples (left panel), tumor samples (middle panel) and non-tumor samples (right panel).
(D) Comparison between tumor and non-tumor samples in human and mouse, respectively (t-test).
Figure S2. EVP Protein Correlation across Samples and Conventional versus Novel EVP Marker Distribution across EVP Sub-populations, Related to Figure 1
(A) Pearson correlation of EVP protein levels among different sample sources. Larger size and darker blue depict a higher correlation between samples. Tumor samples (upper panel) and non-tumor samples (lower panel).
(B) Conventional exosome and novel EVP protein markers in EVP subpopulations. EVP subpopulations comprised of exomeres, <50 nm with an average of 35 nm in diameter; Exo-S, 60–80 nm in diameter and Exo-L, 90–120 nm in diameter, were separated using asymmetric-flow field-flow fractionation (AF4). The absolute expression values of conventional exosome markers (left panel) and 13 novel EVP markers (right panel) are shown. Scale shown is intensity (area) subtracted by mean and divided by row standard deviation (i.e. D [expression-mean]/SD).
(C) Conventional exosome marker positivity across different sample sources in tumor (left panel) and non-tumor (right panel) samples.
(D) Novel EVP marker positivity depicted for tumor (left panel) and non-tumor samples (right panel). The number in each box is the percentage of positive samples for each tissue type.
Figure S3. Gene Set Enrichment Analysis (GSEA) Based on the Complete PaCa Tissue Explant EVP Protein Dataset, Related to Figure 2
(A–I) Top 3 pathways enriched in PaCa TE EVPs compared to matched pancreas AT EVPs using Hallmark, KEGG, and GO gene sets, respectively.
(A–C) Hallmark enrichment plot, heatmap, and protein list for (A) EMT, (B) coagulation, and (C) TGF-beta pathway.
(D–F) KEGG enrichment plot, heatmap, and protein list for (D) cardiac muscle contraction, (E) actin cytoskeleton, and (F) ECM receptor interaction.
(G–I) GO enrichment plot, heatmap, and protein list for (G) endothelial cell apoptosis, (H) actin filament bundle, and (I) peptide cross linking.
Figure S4. Gene Set Enrichment Analysis (GSEA) Based on the Complete LuCa Tissue Explant EVP Protein Dataset, Related to Figure 2
(A-I) Top 3 pathways enriched in LuCa tissue explant EVPs compared to matched lung adjacent tissue EVPs using Hallmark, KEGG and GO genesets, respectively.
(A-C) Hallmark enrichment plot, heatmap and protein list for (A) E2F targets, (B) G2M checkpoint, and (C) MYC targets.
(D-F) KEGG enrichment plot, heatmap and protein list for (D) the spliceosome, (E) RNA degradation, and (F) purine metabolism.
(G-I) GO enrichment plot, heatmap and protein list for (G) RNA processing, (H) mRNA processing, and (I) ncRNA metabolic process.
Figure S5. Identification of Tumor Tissue-Specific EVP Protein Cargo in Tissue Explants Based on the Updated Annotation of the Dataset, Related to Figure 2
(A) Heatmap of tumor (n = 85) and non-tumor (n = 66) samples based on the 16 EVP proteins revealed by random forest algorithm in figure 2D–E.
(B) Classification error matrix using a random forest classifier of 75% training set and 25% test set. The analyses were based on the most recent iteration of the UniProt Complete HUMAN proteome (February, 2020: 74,788 sequences). The number of samples identified are noted in each box.
Figure S6. Identification of EVP Protein Cargo in Cancer Patient Plasma Based on the Updated Annotation of the Dataset, Related to Figures 5 and 6
(A and B) Validation of top (A) PaCa and (B) LuCa plasma EVP markers based on the most recent iteration of the UniProt Complete HUMAN proteome (February, 2020: 74,788 sequences).
(C) Heatmap of the expression of 47 EVP proteins in patient (n = 77) and healthy control (n = 43) plasma samples revealed by random forest algorithm in Figure 6.
(D) Classification error matrix using random forest classifier of 75% training set and 25% test set. The number of samples identified are noted in each box.
Highlights.
Proteomic profiles of extracellular vesicles and particles (EVPs) from 426 human samples
Identification of pan-EVP markers
Characterization of tumor-derived EVP markers in human tissues and plasma
EVP proteins can be useful for cancer detection and determining cancer type
ACKNOWLEDGMENTS
This work is dedicated to Maria de Sousa, Emeritus Professor at the University of Porto, Adjunct Professor in Pediatrics at Weill Cornell Medicine, and cofounder of the prestigious “Graduate Program in Basic and Applied Biology (GABBA)” at the University of Porto, who succumbed to COVID-19 on April 14, 2020. Maria was a noted immunologist who pioneered studies in lymphocyte migration, defining the concept of “ecotaxis.” We honor her last wish: “In your living the hope of my lasting.” We thank the Mesothelioma Research Bank (CDC NIOSH 1-U19-OH009077-01 NMVP) for plasma samples, Medical Illustration & Design Services of Yonsei University College of Medicine for the art, and the Electron Microscopy & Histology services of the Weill Cornell Medicine Microscopy & Image Analysis Core for TEM with NIH Shared Instrumentation Grant (S10RR027699). The authors gratefully acknowledge support from the National Cancer Institute (CA224175 to D.L. and V.P.B., CA210240 to D.L. and M.H., CA232093 to D.L., CA163117 and CA207983 to D.L. and Y.D.C., CA163120 to D.L. and S.K.B., CA169416 to D.L. and H.P., CA169538 to D.L. and M.J.B., CA218513 to D.L. and H.Z., and AI144301 to D.L. and V.P.), the United States Department of Defense (W81XWH-13-1-0425 to D.L. and Y.K., W81XWH-13-1-0427 and W81XWH-13-1-0249 to D.L., and W81XWH-14-1-0199 to D.L. and A.S., the Melanoma Research Alliance (to H.P), the Hartwell Foundation (to D.L. and J.B.), the Thompson Family Foundation (to D.K., A.S., D.L., R.S., Y.Y., I.S., R.S.S., V.P.B., and E.O.), the STARR Consortium (I9-A9-056 to D.L., P.R., K.K., and H.Z. and I8-A8-123 to D.L. and H.P.), the Pediatric Oncology Experimental Therapeutics Investigator’s Consortium (to T.T., D.L., M.H.R., M.B., M.P.L., T.H., and S.A.), Alex’s Lemonade Stand Foundation (to D.L. and P.R.), the Breast Cancer Research Foundation (to D.L., M.J.B., and C.M.G.), the Feldstein Medical Foundation (to D.L. and H.P.), the Tortolani Foundation (to D.L. and J.B.), the Clinical & Translational Science Center (to D.L. and H.Z.), the Mary Kay Ash Charitable Foundation (to D.L. and I.M.), the Malcolm Hewitt Weiner Foundation, the Manning Foundation, the Daniel P. and Nancy C. Paduano Family Foundation, the James Paduano Foundation, the Sohn Foundation, the AHEPA Vth District Cancer Research Foundation, the Daedalus Fund, Selma and Lawrence Ruben Science to Industry Bridge Award, the Children’s Cancer and Blood Foundation (to D.L.), the Scott and Lisa Stuart Family Foundation, the D10 Foundation (to T.T.), Susan G. Komen Postdoctoral Fellowship (PDF15331556, JST PRESTO 30021, and JSPS KAKENHI JP19K23743 to A.H.), the National Research Foundation of Korea (2019R1C1C1006709 and 2018R1A5A2025079 [MSIT]), Severance Hospital Research fund for Clinical Excellence (C-2019-0026), faculty research grant of Yonsei University College of Medicine (6-2019-0090), Daewoong Foundation Research Grant (to H.S.K.), the Swedish Cancer Society Pancreatic Cancer Fellowship (to L.B.), the Lions International Postdoctoral fellowship (to L.B.), the Sweden-America stipend (to L.B.), the Memorial Sloan Kettering Cancer Center Metastasis and Tumor Ecosystems Center fellowship (to C.P.Z.), the Weill Cornell Medical College Clinical and Translational Science Center funded by NIH/NCATS (UL1TR002384 to J.S.J.), the “Little Eric Foundation” (to Y.K.), and the EVAN Foundation (to K.K.).
Footnotes
DECLARATION OF INTERESTS
D.L., A.H., H.S.K., and L.B. have filed a U.S. patent application related to this work.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.07.009.
REFERENCES
- Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, and Lyden D (2016). Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 30, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, and Massagué J (2009). Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, and Bonnerot C (2005). Exosomal-like vesicles are present in human blood plasma. Int. Immunol 17, 879–887. [DOI] [PubMed] [Google Scholar]
- Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, Gascoyne P, Mulu FC, Stephens BM, Huang J, et al. (2018). Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann. Oncol 29, 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Liu L, Ma F, Wong CW, Guo XE, Chacko JV, Farhoodi HP, Zhang SX, Zimak J, Ségaliny A, et al. (2016). Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cell. Mol. Bioeng 9, 509–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Xue L, Hsu CC, Paez JS, Pan L, Andaluz H, Wendt MK, Iliuk AB, Zhu JK, and Tao WA (2017). Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 114, 3175–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Kim DK, Kim YK, and Gho YS (2015). Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrom. Rev 34, 474–490. [DOI] [PubMed] [Google Scholar]
- Colombo M, Raposo G, and Théry C (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol 30, 255–289. [DOI] [PubMed] [Google Scholar]
- Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et al. (2015). Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Munoz W, Man S, Xu P, and Kerbel RS (2008). Development of a preclinical model of spontaneous human melanoma central nervous system metastasis. Cancer Res. 68, 4500–4505. [DOI] [PubMed] [Google Scholar]
- Duruisseaux M, and Esteller M (2018). Lung cancer epigenetics: From knowledge to applications. Semin. Cancer Biol 51, 116–128. [DOI] [PubMed] [Google Scholar]
- Gangoda L, Liem M, Ang CS, Keerthikumar S, Adda CG, Parker BS, and Mathivanan S (2017). Proteomic Profiling of Exosomes Secreted by Breast Cancer Cells with Varying Metastatic Potential. Proteomics 17 10.1002/pmic.201600370. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. (1999). Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537. [DOI] [PubMed] [Google Scholar]
- Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, Yizhak K, Sade-Feldman M, Blando J, Han G, et al. (2020). B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez C, Huebener P, and Schwabe RF (2016). Damage-associated molecular patterns in cancer: a double-edged sword. Oncogene 35, 5931–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, and Tuveson DA (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell 7, 469–483. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, and Meckes DG Jr. (2016). Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget 7, 86999–87015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, et al. (2019). Reassessment of Exosome Composition. Cell 177, 428–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, et al. (2013). Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 13, 1672–1686. [DOI] [PubMed] [Google Scholar]
- Johnstone RM, Adam M, Hammond JR, Orr L, and Turbide C (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem 262, 9412–9420. [PubMed] [Google Scholar]
- Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, et al. (2012). Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, and Massagué J (2003). A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549. [DOI] [PubMed] [Google Scholar]
- Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, Go G, Nhung D, Hong K, Jang SC, et al. (2015). EVpedia: a community web portal for extracellular vesicles research. Bioinformatics 31, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarge MA, Garbe JC, and Stampfer MR (2013). Processing of human reduction mammoplasty and mastectomy tissues for cell culture. J. Vis. Exp (71), 50011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, and Le Pecq JB (2002). Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 270, 211–226. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas SLN, Breakefield XO, and Weaver AM (2017). Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 27, 172–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer M, Steiner M, Mosgoeller W, Prohaska R, and Salzer U (2002). Stomatin is a major lipid-raft component of platelet alpha granules. Blood 100, 897–904. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, and Simpson RJ (2009). ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, and Massagué J (2005). Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel O, Tomas A, Scott CC, and Gruenberg J (2016). Moesin and cortactin control actin-dependent multivesicular endosome biogenesis. Mol. Biol. Cell 27, 3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MF, Mortensen MB, and Detlefsen S (2016). Key players in pancreatic cancer-stroma interaction: Cancer-associated fibroblasts, endothelial and inflammatory cells. World J. Gastroenterol 22, 2678–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll L (2015). Expanding on exosomes and ectosomes in cancer. N. Engl. J. Med 372, 2359–2362. [DOI] [PubMed] [Google Scholar]
- Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med 18, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, and Cottrell JS (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Pizon V, Desjardins M, Bucci C, Parton RG, and Zerial M (1994). Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J. Cell Sci 107, 1661–1670. [DOI] [PubMed] [Google Scholar]
- Procacci P, Moscheni C, Sartori P, Sommariva M, and Gagliano N (2018). Tumor−Stroma Cross-Talk in Human Pancreatic Ductal Adenocarcinoma: A Focus on the Effect of the Extracellular Matrix on Tumor Cell Phenotype and Invasive Potential. Cells 7, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues G, Hoshino A, Kenific CM, Matei IR, Steiner L, Freitas D, Kim HS, Oxley PR, Scandariato I, Casanova-Salas I, et al. (2019). Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol 21, 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr., Carter BS, Krichevsky AM, and Breakefield XO (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol 10, 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyers L, Umlauf E, and Prohaska R (1999). Association of stomatin with lipid-protein complexes in the plasma membrane and the endocytic compartment. Eur. J. Cell Biol 78, 802–812. [DOI] [PubMed] [Google Scholar]
- Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, Greenwood A, Cheng KH, McLaughlin M, Brown D, et al. (2005). Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell 8, 185–195. [DOI] [PubMed] [Google Scholar]
- Stella GM, Senetta R, Cassenti A, Ronco M, and Cassoni P (2012). Cancers of unknown primary origin: current perspectives and future therapeutic strategies. J. Transl. Med 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et al. (2014). Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24, 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, and Clayton A (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Varadhachary GR, and Raber MN (2014). Cancer of unknown primary site. N. Engl. J. Med 371, 757–765. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Lelièvre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, and Bissell MJ (2002). beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell 2, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Weber JS, Hamid O, Lebbé C, Maio M, Schadendorf D, de Pril V, Heller K, Chen TT, Ibrahim R, et al. (2010). Ipilimumab efficacy and safety in patients with advanced melanoma: a retrospective analysis of HLA subtype from four trials. Cancer Immun. 10, 9. [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Williams PJ, Hiraga T, Niewolna M, and Nishimura R (2001). A bone-seeking clone exhibits different biological properties from the MDAMB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J. Bone Miner. Res 16, 1486–1495. [DOI] [PubMed] [Google Scholar]
- Zagorac GB, Mahmutefendić H, Tomaš MI, Kučić N, Le Bouteiller P, and Lučin P (2012). Early endosomal rerouting of major histocompatibility class I conformers. J. Cell. Physiol 227, 2953–2964. [DOI] [PubMed] [Google Scholar]
- Zhang H, and Lyden D (2019). Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc 14, 1027–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, et al. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Higginbotham JN, Jeppesen DK, Yang YP, Li W, McKinley ET, Graves-Deal R, Ping J, Britain CM, Dorsett KA, et al. (2019). Transfer of Functional Cargo in Exomeres. Cell Rep. 27, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing; (Austria: Vienna: ). http://www.R-project.org/. [Google Scholar]
- Thermo-Scientific (2012). Proteome Discoverer version 1.4.1.14.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characterization of EVPs from Murine and Human Cell Lines, Tissue Explants and Bodily Fluids, Related to Figure 1
(A) TEM images for EVPs isolated from a human cell line (Panc-1), human plasma (colon cancer), human explant (melanoma), human lymphatic fluid, human bile duct fluid, a mouse cell line (B16-F10) and mouse tissue explant (lung tissue).
(B) Nanosight profiles showing size distribution for EVPs isolated from a human cell line (HCT116), human plasma (melanoma), human explant (pancreatic cancer), human lymphatic fluid, human bile duct fluid, a mouse cell line (4T1), mouse plasma (B16-F10) and mouse tissue explant (lung tissue).
(C) Average number of EVP proteins detected by MS for each source: all samples (left panel), tumor samples (middle panel) and non-tumor samples (right panel).
(D) Comparison between tumor and non-tumor samples in human and mouse, respectively (t-test).
Figure S2. EVP Protein Correlation across Samples and Conventional versus Novel EVP Marker Distribution across EVP Sub-populations, Related to Figure 1
(A) Pearson correlation of EVP protein levels among different sample sources. Larger size and darker blue depict a higher correlation between samples. Tumor samples (upper panel) and non-tumor samples (lower panel).
(B) Conventional exosome and novel EVP protein markers in EVP subpopulations. EVP subpopulations comprised of exomeres, <50 nm with an average of 35 nm in diameter; Exo-S, 60–80 nm in diameter and Exo-L, 90–120 nm in diameter, were separated using asymmetric-flow field-flow fractionation (AF4). The absolute expression values of conventional exosome markers (left panel) and 13 novel EVP markers (right panel) are shown. Scale shown is intensity (area) subtracted by mean and divided by row standard deviation (i.e. D [expression-mean]/SD).
(C) Conventional exosome marker positivity across different sample sources in tumor (left panel) and non-tumor (right panel) samples.
(D) Novel EVP marker positivity depicted for tumor (left panel) and non-tumor samples (right panel). The number in each box is the percentage of positive samples for each tissue type.
Figure S3. Gene Set Enrichment Analysis (GSEA) Based on the Complete PaCa Tissue Explant EVP Protein Dataset, Related to Figure 2
(A–I) Top 3 pathways enriched in PaCa TE EVPs compared to matched pancreas AT EVPs using Hallmark, KEGG, and GO gene sets, respectively.
(A–C) Hallmark enrichment plot, heatmap, and protein list for (A) EMT, (B) coagulation, and (C) TGF-beta pathway.
(D–F) KEGG enrichment plot, heatmap, and protein list for (D) cardiac muscle contraction, (E) actin cytoskeleton, and (F) ECM receptor interaction.
(G–I) GO enrichment plot, heatmap, and protein list for (G) endothelial cell apoptosis, (H) actin filament bundle, and (I) peptide cross linking.
Figure S4. Gene Set Enrichment Analysis (GSEA) Based on the Complete LuCa Tissue Explant EVP Protein Dataset, Related to Figure 2
(A-I) Top 3 pathways enriched in LuCa tissue explant EVPs compared to matched lung adjacent tissue EVPs using Hallmark, KEGG and GO genesets, respectively.
(A-C) Hallmark enrichment plot, heatmap and protein list for (A) E2F targets, (B) G2M checkpoint, and (C) MYC targets.
(D-F) KEGG enrichment plot, heatmap and protein list for (D) the spliceosome, (E) RNA degradation, and (F) purine metabolism.
(G-I) GO enrichment plot, heatmap and protein list for (G) RNA processing, (H) mRNA processing, and (I) ncRNA metabolic process.
Figure S5. Identification of Tumor Tissue-Specific EVP Protein Cargo in Tissue Explants Based on the Updated Annotation of the Dataset, Related to Figure 2
(A) Heatmap of tumor (n = 85) and non-tumor (n = 66) samples based on the 16 EVP proteins revealed by random forest algorithm in figure 2D–E.
(B) Classification error matrix using a random forest classifier of 75% training set and 25% test set. The analyses were based on the most recent iteration of the UniProt Complete HUMAN proteome (February, 2020: 74,788 sequences). The number of samples identified are noted in each box.
Figure S6. Identification of EVP Protein Cargo in Cancer Patient Plasma Based on the Updated Annotation of the Dataset, Related to Figures 5 and 6
(A and B) Validation of top (A) PaCa and (B) LuCa plasma EVP markers based on the most recent iteration of the UniProt Complete HUMAN proteome (February, 2020: 74,788 sequences).
(C) Heatmap of the expression of 47 EVP proteins in patient (n = 77) and healthy control (n = 43) plasma samples revealed by random forest algorithm in Figure 6.
(D) Classification error matrix using random forest classifier of 75% training set and 25% test set. The number of samples identified are noted in each box.
Data Availability Statement
The MS-based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride) and is available via ProteomeXchange with identifier PXD018301. The code supporting the current study has not been deposited in a public repository as it does not contain newly generated software or custom code, but is available from the corresponding author upon request.