Summary
Skeletal muscle adaptation is mediated by cooperative regulation of metabolism, signal transduction, and gene expression. However, the global regulatory mechanism remains unclear. To address this issue, we performed electrical pulse stimulation (EPS) in differentiated C2C12 myotubes at low and high frequency, carried out metabolome and transcriptome analyses, and investigated phosphorylation status of signaling molecules. EPS triggered extensive and specific changes in metabolites, signaling phosphorylation, and gene expression during and after EPS in a frequency-dependent manner. We constructed trans-omic network by integrating these data and found selective activation of the pentose phosphate pathway including metabolites, upstream signaling molecules, and gene expression of metabolic enzymes after high-frequency EPS. We experimentally validated that activation of these molecules after high-frequency EPS was dependent on reactive oxygen species (ROS). Thus, the trans-omic analysis revealed ROS-dependent activation in signal transduction, metabolome, and transcriptome after high-frequency EPS in C2C12 myotubes, shedding light on possible mechanisms of muscle adaptation.
Subject Areas: Biochemistry, Physiology, Human Metabolism, Omics
Graphical Abstract
Highlights
-
•
We performed electrical pulse stimulation in differentiated C2C12 myotubes
-
•
We constructed trans-omic network after high-frequency electrical pulse stimulation
-
•
Trans-omic network integrates metabolome, transcriptome, and signaling molecules
-
•
We identified ROS-dependent pentose phosphate pathway activation
Biochemistry; Physiology; Human Metabolism; Omics
Introduction
Physiological muscle adaptation, such as muscle hypertrophy that is characteristic of increases in metabolic enzymes and mitochondrial contents and changes in muscle fiber composition, occurs with continuous exercise and frequent muscle contraction. This adaptation is mediated by cooperative regulation of multiple biological processes, including metabolism, signal transduction, and gene expression (Egan and Zierath, 2013; Hawley et al., 2014). Muscle contraction activates glycolysis and mitochondrial oxidation to maintain energy metabolism. Muscle contraction also activates signaling molecules such as AMP-activated protein kinase (AMPK), Ca2+/calmodulin-dependent protein kinase II (CaMKII), mitogen-activated protein kinases (MAPKs), and mammalian target of rapamycin (mTOR) by changing creatine/phosphocreatine, NAD/NADH, AMP/ATP, Ca2+, reactive oxygen species (ROS), and mechanical stress. Activation of signaling molecules further induces gene expression via transcription factors (TFs). Therefore, skeletal muscle contraction elicits cooperative changes across a multi-layer network, including metabolism, signal transduction, and gene expression, leading to muscle adaptation.
Changes in metabolites, signaling molecule activities, and gene expression in response to muscle contraction depend on the intensity of muscle contraction. In high-intensity exercise, the energy supply by glycogen is dominant and lactate concentration increases, because of activation of glycolysis and glycogenolysis, whereas in low-intensity exercise, the energy supply from fatty acids is dominant and the glycolysis and glycogenolysis rates decrease (van Loon et al., 2001; Romijn et al., 1993). Activation of signaling molecules such as AMPK, p38, and CaMKII also depends on the intensity of muscle contraction (Chen et al., 2003; Egan et al., 2010). Intensity of contraction in the isolated intact muscles is controlled by frequency of electrical stimulation (Cairns et al., 2007). Activation of signaling molecules dependent on frequency of electrical stimulation has thus far been studied using excised muscles and primary myotubes (Atherton et al., 2005; Scheler et al., 2013). Moreover, metabolism-related gene expression is regulated by contraction intensity (Egan et al., 2010; Hildebrandt et al., 2003).
Muscle contraction-induced changes in signal transduction, gene expression, and metabolism have been reported to last for hours to days in animals and humans (Chen et al., 2002; Mahoney et al., 2008; Neubauer et al., 2014). Genes induced by muscle contraction, especially those related to energy metabolism, are responsible for long-term effects of exercise such as increase in protein levels of metabolic enzymes and improvement in exercise capacity (Egan and Zierath, 2013; Perry et al., 2010). Metabolites of glycolysis and amino acids change during exercise (Gibala et al., 1997), whereas amino acids and fatty acids in the blood continue to increase for hours after exercise (Peake et al., 2014). Signaling molecules such as phosphorylated Akt (pAkt), mTOR, and S6K also continue to increase after exercise in human skeletal muscle (Camera et al., 2010). However, the interplay among signaling molecules, gene expression, and metabolites after muscle contraction remains to be elucidated.
Quantitative and global measurements using single-omic technology are becoming more available. Metabolome (Miyamoto et al., 2013; Peake et al., 2014), transcriptome (Chen et al., 2002; Mahoney et al., 2008; Neubauer et al., 2014), and phosphoproteome (Camera et al., 2017; Hoffman et al., 2015) have been used to examine the biological mechanisms of exercise in skeletal muscle of rodents and humans. However, despite the progress of single-omic studies, the interplay between different omic layers during exercise has not been reported. Here, we propose using “trans-omics” to construct biochemical interaction networks from multiple omic datasets (Yugi et al., 2016). In our previous studies, we developed a method to construct trans-omic network from several omic datasets and found a novel insulin action for metabolic regulation in hepatoma cells (Kawata et al., 2018; Yugi et al., 2014). However, a trans-omic analysis for muscle contraction has not been explored thus far.
In this study, we performed metabolome and transcriptome analyses and investigated phosphorylation status of signaling molecules in C2C12 myotubes during and after high- or low-frequency electrical pulse stimulation (EPS). We found that extensive changes in metabolites, gene expression, and phosphorylation of signaling molecules selectively occur after high-frequency EPS. We constructed the trans-omic network across signaling molecules, TFs, gene expression, metabolic enzymes, and metabolites induced by high-frequency EPS. The trans-omic analysis revealed that the signaling molecules, gene expression, and metabolites involved in the pentose phosphate pathway increased in a ROS-dependent manner.
Results
Analysis of Metabolome, Signaling Molecules, and Transcriptome Induced by EPS
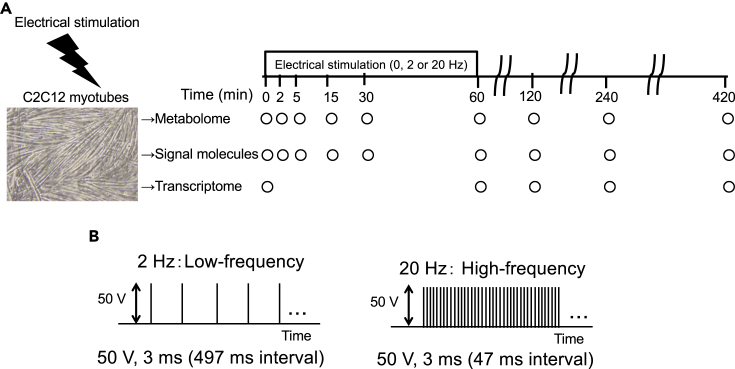
We performed low- and high-frequency EPS on C2C12 myotubes for 60 min (Manabe et al., 2012) and measured metabolome, signaling molecules, and transcriptome during and after EPS (Figures 1A and 1B).
Figure 1.
Experimental Design of Metabolome, Signaling Molecules, and Transcriptome Analyses in EPS-Stimulated Differentiated C2C12 Myotubes
(A) Metabolome, signaling molecules, and transcriptome were measured during (0, 2, 5, 15, 30, and 60 min) and after (120, 240, and 420 min) electrical pulse stimulation (EPS) at 0, 2, and 20 Hz in differentiated C2C12 myotubes.
(B) Stimulation protocol of EPS.
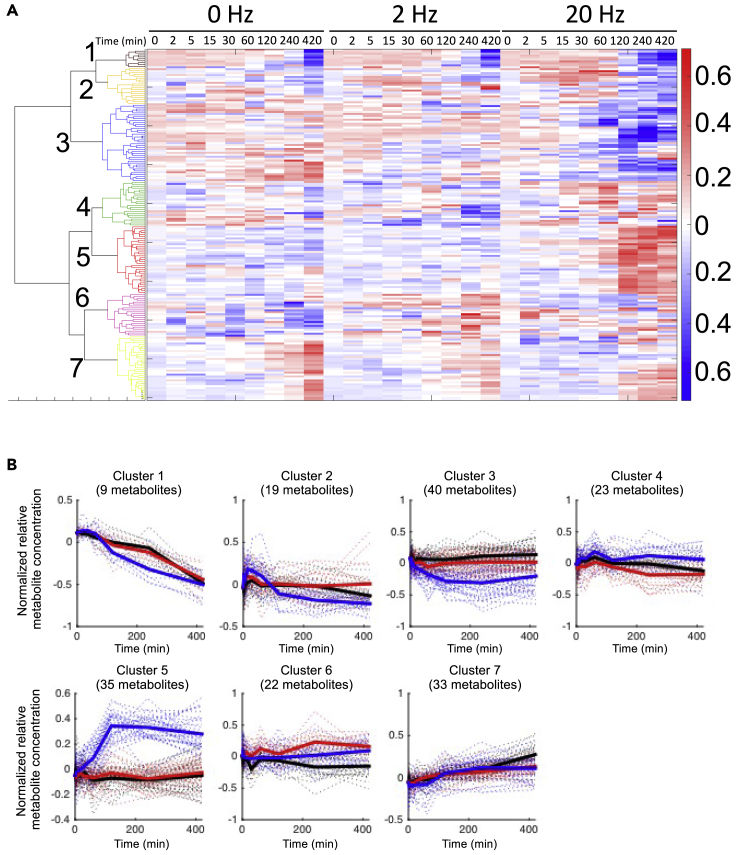
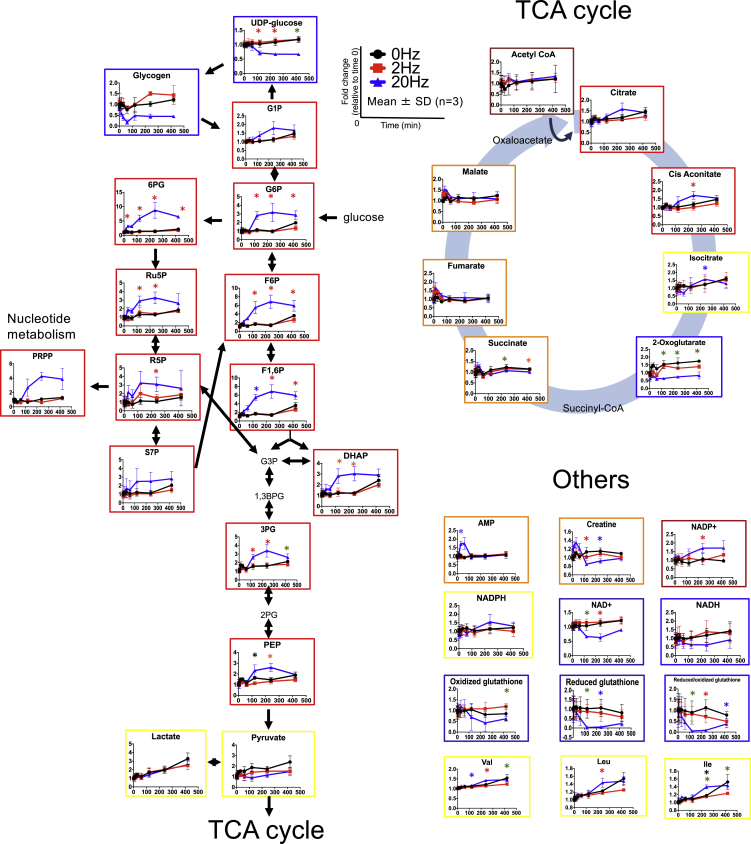
For metabolome, 179 metabolites were detected at more than one time point. The metabolites were grouped into seven clusters by cluster analysis using the time series data (Figures 2A, 2B, and S1 and Table S1). The metabolites involved in carbohydrate metabolism, glycolysis, and pentose phosphate pathway were in cluster 5, which showed continuous increase after 20-Hz EPS. Metabolites in TCA cycle (malate, fumarate, and succinate) and nucleotide metabolism (AMP, adenosine, UTP, UDP, and guanosine) were in cluster 2, which showed transient increase during 20-Hz EPS and then returned to the basal level after the EPS. Metabolites in amino acid metabolism were in cluster 3, which showed continuous decrease after 20-Hz EPS. Next, we examined the metabolites that significantly changed with EPS. One single metabolite (6PG) significantly changed during 20-Hz EPS (0–60 min), and 32 metabolites significantly changed after 20-Hz EPS but not during or after 2-Hz EPS (Table S2). These results indicate that extensive metabolic changes occurred after 20-Hz EPS. Among 32 significantly changed metabolites, 17 metabolites increased, 14 metabolites decreased, and one metabolite increased and decreased. G6P, F6P, F1, 6P, DHAP, 3PG, and PEP in glycolysis and 6PG, Ru5P, and R5P in pentose phosphate pathway increased significantly after 20-Hz EPS (Figure 3). UDP-glucose, reduced/oxidized glutathione, and 2-oxoglutarate significantly decreased after 20-Hz EPS. We confirmed that Ca2+ signals were induced by 2- and 20-Hz EPS but the Ca2+ signals were not maintained during 60 min of 20-Hz EPS. At the same time, the number of myotubes were not reduced (Figure S2A) and ATP did not change significantly after 20-Hz EPS (Figure S2B). Calculated energy charge, an index of energy status of biological cells, showed a slight decrease owing to the changes in ADP (Figure S2C) and AMP levels during 20-Hz EPS and then returned to the basal level after the 20-Hz EPS (Figure S2D). In summary, many metabolites changed only after 20-Hz EPS but not during 20-Hz EPS, and no metabolites changed significantly during or after 2-Hz EPS.
Figure 2.
Hierarchal Clustering Analysis of Metabolomic Time Series Data
(A) Heatmap of changes in metabolite concentration at each time point during and after 0, 2, and 20-Hz EPS. Hierarchical clustering was performed using Ward's method with the normalized average value of each metabolite concentration. This average value was calculated with data from three biological replicates.
See also Figure S1 and Table S1.
(B) Time course of metabolites in each cluster. Dashed lines indicate the time series of each metabolite. Black, red, and blue lines indicate values during and after 0, 2, and 20-Hz EPS, respectively. Bold line represents the average value at each condition in each cluster.
Figure 3.
Time Course of Metabolites in Central Carbon Metabolism
Black, red, and blue lines represent 0, 2, and 20 Hz, respectively. Orange, blue, red, and yellow frames correspond to clusters 2, 3, 5, and 7 in Figure 2, respectively. Average concentrations were compared between different stimulations at each time point using the Welch's t test with the Storey's multiple testing correction. At time points where FDR q < 0.2, ∗red indicates significant difference between 0 and 20 Hz and 2 and 20 Hz, ∗blue indicates significant difference between 0 and 20 Hz, ∗green indicates significant difference between 2 and 20 Hz, and ∗black indicates significant difference between 0 and 2 Hz. The means and SDs of three independent experiments each with three biological replicates are shown.
See also Figures S1 and S2 and Table S2.
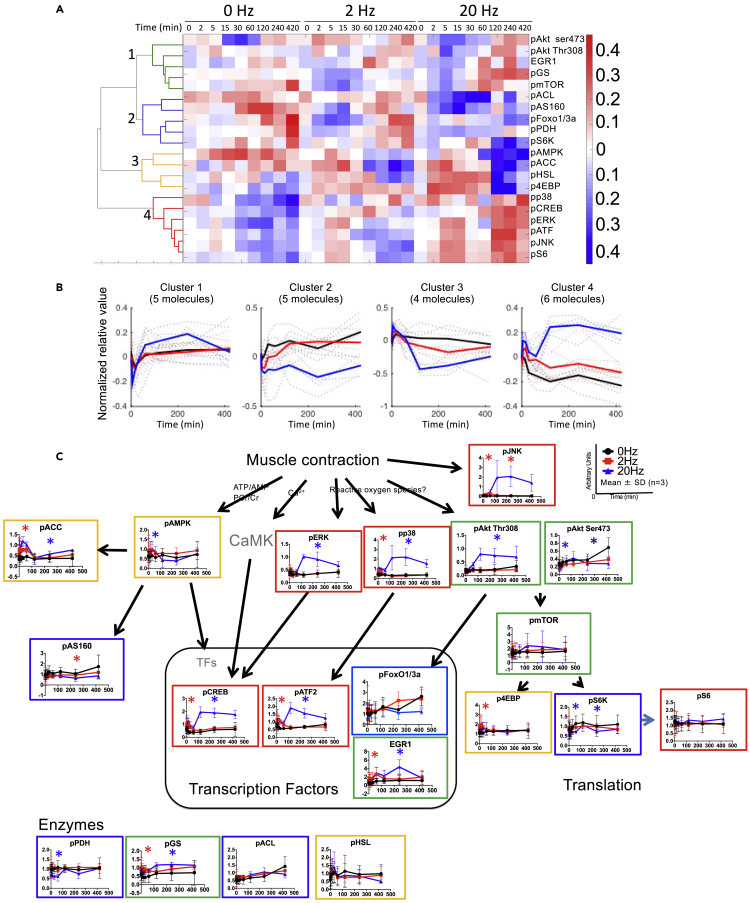
We measured protein levels of phosphorylated signaling molecules, including TFs, protein kinases, and metabolic enzymes, which are known to be regulated by muscle contraction (Egan and Zierath, 2013), using western blot analysis (Figure S3). We performed cluster analysis that resulted in four clusters (Figures 4A–4C and Table S3). pAkt (Ser308 and Thr473), phosphorylated mammalian target of rapamycin (pmTOR), and early growth response protein 1 (EGR1) were in cluster 1, which showed an increase after 60 min of 20-Hz EPS. Phosphorylated pyruvate dehydrogenase (pPDH), phosphorylated ATP citrate lyase (pACL), and phosphorylated S6K (pS6K) were in cluster 2, which showed decrease during and after 20-Hz EPS. Phosphorylated AMPK (pAMPK) and phosphorylated acetyl-CoA carboxylase (pACC) were in cluster 3, which showed a transient increase during 2- and 20-Hz EPS. MAPK such as phosphorylated p38 (pp38), phosphorylated ERK1/2 (pERK), phosphorylated JNK (pJNK), and TFs such as phosphorylated cAMP response element-binding protein (pCREB) and phosphorylated activating TF 2 (pATF2) were in cluster 4, which showed an increase during 2- and 20-Hz EPS and increased after 20-Hz EPS but not after 2-Hz EPS. pERK, p38, JNK, pAkt, pATF2, and pCREB increased significantly after 20-Hz EPS. These results indicate that phosphorylation of a subset of signaling molecules, such as MAPKs and Akt, and TFs, such as pCREB and pATF2, increased continuously after 20-Hz EPS.
Figure 4.
Hierarchal Clustering Analysis Using Time Series Data of Signaling Molecules
(A) Heatmap of changes in signaling molecules at each time point during and after 0, 2, and 20-Hz EPS. Hierarchical clustering was performed using the Ward's method with the normalized average value of each signaling molecule. This average value was calculated with data from three biological replicates.
(B) Time course of signaling molecules in each cluster. Dashed lines indicate the value of each signaling molecule. Black, red, and blue lines indicate values during and after 0, 2, and 20-Hz EPS, respectively. Bold line represents the average value at each EPS frequency in each cluster.
(C) Time course of signaling molecules. Black, red, and blue lines represent 0, 2, and 20 Hz, respectively. Green, blue, yellow, and red frames indicate clusters 1, 2, 3, and 4, respectively. Statistical comparison of the level of each molecule among 0, 2, and 20 Hz at each time point was performed using two-way ANOVA. The calculated p value for the electrical stimulation was corrected for multiple testing using the Benjamini-Hochberg method. FDR q < 0.1 was regarded as a significant difference. ∗Red indicates significant difference between 0 and 20 Hz and 0 and 2 Hz, ∗blue indicates significant difference between 0 and 20 Hz, and ∗black indicates the difference between 0 and 2 Hz. The means and SDs of three independent experiments each with three biological replicates are shown.
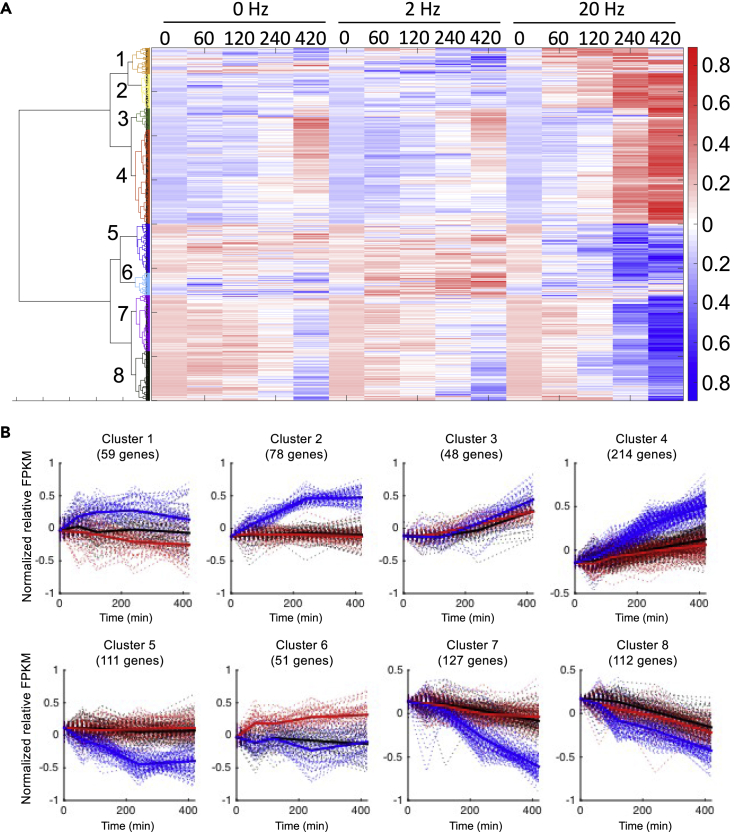
We performed mRNA sequencing of the myotubes, mapped the sequences to the mouse genome using TopHat (Trapnell et al., 2009), and calculated mRNA expression level as fragments per kilobase of exon per million fragments mapped (FPKM) using Cufflinks (Trapnell et al., 2010). We thus obtained expression data of 11,185 genes throughout the time course (Figure S4). To identify differentially expressed genes (DEGs) in response to EPS, we calculated fold-change of FPKMs at each time point compared with those at time 0 at 2- and 20-Hz EPS and defined genes with changes of 2.0-fold or more and of 0.5-fold or less as DEGs. We identified 800 DEGs. To clarify the pattern of time-dependent changes of the DEGs, we performed hierarchical cluster analysis using the time series data of the DEGs and obtained eight clusters (Figure 5). Next, we examined biological functions of genes in each cluster using KEGG pathway analysis in the DAVID online tool (Huang et al., 2008, Table S4). Genes in MAPK and p53 signaling pathways were enriched in cluster 2, and genes in glutathione metabolism and pentose phosphate pathway were enriched in cluster 4. Clusters 2 and 4 showed continuous increase after 20-Hz EPS. Genes in phosphatidylinositol signaling were enriched in cluster 5, and genes in focal adhesion and TGF-beta signaling pathways were enriched in cluster 7. The clusters 5 and 7 showed continuous decrease after 20-Hz EPS. Taken together, there were widespread and persistent changes in metabolite, phosphorylation of signaling molecules, and gene expression after 20-Hz EPS.
Figure 5.
Hierarchal Clustering Analysis Using Time Series of DEGs from Transcriptomic Data
(A) Heatmap of changes in DEGs at each time point during and after 0, 2, and 20-Hz EPS. Hierarchical clustering was performed using Ward's method with normalized value of each DEG (one independent biological experiment at each time point).
(B) Time course of DEGs in each cluster. Dashed lines indicate the time series of each DEG. Black, red, and blue lines indicate values during and after 0, 2, and 20-Hz EPS. Bold line represents the average value at each EPS frequency in each cluster.
See also Figure S4 and Tables S4–S7.
Connection of the Transcriptomic Layer to the Signaling Layer
Having observed changes in both phosphorylation of signaling molecules and gene expression after 20-Hz EPS, we set out to identify TFs that connect these two layers. The analysis was conducted in two steps: (1) prediction of TFs and (2) identification of signaling molecules regulating the predicted TFs using KEGG database.
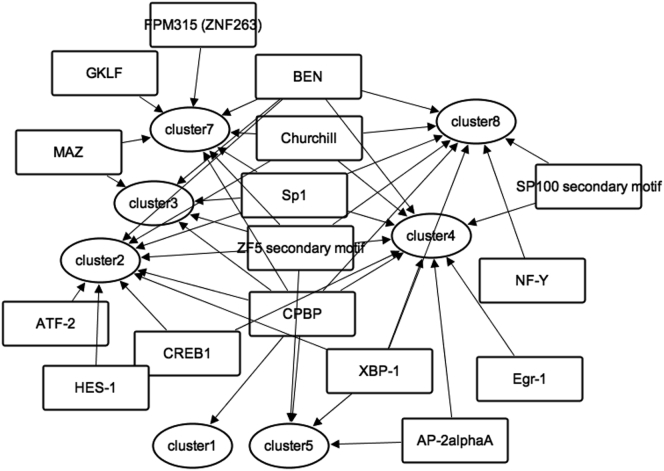
To identify TFs that regulate DEGs, we identified putative binding motifs for TFs in the promoter region of each DEG (−300 bp to +100 bp around the consensus transcription start site [Arner et al., 2015; Kinsella et al., 2011]) using TRANSFAC (Matys et al., 2006) and Match (Kawata et al., 2018; Kel et al., 2003). Subsequently, we used a modified motif enrichment analysis to predict TFs that bind to promoters of DEGs in each cluster (Mina et al., 2015). We thus predicted 16 TFs for the DEG clusters (Figure 6), with some of them shared by multiple clusters.
Figure 6.
Prediction of Transcription Factors from Clusters of DEGs Using Motif Enrichment Analysis
Squares indicate predicted TFs. Circles indicate clusters resulting from hierarchical clustering. Arrows indicate from which cluster the TF was predicted.
See also Figure S4.
Next, we identified the signaling molecules that regulate these predicted TFs, using KEGG pathway analysis. We focused on MAPK, PI3K-Akt, AMPK, and calcium signaling pathways (Figure S5), which are known to be regulated by muscle contraction (Egan and Zierath, 2013). Twelve signaling molecules were identified as the regulators of the predicted TFs: Akt, ERK, PKA, PKC, JNK, p38, mTORC2, MSK1/2, CAMK, GSK3, CREB, and EGR1. Among them, phosphorylation of ERK, Akt, CREB, JNK, p38, and EGR1 increased significantly after 20-Hz EPS (Figure 4C). These results indicate that the signaling molecules upstream of TFs and their target DEGs are activated after 20-Hz EPS.
Connection of the Transcriptomic Layer to the Metabolic Layer
We connected the metabolomic layer with the transcriptomic layer by identifying metabolic enzymes responsible for the changes in the metabolites. We identified responsible metabolic enzymes using the method developed in our previous study (Yugi et al., 2014). First, we measured levels of 32 metabolites that significantly changed after 20-Hz EPS. Second, we searched for metabolic enzymes that have these 32 metabolites as substrates or products using KEGG database. Third, the metabolic enzymes encoded by DEGs were defined as responsible metabolic enzymes. As a result, 27 responsible metabolic enzymes were identified (Table S5). KEGG pathway enrichment analysis showed that the 27 responsible metabolic enzymes are involved in carbon metabolism, pentose phosphate pathway, and biosynthesis of amino acids (Table S6). Given that 27 metabolic enzymes were encoded by DEGs in cluster 4 in Figure 5, which continuously increased after 20 Hz, we performed KEGG pathway enrichment analysis for cluster 4. Among enzymes involved in central carbon metabolism, genes encoding metabolic enzymes in the pentose phosphate pathway were the largest gene group that was significantly enriched (Table S7). These results indicate that both the metabolites and the genes encoding the metabolic enzymes in the pentose phosphate pathway increased after 20 Hz stimulation (Figure S6).
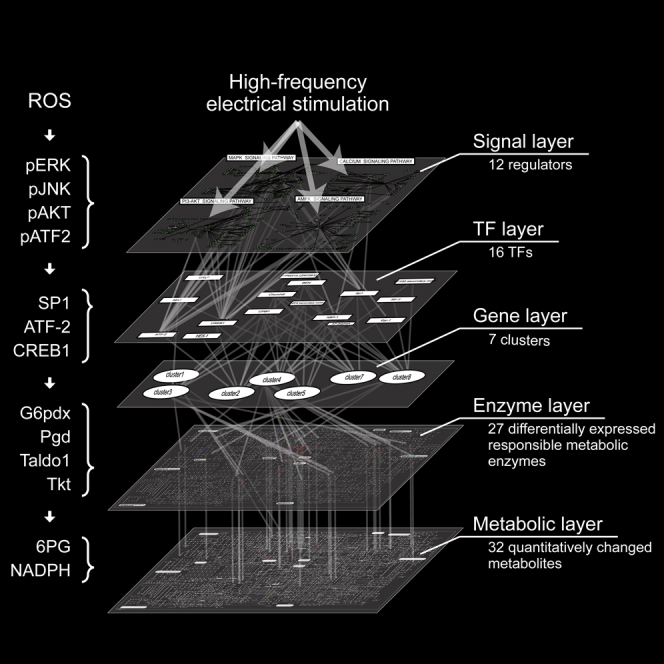
Construction of the Trans-omic Network Induced by EPS
Our analysis showed that, after 20-Hz EPS, extensive changes occur in multiple layers of metabolites, signal molecules, and gene expression. Therefore, we constructed a trans-omic network of metabolites, metabolic enzymes, gene expression, TFs, and signal molecules after 20-Hz EPS (Figure 7). Thirty-two metabolites significantly changed after 20-Hz EPS. Twenty-seven DEGs encoding metabolic enzymes use these 32 metabolites as substrates or products. We performed hierarchical clustering analysis using 800 DEGs and predicted 16 TFs for the DEG clusters. Twelve signaling molecules presumably interact with these 16 TFs, as they are involved in the same KEGG pathways (MAPK, AMPK, calcium, and PI3K-Akt pathways).
Figure 7.
Trans-omic Network after High-Frequency Electrical Stimulation of C2C12 Myotubes
The diagram connects five layers of signaling molecules, transcriptional factors, gene expression, metabolic enzymes, and metabolites. In the signaling molecules layer, the 12 regulators were identified in the four KEGG signaling pathways and were shown in red, and the others were shown in green. In the transcriptional factor and gene expression layers, the 16 TFs were predicted from seven clusters of DEGs. In the metabolic enzyme layer, 27 differentially expressed responsible metabolic enzymes were identified, and in the metabolite layer, 32 metabolites were identified as quantitatively changed metabolites.
See also Figures S5 and S6 and Tables S5–S7.
ROS-Dependent Metabolic Network after High-Frequency EPS
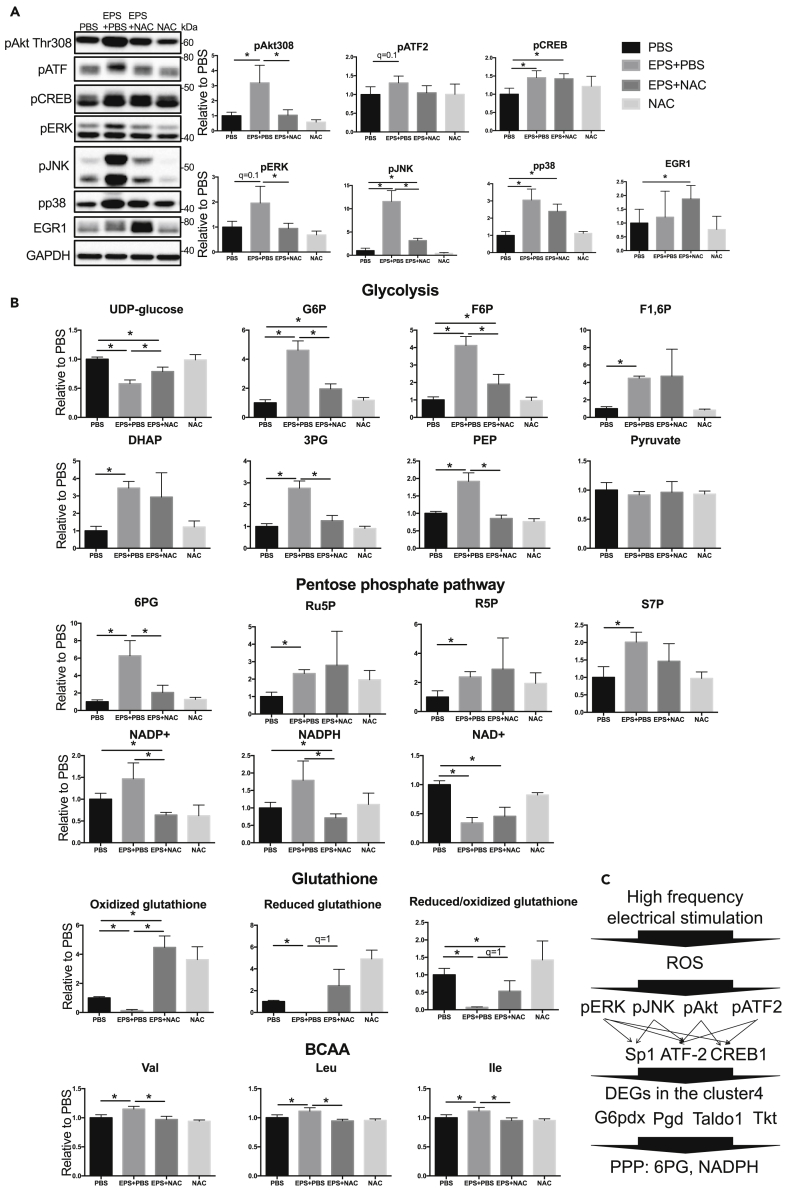
We found that ROS were involved in the multi-layer responses after 20-Hz EPS because our results showed that reduced/oxidized glutathione, which is a marker for accumulation of ROS, greatly and continuously changed after 20-Hz stimulation (Figure 3A and Table S2). Consistently, p38 and JNK phosphorylation, which are known to be regulated by ROS, increased after 20-Hz EPS (Figure 4C). To experimentally validate our findings, we added an antioxidant, N-acetylcysteine (NAC), immediately after 1 h of 20-Hz EPS and examined changes of signaling molecules and metabolites 180 min time point after of the 1 h 20-Hz EPS. The addition of NAC attenuated the changes in reduced/oxidized glutathione after 20-Hz EPS. The addition of NAC after 20-Hz EPS also suppressed phosphorylation of JNK, ERK, Akt Ser308, and ATF2 after 20-Hz EPS (Figure 8A) and the increase of 6PG, S7P, and NADPH in the pentose phosphate pathway (Figure 8B). These results indicate the pivotal role of ROS in inducing changes of signal molecules and metabolites involved with the pentose phosphate pathway after 20-Hz EPS (Figure 8C).
Figure 8.
Effects of NAC on Signaling Molecules and Metabolites after 20-Hz EPS
(A and B) Signaling molecules and metabolite concentrations. Welch's t test was performed on each signaling molecule and each metabolite, and multiple comparisons were performed based on FDR (BH method). ∗q < 1 was defined as a significant difference. The means and SDs of four independent experiments each with four biological replicates are shown.
(C) ROS-dependent activation of signaling molecules, enzymes, and metabolites involved with the pentose phosphate pathway (PPP) after high-frequency electrical stimulation.
Discussion
In this study, we characterized metabolome, transcriptome, and phosphorylation and protein of signal molecules in the EPS-stimulated C2C12 myotubes and constructed a trans-omic network by integrating the multi-omic data. The trans-omic analysis and experimental validation revealed that ROS-mediated phosphorylation of signaling molecules, gene expression of metabolic enzymes, and metabolites in the pentose phosphate pathway were upregulated after 20-Hz EPS.
In this study, we used an electrical stimulation system for C2C12 myotubes to that has been shown to mimic muscle contraction in vitro in our laboratory (Furuichi et al., 2018; Manabe et al., 2012; Matsuda et al., 2020; Wada et al., 2020). We used 2 Hz for low-frequency and 20 Hz for high-frequency stimulation. Previous studies confirmed that 20 Hz is high frequency for C2C12 myotubes (Fujita et al., 2007; Yamasaki et al., 2009) but 20 Hz is not considered high-frequency stimulation for skeletal muscle contraction in vivo (Cairns et al., 2007). Creatine and AMP increased transiently during 20 Hz stimulation, suggesting activation of ATP hydrolysis and PCr breakdown. These results are consistent with electrical stimulated rat skeletal muscles (Spriet, 1989) and exercised human skeletal muscles (Parolin et al., 1999; Perry et al., 2008). However, the increase in ADP and AMP levels of C2C12 by EPS was smaller than that of in vivo muscle contraction, suggesting the difference between C2C12 EPS and in vivo muscle contraction. In the TCA cycle intermediates, Fumarate and Malate increased and returned to basal rapidly during 20 Hz, and 2-oxoglutarate decreased due to 20-Hz EPS. These results are consistently seen in human skeletal muscles during 60-min exercise (Gibala et al., 1997). In signaling molecules, pACC, pJNK and pp38 increased during 2- and 20-Hz EPS. This result is similar to the previous study using EPS in C2C12 myotubes (Manabe et al., 2012). pERK, pp38, pJNK, pAkt, pATF2, and pCREB increased continuously after 20-Hz EPS. Previous studies have reported that pAkt and pp38 increased continuously in skeletal muscles after exercise (Camera et al., 2010; Williamson et al., 2006). Gene expression results show that EGR1, IL6, FOS, HSPB1, and mdm2 increased after 20-Hz EPS. These genes were known to increase after exercise or EPS (Furuichi et al., 2018; Irrcher and Hood, 2004; Neubauer et al., 2014; Roudier et al., 2012). Gene expression of dgkq, Pik3r1, PIK3C2B, and Thbs1 decreased after 20-Hz EPS. These reductions after EPS have never been reported but may be important for an increase in insulin sensitivity in skeletal muscles because these genes are associated with insulin resistance (Alliouachene et al., 2015; Inoue et al., 2013; Mannerås-Holm et al., 2015; McCurdy et al., 2012). We confirmed that Ca2+ signals were induced by 2- and 20-Hz EPS but these were not maintained during 60 min of 20-Hz EPS. This result suggests fatigue state of myotubes after 60 min of 20-Hz EPS; 20-Hz EPS was considered high-frequency stimulation because Ca2+ signal and contraction responses against frequency are different between C2C12 myotubes and skeletal muscle. A sustained increase in Ca2+ signal and tetanus contraction by 10-Hz EPS in C2C12 myotubes were shown previously (Fujita et al., 2007; Yamasaki et al., 2009). However, the number of cells and ATP were not reduced during and after 20-Hz EPS. Calculated energy charge reduced slightly during 20 Hz but returned to the basal level after the 20-Hz EPS. We believe that EPS-stimulated biological alterations involved with metabolome, signaling molecules, and gene expression were not attributable to cell death.
Because phosphorylation of signaling molecules in response to ROS and markers for ROS accumulation increased after 20-Hz EPS, we hypothesized that ROS accumulation after 20-Hz EPS is involved in activation of the pentose phosphate pathway. ROS accumulation in muscle by in vitro EPS has also been reported in the previous study (Horie et al., 2015). Although phosphorylation of ERK, p38, JNK, Akt, ATF2, and CREB increased significantly after 20-Hz EPS, addition of NAC, an antioxidant, after 20-Hz EPS inhibited phosphorylation of Akt, ERK, and JNK but not the other signaling factors. Previous studies suggest that activation of Akt signaling upregulates expression of genes encoding the metabolic enzymes and corresponding metabolites in the pentose phosphate pathway (Wagle et al., 1998; Wu et al., 2017). ERK and JNK have also been shown to mediate activation of the pentose phosphate pathway in cancer cells (Papa et al., 2019). Since NAC was added after 20-Hz EPS to clarify the effect of ROS accumulation after 20-Hz EPS, ROS production and accumulation during EPS could not be suppressed. It is possible that ROS during 20-Hz EPS affected the activation of signaling molecule. Indeed, EGR1 was not attenuated by NAC. This result may be associated with ROS production and/or accumulation during 20-Hz EPS because EGR1 increased rapidly in C2C12 myotubes after electrical stimulation (Irrcher and Hood, 2004). These results suggest that accumulation of ROS after 20-Hz EPS regulates activation of the pentose phosphate pathway via Akt, ERK, and JNK signaling.
We identified responsible metabolic enzymes, such as Pgd, Tkt, Taldo1, and G6pdx, which are involved in the pentose phosphate pathway, using gene expression data. These metabolic enzymes are encoded by DEGs in cluster 4, which continuously increased after 20-Hz EPS. These results indicate that the pentose phosphate pathway was activated after 20-Hz EPS. To the best of our knowledge, this is the first study reporting that expression levels of genes encoding enzymes of the pentose phosphate pathway increased after high-frequency electrical stimulation. We estimate that gene expression of enzymes related in the pentose phosphate pathway was also inhibited by NAC because phosphorylation of Akt, ERK, and JNK was inhibited. Previous studies reported that these signaling molecules regulate gene expression of enzymes involved in pentose phosphate pathway (Ham et al., 2013; Wang et al., 2019; Wu et al., 2017). Therefore, our data-driven trans-omic analysis revealed activation of the pentose phosphate pathway after high-frequency electrical stimulation.
Along with increase in gene expression related to the pentose phosphate pathway, metabolites in the same pathway, such as Ru5P, 6PG, and R5P, increased significantly after 20-Hz EPS. It has been shown that insulin sensitivity and glucose uptake increased for up to 3 h after muscle contraction (Cartee et al., 1989). Transported glucose has been thought to be used primarily for glycogen repletion (Jentjens and Jeukendrup, 2003). Flux of glucose into the alternative pathway, the pentose phosphate pathway, has received little attention because expression levels of genes and proteins for the enzymes are relatively low in skeletal muscle (Battistuzzi et al., 1985). Our findings suggest that the pentose phosphate pathway, another glucose metabolic pathway in addition to glycogen synthesis, is also triggered after high-frequency electrical stimulation for physiological adaptation. Furthermore, addition of NAC suppressed the increase in metabolites from the pentose phosphate pathway such as 6PG and NADPH. There are two possible explanations for the ROS-dependent activation of the pentose phosphate pathway. First, ROS is known to enhance insulin sensitivity and glucose uptake (Loh et al., 2009). Alternatively, protein levels of the enzymes involved in the pentose phosphate pathway were increased, although we did not measure protein abundance and protein phosphorylation in large scale in this study.
Interestingly, the concentration of metabolites in the glycolysis pathway was also increased after 20-Hz EPS. It is unclear why the concentrations of these metabolites increased after 20-Hz EPS because the flux of the glycolytic system increases rapidly owing to exercise and decreases rapidly at basal state after exercise (Crowther et al., 2002; Spriet et al., 1985). We estimate that increases in these intermediate metabolites did not reflect an increase in glycolytic flux. The activity of pyruvate kinase may be associated with increases in these metabolite concentrations because the pyruvate did not increase in spite of increasing from G6P to PEP. It is necessary to examine metabolic flux because the concentration is a balance between production and degradation of the metabolite.
The ROS-dependent activation of the pentose phosphate pathway may be associated with physiological adaptation of skeletal muscle. The pentose phosphate pathway plays a role in producing substrates for nucleic acid synthesis and lipid synthesis. Since high-frequency stimulation is accompanied by muscle damage, it is physiologically important that nucleic acid synthesis and lipid synthesis are activated for muscle hypertrophy and membrane recovery. We measured NADPH, which is a source of lipid synthesis, and PRPP, which is a source of nucleic acid synthesis. NADPH and PRPP of 20-Hz EPS at 240 min were about 1.5- and 4-fold, respectively, higher than those of 0-Hz EPS at 240 min, although these differences were not significant. These results suggest the activation of lipid synthesis and nucleotide synthesis after 20-Hz EPS to lead to physiological adaptation. Indeed, it has been shown that regeneration from drug-induced muscle damage requires activation of the pentose phosphate pathway (Wagner et al., 1978). Furthermore, ROS has been shown to be associated with repair of membrane damage after eccentric muscle contraction (Horn et al., 2017). Taken together, our findings of ROS-dependent activation of the pentose phosphate pathway provide new insights into muscle adaptation under physiological conditions.
Limitations of the Study
Trans-omic analysis revealed ROS-dependent pentose phosphate pathway activation after 20-Hz EPS in C2C12 myotubes. We could not construct low-frequency stimulation-induced trans-omic network because of weak responses in each multi-layer in this study. Clarifying the difference in trans-omic network between low-frequency and high-frequency stimulation will further deepen the understanding of mechanisms of frequency-dependent muscle adaptation. Furthermore, it is not clear whether exercise in animals stimulates the signaling molecules, genes, and metabolites in the pentose phosphate pathway in vivo. In the future, it is necessary to examine the validation of trans-omic network we constructed and activation of pentose phosphate pathway using an animal experiment. We also could not verify the physiological significance of activation of the pentose phosphate pathway. Exercise has a wide variety of modes such as endurance exercise and resistance exercise; cellular responses are different between each exercise mode (Atherton et al., 2005). Therefore, clarifying the relationship between exercise modes and activation of the pentose phosphate pathway enables clarification of the physiological significance in vivo.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shinya Kuroda (skuroda@bs.s.u-tokyo.ac.jp).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The RNA-seq data in this study are available at DDBJ (Accession number DRA010400).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank our laboratory members for critically reading this manuscript and for their technical assistance with the experiments. This work was supported by the Creation of Fundamental Technologies for Understanding and Control of Biosystem Dynamics, CREST (JPMJCR12W3) from the Japan Science and Technology Agency (JST) and by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 17H06299, 17H06300. D.H. received a Grant-in-Aid for JSPS Research Fellow (14J05223). K. Kawata receives funding from JSPS KAKENHI (19K16635) and the Takeda Life Science Foundation. K. Kunida receives funding from a Grant-in-Aid for Young Scientists (B) (16K19028) from JSPS. K.Y. receives funding from the JSPS (KAKENHI grant numbers JP15H05582 and JP18H05431 and Creation of Innovative Technology for Medical Applications Based on the Global Analyses and Regulation of Disease-Related Metabolites) and PRESTO (JPMJPR1538) from the JST. Y.S. was supported by the JSPS KAKENHI Grant Number 17H06306. T. Soga receives funding from the AMED-CREST (JP18gm0710003) from the Japan Agency for Medical Research and Development, AMED.
Author Contributions
Conception and design of research: D.H. and S.K. Conception and design of the experiments: D.H., K. Kunida, A.H., K.Y., T.W., Y.F., Y.M., N.L.F., and S.K. Collection, analysis, and interpretation of data: D.H., K. Kawata, K. Kunida, A.H., K.Y., T.W., M.F., T. Sano, Y.I., Y.F., Y.M., Y.S., N.L.F., T. Soga, and S.K. Drafting the article or revising it critically for important intellectual content: D.H., K. Kawata, A.H, Y.K., T.W., and S.K. All authors approved the final version of the manuscript and listed qualify for authorship.
Declaration of Interests
The authors declare no conflicts of interest.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101558.
Supplemental Information
References
- Alliouachene S., Bilanges B., Chicanne G., Anderson K.E., Pearce W., Ali K., Valet C., Posor Y., Low P.C., Chaussade C. Inactivation of the class II PI3K-C2β potentiates insulin signaling and sensitivity. Cell Rep. 2015;13:1881–1894. doi: 10.1016/j.celrep.2015.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drablos F., Lennartsson A., Ronnerblad M., Hrydziuszko O., Vitezic M. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton P.J., Babraj J., Smith K., Singh J., Rennie M.J., Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- Battistuzzi G., D’Urso M., Toniolo D., Persico G.M., Luzzatto L. Tissue-specific levels of human glucose-6-phosphate dehydrogenase correlate with methylation of specific sites at the 3’ end of the gene. Proc. Natl. Acad. Sci. U S A. 1985;82:1465–1469. doi: 10.1073/pnas.82.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns S.P., Chin E.R., Renaud J.-M. Stimulation pulse characteristics and electrode configuration determine site of excitation in isolated mammalian skeletal muscle: implications for fatigue. J. Appl. Physiol. 2007;103:359–368. doi: 10.1152/japplphysiol.01267.2006. [DOI] [PubMed] [Google Scholar]
- Camera D.M., Edge J., Short M.J., Hawley J.A., Coffey V.G. Early time course of akt phosphorylation after endurance and resistance exercise. Med. Sci. Sports Exerc. 2010;42:1843–1852. doi: 10.1249/MSS.0b013e3181d964e4. [DOI] [PubMed] [Google Scholar]
- Camera D.M., Burniston J.G., Pogson M.A., Smiles W.J., Hawley J.A. Dynamic proteome profiling of individual proteins in human skeletal muscle after a high-fat diet and resistance exercise. FASEB J. 2017;31:5478–5494. doi: 10.1096/fj.201700531R. [DOI] [PubMed] [Google Scholar]
- Cartee G.D., Young D.A., Sleeper M.D., Zierath J., Wallberg-Henriksson H., Holloszy J.O. Prolonged increase in insulin-stimulated glucose transport in muscle after exercise. Am. J. Physiol. 1989;256:E494–E499. doi: 10.1152/ajpendo.1989.256.4.E494. [DOI] [PubMed] [Google Scholar]
- Chen Y.-W., Nader G.A., Baar K.R., Fedele M.J., Hoffman E.P., Esser K.A. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J. Physiol. 2002;545:27–41. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-P., Stephens T.J., Murthy S., Canny B.J., Hargreaves M., Witters L.A., Kemp B.E., McConell G.K. Effect of exercise intensity on skeletal muscle AMPK signaling in humans. Diabetes. 2003;52:2205–2212. doi: 10.2337/diabetes.52.9.2205. [DOI] [PubMed] [Google Scholar]
- Crowther G.J., Kemper W.F., Carey M.F., Conley K.E. Control of glycolysis in contracting skeletal muscle. II. Turning it off. Am. J. Physiol. Endocrinol. Metab. 2002;282:E74–E79. doi: 10.1152/ajpendo.2002.282.1.E74. [DOI] [PubMed] [Google Scholar]
- Egan B., Zierath J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Egan B., Carson B.P., Garcia-Roves P.M., Chibalin A.V., Sarsfield F.M., Barron N., McCaffrey N., Moyna N.M., Zierath J.R., O’Gorman D.J. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J. Physiol. 2010;588:1779–1790. doi: 10.1113/jphysiol.2010.188011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Nedachi T., Kanzaki M. Accelerated de novo sarcomere assembly by electric pulse stimulation in C2C12 myotubes. Exp. Cell Res. 2007;313:1853–1865. doi: 10.1016/j.yexcr.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Manabe Y., Takagi M., Aoki M., Fujii N.L. Evidence for acute contraction-induced myokine secretion by C2C12 myotubes. PLoS One. 2018;13:e0206146. doi: 10.1371/journal.pone.0206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibala M.J., Tarnopolsky M.A., Graham T.E. Tricarboxylic acid cycle intermediates in human muscle at rest and during prolonged cycling. Am. J. Physiol. 1997;272:E239–E244. doi: 10.1152/ajpendo.1997.272.2.E239. [DOI] [PubMed] [Google Scholar]
- Ham M., Lee J.-W., Choi A.H., Jang H., Choi G., Park J., Kozuka C., Sears D.D., Masuzaki H., Kim J.B. Macrophage glucose-6-phosphate dehydrogenase stimulates proinflammatory responses with oxidative stress. Mol. Cell Biol. 2013;33:2425–2435. doi: 10.1128/MCB.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley J.A., Hargreaves M., Joyner M.J., Zierath J.R. Integrative biology of exercise. Cell. 2014;159:738–749. doi: 10.1016/j.cell.2014.10.029. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A.L., Pilegaard H., Neufer P.D. Differential transcriptional activation of select metabolic genes in response to variations in exercise intensity and duration. Am. J. Physiol. Endocrinol. Metab. 2003;285:E1021–E1027. doi: 10.1152/ajpendo.00234.2003. [DOI] [PubMed] [Google Scholar]
- Hoffman N.J., Parker B.L., Chaudhuri R., Fisher-Wellman K.H., Kleinert M., Humphrey S.J., Yang P., Holliday M., Trefely S., Fazakerley D.J. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 2015;22:922–935. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M., Warabi E., Komine S., Oh S., Shoda J. Cytoprotective role of Nrf2 in electrical pulse stimulated C2C12 myotube. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0144835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A., Van Der Meulen J.H., Defour A., Hogarth M., Sreetama S.C., Reed A., Scheffer L., Chandel N.S., Jaiswal J.K. Mitochondrial redox signaling enables repair of injured skeletal muscle cells. Sci. Signal. 2017;10:1–12. doi: 10.1126/scisignal.aaj1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Inoue M., Jiang Y., Barnes R.H., Tokunaga M., Martinez-Santibañez G., Geletka L., Lumeng C.N., Buchner D.A., Chun T.-H. Thrombospondin 1 mediates high-fat diet-induced muscle fibrosis and insulin resistance in male mice. Endocrinology. 2013;154:4548–4559. doi: 10.1210/en.2013-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I., Hood D.A. Regulation of Egr-1, SRF, and Sp1 mRNA expression in contracting skeletal muscle cells. J. Appl. Physiol. 2004;97:2207–2213. doi: 10.1152/japplphysiol.00388.2004. [DOI] [PubMed] [Google Scholar]
- Jentjens R., Jeukendrup A. Determinants of post-exercise glycogen synthesis during short-term recovery. Sports Med. 2003;33:117–144. doi: 10.2165/00007256-200333020-00004. [DOI] [PubMed] [Google Scholar]
- Kawata K., Hatano A., Yugi K., Kubota H., Sano T., Fujii M., Tomizawa Y., Kokaji T., Tanaka K.Y., Uda S. Trans-omic analysis reveals selective responses to induced and basal insulin across signaling, transcriptional, and metabolic networks. iScience. 2018;7:212–229. doi: 10.1016/j.isci.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel A.E., Gössling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella R.J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K., Deng H., Fukushima A., Cai X., Boivin B., Galic S., Bruce C., Shields B.J., Skiba B., Ooms L.M. Reactive oxygen species enhance insulin sensitivity. Cell Metab. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney D.J., Safdar A., Parise G., Melov S., Fu M., MacNeil L., Kaczor J., Payne E.T., Tarnopolsky M.A. Gene expression profiling in human skeletal muscle during recovery from eccentric exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1901–R1910. doi: 10.1152/ajpregu.00847.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe Y., Miyatake S., Takagi M., Nakamura M., Okeda A., Nakano T., Hirshman M.F., Goodyear L.J., Fujii N.L. Characterization of an acute muscle contraction model using cultured C2C12 myotubes. PLoS One. 2012;7:e52592. doi: 10.1371/journal.pone.0052592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannerås-Holm L., Kirchner H., Björnholm M., Chibalin A.V., Zierath J.R. mRNA expression of diacylglycerol kinase isoforms in insulin-sensitive tissues: effects of obesity and insulin resistance. Physiol. Rep. 2015;3:e12372. doi: 10.14814/phy2.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Hironaka K.I., Fujii M., Wada T., Kunida K., Inoue H., Eto M., Hoshino D., Furuichi Y., Manabe Y. Monitoring and mathematical modeling of mitochondrial ATP in myotubes at single-cell level reveals two distinct population with different kinetics. Quant. Biol. 2020:1–10. doi: 10.1007/s40484-020-0211-8. [DOI] [Google Scholar]
- Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. TRANSFAC(R) and its module TRANSCompel(R): transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy C.E., Schenk S., Holliday M.J., Philp A., Houck J.A., Patsouris D., MacLean P.S., Majka S.M., Klemm D.J., Friedman J.E. Attenuated Pik3r1 expression prevents insulin resistance and adipose tissue macrophage accumulation in diet-induced obese mice. Diabetes. 2012;61:2495–2505. doi: 10.2337/db11-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina M., Magi S., Jurman G., Itoh M., Kawaji H., Lassmann T., Arner E., Forrest A.R.R., Carninci P., Hayashizaki Y. Promoter-level expression clustering identifies time development of transcriptional regulatory cascades initiated by ErbB receptors in breast cancer cells. Sci. Rep. 2015;5:11999. doi: 10.1038/srep11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto L., Egawa T., Oshima R., Kurogi E., Tomida Y., Tsuchiya K., Hayashi T. AICAR stimulation metabolome widely mimics electrical contraction in isolated rat epitrochlearis muscle. Am. J. Physiol. Cell Physiol. 2013;305:C1214–C1222. doi: 10.1152/ajpcell.00162.2013. [DOI] [PubMed] [Google Scholar]
- Neubauer O., Sabapathy S., Ashton K.J., Desbrow B., Peake J.M., Lazarus R., Wessner B., Cameron-Smith D., Wagner K.-H., Haseler L.J. Time course-dependent changes in the transcriptome of human skeletal muscle during recovery from endurance exercise: from inflammation to adaptive remodeling. J. Appl. Physiol. 2014;116:274–287. doi: 10.1152/japplphysiol.00909.2013. [DOI] [PubMed] [Google Scholar]
- Papa S., Choy P.M., Bubici C. The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene. 2019;38:2223–2240. doi: 10.1038/s41388-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolin M.L., Chesley A., Matsos M.P., Spriet L.L., Jones N.L., Heigenhauser G.J. Regulation of skeletal muscle glycogen phosphorylase and PDH during maximal intermittent exercise. Am. J. Physiol. 1999;277:E890–E900. doi: 10.1152/ajpendo.1999.277.5.E890. [DOI] [PubMed] [Google Scholar]
- Peake J.M., Tan S.J., Markworth J.F., Broadbent J.A., Skinner T.L., Cameron-Smith D. Metabolic and hormonal responses to isoenergetic high-intensity interval exercise and continuous moderate-intensity exercise. Am. J. Physiol. Endocrinol. Metab. 2014;307:E539–E552. doi: 10.1152/ajpendo.00276.2014. [DOI] [PubMed] [Google Scholar]
- Perry C.G.R., Heigenhauser G.J.F., Bonen A., Spriet L.L. High-intensity aerobic interval training increases fat and carbohydrate metabolic capacities in human skeletal muscle. Appl. Physiol. Nutr. Metab. 2008;33:1112–1123. doi: 10.1139/H08-097. [DOI] [PubMed] [Google Scholar]
- Perry C.G.R., Lally J., Holloway G.P., Heigenhauser G.J.F., Bonen A., Spriet L.L. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J. Physiol. 2010;588:4795–4810. doi: 10.1113/jphysiol.2010.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn J.A., Coyle E.F., Sidossis L.S., Gastaldelli A., Horowitz J.F., Endert E., Wolfe R.R. Regulation of endogenous fat and carbohydrate metabolism in relation to exercise intensity and duration. Am. J. Physiol. 1993;265:E380–E391. doi: 10.1152/ajpendo.1993.265.3.E380. [DOI] [PubMed] [Google Scholar]
- Roudier E., Forn P., Perry M.E., Birot O. Murine double minute-2 expression is required for capillary maintenance and exercise-induced angiogenesis in skeletal muscle. FASEB J. 2012;26:4530–4539. doi: 10.1096/fj.12-212720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler M., Irmler M., Lehr S., Hartwig S., Staiger H., Al-Hasani H., Beckers J., de Angelis M.H., Häring H.-U., Weigert C. Cytokine response of primary human myotubes in an in vitro exercise model. Am. J. Physiol. Cell Physiol. 2013;305:C877–C886. doi: 10.1152/ajpcell.00043.2013. [DOI] [PubMed] [Google Scholar]
- Spriet L.L. ATP utilization and provision in fast-twitch skeletal muscle during tetanic contractions. Am. J. Physiol. 1989;257:E595–E605. doi: 10.1152/ajpendo.1989.257.4.E595. [DOI] [PubMed] [Google Scholar]
- Spriet L.L., Matsos C.G., Peters S.J. Muscle metabolism and performance in perfused rat hindquarter during heavy exercise. Am. J. Physiol. 1985;248 doi: 10.1152/ajpcell.1985.248.1.C109. C109–18. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon L.J.C., Greenhaff P.L., Constantin-Teodosiu D., Saris W.H.M., Wagenmakers A.J.M. The effects of increasing exercise intensity on muscle fuel utilisation in humans. J. Physiol. 2001;536:295–304. doi: 10.1111/j.1469-7793.2001.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Hironaka K., Wataya M., Fujii M., Eto M., Uda S., Hoshino D., Kunida K., Inoue H., Kubota H. Single-cell information analysis reveals that skeletal muscles incorporate cell-to-cell variability as information not noise. Cell Rep. 2020;32:108051. doi: 10.1016/j.celrep.2020.108051. [DOI] [PubMed] [Google Scholar]
- Wagle A., Jivraj S., Garlock G.L., Stapleton S.R. Insulin regulation of glucose-6-phosphate dehydrogenase gene expression is rapamycin-sensitive and requires phosphatidylinositol 3-kinase. J. Biol. Chem. 1998;273:14968–14974. doi: 10.1074/jbc.273.24.14968. [DOI] [PubMed] [Google Scholar]
- Wagner K.R., Kauffman F.C., Max S.R. The pentose phosphate pathway in regenerating skeletal muscle. Biochem. J. 1978;170:17–22. doi: 10.1042/bj1700017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Davis S.S., Borch Jensen M., Rodriguez-Fernandez I.A., Apaydin C., Juhasz G., Gibson B.W., Schilling B., Ramanathan A., Ghaemmaghami S. JNK modifies neuronal metabolism to promote proteostasis and longevity. Aging Cell. 2019;18:e12849. doi: 10.1111/acel.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D.L., Bolster D.R., Kimball S.R., Jefferson L.S., David L. Time course changes in signaling pathways and protein synthesis in C2 C12 myotubes following AMPK activation by AICAR. Am. J. Physiol. Endocrinol. Metab. 2006;17033:80–89. doi: 10.1152/ajpendo.00566.2005. [DOI] [PubMed] [Google Scholar]
- Wu C.L., Satomi Y., Walsh K. RNA-seq and metabolomic analyses of Akt1-mediated muscle growth reveals regulation of regenerative pathways and changes in the muscle secretome. BMC Genomics. 2017;18:1–18. doi: 10.1186/s12864-017-3548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K.I., Hayashi H., Nishiyama K., Kobayashi H., Uto S., Kondo H., Hashimoto S., Fujisato T. Control of myotube contraction using electrical pulse stimulation for bio-actuator. J. Artif. Organs. 2009;12:131–137. doi: 10.1007/s10047-009-0457-4. [DOI] [PubMed] [Google Scholar]
- Yugi K., Kubota H., Toyoshima Y., Noguchi R., Kawata K., Komori Y., Uda S., Kunida K., Tomizawa Y., Funato Y. Reconstruction of insulin signal flow from phosphoproteome and metabolome data. Cell Rep. 2014;8:1171–1183. doi: 10.1016/j.celrep.2014.07.021. [DOI] [PubMed] [Google Scholar]
- Yugi K., Kubota H., Hatano A., Kuroda S. Trans-omics: how to reconstruct biochemical networks across multiple ‘omic’ layers. Trends Biotechnol. 2016;34:276–290. doi: 10.1016/j.tibtech.2015.12.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data in this study are available at DDBJ (Accession number DRA010400).