Abstract
Background and aims:
The genotypic method could significantly shorten the time needed to obtain antibiotic susceptibility data for Helicobacter pylori. The aim of this study was to explore the profile of H. pylori from gastric biopsies and strains with antibiotic-induced resistance.
Methods:
A total of 124 gastric biopsies were used to perform gene sequencing and to perform bacterial culture and susceptibility testing. Seven susceptible strains were selected to develop resistance to clarithromycin, levofloxacin, and metronidazole. Four susceptible strains were selected to transfer candidate mutations. The genotype profiles of these groups were analyzed by sequencing analysis. The antibiotic susceptibility of these strains was detected using the E-test method.
Results:
Phenotypic resistance to clarithromycin, levofloxacin, and metronidazole was observed in 35.5%, 40.0%, and 79.8% strains, respectively. Point mutations in 23 S rRNA, gyrA, and rdxA genes were observed in 39.5%, 38.7%, and 86.3% of gastric biopsies, respectively. The A2143G mutation in the 23S rRNA occurs in most clarithromycin-resistant samples. The A2142C point mutation showed a higher efficacy than A2142G and A2143G for inducing clarithromycin resistance. The D91N and N87K mutations in gyrA occurs in most levofloxacin-resistant samples, and double point mutations showed a higher efficacy than single mutations for inducing levofloxacin resistance. Phenotypic resistance and mutations in rdxA lacked consistency.
Conclusion:
Genotype-based gastric biopsy analysis was reliable for determining clarithromycin and levofloxacin resistance. A2143G in 23S rRNA and N87K/D91N in the gyrA gene occurred in most resistant strains. Mutations in the rdxA gene were not good indicators of metronidazole resistance.
Keywords: antibiotic, genotype, Helicobacter pylori, resistance
Introduction
Helicobacter pylori is a gastric pathogen that colonizes the human stomach, leading to gastrointestinal diseases such as chronic gastritis, peptic ulcer disease, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma.1 More than half of the world’s population is infected by this bacterium. Therefore, anti-H. pylori treatment has been widely suggested for H. pylori-infected patients.1–3 Antibiotic susceptibility testing is a reliable assay that provides antibiotic susceptibility information for clinicians to prescribe anti-H. pylori regimens. Traditional phenotypic detection methods, such as agar dilution experiments and epsilometer tests (E-tests) based on isolated strains, are time consuming and difficult to apply in most hospitals due to the strict conditions required to culture the bacteria.4,5 In addition, as a standard method to detect antibiotic susceptibility, the E-test method cannot distinguish heteroresistant strains, which may provide incorrect information for clinical treatment.6 The genotyping method could significantly shorten the time needed to obtain antibiotic susceptibility data for H. pylori. This method has been explored since 1996.7 Mutations in the clarithromycin resistance gene 23S rRNA and the levofloxacin resistance gene gyrA show a good relationship with phenotypic antibiotic susceptibility in H. pylori clinical strains.8,9 Genotypic methods based on gastric biopsy samples and stool samples have also been used in several studies; some of these studies showed excellent consistency, while others did not.10–12 On the other hand, metronidazole, which shows a resistance rate of more than 50% in many countries or areas,13–15 has a resistance mechanism that is still obscure.16,17 The results of some studies have suggested that rdxA and frxA mutations play a crucial roles in metronidazole resistance as well as the outcomes of eradication therapy.17–19 However, some studies have also provided evidence that rdxA or frxA mutations are unable to explain metronidazole resistance in H. pylori.20,21 Some new potential mechanisms, such as the D85N mutation in the inner-membrane protein RclC or efflux pump also been suggest involved in metronidazole resistance.16,17 However, these mutations still need more evidence to prove their clinical value.
Currently, most studies about antibiotic resistance and gene mutations are based on clinically isolated resistant strains and susceptible strains, but less research has focused on the process by which susceptible strains become resistant, which may provide some different information. The aim of this research was to explore the profile of resistance to clarithromycin, levofloxacin, and metronidazole in H. pylori strains from gastric biopsy samples and strains with antibiotic-induced resistance.
Methods
Patients and biopsy
Adult outpatients referred for gastroscopy at The First Affiliated Hospital of Nanchang University between January 2018 and August 2019 who were positive for the urea breath test or histopathology were enrolled. Written informed consent was obtained from each participating patient before enrollment in the study. The research protocol was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (IRB 2018-019-1). Patients were excluded if they were taking PPI or H2-receptor antagonists 4 weeks prior to enrollment. Biopsies from the antrum and body of the stomach underwent bacterial culture, susceptibility testing, and DNA extraction.
H. pylori culture
Briefly, the gastric biopsy specimen was stored in brain heart infusion broth (Oxoid, Basingstoke, UK) with 20% glycerin at –80°C before use. After homogenization, part of the biopsy was cultured on Campylobacter agar (Oxoid, Basingstoke, UK) plates supplemented with 5% defibrinated sheep blood (Bio-kont, Zhejiang, China), 2.5 mg/l vancomycin, 3 mg/l trimethoprim, 2 mg/l polymyxin B, and 2 mg/l amphotericin B (Duly Biotech, Nanjing, China). The plates were incubated under microaerobic atmosphere conditions (10% CO2, 5% O2, and 85% N2) at 37°C for up to 5 days.
E-test method
The antibiotic susceptibility of the clinically isolated strains was detected using the E-test following the protocols of the Clinical and Laboratory Standards Institute. Briefly, bacteria were subcultured for 2 days on Mueller-Hinton agar plates supplemented with 5% defibrinated sheep blood. The bacterial suspension was adjusted to an optical density (OD) of 0.2 and inoculated onto the plates under microaerobic atmosphere conditions. The minimum inhibitory concentrations (MICs) of antibiotics were determined after 3–5 days of incubation. The resistance break points to metronidazole, clarithromycin, and levofloxacin were set at >8, >0.5, and >1 mg/l, respectively, which were selected using breakpoint tables for interpretation of MICs and zone diameters provided by the European Committee on Antimicrobial Susceptibility Testing Version 9.0, 2019 (http://www.eucast.org). ATCC43504 was used as the control strain.
Agar dilution test
The antibiotic susceptibility of the strain used for further development of the resistant strain was detected using the agar dilution assay to determine the MIC of the antibiotic. Clarithromycin, levofloxacin, and metronidazole (China) were used in this experiment. Briefly, bacteria were subcultured for 2 days on Mueller-Hinton agar plates supplemented with 5% sheep blood and two-fold serial dilutions of antibiotics. The bacterial suspension was adjusted to an OD or 0.2 and inoculated onto the plates under microaerobic atmosphere conditions. The MICs of antibiotics were determined after 3–5 days of incubation.
Genotypic analysis of antibiotic susceptibility
DNA was extracted from biopsy samples using a QIAamp1 DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. PCR for 23S rRNA, gyrA, and rdxA was performed with previously described primers.22,23 The primer sequences were 23S rRNA_F (5'-CCAC AGCGATGTGGTCTCAG-3'), 23S rRNA_R (5'-CTCCATAAGAGCCAAAGCCC-3'), (product 425 bp); gyrA_F (5'-AGCTTATTCCATGAGCGTGA-3'), gyrA _R (5'-TCAGGCCC TTTGACAAATTC-3'), (product 582 bp); and rdxA_F (5'-TTACAGAGAGCCAGATAGCC-3'), rdxA_R (5'-CACAACCAAGTAATCGCATC-3') (product 780 bp). The thermal profile used for amplification of the encoded sequences consisted of an initial step at 96°C for 5 min; 35 cycles at 96°C for 30 s, 56°C for 25 s and 72°C for 1 min; and a final step at 72°C for 5 min.
DNA sequencing and analysis
Sequence data were analyzed using the software DNAMAN (2005, Lynnon) and MEGA (version 10.0.4, Glen Stecher), and heteroresistance status was evaluated with ContigExpress (2000, InforMax). Following alignment with a reference sequence (H. pylori 26695), sequence data were examined in terms of codons, and comparisons of amino acids were performed.
Antibiotic-resistant strain development
Seven susceptible H. pylori strains were inoculated onto Columbia agar plates, and the agar dilution method was performed to obtain the MICori values for each strain. After obtaining the MICori values, the H. pylori strains were stimulated with antibiotics to induce antibiotic resistance individually. The process was performed as follows: H. pylori strains were collected from Columbia agar plates, the concentration was adjusted to an OD of 0.2 in Brucella broth medium containing 5% FBS, and the suspension was cultured at 37°C with shaking under microaerobic atmosphere conditions overnight. Antibiotics were added to Brucella broth medium to obtain a concentration of 1/2 MICori. After culturing for 6 h, the medium was transferred to Columbia agar plates containing 1/2 MICori and incubated for 2–3 days. Single colonies grown in this plate were labeled with Hp_1/2 × MICori and selected for the next step of resistance development. The process was repeated to obtain Hp_1 × MICori, Hp_2 × MICori, Hp_4 × MICori . . . Hp_n × MICori. The process stopped when the antibiotic concentration was 32 mg/l or induction of resistance failed three times.
Natural transformation of the candidate mutation
The amplified PCR products containing either wild-type sequences or candidate mutations were separately introduced into four susceptible strains through natural transformation, as previously described.24,25 Briefly, recipient cells were inoculated onto Mueller-Hinton agar plates and were grown for 5 h, after which 1.0 g of PCR fragments diluted in TE [10 mM Tris-HCl (pH 8.0) and 1 mM EDTA] was added directly onto the bacterial lawn. After incubation for 24 h under microaerophilic conditions, the transformed cells were streaked onto Mueller-Hinton II agar plates containing clarithromycin/levofloxacin/metronidazole (0.25, 0.5, 1.0, 2.0, 4.0, 8.0, 16, and 32 mg/liter), and several single colonies were separately collected from the lowest to the highest concentrations on the antibiotic-containing plates and spread onto antibiotic-free defibrinated sheep blood agar plates. Successful transformations and mutations were confirmed with PCR, followed by DNA sequencing analysis.
Statistical analyses
SPSS Statistics for Windows (version 21.0, IBM Corp, Armonk, NY, USA) was used to perform all statistical analyses. Chi-square and Fisher’s exact tests were used to determine the statistical significance of differences between categorical variables. A p-value ⩽ 0.05 was considered statistically significant.
Results
Genotype profiles of H. pylori from gastric biopsy
Patients and bacterial isolates
A total of 124 patients were ultimately recruited for this study during 2018–2019. They were positive for both H. pylori culture and PCR.
According to the E-test method results, clarithromycin, levofloxacin, and metronidazole resistance was observed in 44 (35.5%), 50 (40.0%), and 99 (79.8%) strains, respectively.
Profile of clarithromycin-related resistance genotype in biopsies
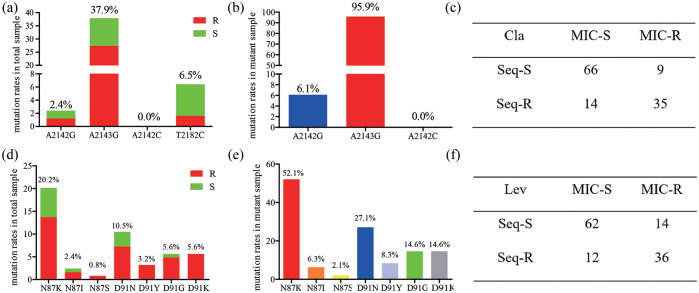
Among 124 H. pylori-positive biopsy samples, the 23S rRNA gene sequence classified 49 biopsies as having at least one of three point mutations responsible for clarithromycin resistance. The A2143G point mutation was observed in 47 (37.9%) biopsies (34 cases for resistant biopsies, 13 for susceptible biopsies); the A2142G point mutation was observed in three cases (2.4%) biopsies (two for resistant biopsies, one for susceptible biopsy). The A2142C mutation was not observed in any patient. In addition, we also noted that eight (6.45%) biopsies showed a T2182C point mutation (two for resistant biopsies, six for susceptible strains) (Figure 1a). For these strains with A2143G and/or A2142G point mutations, the A2143G mutation was predominantly observed (95.9%) (Figure 1b), A mixture of resistant and susceptible strains (heteroresistance status) were observed in 23 (18.5%) biopsies.
Figure 1.
Profile of clarithromycin- and levofloxacin-related resistance genotypes in biopsies. (a) Prevalence of point mutations in 23S rRNA in total samples; (b) prevalence of point mutations in 23S rRNA in mutant samples; (c) consistency between the 23S rRNA gene sequence and E-test result; (d) prevalence of amino mutations in gyrA in total samples; (e) prevalence of amino mutations in gyrA in mutant samples; (f) consistency between the gyrA gene sequence and E-test results.
Cla, clarithromycin; Lev, levofloxacin; MIC, minimum inhibitory concentration; R, resistant; S, susceptible; Seq-R, sequence result contain a point mutation (A2142G, A2143G, and/or A2142C point mutations for the 23S rRNA gene; point mutations resulting in the amino mutations N87K, N87I, N87S, D91N, D91Y, D91G, and/or D91K for the gyrA gene sequence); Seq-S, sequence result was the same as the reference sequence.
When comparing the genotypic and phenotypic methods for clarithromycin resistance detection, the 23S rRNA gene sequence and E-test showed 101 (81.5%) concordant susceptible or resistant results. For the 80 clarithromycin-susceptible strains, 66 corresponding biopsies contained a wild-type genotype, and 14 contained point mutations. For the 44 clarithromycin-resistant isolates, point mutations were detected in 35 biopsies, and the wild-type genotype was observed in 9 biopsies by 23S rRNA gene sequencing.
Profile of the levofloxacin-related resistance genotype in biopsies
Among 124 H. pylori-positive biopsy samples, gyrA amino acid sequences revealed 48 biopsies with mutations responsible for levofloxacin resistance. The N87K point mutation was observed in 25 (20.2%) biopsies (17 cases for resistant biopsies, 8 for susceptible biopsies); the N87I point mutation was observed in 3 (2.4%) biopsies (2 cases for resistant biopsies, 1 for susceptible biopsy); the N87S point mutation was observed in 1 (0.8%) biopsy (1 case for resistant biopsy); the D91N point mutation was observed in 13 (10.5%) biopsies (9 cases for resistant biopsies, 4 for susceptible biopsies); the D91Y point mutation was observed in 4 (3.2%) biopsies (4 cases for resistant biopsies); the D91G point mutation was observed in 7 (5.6%) biopsies (6 cases for resistant biopsies, 1 for susceptible biopsy); the D91K point mutation was observed in 7 (5.6%) biopsies (7 cases for resistant biopsies) (Figure 1d). For these strains with gyrA point mutations, the N87K mutation was predominantly detected (52.1%) (Figure 1e). A mixture of resistant and susceptible strains (heteroresistance status) were observed in 25 biopsies (20.2%).
Comparing the genotypic and phenotypic methods for levofloxacin resistance detection, gyrA amino acid sequences and E-tests showed 98 (79.0%) concordant susceptible or resistant results. For the 74 levofloxacin-susceptible strains, 62 corresponding biopsies contained a wild-type genotype, and 12 contained an amino acid mutation. For the 50 levofloxacin-resistant isolates, amino acid mutations were detected in 36 biopsies, and the wild-type genotype was found in 14 biopsies by gyrA amino acid sequencing (Figure 1f).
Profile of metronidazole-related resistance genotypes in biopsies
Among 124 H. pylori-positive biopsy samples, RdxA amino acid sequences revealed 107 biopsies with mutations in the rdxA gene. As there are no known point mutations that are responsible for metronidazole resistance, we analyzed mutations that had an occurrence rate of more than 5% in our biopsies. END (stop codon mutations), frameshift, R16H/C, M21A, R53H/Y, V62L/I, A68T/V, P91S, S108A/P, and A118S mutations were observed in 15 (12.1%), 25 (20.2%), 14 (11.3%), 16 (12.9%), 12 (9.7%), 23 (18.5%), 16 (12.9%), 7 (5.6%), 8 (6.5%), and 8 (6.5%) biopsies, respectively (Table 1). We did not count double or multiple point mutations in this part of the experiment, as RdxA amino acid sequences were mutable.
Table 1.
Profile of metronidazole-related resistance genotypes in biopsies.
| END | FS | 16re | 21re | 53re | 62re | 68re | 91re | 108re | 118re | WT | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S (25) | 3 (12.0%) | 3 (12.0%) | 3 (12.0%) | 3 (12.0%) | 2 (8.0%) | 6 (24.0%) | 4 (16.0%) | 0 (0.0%) | 1 (4.0%) | 3 (12.0%) | 3 (12.0%) |
| R (99) | 12 (12.1%) | 22 (22.2%) | 11 (11.1%) | 13 (13.1%) | 10 (10.1%) | 17 (17.2%) | 12 (12.1%) | 7 (7.1%) | 7 (7.1%) | 5 (5.1%) | 14 (14.1%) |
| Tot (124) | 15 (12.1%) | 25 (20.2%) | 14 (11.3%) | 16 (12.9%) | 12 (9.7%) | 23 (18.5%) | 16 (12.9%) | 7 (5.6%) | 8 (6.5%) | 8 (6.5%) | 17 (13.7%) |
16re, the 16th amino acid was replaced by another amino acid.
END, stop codon mutations, FS, frameshift; R, resistant; S, susceptible; Tot, total; WT, wild type.
When we tried to determine which mutation was responsible for metronidazole resistance, we analyzed the distribution of amino acid mutations in metronidazole-resistant and metronidazole-susceptible cases. To our surprise, END, frameshift, and nonsense mutations showed no significant differences (p > 0.05), which means that single amino acid mutations in H. pylori rdxA gene sequences may not be able to indicate a resistance phenotype for metronidazole. When we combined these amino acid mutations together, for the 99 metronidazole-resistant strains, 85 corresponding biopsies contained amino acid mutations, and the wild-type genotype was found in 14 cases by the rdxA amino acid sequence. For the 25 metronidazole-susceptible strains, 3 cases showed the wild-type genotype, and 22 showed amino acid mutations, which means that combining all these amino acid mutations to diagnose resistance is also not effective.
Profile of mutations during the process of antibiotic-induced resistance
After investigating the difference between clinically isolated resistant and susceptible strains, we aimed to determine the profiles of mutations that occurred after the H. pylori strain was stimulated with antibiotics. Seven susceptible H. pylori strains were selected to develop antibiotic resistance. The results showed that six individual strains successfully developed clarithromycin, levofloxacin, and metronidazole resistance, and one strain successfully developed levofloxacin and metronidazole resistance.
Profile of mutations during the process of clarithromycin-induced resistance
Among the six strains that developed clarithromycin resistance, one showed an A2142C point mutation after being stimulated with a 1 × MICori concentration of clarithromycin, and one showed an A2142G point mutation after being stimulated with a 1 × MICori concentration of clarithromycin. For these two strains, no additional point mutations were found in the further development process. The remaining four strains all showed A2143G point mutations after being stimulated with different concentrations of clarithromycin. Two strains subsequently converted the A2143G point mutation to the A2142G point mutation, and two strains remained the A2143G mutation throughout the entire process (Table 2). These results suggest that A2142G was more effective than the A2143G point mutation. In addition, we did not detect the point mutation T2182C in these strains.
Table 2.
Profile of mutations during the process of antibiotic-induced resistance.
| 0 × MICori | 0.5 × MICori | 1 × MICori | 2 × MICori | 4 × MICori | 8 × MICori | 16 × MICori | 32 × MICori | 64 × MICori | 128 × MICori | 256 × MICori | 512 × MICori | 1024 × MICori | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cli1 | WTcla | – | – | – | A2143G | – | – | – | G2143A A2142G |
– | – | ||
| Cli2 | WTcla | – | A2142G | – | – | – | – | – | – | – | – | ||
| Cli3 | WTcla | – | – | A2143G | – | – | – | – | G2143A A2142G |
– | – | – | |
| Cli4 | WTcla | – | failure | ||||||||||
| Cli5 | WTcla | – | – | – | A2143G | – | – | – | – | – | – | – | – |
| Cli6 | WTcla | – | A2142C | – | – | – | – | – | – | – | – | – | – |
| Cli7 | WTcla | – | A2143G | – | – | – | – | – | – | – | – | – | – |
| Cli1 | WTlev | – | 87N, D91N |
– | – | – | – | – | N87K, 91N |
– | |||
| Cli2 | WTlev | 87N, D91G |
– | – | N87K, G91D |
87K, D91G |
– | – | – | ||||
| Cli3 | WTlev | – | – | 87N, D91N |
N87K, N91D |
– | 87K, D91N |
– | – | – | – | ||
| Cli4 | WTlev | – | 87N, D91G |
87N, G91Y |
– | – | – | – | – | – | |||
| Cli5 | WTlev | – | – | N87K, 91D |
– | 87K, D91G |
– | – | |||||
| Cli6 | WTlev | – | 87N, D91G |
– | – | – | – | N87K, 91G |
– | ||||
| Cli7 | WTlev | – | 87N, D91G |
– | N87K, 91G |
– | – | – | – | – | |||
| Cli1 | WTmet | – | S29F S43F T58I |
– | – | 26FS | – | V14L I26L |
– | ||||
| Cli2 | WTmet | – | 65FS | – | T68S T79S87 |
– | – | – | – | ||||
| Cli3 | WTmet | – | E133K I172N |
E75K E138K |
N172I | K75E K133E K138E |
– | – | – | ||||
| Cli4 | WTmet | – | – | – | – | – | – | – | – | ||||
| Cli5 | WTmet | – | – | – | – | E133K | |||||||
| Cli6 | WTmet | – | – | – | R16C | – | – | – | C16H E73G N98S A118S S150G |
– | |||
| Cli7 | WTmet | – | – | N64K E74G |
H17Y P51T G74E |
– | – | – | – | – |
Profile of 23S rRNA/gyrA amino acid/rdxA amino acid mutations during the process of antibiotics-induced resistance.
–, same as the former genotype; MICori, the original MIC for clinical strain; WTcla, wild-type genotype for 23Sr RNA; WTlev, wild-type genotype for gyrA; WTmet, wild-type genotype for rdxA.
Profile of mutations during the process of levofloxacin-induced resistance
Among the seven strains that developed levofloxacin resistance, six developed the D91N/G amino acid mutation as their first mutation after being stimulated with levofloxacin. Five of these strains showed the N87K amino acid mutation after being further stimulated with a higher concentration of levofloxacin. Interestingly, two of these strains showed N/G91D when N87K was occurring, which means that mutations at amino acid 87 would be more efficacious than those at amino acid 91, and the latter mutations were reversible. Of the seven strains in this process, four showed 87K and 91G in their final status, two showed 87K and 91N in their final status, and the rest showed 87N and 91Y in their final status (Table 2). These results suggest that the single amino acid mutation N87K was more efficacious than the D91G/N mutation. Furthermore, 87K and 91G/N combined were more efficacious than 87K or 91G/N alone. We did not find other amino acid mutations, such as N87I, N87S, and D91K, in these strains.
Profile of mutations during the process of metronidazole-induced resistance
Among the seven strains that developed metronidazole resistance, six showed point mutations during the process of metronidazole-induced resistance, and amino acid mutations such as S29F, S43F, T58I, T67S, K73R, T79S, K81N, K127R, E133K, I172N, E75K, E138K, R16C, N98R, A118S, S150G, N84K, E74G, H17Y, P51T, and G74E and frameshift mutations occurred (Table 2). These mutations showed a wide range of rdxA genes, and few of them showed repeated mutations in different strains. In addition, one strain did not show an amino acid mutation during the process of metronidazole-induced resistance, and all these results suggest that the rdxA point mutation was not a good molecular marker to indicate a resistance phenotype for metronidazole.
Profile of point mutations in transformed H. pylori strains
To determine whether these mutations in 23S rRNA, gyrA, and rdxA were necessary and sufficient to mediate antibiotic resistance, the mutated PCR products were transformed into the susceptible H. pylori strain 26695 and three susceptible clinically isolated strains using natural transformation.
Profile of strains transformed with clarithromycin-related mutations
Three mutations (A2142C, A2142G, and A2143G) in 23S rRNA were transferred to these four susceptible strains, and the MICs of these strains are shown in Table 3. As the data show, compared with the original strains, the MICs for these strains increased considerably after transfer of these point mutations. Strains with the A2142C point mutation showed the highest MIC (>256 µg/ml), the MIC for strains with A2142G ranged from 32 to 128 µg/ml, and the MIC for strains with A2143G ranged from 3 to 12 µg/ml. These results suggest that the point mutations A2142C, A2142G, and A2143G in 23S rRNA were sufficient to mediate antibiotic resistance and once again proved that the point mutation A2142G was more efficacious than A2143G and that A2142C was more efficacious than A2142G.
Table 3.
MIC changes for strains transformed with clarithromycin-related mutations.
| Strain | Ori | A2142G | A2142C | A2143G |
|---|---|---|---|---|
| 26695 | 0.008 | 64 | >256 | 12 |
| Cli1 | 0.032 | 32 | >256 | 6 |
| Cli3 | 0.016 | 32 | > 256 | 3 |
| Cli5 | 0.008 | 128 | > 256 | 12 |
26695, H. pylori strain 26695; cli, clinical isolated strain; MIC, minimal inhibitory concentration; ori, original.
Profile of strains transformed with levofloxacin-related mutations
Six amino acid mutations (91N, 91G, 91Y, 87K, 87K and 91N) and 87K and 91D in gyrA were transferred to these four susceptible strains, and the MICs of these strains are shown in Table 4. The data show that, compared with those of the original strains, the MICs for these strains increased to some degree after transfer of these point mutations. However, unlike transfer of mutations in 23S rRNA, strains with a mutated gyrA gene could not improve their MICs to a very high level, which suggests that, during the process of levofloxacin-induced resistance, gyrA mutation is a very important part of overcoming antibiotic pressure; however, the mechanisms involved in this process remain unclear.
Table 4.
MIC changes for strains transformed with levofloxacin-related mutations.
| Strain | Ori | 87N,91N | 87K,91N | 87N,91G | 87K,91D | 87K,91G | 87N,91Y |
|---|---|---|---|---|---|---|---|
| 26695 | 0.125 | 0.380 | 1.500 | 0.750 | 1.000 | >32.000 | 0.750 |
| Cli1 | 0.250 | 1.000 | 1.500 | 1.000 | 4.000 | 16.000 | 0.750 |
| Cli3 | 0.030 | 3.000 | 3.000 | 0.750 | 1.500 | 8.000 | 1.500 |
| Cli5 | 0.250 | 1.000 | 4.000 | 1.500 | 3.000 | >32.000 | 16 |
26695, H. pylori strain 26695; cli, clinical isolated strain; MIC, minimal inhibitory concentration; ori, original.
Discussion
In recent decades, the problem of increasing antibiotic resistance rates has greatly decreased the efficacy of antibiotic-based methods. Tailored treatment based on individual susceptibility data can achieved a satisfactory H. pylori eradication rate.26–28 The E-test method could provide several kinds of antibiotic susceptibility information for the bacteria, but the strict conditions required to culture the bacteria make it difficult to apply in most hospitals. Genotypic methods based on gastric biopsy, gastric juice, or perhaps stool samples deserve more attention. As the results show, the genotypic method based on the 23S rRNA point mutation was reliable for the diagnosis of clarithromycin resistance, and the point mutation A2143G was found in most clarithromycin-resistant strains but was less efficacious than A2142G in inducing clarithromycin resistance. The point mutation A2142C was not found in these clinical biopsies but was detected in the antibiotic-induced resistant strain, and natural transformation also proved that this mutation was more efficacious than the A2142G and A2143G point mutations. Some studies suggest that T2182C is also involved in clarithromycin resistance, but our data do not support this hypothesis. Indeed, the results of our previous work also showed that T2182C could not improve the specificity and sensitivity of the genotypic method.4 One inconsistency was that the point mutation A2142G was found in two clinical strains, while only one of these strains was susceptible to clarithromycin, which may be explained by heteroresistance status. When the wild-type strain and mutant strain coexist in one patient, the wild-type strain may act as the dominant group with no antibiotic stimulation. This leads to a susceptible phenotype.
Mutations in the gyrA and gyrB gene were initially suggested to indicate quinolone resistance, but recent studies have indicated an inconsistent association between gyrB mutations and quinolone resistance;22,29 therefore, only gyrA mutations were detected in levofloxacin-resistant strains in this work. These results indicate that the genotypic method based on gyrA amino acid mutations was reliable for the diagnosis of levofloxacin as well. The amino acid mutations N87K and D91N were most common in the levofloxacin-resistant strain, and N87K was more efficacious than D91N. Interestingly, when we transferred amino acid mutations that may participate in levofloxacin resistance to four susceptible strains, their MIC values did not improve much, which means that, although these amino acid mutations may be a good indicator for diagnosing levofloxacin resistance, some other mechanism may also be involved in resistance.
Metronidazole is still widely used in anti-H. pylori treatment, although it had the highest resistance rate among the investigated antibiotics. Ji et al. suggested that a high dose of metronidazole could increase the eradication rate of anti-H. pylori treatment in populations with high metronidazole resistance but could cause more side effects.30 In some areas, such as Japan, the metronidazole resistance rate is still low, and they could achieve a satisfactory eradication rate based on metronidazole treatment.31,32 We also explored the genotype method for metronidazole; however, unlike clarithromycin and levofloxacin, the mechanism of metronidazole resistance remains obscure, and the most widely accepted mechanism is mutation in the rdxA gene.33,34 Although the reliability of this gene has also been frequently questioned,9,20,21 we chose the whole rdxA gene as our target. Similar to other research, we did not find any amino acid mutation of rdxA that could explain metronidazole resistance well. There are diverse mutations in the rdxA gene, but the distribution and repeatability of these mutations were not sufficient to indicate metronidazole resistance. In addition, we found some cases that possessed a resistance phenotype but had no point mutation in the rdxA gene. During the process of developing metronidazole-induced resistance, one strain also showed no point mutation, which again suggests that mutations in rdxA may not always be essential for metronidazole resistance. On the other hand, both the E-test results and the genotypic method show a high resistance rate for metronidazole, and there may be no need to perform a susceptibility test when metronidazole is used in anti-H. pylori treatment, as a higher concentration can be selected when necessary.
Tailored treatment based on individual susceptibility data has been shown to achieved satisfactory H. pylori eradication rates.26–28 The Maastricht V/Florence Consensus Report have mentioned that the value of culture is primarily to perform antimicrobial susceptibility testing for clarithromycin, levofloxacin, metronidazole, amoxicillin, and tetracycline.1 However, susceptibility testing based on the culture method is time consuming and difficult to apply in most hospitals. The development of genotypic methods based on gastric biopsies that could obtain similar results as those observed with the E-test method would overcome the limitation of antimicrobial susceptibility testing and allow the tailored method to be applied more easily in most hospitals in the future. Our results showed that the detection of clarithromycin and levofloxacin resistance with genotypic methods using gastric biopsies is effective, suggesting that tailored treatment based on genotypic method with gastric biopsy deserve further consideration.
Conclusion
Genotypic method-based gastric biopsy was reliable for inferring clarithromycin and levofloxacin resistance. A2143G in 23S rRNA and 87K and 91N occurred in most resistant strains. Mutations in the rdxA gene were not good indicators of metronidazole resistance.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Key Research and Development Program of China (2016YFC1302201), the National Natural Science Foundation of China (81970502, 81860107, 81260076), and the Leading Talent Training Plan of the Ganpo Outstanding Talents 555 Project of Jiangxi Province (2010-3-61).
ORCID iD: Yong Xie  https://orcid.org/0000-0002-5290-5579
https://orcid.org/0000-0002-5290-5579
Contributor Information
You-hua Wang, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Fang-fei Wang, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Xiao-ling Gong, Department of Blood Transfusion, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Li-li Yan, Department of Medical College, Nanchang University, Nanchang, Jiangxi Province, China.
Qiao-yun Zhao, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Yan-ping Song, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Ru-lin Zhao, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Ya-jing He, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, China.
Linfu Zhou, Department of Biochemistry and Molecular Biology, Zhejiang University School of Medicine, Hangzhou, Zhejiang Province, China.
Dong-sheng Liu, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, 17 Yongwai Zheng Street, Nanchang, Jiangxi Province, 330000, China.
Yong Xie, Department of Gastroenterology, The First Affiliated Hospital of Nanchang University, 17 Yongwai Zheng Street, Nanchang, Jiangxi Province, 330000, China.
References
- 1. Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection-the Maastricht V/florence consensus report. Gut 2017; 66: 6–30. [DOI] [PubMed] [Google Scholar]
- 2. Liu WZ, Xie Y, Lu H, et al. Fifth Chinese national consensus report on the management of Helicobacter pylori infection. Helicobacter 2018; 23: e12475. [DOI] [PubMed] [Google Scholar]
- 3. Sugano K, Tack J, Kuipers EJ, et al. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 2015; 64: 1353–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Y-H, Li Z, Wang L, et al. A systematic review and meta-analysis of genotypic methods for detecting antibiotic resistance in Helicobacter pylori. Helicobacter 2018; 23: e12467. [DOI] [PubMed] [Google Scholar]
- 5. Giorgio F, Ierardi E, Sorrentino C, et al. Helicobacter pylori DNA isolation in the stool: an essential pre-requisite for bacterial noninvasive molecular analysis. Scand J Gastroenterol 2016; 51: 1429–1432. [DOI] [PubMed] [Google Scholar]
- 6. Farzi N, Behzad C, Hasani Z, et al. Characterization of clarithromycin heteroresistance among Helicobacter pylori strains isolated from the antrum and corpus of the stomach. Folia Microbiol 2019; 64: 143–151. [DOI] [PubMed] [Google Scholar]
- 7. Versalovic J, Shortridge D, Kibler K, et al. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother 1996; 40: 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tuan VP, Narith D, Tshibangu-Kabamba E, et al. A next-generation sequencing-based approach to identify genetic determinants of antibiotic resistance in Cambodian Helicobacter pylori clinical isolates. J Clin Med 2019; 8: 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lauener FN, Imkamp F, Lehours P, et al. Genetic determinants and prediction of antibiotic resistance phenotypes in Helicobacter pylori. J Clin Med 2019; 8: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Phan TN, Santona A, Tran VH, et al. High rate of levofloxacin resistance in a background of clarithromycin- and metronidazole-resistant Helicobacter pylori in Vietnam. Int J Antimicrob Agents 2015; 45: 244–248. [DOI] [PubMed] [Google Scholar]
- 11. Trespalacios AA, Rimbara E, Otero W, et al. Improved allele-specific PCR assays for detection of clarithromycin and fluoroquinolone resistant of Helicobacter pylori in gastric biopsies: identification of N87I mutation in GyrA. Diagn Microbiol Infect Dis 2015; 81: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Francesco V, Zullo A, Giorgio F, et al. Change of point mutations in Helicobacter pylori rRNA associated with clarithromycin resistance in Italy. J Med Microbiol 2014; 63: 453–457. [DOI] [PubMed] [Google Scholar]
- 13. Liu DS, Wang YH, Zeng ZR, et al. Primary antibiotic resistance of Helicobacter pylori in Chinese patients: a multiregion prospective 7-year study. Clin Microbiol Infect 2018; 24: 780–785. [DOI] [PubMed] [Google Scholar]
- 14. Khien VV, Thang DM, Hai TM, et al. Management of antibiotic-resistant Helicobacter pylori infection: perspectives from Vietnam. Gut Liver 2019; 13: 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kageyama C, Sato M, Sakae H, et al. Increase in antibiotic resistant Helicobacter pylori in a University Hospital in Japan. Infect Drug Resist 2019; 12: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chua EG, Debowski AW, Webberley KM, et al. Analysis of core protein clusters identifies candidate variable sites conferring metronidazole resistance in Helicobacter pylori. Gastroenterol Rep (Oxf) 2019; 7: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee SM, Kim N, Kwon YH, et al. rdxA, frxA, and efflux pump in metronidazole-resistant Helicobacter pylori: their relation to clinical outcomes. J Gastroenterol Hepatol 2018; 33: 681–688. [DOI] [PubMed] [Google Scholar]
- 18. Hashemi SJ, Sheikh AF, Goodarzi H, et al. Genetic basis for metronidazole and clarithromycin resistance in Helicobacter pylori strains isolated from patients with gastroduodenal disorders. Infect Drug Resist 2019; 12: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pourakbari B, Mahmoudi S, Parhiz J, et al. High frequency of metronidazole and clarithromycin-resistant Helicobacter pylori in formalin-fixed, paraffin-embedded gastric biopsies. Br J Biomed Sci 2018; 75: 61–65. [DOI] [PubMed] [Google Scholar]
- 20. Marques B, Donato MM, Cardoso O, et al. Study of rdxA and frxA genes mutations in metronidazole-resistant and -susceptible Helicobacter pylori clinical isolates from the central region of Portugal. J Glob Antimicrob Resist 2019; 17: 300–304. [DOI] [PubMed] [Google Scholar]
- 21. Miftahussurur M, Shrestha PK, Subsomwong P, et al. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol 2016; 16: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Binkowska A, Biernat MM, Laczmanski L, et al. Molecular patterns of resistance among Helicobacter pylori strains in South-Western Poland. Front Microbiol 2018; 9: 3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Farzi N, Yadegar A, Sadeghi A, et al. High prevalence of antibiotic resistance in Iranian Helicobacter pylori isolates: importance of functional and mutational analysis of resistance genes and virulence genotyping. J Clin Med 2019; 8: 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Binh TT, Suzuki R, Trang TT, et al. Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob Agents Chemother 2015; 59: 2343–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Binh TT, Shiota S, Suzuki R, et al. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J Antimicrob Chemother 2014; 69: 1796–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ierardi E, Giorgio F, Iannone A, et al. Noninvasive molecular analysis of Helicobacter pylori: Is it time for tailored first-line therapy? World J Gastroenterol 2017; 23: 2453–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Butenko T, Jeverica S, Orel R, et al. Antibacterial resistance and the success of tailored triple therapy in Helicobacter pylori strains isolated from Slovenian children. Helicobacter 2017; 22: e12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen H, Dang Y, Zhou X, et al. Tailored therapy versus empiric chosen treatment for Helicobacter pylori eradication: a meta-analysis. Medicine 2016; 95: e2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miftahussurur M, Waskito LA, Syam AF, et al. Alternative eradication regimens for Helicobacter pylori infection in Indonesian regions with high metronidazole and levofloxacin resistance. Infect Drug Resist 2019; 12: 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ji Y, Lu H. Meta-analysis: high-dose vs. low-dose metronidazole-containing therapies for Helicobacter pylori eradication treatment. PLoS One 2018; 13: e189888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Okuda M, Lin Y, Wang C, et al. Metronidazole for Helicobacter pylori eradication therapy among children and adolescents in Japan: overcoming controversies and concerns. Helicobacter 2019; 24: e12575. [DOI] [PubMed] [Google Scholar]
- 32. Mabe K, Okuda M, Kikuchi S, et al. Randomized controlled trial: PPI-based triple therapy containing metronidazole versus clarithromycin as first-line treatment for Helicobacter pylori in adolescents and young adults in Japan. J Infect Chemother 2018; 24: 538–543. [DOI] [PubMed] [Google Scholar]
- 33. Ramzy I, Elgarem H, Hamza I, et al. Genetic mutations affecting the first line eradication therapy of Helicobacter pylori-infected Egyptian patients. Rev Inst Med Trop Sao Paulo 2016; 58: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rasheed F, Campbell BJ, Alfizah H, et al. Analysis of clinical isolates of Helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter 2014; 19: 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]