Abstract
Objective
Although N6-methyladenosine (m6A) RNA methylation is the most common mRNA modification process, few studies have examined the role of m6A in stomach adenocarcinomas (STADs).
Methods
In this retrospective study, we analyzed 293 STAD samples from The Cancer Genome Atlas with complete clinicopathological feature profiles. The m6A methylation risk signature was derived from LASSO–Cox regression analyses with 15 m6A regulators. Statistical analysis was performed and figures were prepared using R software (https://www.R-project.org/).
Results
The m6A signature was established as follows: risk score = FTO × 0.127 + YTHDF1 × 0.004 + KIAA1429 × 0.044 + YTHDC2 × 0.112 − RBM15 × 0.135 − ALKBH5 × 0.019 − YTHDF2 × 0.028, which was confirmed as an independent prognostic indicator to predict overall survival of patients with STAD. Risk scores and tumor grades were closely associated. Cell cycle, p53 signaling pathways, DNA mismatch repair, and RNA degradation were enriched in the low-risk subgroup. This subgroup showed significantly higher expression of immune checkpoint molecules including PD-1 (programmed death 1), PD-L1 (programmed death-ligand 1), and CTLA-4 (cytotoxic T-lymphocyte–associated antigen 4), suggesting that the signature may be a useful immunotherapy predictor.
Conclusions
We established an m6A methylation signature as an independent prognostic tool to predict overall survival, which may also be useful as an immunotherapy predictor.
Keywords: N6-methyladenosine, mRNA modification, signature, stomach carcinoma, prognosis, immune checkpoint
Introduction
Stomach carcinoma is a lethal cancer of the digestive system, with approximately 1,033,701 new cases diagnosed in 185 countries worldwide, resulting in 782,685 deaths in 2018.1 Although immune and target therapies have developed rapidly in recent years,2,3 the mechanisms underlying the pathogenesis and progression of stomach adenocarcinoma (STAD) require further investigation to discover new prognostic tools and improve the efficacy of current therapies.
Epigenetic changes were initially thought to be limited to DNA and histone layers; however, accumulating evidence shows that epigenetic modification of RNA plays a critical role in regulating RNA function and metabolism.4,5 Various types of RNAs have shown these modifications, including mRNAs, rRNAs, tRNAs, long noncoding RNAs, and microRNAs.5,6 Among these, N6-methyladenosine (m6A) modification is the most extensive form of mRNA modification in eukaryotes;7,8 it is located in the 3′-untranslated region (UTR) and regulates the expression of more than 7000 genes.9 m6A modification acts as an inhibitory regulator to mediate the translation of target mRNA, such that a reduction in m6A modification increases protein expression.10 m6A modification regulates cellular processes, including cell proliferation, impairment, and death, thereby influencing tissue development (e.g., nervous system tissue)11 and the progression and clinical outcome of several types of tumor.8,12 The detailed molecular mechanisms of these processes remain unknown.
m6A RNA modification is dynamically mediated by three categories of regulating genes (writers, readers, and erasers), encoding methyltransferases, binding proteins, and demethylases, respectively.13 Prominent regulators classified as writers include KIAA1429, RNA binding motif protein 15 (RBM15), the methyltransferase-like family (e.g., METTL3 and METTL14), zinc finger CCCH domain-containing protein 13 (ZC3H13), and WT1-associated protein (WTAP). Readers include the YTH domain-containing family (e.g., YTHDC1 and YTHDC2), the YTH N6-methyladenosine RNA-binding protein family (e.g., YTHDF1, YTHDF2, and YTHDF3), and the heterogeneous nuclear ribonucleoprotein family (e.g., HNRNPC and HNRNPA2B1). Erasers include fat mass and obesity associated protein (FTO) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5).14–20 Recent studies have revealed that changes in expression among these regulators are associated with malignant biological processes in several types of tumors, including gastric cancer.21–25 However, because these studies were limited to single regulators, the relationship between STAD and m6A RNA methylation has not been comprehensively analyzed. m6A RNA methylation also regulates the synthesis of type I interferon, which is associated with tumor immune checkpoints and immunotherapy efficacy.26,27 Therefore, we analyzed the expression of the 15 regulators mentioned above in STAD samples from a dataset extracted from The Cancer Genome Atlas (TCGA) and constructed m6A risk scores to estimate its association with clinical characteristics and immune checkpoints.
Materials and methods
Ethics statement
The ethical review board provided an exemption for this study.
Data processing
All RNA-seq transcriptome and corresponding clinicopathological profiles were obtained from the publicly available TCGA database (http://cancergenome.nih.gov). In this retrospective study, we included 293 STAD samples from TCGA including complete age; sex; pathological stage; histological grade; and tumor, node, and metastasis (TNM) stage information. We included only samples with an overall survival (OS) time >1 month (Table 1). Normalized mRNA expression matrix data (fragments per kilobase of transcript per million mapped reads, FPKM) were downloaded from TCGA.
Table 1.
Clinicopathological characteristics of TCGA samples included in our study.
| Variable | TCGA dataset (n = 293) |
Total | ||
|---|---|---|---|---|
| Training cohort(n = 147) | Testing cohort(n = 146) | p-value | ||
| Age | <0.01 | |||
| ≤65 years | 64 | 70 | 134 (45.7%) | |
| >65 years | 83 | 76 | 159 (54.3%) | |
| Gender | 0.42 | |||
| Female | 58 | 50 | 108 (36.9%) | |
| Male | 89 | 96 | 185 (63.1%) | |
| Grade | 0.85 | |||
| G1 | 3 | 3 | 6 (2.0%) | |
| G2 | 53 | 48 | 101 (34.5%) | |
| G3 | 91 | 95 | 186 (63.5%) | |
| Stage | 0.45 | |||
| I | 23 | 14 | 37 (12.6%) | |
| II | 48 | 49 | 97 (33.1%) | |
| III | 63 | 67 | 130 (44.4%) | |
| IV | 13 | 16 | 29 (9.9%) | |
| T | 0.28 | |||
| T1 | 9 | 4 | 13 (4.4%) | |
| T2 | 33 | 27 | 60 (20.5%) | |
| T3 | 72 | 72 | 144 (49.2%) | |
| T4 | 33 | 43 | 76 (25.9%) | |
| M | 0.99 | |||
| M0 | 139 | 137 | 276 (94.2%) | |
| M1 | 8 | 9 | 17 (5.8%) | |
| N | 0.85 | |||
| N0 | 46 | 42 | 88 (30%) | |
| N1 | 38 | 42 | 80 (27.3%) | |
| N2 | 30 | 33 | 63 (21.5%) | |
| N3 | 33 | 29 | 62 (21.2%) | |
| Pathology | 0.99 | |||
| Signet ring | 4 | 4 | 8 (2.73%) | |
| Others | 141 | 143 | 284 (96.9%) | |
| Localization | 0.08 | |||
| Antrum | 51 | 59 | 110 (37.5%) | |
| Cardia | 24 | 16 | 40 (13.7%) | |
| Fundus | 55 | 47 | 102 (34.8%) | |
| GEJ | 10 | 21 | 31 (10.6%) | |
TCGA, The Cancer Genome Atlas; T, N, M, tumor, node, metastasis; GEJ, gastroesophageal junction.
Prognostic model construction and validation
We selected 15 m6A RNA methylation regulators from previous studies, including METTL3, METTL14, WTAP, KIAA1429, RBM15, ZC3H13, YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, HNRNPC, HNRNPA2B1, ALKBH5, and FTO; expression data for these regulators were available from TCGA. To calculate risk scores, we divided the 293 samples into training (147 cases) and testing (146 cases) cohorts using a randomization method. Coefficients for selected regulators were calculated by applying LASSO (least absolute shrinkage and selection operator)–Cox regression to the training cohort, based on minimum partial likelihood deviance. Risk scores were calculated using the following formula:28
where coef(x) is the coefficient and gene(x) is the standardized gene expression profile. The testing cohort was validated using the same formula.
Gene set enrichment analysis
To identify enriched biological pathways between subgroups with high and low risk scores according to Kyoto Encyclopedia of Genes and Genomes (KEGG),29 we performed gene set enrichment analysis (GSEA) using software provided by the Broad Institute (Cambridge, MA, USA). We used the GSEA canonical pathways gene set (c2.cp.kegg.v7.0.symbols.gmt) with GSEA software 4.0.2 (www.gsea-msigdb.org); significance was determined using normalized p-values (<0.05) and false discovery rate (<0.25).
Statistical analyses
Statistical analyses were performed using R software, version 3.5.2 (http://www.r-project.org). LASSO-Cox regression was performed using the glmnet package. Kaplan–Meier plots of patient OS and disease-free survival (DFS) were created using the survival and ggplot packages, with p-values calculated using the log-rank test. The stability of the m6A model was tested with receiver-operating characteristic (ROC) curves created using the survivalROC package. Differences in gene expression, risk scores, and histological grades among subgroups were analyzed using the Wilcoxon test. Significance was determined at a level of p < 0.05.
Results
STAD prognostic value of m6A RNA methylation regulators
We first compared clinical outcomes among the 15 m6A RNA methylation regulators. Univariate Cox regression analyses showed that expression levels of RBM15 and ALKBH5 were significantly associated with OS among the 293 STAD samples acquired from TCGA (Table 2). Because of the limited number of prognosis-related regulators, all 15 m6A regulators were included in our subsequent analyses.
Table 2.
Univariable Cox regression analysis to evaluate the prognosis value of 15 m6A regulators.
| Regulators | HRa | 95% CI |
p-valueb | |
|---|---|---|---|---|
| Low | High | |||
| Writers | ||||
| KIAA1429 | 1.01 | 0.932 | 1.095 | 0.808 |
| RBM15 | 0.839 | 0.738 | 0.954 | 0.008 |
| METTL14 | 0.811 | 0.623 | 1.058 | 0.122 |
| METTL3 | 0.928 | 0.813 | 1.059 | 0.269 |
| ZC3H13 | 0.996 | 0.969 | 1.025 | 0.799 |
| WTAP | 0.949 | 0.886 | 1.017 | 0.139 |
| Readers | ||||
| YTHDC1 | 0.942 | 0.858 | 1.034 | 0.208 |
| YTHDC2 | 1.016 | 0.889 | 1.162 | 0.812 |
| YTHDF1 | 0.996 | 0.978 | 1.014 | 0.627 |
| YTHDF2 | 0.961 | 0.922 | 1.001 | 0.055 |
| YTHDF3 | 0.994 | 0.961 | 1.028 | 0.727 |
| HNRNPC | 0.994 | 0.976 | 1.012 | 0.511 |
| HNRNPA2B1 | 0.993 | 0.986 | 1.001 | 0.069 |
| Erasers | ||||
| FTO | 1.116 | 0.976 | 1.276 | 0.108 |
| ALKBH5 | 0.97 | 0.943 | 0.999 | 0.045 |
m6A, N6-methyladenosine; STAD, stomach adenocarcinoma; HR, hazard ratio; CI, confidence interval.
aValues >1.0 indicate that expression is positively associated with poor survival.
bLikelihood ratio test p-value; significant values < 0.05.
Construction and validation of m6A risk scores
We next constructed risk scores using the training cohort by integrating 7 of the 15 m6A regulators into a formula, in which the 7 regulators were chosen based on the LASSO–Cox regression results as follows: FTO × 0.127 + YTHDF1 × 0.004 + KIAA1429 × 0.044 + YTHDC2 × 0.112 − RBM15 × 0.135 − ALKBH5 × 0.019 − YTHDF2 × 0.028 (Figure 1A–C). To validate the prognostic value of this model, we divided the training (n = 147) and testing (n = 146) cohorts into high- and low-risk groups based on significant differences in OS determined by Kaplan–Meier curves (ptraining < 0.001, ptesting < 0.05; Figure 1D,E). Based on area under the curve (AUC) values, the model adequately predicted OS rates for STAD patients in both cohorts (AUCtraining = 62.6%, AUCtesting = 65.9%; Figure 1F,G).
Figure 1.
Risk scores constructed using m6A RNA methylation regulators. (a) Partial likelihood deviance determined by LASSO–Cox regression of OS rates among patients in the training cohort (n = 147). Dashed vertical line indicates logLambda = −3.55 (minimum partial likelihood deviance). (b, c) LASSO–Cox coefficients of 7 selected m6A regulators in the following model: risk score = FTO × 0.127 + YTHDF1 × 0.004 + KIAA1429 × 0.044 + YTHDC2 × 0.112 − RBM15 × 0.135 − ALKBH5 × 0.019 − YTHDF2 × 0.028. (d, e) Kaplan–Meier curves for OS rates among patients in the training cohort (d) and testing cohort (n = 146) (e). Patients in both cohorts were assigned to low- and high-risk categories based on a risk score cut-off value. (f, g) ROC curves indicated adequate predictive ability of the m6A risk score for the training (F) and testing (g) cohorts. m6A, N6-methyladenosine; OS, overall survival; ROC, receiver operating characteristic.
m6A risk scores as independent prognostic indicators
Because the training and testing cohorts were derived from the same datasets and the sample size was limited, we subsequently merged all samples to increase the sample size. To expand the sample size, we merged all 293 samples into two risk subgroups according to risk scores (Figure 2A,B). The 1-, 3-, and 5-year OS and DFS rates showed significant differences between the two risk categories (log-rank, p < 0.001; Table 3). To further evaluate the prognostic value of the m6A risk signature, factors including risk score, age, TMN stage, pathological stage, and histological grade were successively included in univariate and multivariate Cox regression models. Only risk score was significantly related to OS in both Cox analyses (p < 0.001; Figure 2C, Table 4), indicating that the signature may be an independent prognostic tool.
Figure 2.
Relationships between risk scores and clinicopathological features. (a) Kaplan–Meier curves for OS rates among all patients (n = 293). (b) ROC curves validated the predictive ability of the merged cohort. (c) Univariate and multivariate Cox regression analyses of the relationship between clinicopathological characteristics (including risk scores) and OS rates of patients in the merged cohort. The risk score was found to be an independent prognostic indicator. (d) A heatmap showing the relationship between expression levels of the seven selected m6A RNA methylation regulators and clinicopathological features for the low- and high-risk categories. (e) Distribution of risk scores stratified by histological grades. Significant differences in risk score were not detected among other clinicopathological features. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001. OS, overall survival; ROC, receiver operating characteristic; m6A, N6-methyladenosine.
Table 3.
Overall survival and disease-free survival of 293 STAD patient samples.
| Group | Survival rate (%) |
p-valuea | ||
|---|---|---|---|---|
| 1 year | 3 year | 5 year | ||
| Overall survival | ||||
| Low risk | 84.7 | 60.3 | 56.8 | <0.001 |
| High risk | 72.6 | 38.0 | 22.3 | |
| Disease-free survival | ||||
| Low risk | 87.3 | 68.4 | 68.4 | <0.001 |
| High risk | 67.5 | 48.5 | 31.2 | |
STAD, stomach adenocarcinoma.
aLog-rank p-value; significant values <0.05.
Table 4.
Univariate and multivariate Cox regression analysis of m6A regulators for overall survival in patients with STAD.
| Variable | Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| HRa | 95% CI | p-valueb | HRa | 95% CI | p-valueb | |
| Age (>65 vs ≤65) | 0.998 | 0.993–1.004 | 0.606 | |||
| Gender | 1.454 | 0.952–2.219 | 0.083 | |||
| Grade | 1.342 | 0.923–1.951 | 0.124 | |||
| Stage | 1.516 | 1.200–1.915 | <0.001 | 1.33 | 0.927–1.908 | 0.122 |
| T | 1.311 | 1.028–1.672 | 0.029 | 1.075 | 0.778–1.485 | 0.661 |
| M | 1.751 | 0.882–3.475 | 0.109 | |||
| N | 1.268 | 1.065–1.510 | 0.008 | 1.134 | 0.899–1.430 | 0.288 |
| Risk score | 2.949 | 1.720–5.055 | <0.001 | 3.283 | 1.866–5.778 | <0.001 |
m6A, N6-methyladenosine; STAD, stomach adenocarcinoma; HR, hazard ratio; CI, confidence interval; T, N, M, tumor, node, metastasis.
aValues >1.0 indicate that expression is positively associated with poor survival.
bLikelihood ratio test p-value; significant values <0.05.
Association between m6A scores and clinicopathological characteristics
Next, we evaluated the association between risk scores and clinicopathological features by producing a heatmap of clinical characteristics including TNM stage, histological grade, and pathological stage, associated with expression levels of the seven selected regulators (Figure 2D). Patients with grade 3 tumors (poor differentiation) had higher scores than those with grade 2 tumors (moderate differentiation) (p < 0.05; Figure 2E); no significant differences were detected among other clinical characteristics (data not shown).
Association between m6A RNA methylation scores and GSEA results
Next, given the tight association between m6A scores and both prognosis and histological grade, we identified the genes and signaling pathways involved in m6A RNA methylation that affected clinical outcomes. We applied GSEA using the KEGG database to examine enriched gene sets among samples from the two risk categories. Several biological processes were enriched among the low-risk subgroup (Figure 3A), including cell cycle (normalized enrichment score (NES) = 2.04, normalized p < 0.001), p53 signaling pathways (NES = 1.96, normalized p < 0.003), DNA mismatch repair (NES = 1.80, normalized p < 0.005), and RNA degradation (NES = 1.81, normalized p < 0.007). Some of these gene sets were previously identified as being related to m6A modification; however, such a relationship has not been reported for DNA mismatch repair, providing an avenue for future STAD research. We also examined the expression of p53 (also called TP53) to validate these GSEA findings; we detected a significant difference between the two risk categories (p < 0.001; Figure 3B). Interestingly, the low-risk cohort (i.e., better prognosis) was related to higher p53 expression, contradicting the findings of several previous studies and suggesting an unusual association between m6A modification and p53 activity.
Figure 3.
GSEA results for low- and high-risk categories in cell cycle (a), p53 (TP53) signaling pathway (b), DNA mismatch repair (c), and RNA degradation (d) determined using the KEGG database. (e) Evaluation of p53 (TP53) expression stratified among risk subgroups. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001. GSEA, gene set enrichment analyses; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Identification of m6A risk scores associated with immune checkpoint molecules
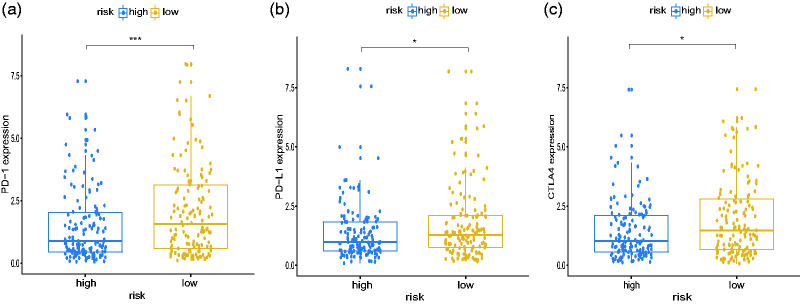
Next, we analyzed the effects of m6A modification on immune responses in STAD patients and found that the m6A-related low-risk subgroup was associated with significantly higher expression of several immune checkpoints, including PD-1 (programmed death 1), PD-L1 (programmed death-ligand 1), and CTLA-4 (cytotoxic T-lymphocyte–associated antigen 4) (Figure 4). These findings suggest that our score may predict the efficacy of immunotherapy in STAD patients; however, this finding must be verified clinically.
Figure 4.
GSEA results for low- and high-risk categories. Expression of immune checkpoint molecules PD-1 (a), PD-L1 (b), and CTLA4 (c) among different risk subgroups. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001. GSEA, gene set enrichment analyses; PD-1, programmed death 1; PD-L1, programmed death-ligand 1; CTLA4, cytotoxic T-lymphocyte–associated antigen 4.
Discussion
m6A RNA methylation is a reversible RNA modification process that has recently received much attention. Because of the limited technology for detecting m6A modification levels, several studies have applied indirect methods to detect changes in the expression of m6A regulatory genes to evaluate relationships between m6A status and human diseases.
In this study, risk scores were constructed for 15 selected m6A regulators, which were used to divide patients into low- and high-risk categories according to OS. The two categories represented different m6A modification statuses, which were related to distinct clinical patient outcomes. The mechanism of m6A methylation is complex; for example, FTO and ALKBH5 (erasers) were positively and negatively related, respectively, to our risk score. Previous studies have shown that FTO regulates the demethylation of both N6, 2-O-dimethyladenosine (m6Am) and m6A (the former being preferred), whereas AKLBH5 prefers m6A regulation.30 METTL14 and METTL3 (writers) have been shown to promote tumor progression in acute myelocytic leukemia (AML) but act as suppressor genes in glioblastoma multiforme (GBM) cancer.21,22 These findings suggest that genetic changes to individual m6A regulators are linked to different dysregulated genes and play distinct roles in different tumor types. Because the coefficients of regulators within the same groups (i.e., writers, readers, and erasers) showed opposite signs within the risk score model, the score represents the collective contribution of individual genes mediated by specific m6A regulators. Differences between the two risk categories therefore include simultaneous effects of upregulated and downregulated m6A genes, jointly affecting STAD clinical outcomes.
The molecular mechanisms of RNA m6A methylation in cancers have only recently begun to be elucidated. We performed GSEA using the KEGG database to examine the effects of genes regulated by m6A methylation on OS within our dataset. In consideration of the signature, we speculated that GSEA biological processes related to low risk score, including cell cycle, p53 signaling pathways, DNA mismatch repair, and RNA degradation, are mainly related to the seven selected regulators. In a previous study, more than 7000 human genes were analyzed using m6A sequencing; silencing m6A methyltransferase was found to regulate p53 signaling pathway-mediated cell apoptosis.31 As possible effectors of p53 signaling pathway, FTO was confirmed to reduce p53 expression, whereas ALKBH5 could activate the p53 signaling pathway by m6A RNA demethylation in a YTHDF2-dependent manner.32,33 Our results are in line with these findings, because ALKBH5 and YTHDF2 were related to low risk score, whereas FTO was related to high risk score. Effectors including RBM15, ALKBH5, and YTHDF2 can regulate the cell cycle biological process.34–36 Another study similarly reported that YTHDF2 mediated the degradation of RNAs containing m6A in mammals.37 However, the involvement of m6A in DNA mismatch repair has not previously been reported; this relationship is a potential avenue for future research. The relationship between p53 expression and gastric cancer prognosis is also controversial,38,39 suggesting that a similar mechanism underlies m6A modification and p53 signaling pathways.
PD-1 and CTLA-4 are the most effective immune checkpoint molecules targeted by treatments for various cancers, including gastric cancer.40 However, significant differences among individual effects have been reported in many patients.41 Therefore, it is important to identify patients who might benefit from improved immune target therapy. Because the PD1/PD-L1 checkpoint blockade is regulated by YTHDF1 (an m6A reader) and FTO (an eraser), we conclude that m6A regulators may be potential anticancer immunotherapy targets.42,43 Thus, the mechanisms and effects of m6A on immune checkpoints in gastric cancer deserve further exploration. In this study, patients (samples) with low m6A-related risk had higher expression of immune checkpoint molecules, suggesting that immune checkpoint inhibitor therapy may be more effective for this subgroup. Therefore, m6A risk signatures may be useful in the clinical setting as immunotherapy predictors of STAD outcome. The underlying mechanisms of the relationship between checkpoint molecules and m6A RNA methylation require further investigation.
This signature might be important clinically as an independent prognostic tool to predict prognosis. Our results suggested a possible relationship between m6A-related prognosis and immune checkpoints, which is a fascinating area for further research. In addition, our data inclusion criteria were much stricter than that in other studies because we removed samples that lacked complete clinicopathological characteristics from the total of 375 STAD samples with RNA-seq data from TCGA. However, there were also some limitations in our study. The research was based on bioinformatics analysis and lacks confirmation from experiments and clinical trials. The correlation analysis of m6A RNA methylation and immune checkpoints remains superficial and needs further research.
In summary, we established an m6A modification risk score that may be applied as an independent prognostic tool and that could predict clinical immunotherapy outcomes in patients with STAD. We found a significant relationship between m6A risk scores and histological tumor grades. Gene sets involved in the cell cycle, p53 signaling pathway, DNA mismatch repair, and RNA degradation were involved in m6A regulation, affecting prognosis among patients with STAD. Future studies using m6A-related methods should be conducted to confirm these findings.
Authors’ contributions
JM designed and conceived this study; PM performed most of the data analysis and wrote the manuscript; SX, WC, QD, JY, and JR downloaded data from the TCGA database and performed the rest of the data analysis. All authors read and approved the final manuscript.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the TCGA repository, http://cancergenome.nih.gov/. We have de-identified the details such that the identity of the patients may not be ascertained in any way.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
The authors disclosed receipt of the following financial support for the research, authorship and/or publication of this article: The work was supported by grants from the National Natural Science Foundation of China (No. 81372348, No. 81773065) and the Natural Science Foundation of Zhejiang Province (No. LY16H160030).
ORCID iD
Jianshan Mao https://orcid.org/0000-0003-3308-3132
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Dyck L, Mills KHG. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur J Immunol 2017; 47: 765–779. [DOI] [PubMed] [Google Scholar]
- 3.Ruschoff J, Hanna W, Bilous M, et al. HER2 testing in gastric cancer: a practical approach. Mod Pathol 2012; 25: 637–650. [DOI] [PubMed] [Google Scholar]
- 4.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol 2010; 6: 863–865. [DOI] [PubMed] [Google Scholar]
- 5.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell 2017; 169: 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 2011; 39: D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu N, Pan T. N-6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol 2016; 23: 98–102. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N-6-methyladenosine (m(6)A) in human cancers. Cancer Lett 2017; 408: 112–120. [DOI] [PubMed] [Google Scholar]
- 9.Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015; 347: 1002–1006. [DOI] [PubMed] [Google Scholar]
- 11.Weng YL, Wang X, An R, et al. Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 2018; 97: 313–325.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Y, Ma P, Liu Y, et al. Multiple functions of m(6)A RNA methylation in cancer. J Hematol Oncol 2018; 11: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018; 28: 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li A, Chen YS, Ping XL, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res 2017; 27: 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alarcon CR, Goodarzi H, Lee H, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 2015; 162: 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer 2019; 18: 103 DOI: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholler E, Weichmann F, Treiber T, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 2018; 24: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang C, Klukovich R, Peng H, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A 2018; 115: E325–E333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojtas MN, Pandey RR, Mendel M, et al. Regulation of m(6)A transcripts by the 3′ -> 5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell 2017; 68: 374–387.e12. [DOI] [PubMed] [Google Scholar]
- 20.Wu RF, Yao YX, Jiang Q, et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m(6)A-YTHDF2-dependent manner. Int J Obesity 2018; 42: 1378–1388. [DOI] [PubMed] [Google Scholar]
- 21.Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep 2017; 18: 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok CT, Marshall AD, Rasko JEJ, et al. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol 2017; 10: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, Zhang M, Ge S, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med 2019; 8: 4766–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zheng DY, Wang F, et al. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Digest Dis Sci 2019; 64: 1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin S, Liu JN, Jiang W, et al. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Wars) 2019; 14: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minn AJ, Wherry EJ. Combination cancer therapies with immune checkpoint blockade: convergence on interferon signaling. Cell 2016; 165: 272–275. [DOI] [PubMed] [Google Scholar]
- 27.Winkler R, Gillis E, Lasman L, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 2019; 20: 173–182. [DOI] [PubMed] [Google Scholar]
- 28.Bovelstad HM, Nygard S, Storvold HL, et al. Predicting survival from microarray data–a comparative study. Bioinformatics 2007; 23: 2080–2087. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei J, Liu F, Lu Z, et al. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 2018; 71: 973–985.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature 2012; 485: 201-U284. [DOI] [PubMed] [Google Scholar]
- 32.Zhou P, Wu M, Ye C, et al. Meclofenamic acid promotes cisplatin-induced acute kidney injury by inhibiting fat mass and obesity-associated protein-mediated mA abrogation in RNA. J Biol Chem 2019; 294: 16908–16917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X, Li K, Jiang W, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer 2020; 19: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaki Y, Motoyama S, Yamaguchi T, et al. m6A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells 2020; 25: 547--561. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Wang S, Zhang Y, et al. Biological effects of decreasing RBM15 on chronic myelogenous leukemia cells. Leuk Lymphoma 2012; 53: 2237–2244. [DOI] [PubMed] [Google Scholar]
- 36.Fei Q, Zou Z, Roundtree IA, et al. YTHDF2 promotes mitotic entry and is regulated by cell cycle mediators. PLoS Biol 2020; 18: e3000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 2016; 7: 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabbert HE, Muller W, Schneiders A, et al. The relationship of p53 expression to the prognosis of 418 patients with gastric carcinoma. Cancer 1995; 76: 720–726. [DOI] [PubMed] [Google Scholar]
- 39.Martin HM, Filipe MI, Morris RW, et al. p53 expression and prognosis in gastric carcinoma. Int J Cancer 1992; 50: 859–862. [DOI] [PubMed] [Google Scholar]
- 40.Janjigian YY, Bendell J, Calvo E, et al. CheckMate-032 Study: Efficacy and safety of nivolumab and nivolumab plus ipilimumab in patients with metastatic esophagogastric cancer. J Clin Oncol 2018; 36: 2836–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akin Telli T, Bregni G, Camera S, et al. PD-1 and PD-L1 inhibitors in oesophago-gastric cancers. Cancer Lett 2020; 469: 142–150. [DOI] [PubMed] [Google Scholar]
- 42.Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA mA methylation and YTHDF1 in dendritic cells. Nature 2019; 566: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S, Wei J, Cui YH, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun 2019; 10: 2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the TCGA repository, http://cancergenome.nih.gov/. We have de-identified the details such that the identity of the patients may not be ascertained in any way.