Abstract
For more effective early-stage cancer diagnostics, there is a need to develop sensitive and specific, non- or minimally invasive, and cost-effective methods for identifying circulating nanoscale extracellular vesicles (EVs). Here, we report the utilization of a simple plasmonic scaffold composed of a microscale biosilicate substrate embedded with silver nanoparticles for surface-enhanced Raman scattering (SERS) analysis of ovarian and endometrial cancer EVs. These substrates are rapidly and inexpensively produced without any complex equipment or lithography. We extensively characterize the substrates with electron microscopy and outline a reproducible methodology for their use in analyzing EVs from in vitro and in vivo biofluids. We report effective chemical treatments for (i) decoration of metal surfaces with cysteamine to nonspecifically pull down EVs to SERS hotspots and (ii) enzymatic cleavage of extraluminal moieties at the surface of EVs that prevent localization of complementary chemical features (lipids/proteins) to the vicinity of the metal-enhanced fields. We observe a major loss of sensitivity for ovarian and endometrial cancer following enzymatic cleavage of EVs’ extraluminal domain, suggesting its critical significance for diagnostic platforms. We demonstrate that the SERS technique represents an ideal tool to assess and measure the high heterogeneity of EVs isolated from clinical samples in an inexpensive, rapid, and label-free assay.
Keywords: cancer, exosomes, nanomaterials, liquid biopsy, biophotonics, Raman spectroscopy, SERS
It is estimated that by 2025, over 20 million new cancer cases will be annually diagnosed.1,2 While early-stage diagnosis leads to measurably improved patient outcomes, tumor heterogeneity and transformability are large obstacles. Tissue biopsies are the current gold standard for cancer diagnosis yet are invasive, often fail to capture tumor heterogeneity, are incapable of assessing small and hard-to-reach tumors or ones that have metastatically spread, and risk propagating the tumor to adjacent tissues.3,4 Liquid biopsy entails the detection of tumor-associated biological materials in circulating biofluids, including circulating tumor cells (CTCs), circulating tumor nucleic acids (ctDNA/ctRNA), tumor-educated platelets (TEPs), small molecular products of tumor metabolism, circulating tumor-derived proteins, and more recently extracellular vesicles (EVs).5−9
EVs are lipid-bilayer enveloped nanoscale assemblies (from ∼30 to hundreds of nanometers in diameter) that traffic bioactive molecules including nucleic acids, proteins, lipids, carbohydrates, and metabolites and related small molecules.10,11 They are heavily implicated as mediators of cardiovascular and metabolic diseases, inflammatory diseases, neurological disorders (e.g., Alzheimer’s disease), and cancer.12,13 While EVs are released from cells in both normal and diseased states, it is clear that cancer cells exploit these signaling routes to dispatch EVs that promote tumor progression in the local microenvironment as well as for metastatic purposes, including via the formation of a pre-metastatic niche.14−16 Tracking tumor-associated EV aberrations in the biochemical landscape of body fluids can unveil the presence of cancer, recurrence, relapse, and drug resistance.17 In ovarian cancer (OvCa), a significant number of EV-trafficked proteins have been reported to correlate with cancer type and staging.18−20
Despite this limited success, liquid-biopsy-based EV diagnostics are largely impeded by the high number of off-target healthy EVs present in all biofluids. Many analytical tools have been applied to EV-based liquid biopsies including Western blots, nanoparticle tracking analysis (NTA), flow cytometry, and direct fluorescence imaging techniques via antibody or aptamer labeling. However, these approaches often require tedious preanalytical isolation and large sample volumes, are lacking in throughput and multiplexibility, and are not cost-effective. An attractive solution to overcome these limitations is label-free analysis using chemical spectroscopy, such as Raman scattering. This technique provides the global chemical composition of EVs and has been recently used to elucidate key differences between cancerous and noncancerous EVs.19,21−26 To address the weak nature of Raman scattering, surface-enhanced Raman scattering (SERS) can be employed via the use of plasmonic substrates or particles.27−29 We have recently reviewed the application of nanoplasmonics to detect and analyze EVs.30 SERS in particular is very attractive for liquid biopsies, given that it is label-free, exhibits unprecedented and highly tunable sensitivity and specificity, and has the potential for automation, miniaturization, and integration with microfluidics and machine learning.30−32
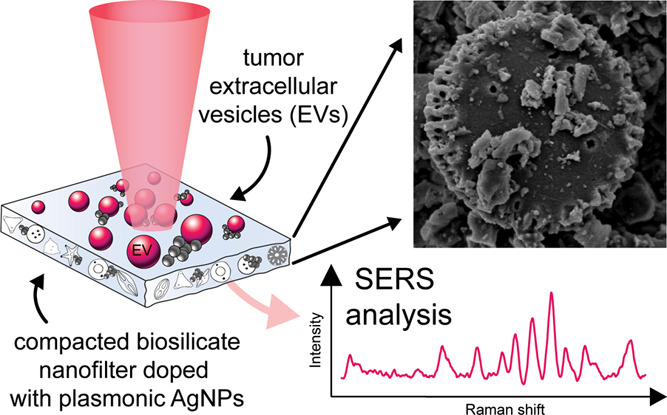
In this report, we employ a new type of inexpensive, biocompatible plasmonic substrate integrating the benefits of a novel biosilicate material embedded with silver NPs (AgNPs) for use in label-free liquid biopsy of tumor EVs (schematized in Figure 1). We use multivariate data analysis to extract relevant features among spectral sets that distinguish tumor samples from controls in in vitro cell-derived EVs and also clinical human serum samples from patients suspected of gynecologic malignancy. We further explore cysteamine functionalization of AgNPs for nonspecific recruitment of EVs to the substrate and chemical treatment of EVs to modulate their localization at the plasmonic AgNPs.
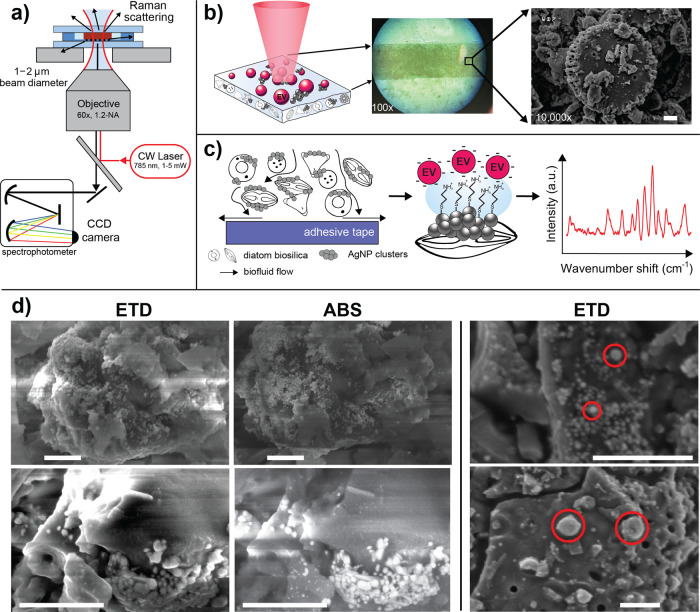
Figure 1.
Overview of the nanoplasmonic substrate and SERS imaging process. (a) Schematic of the SERS optical setup, where the substrate is sandwiched between quartz windows for analysis using an inverted confocal Raman scanning instrument. (b) The biosilicate SERS substrate is irradiated by laser light to instigate Raman scattering. The insets show the heterogeneous surface structure of the compacted diatom mesh at 100× and then under SEM at 10k×, where single diatoms are visible. (c) The substrate allows for transport of EVs from solution to the proximity of AgNP clusters adsorbed to the compacted silicate scaffold. When functionalized with cysteamine, thiol bonds anchor to the AgNPs, enabling anionic EVs to adhere electrostatically to cysteamine’s terminal amine groups. Spectral SERS fingerprints can be acquired from EVs adjacent to AgNPs. (d) SEM micrographs of hybrid biosilicate mesh with AgNP clusters. An Everhart–Thornley detector (ETD) records the secondary electron scattering from the surface, whereas the annular backscattering detector (ABS) collects electrons more sensitive to atomic weight, highlighting the AgNP clusters. The images on the right show likely EV candidates localized in the vicinity of AgNP clusters throughout the hybrid material. The approximate starting concentration of EVs was ∼5 × 108 EV/mL. All scale bars are 1 μm.
Materials and Methods
EV Isolation and Preparation
The EVs studied in this work were isolated from clinical serum samples provided by the UC Davis Comprehensive Cancer Center (UCDCCC) Pathology Biorepository Resource (IRB ID: 1314848-1). Patient serum samples were obtained as deidentified remnants following a clinician-ordered CA125 assay. Histopathology analysis by trained clinicians identified sample types as either endometrial cancer (EnCa), ovarian cancer (OvCa), or benign (control). Therefore, some patients are not diagnosed with cancer even after the CA125 assay; thus, negative control samples can be obtained. Approximately 1 mL of serum was obtained per patient, from which EVs were isolated using differential ultracentrifugation (UC) methods.33 First, 1 mL of serum was centrifuged at 300g for 10 min at 4 °C to clear any remaining whole cells. The resulting supernatant was then centrifuged at 2000g for 15 min at 4 °C followed by a 10,000g spin for 30 min at 4 °C to pellet any dead cells and cell debris/larger microvesicles, respectively. All low speed spins were performed using a Beckman Coulter Microfuge 20R centrifuge with an FA361.5 Biosafe rotor. Finally, the samples were ultracentrifuged twice at 120,000g for 70 min at 4 °C to pellet EVs, dispersing in ultrapure water between spins. Ultracentrifugation was carried out using a Beckman Optima TLX Ultracentrifuge with a TLA 100.1 fixed angle rotor. Resulting pellets were resuspended in up to 100 μL of ultrapure water and stored at −80 °C for up to a few weeks until use. Samples were aliquoted to reduce freeze–thaw cycling.
For the method development stage, two cell lines were cultured to model OvCa and EnCa: SKOV-3 and HEC-A-1 cells, respectively (ATCC, Manassas, VA, USA). Cells were first cultured in T-75 flasks with Gibco McCoy’s 5A medium (ThermoFisher Scientific, Waltham, MA, USA) and 10% (v/v) FBS and 100 units/mL penicillin–streptomycin. From here, 25 × 106 cells were used to seed high-yield EV collection CELLine 1000 AD (DWK Life Sciences, Millville, NJ, USA) bioreactor flasks.34 The bioreactor’s upper compartment was supplied with 950 mL of Gibco McCoy’s 5A medium complemented with 10% (v/v) FBS and 100 units/mL penicillin–streptomycin. The bioreactor’s cell compartment was supplied with 15 mL of Gibco McCoy’s 5A medium with 10% (v/v) Gibco Exosome-Depleted FBS and 100 units/mL penicillin–streptomycin. The T-75 flask and bioreactor cell cultures were propagated and cultured at 37 °C, 95% relative humidity, and 5% CO2 atmosphere. For EV isolation, the cell compartment medium (15 mL) was collected once a week from the bioreactor, and 15 mL of fresh medium was added. Concurrently, the upper compartment medium (950 mL) was discarded and replaced with the same amount of fresh medium. EV isolation was performed by UC as described above.
EV Characterization
Nanoparticle Tracking Analysis (NTA)
NTA was performed using a NanoSight model LM10 (Malvern Panalytical Ltd., UK), equipped with a violet (405 nm) laser and sCMOS camera. Aliquoted EVs were thawed to room temperature and typically diluted 1000-fold in filtered ultrapure water, which was also used to copiously flush the NTA lines to confirm background to be free of particulates prior to sample addition. 1 mL of sample was loaded into a syringe and fit into an automated syringe pump (Harvard Bioscience, MA, USA) for injection. Three consecutive 30 s videos of each sample in flow conditions with at least 200 particle tracks present per video were recorded at camera level 13. The data was analyzed using NanoSight NTA 3.1 software with the detection threshold set to 5 and screen gain 10 to track the statistically relevant number of particles, simultaneously minimizing the distorting background artifacts. A representative video of the NTA analysis, a typical concentration and size distribution charts for in vitro SKOV-3 EVs, and a clinical ovarian cancer patient EV sample are provided in Figure S1.
Bicinchoninic Acid (BCA) Assay
Total protein was quantified using a BCA assay (Thermo Scientific). 15 μL of EVs was mixed in an Eppendorf tube with 8 μL of 10× RIPA buffer and 57 μL of ultrapure water and vortexed briefly for ∼5 s. The tubes were then incubated on ice for 30 min. The working reagent and protein standards were prepared according to the manufacturer’s instructions. 25 μL of each standard or sample was pipetted in triplicate into a 96-well microplate. 200 μL of working reagent was added to each well, and the plate was wrapped in tin foil and then incubated at 37 °C for 30 min. After incubation, the plate was allowed to cool to room temperature and the absorbance at 562 nm was measured. Representative total protein estimates for in vitro samples are presented in Figure S2.
SERS Substrate Preparation, Characterization, and Measurement
Preparation of Plasmonic Composite Substrates
The biosilica/AgNP composite substrates used in this study were prepared as described in a recent report by Korkmaz et al.35 Silver nitrate, sodium citrate, and diatoms (Celite 209) were purchased from Sigma-Aldrich. Silver nanoparticles were synthesized from silver nitrate based on the protocol reported by Lee and Meisel.36 Briefly, 90 mg of silver nitrite was dissolved in 500 mL of water. The solution was then heated and stirred on a hot plate at 300 °C with a speed of 300 rpm until it was boiling. Once boiling, 10 mL of 1% sodium citrate solution was added to the solution drop by drop. The solution was then kept boiling for 1 h. The AgNP solution was obtained. Characterization data of the AgNPs and the diatom surface are presented in Figure S3. An optimized ratio of 1 g of diatom powder and 240 mL of 50 nm (±20 nm) AgNP suspension were mixed and heated until all suspension water had evaporated. The composite was further dried in an oven for 0.5 h at 120 °C. Next, a 5 cm × 0.2 cm substrate template was filled with the prepared diatom/AgNP composite. Finally, the SERS substrates were obtained by peeling off the composites from the template using office-grade adhesive tape.
Electron Microscopy Imaging of Substrates
Plasmonic substrates were imaged using a scanning electron microscope (SEM) before and after EV adsorption. SEM images were obtained using a ThermoFisher Quattro S (ThermoFisher Scientific, Waltham, MA, USA) to visualize surface attachment. The instrument is equipped with an Everhart–Thornley detector (ETD) for secondary electron imaging and a dedicated annular backscattered (ABS) detector for backscattered electron imaging. Prior to imaging, substrates were treated with cysteamine and EVs as detailed in EV–Substrate Incubation. They were then washed 1–2 times with sodium phosphate buffered solution (PBS) and fully submerged for 15 min each in glutaraldehyde and osmium tetroxide. Following, substrates were gradually dehydrated using a graded series of ethanol (EtOH) solutions: 30 (5 min), 50 (5 min), 70 (5 min), 95 (2 × 5 min), and 100% (2 × 5 min). To finalize the preparation, substrates were allowed to air dry overnight in a fume hood. Substrates were then mounted on metal studs using two-sided black carbon tape, and the following typical imaging parameters were applied: working distance 10.0–13.4 mm, spot size 2.5 or 5.0, accelerating voltage 10.0 kV, and chamber pressure 50 Pa or 100 Pa (100 Pa while using ETD + ABS detectors in combination).
EV–Substrate Incubation
Small sensor pieces (∼2 mm by 5 mm) were cut out from the prepared parallel (0.2 × 5 cm) stripe-shaped plasmonic sensor substrates. One 2 × 5 mm piece was used per measured sample. Before EV incubation, the sensors were pretreated at pH 6.5 with 10 μL of 20 mM (in 95% EtOH) cysteamine, which binds to the AgNPs via thiol bonds. The free −NH3 groups of cysteamine generate a positive charge around the AgNPs. After 1 h, the substrates were washed 2–3 times by dipping into Eppendorf tubes of pH 6.4 buffer. EV samples were diluted 1:100 in the same buffer, and 30 μL drops were pipetted onto the substrates, covered, and incubated at room temperature for 2 h. This volume was sufficient as to not allow the substrates to dry out during this incubation period. The substrates were then washed again and inverted onto a #1.5 glass coverslip for SERS measurements. This procedure was used to ensure that the inherently anionic EVs in solution adhere and concentrate to the vicinity of the AgNPs. Thus, the EVs were ubiquitously located at the plasmonic hotspots allowing for effective SERS measurements. All steps were performed in liquid conditions, including the EV measurements. Since EVs in biological systems always appear in solutions, the primary motivation for this approach was to minimize experimental artifacts potentially induced by drying the EVs, thus possibly biasing the observations. For EV treatment with trypsin, prior to measurement, 5 μL of EV sample was mixed with 500 μL of 0.25% w/v trypsin, pipetted rapidly up and down, and then incubated at 37 °C for 1 h to allow for cleavage of extraluminal surface moieties. Samples were washed thrice with ultrapure water in 10 kDa Amicon Ultra (0.5 mL) regenerated cellulose centrifugal filters according to the manufacturer’s instructions.
SERS Spectral Acquisition and Data Processing
SERS spectra were acquired using a custom-built inverted Raman scanning confocal microscope with an excitation wavelength of 785 nm and a 60×, 1.2 NA water immersion objective on an inverted IX73 Olympus microscope. Raman spectra were captured via an Andor Kymera-3281-C spectrophotometer and Newton DU920P-BR-DD CCD camera and initially processed using Solis v4.31.30005.0 software. Exposure time was set to 1 s per scan with a laser power of ∼1–5 mW unless otherwise specified. Across all samples, SERS spectra were sampled randomly in 5–10 different spatial locations with a 20-spectra kinetic series collected. Representative spots were chosen for data collection when noticeable SERS peaks became visible and stable during randomly traversing across the substrate, so there is a positive bias in each spot for containing EV materials.
SERS spectral analysis was performed using custom scripts written in MATLAB v2019b (MathWorks, MA, USA). Spectral preprocessing included cosmic ray removal, penalized least-squares (PLS) background correction, smoothing, and normalization. Preprocessed spectral sets were further subjected to principal component analysis (PCA) and hierarchical clustering analysis based on the corresponding MATLAB built-in functions. These procedures are described in further detail in our previous works.19,24 LDA and subsequent calculation of the confusion matrix, accuracy, sensitivity, and specificity of the assay were also carried out in MATLAB using built-in functions and an in-house written code that can be provided on request. Tests were performed to choose an adequate number of PCs for the LDA, simultaneously avoiding overfitting. Thus, an LDA analysis using the two first PCs (PC1 and PC2) was determined to be the most suitable for the analysis, consistent with the data presented throughout this study.
Results and Discussion
AgNP-Embedded Plasmonic Biosilicate Scaffold for SERS-Based Diagnostics Can Filter and Trap EVs from Solution
At the core of plasmonic sensing are nanostructured metal materials, and cost-effective, highly sensitive, and flexible nanomaterials are of great interest,37 particularly for low-resource settings. The main function of plasmonic SERS scaffolds is to boost weak spontaneous Raman scattering upon irradiation with light. We have recently introduced the fabrication of a simple, robust, and flexible SERS substrate that has an increased enhancement factor (EF) of ∼1.0 × 105 with a high level of reproducibility for the development of inexpensive label-free biosensors.35 In these substrates, enhancement is achieved via decoration of AgNPs throughout compacted biosilicate diatoms (Figure 1). Diatoms are unicellular photosynthetic biomineralized marine microalgae that excrete an amorphous silica cell wall, or frustule. These frustules serve as inexpensive photonic crystals38,39 without the need for labor-intensive and costly lithography or etching techniques. When AgNPs are embedded within the diatom frustules, a dual plasmonic effect can be accomplished stemming from the coupling between LSPRs of metal NPs and guided-mode resonances of the frustules.40,41 There are a few examples of utilizing gold or silver NPs as diatom-based hybrid plasmonic biosilicate SERS substrates, wherein the diatom–metal composites are typically achieved via in situ growth or self-assembly of metal NPs.42−44 These novel materials represent an inexpensive, easily fabricated SERS sensing alternative for a broad range of applications in life and materials sciences. Here, we demonstrate the proof-of-concept use of these substrates for EV-based cancer diagnostic application.
Prior to incubation with biological materials, we used SEM to microscopically evaluate the surface of the substrate and the adherence of AgNPs to the diatom surfaces (Figure 1d). The ETD provides micrographs with contrast stemming from surface features, whereas the ABS detector is well suited for establishing contrast sensitive to atomic weight. Hence, in the ABS images, AgNP clusters are clearly separable as bright structures compared to the biosilica material. We also point out that several candidates of EVs are adsorbed to the frustules at the vicinity of the AgNPs. Although the isolation procedure yields predominantly EVs in the size range of 50–200 nm as seen in Figure 1d in the upper right corner SEM image, it inevitably co-isolates proportions of bigger (>200 nm) EV subpopulations, some of which may be readily apparent in the lower right corner image. These EV populations are traditionally called “microvesicles”, but due to the manifold nomenclature in the EV field, we chose to use a collective term “EVs”. Comparably similar EV structures as visualized by SEM have been demonstrated previously.45
Recently, we have shown that the porous nature of the diatoms allows smaller molecules and nanostructures to disperse within the substrate, filtering through the biosilicate material.35 The small average hydrodynamic diameter of EVs allows them to enter and distribute throughout the three-dimensional biosilica mesh, with smaller contaminants exiting through the bottom of the substrate (theoretical calculations for this approximation can be found in the Supporting Information section Theoretical Calculations of the SERS Surface Coverage). To utilize these capabilities and immense surface area of the biosilica microstructure, we analyzed EVs isolated from SKOV-3 and HEC-A-1 cells, common in vitro human ovarian and endometrial cancer cell lines, respectively, as well as EVs isolated from various clinical serum samples. We chose to call 120,000g pelleted materials as EVs for the purpose of this study unless otherwise explicitly noted.46
To better localize the anionic EVs close to the AgNP clusters throughout the heterogeneous biosilica substrates, we pretreated them with cysteamine (H2NCH2CH2SH), a practical functionalizing agent to couple anionic species to metal surfaces.47−49 The sulfhydryl group (−SH) of cysteamine binds to Ag/Au, while the opposite terminal amine group (−NH2) is freely exposed. A representative example of SERS measurements following cysteamine treatment is shown in Figure S3, where peaks in the substrate are comparable to the spontaneous Raman of bulk cysteamine. Even after thorough washing, cysteamine remains present throughout the material. The strong peak at 650 cm–1 and the moderately fainter peak at 735 cm–1 both attributed to the C–S stretching are practically absent in the spontaneous Raman spectrum of cysteamine (full detailed peak assignments are displayed in Table 1). This highlights that cysteamine forms thiol bonds with the AgNPs, such that these bonds undergo significant enhancements due to their proximity to the strong electromagnetic SERS fields.
Table 1. Chemical Assignments for the Relevant Spectral Peaks or Bands Identified in This Work.
| peak/band (cm–1) | chemical assignment |
|---|---|
| 643 | amino acids in proteins, e.g., tyrosine54 |
| 650 | C–S stretching47 |
| 735 | C–S stretching47 |
| 789–795 | vibrations in nucleic acids22,24,74 |
| 805 | Si–O stretching; predominantly silicon motion, e.g., within Si–O–Si units60,61 |
| 903 | carbohydrate-related SERS vibrations62 |
| 931 | C–C ring stretching in, e.g., proline68 |
| 940 | C–C stretching vibration possibly coupled to C–N stretching vibration47 |
| 960 | protein vibrational modes, e.g., C=C deformation or C–N stretching54,71,72 |
| 1010, 1050, 1090 | Si–O stretching; oxygen vibrating between silicon in the Si–O–Si bond60,61 |
| 1015 | C–C stretching vibration possibly coupled to C–N stretching vibration47 |
| 1095 | PO2– stretching, C–C stretching, C–O–C stretching, glycosidic link in DNA/RNA68 |
| 1110 | Cα–N, Cα–C, C–N stretching in the protein backbone, C–C stretching in acyl chains of lipids24,68 |
| 1160–1170 | carbohydrate-related SERS vibrations56 |
| 1175 | nucleic acid vibrations in DNA/RNA, phenylalanine, or tyrosine vibrations in proteins24,68 |
| 1240 | C–N stretching + N–H deformation; amide III in proteins70 |
| 1287 | CH2, CH3 deformation/C–N stretching + N–H deformation; amide III in proteins24,68 |
| 1290 | CH2 deformation in acyl chains of lipids24,25,68 |
| 1310–1340 | carbohydrate-related SERS vibrations62 |
| 1336 | backbone deformation Cα–H/Cα–C stretching/CH2, CH3 twisting or wagging in proteins24,68 |
| 1360 | CH2, CH3 wagging in proteins68 |
| 1386–1390 | symmetrical CH3 deformation in DNA/RNA, proteins, or lipids68,70 |
| 1400 | protein vibrational modes, e.g., CH2 deformations54 |
| 1445–1460 | CH2 and CH3 deformations in proteins and lipids23,24 |
| 1500 | conjugated −C=C– vibrations in nucleic acids24,68,73 |
| 1545 | protein vibrational modes, e.g., amide II vibrations68,70 |
| 1590 | C–C ring vibration in aromatic groups69 |
| 1595 | vibrations in nucleic acids24,68,71 |
| 1620 | C=C vibration in, e.g., proteins70 |
| 1630 | amide I C=O stretching vibrations in proteins24,74 |
| 1650 | amide I vibrations in proteins or C=C stretching in lipids23,24 |
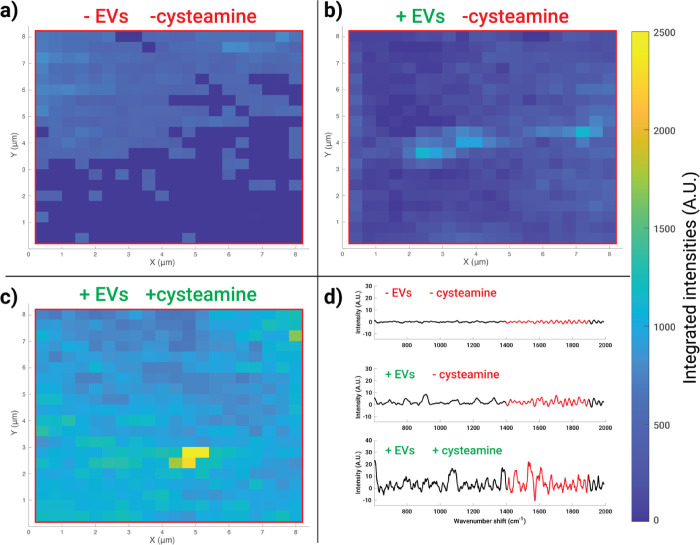
Following substrate treatment with cysteamine to functionalize AgNP clusters, we used a slightly acidic (pH = 6.4) buffer to wash the cysteamine-treated substrates in order to facilitate EV adsorption since at this pH the amine groups are pushed toward cationicity (NH3+). On the basis of the preliminary testing with various types of EVs and concentrations, it was significantly more difficult to find SERS hotspots with EV signatures when the surface was not pretreated with cysteamine. To highlight the effectiveness of cysteamine modification, in vitro SKOV-3 EVs were incubated on noncysteamine-treated and cysteamine-functionalized substrates. An area of 64 μm2 with a step size of 400 nm (i.e., 20 × 20 pixels) was raster-scanned (Figure 2). A section of the fingerprint region (1400–1900 cm–1) was integrated over, given that this region contains many peaks arising from biomolecules (Table 1) and is a quiet region for the blank substrates. Heatmaps were generated from the scans, and it is apparent that the cysteamine-functionalized surfaces exhibit more “hot” areas than the substrates without cysteamine. We concluded that without cysteamine, the EVs do not strongly adhere to the surface easily and are mostly rinsed away during subsequent washing steps. Cysteamine functionalization allows us to achieve reasonable measurement reproducibility via nonspecific binding of EVs.
Figure 2.
Spectral maps show the increased signal of biosilicate SERS substrates upon cysteamine treatment. Maps of dimension 8 × 8 μm with 400 nm spacing between spectra were collected from the (a) control substrate, (b) substrate with SKOV-3 EVs, and (c) cysteamine-pretreated substrate with SKOV-3 EVs. Representative spectra for the maps are shown in panel (d). The red highlighted portion of each spectrum in panel (d) represents the portion integrated under to generate the maps, chosen due to its coverage of protein and lipid peaks (Table 1), thus used as a surrogate for the biomaterial. It is apparent that cysteamine pretreatment enabled increased coverage of the biomaterial in the substrate.
EVs Treated with and without Trypsin Provide Complementary Biomolecular Analysis
Method development for EV characterization and identification was initiated using in vitro SKOV-3 and HEC-A-1 EVs produced in bioreactor flasks, which allow for cost-effective high-yield EV production.34,50 Particle concentrations as measured by NTA were used to normalize EV additions. In early experiments, we noticed that the majority of chemical components in SERS spectra of EVs could be attributed mainly to sugars with some small extent of protein features, but no nucleic acids, in contrast to our previous report of spontaneous Raman scattering from optically trapped whole EVs.24 We suspected that this was due the distance dependence of SERS; thus, we used enyzmatic treatment of EVs to modulate the portion in contact with the plasmonic substrate. We previously demonstrated that EV surface proteins residing on a hydration layer can be modified by trypsin treatment.24 Trypsin nonspecifically cleaves extraluminal domains of surface proteins and a thick layer of carbohydrates (i.e., the glycocalyx),51 which has been demonstrated to be a part of the EV structure.52 Our assumption was that this corona prevents EVs from getting close enough to the plasmonically active AgNPs (Figure 3). In turn, removing this layer may help to (i) expose complementary biomolecules for plasmonic signal amplification that takes place approximately within 5–10 nm distance53,54 from the AgNPs and (ii) eliminate a major source of heterogeneity of EVs present in the glycocalyx. It is worthwhile to stress that the SERS distance is an estimate. It can slightly vary depending on the plasmonically active metal used (i.e., whether the metal is, for example, Ag, Au, Pt, or Al) and whether the measurements are performed on a film or planar nanoroughened surface or a colloidal metal nanoparticle solution.53,54 The size and shape of nanostructures also contribute; in the case of films or planar surfaces, the dimensions of nanoroughened surfaces dominate, while when using colloidal metal nanoparticle solutions, the size of nanoparticles is essential.53,54
Figure 3.
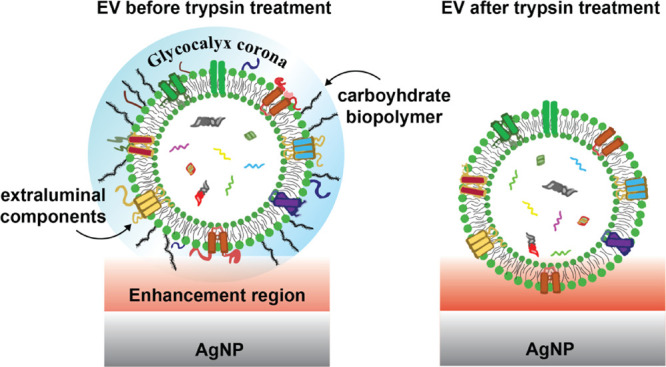
Potential effect of trypsin treatment on the glycocalyx and protein corona of EVs. Prior to trypsinization, the chemical components comprising the corona and near the outer shell of the EV are mainly exposed to the electromagnetic SERS amplification field (red). Trypsin cleaves off extraluminal domains of surface proteins and sugars that extend outside the vesicle’s phospholipid shell, placing the EVs in closer contact with the AgNP with different parts, including some intraluminal components, experiencing stronger signal amplification.
Furthermore, we acknowledge the potential of the glycocalyx for diagnostic purposes; therefore, we hypothesized that a portion of the EVs’ diagnostic value may be lost by enzymatic treatment. As meticulously demonstrated by Shurer et al., the overall composition and physical nature of glycocalyx appear to possess a remarkable role in cell membrane shape regulation, EV biogenesis, and thus on the surface composition of secreted EVs – including their glycocalyx corona.55 The level of glycosylation of EVs is reported to affect their in vivo biodistribution,56 and simultaneously, EVs can carry enzymes on their shell that shape their own glycocalyx as well as similar glycocalyx structures in the extracellular matrix and surrounding cells.57 Furthermore, given that tumor cells typically secrete high numbers of EVs58 and their cell surfaces are abundantly crowded by mucins and hyaluronan,59 we have a good reason to believe that (i) the EVs secreted by tumor cells reflect similar glycocalyx membrane composition to their parent cells and therefore (ii) investigating the different constituents of the EV-associated glycocalyx corona indeed holds great uncharted potential in the context of liquid biopsies and cancer diagnostics.
To test that trypsin did not affect the cysteamine pretreatment, we incubated substrates with trypsin prior to any addition of EVs and rinsed thoroughly. We also measured the SERS spectrum of a nontreated empty substrate in order to demonstrate that the weak spectral features stemming from the substrate have no to minimal interference with the measured biological specimens. Figure S4a–c shows representative spectra of these controls, including solid cysteamine and cysteamine-functionalized substrates treated without and with 0.25% w/v trypsin solution, respectively. Characteristic cysteamine SERS peaks remained, suggesting that even harsh trypsin treatment did not significantly remove cysteamine nor block its ability for EV enrichment. Figure S4d shows the representative spectrum of a blank substrate, with discernible peaks/bands that are well characterized and known to relate to various vibrational modes of Si compositions.60,61 In particular, features at 805 (Si–O stretching; predominantly silicon motion, e.g., within Si–O–Si units), 1010, 1050, and 1090 cm–1 (Si–O stretching; oxygen vibrating between silicon in the Si–O–Si bond) are prevalent in the measured spectrum (Table 1). On this note, these spectral regions do not overlap and therefore interfere with our analyses of distinguishing noncancerous and cancerous EVs.
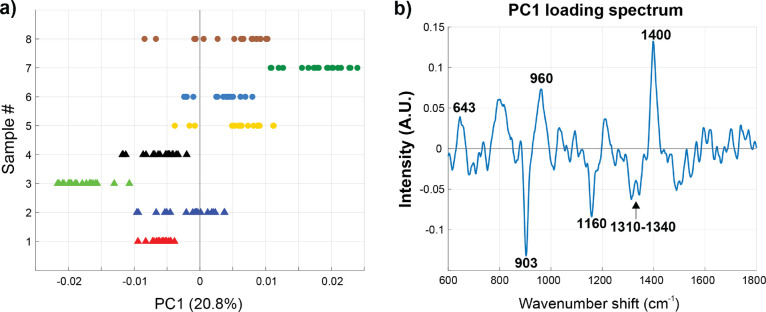
Subsequent analyses comprised stages where the EVs were treated with trypsin followed by thorough washing. As shown in Figure 4a, a clear separation between native nontreated (triangle markers) and trypsin-treated (circular markers) SKOV-3 in vitro EVs was observed in the one-dimensional principal component (PC) space, where each marker represents one individual spectrum. The PC loadings report the variables, i.e., wavenumber regions, in the spectra that are pertinent for the group separation and thus provide information on the chemical differences between the measured groups. Figure 4b displays a PC1 loading spectrum that captures ∼20.8% of the total variation between the SERS spectra acquired from native and trypsin-treated EVs. Although the other PCs also have their contribution to the observed differences throughout the analyses, for ease of interpretation, we display these results on the basis of PC1 since that dimension adequately and consistently identifies the relevant biochemical contributors that differentiate these analyzed specimens from each other. In any case, all raw SERS spectral data are available for viewing or independent analysis (Supporting Information).
Figure 4.
Trypsin treatment removes carbohydrates from EVs. (a) PC1 score plot of native SKOV-3 EVs (triangle markers) and trypsin-treated SKOV-3 EVs (circular markers) measured on the substrate. (b) PC1 loading spectrum with six spectral regions identified, three assigned to protein vibrational modes (643, 960, and 1400 cm–1) and three assigned to carbohydrates (903, 1160, and 1310–1340 cm–1). The scores on PC1 for trypsin-treated EVs correspond to carbohydrates, indicating that the treatment effectively cleaves the extraluminal domain of EVs, exposing complementary biomolecules.
As the PC score values (Figure 4a) are primarily negative for the native SKOV-3 EVs and positive for the trypsin-treated SKOV-3 EVs, this indicates that in the SERS data, the wavenumber regions 643 (amino acids in proteins, e.g., tyrosine), 960 (protein vibrational modes, e.g., C=C deformation or C–N stretching), and 1400 cm–1 (protein vibrational modes, e.g., CH2 deformations)62 are pronouncedly present in the trypsinized EVs. Simultaneously, the regions at around 903, 1160, and 1310–1340 cm–1 are less represented. Intriguingly, these three bands are potentially attributed to carbohydrate-related SERS vibrations61 (Table 1). Trypsin treatment also influences the total protein content of EVs as investigated by a BCA assay using native and trypsinized in vitro SKOV-3 EVs (Figure S2). Trypsinized EVs had clearly reduced protein contents in comparison to native EVs, while particle count by NTA remained in a similar range. Taken together, these findings imply that the EV glycocalyx/corona is indeed affected by trypsin treatment and, as hypothesized, complementary EV surface structures are exposed to SERS amplification. This may be a generalizable treatment that can be applied to a variety of SERS substrates.
Limit of Detection (LOD) for the Biosilicate SERS Scaffold
We determined the limit of detection (LOD) for sample concentration and laser power. SKOV-3 EVs were chosen for both of these experiments to promote consistency over the course of the testing. For the concentration LOD, SKOV-3 EVs were tested starting with an initial concentration of ∼5 × 1012 particles/mL. After diluting 106-fold, the signal was still easily located in different areas on the substrate. After another 104-fold dilution (bringing the total dilution to 1010-fold), we were still able to locate a few spots that produced signals, indicating that the LOD for EV concentration is less than 600 particles/mL. Many plasmonics studies demonstrate LODs down to hundreds or a few thousands of EVs.45,64 Thus, the performance of our substrate resides in a similar range demonstrating its comparability and feasibility for EV characterization. We additionally determined the LOD for our laser power, an appropriate measure, since it would be useful for point-of-care diagnostics, particularly in low resource settings, to minimize the laser power. A lower laser power may also help preserve the sample condition to ensure platform reliability and repeatability. The substrates were prepared once again with SKOV-3 in a typical 100-fold dilution. Spots were interrogated across the substrate at decreasing levels of laser power. Spots were fairly easy to locate down to 800 μW, with even an occasional signal being seen as low as ∼500 μW. The testing was eventually stopped because the laser power could not be decreased any further. We previously established the SERS enhancement factor for these novel substrates.35
High Inherent Chemical Heterogeneity of EVs Isolated from a Single Patient Sample
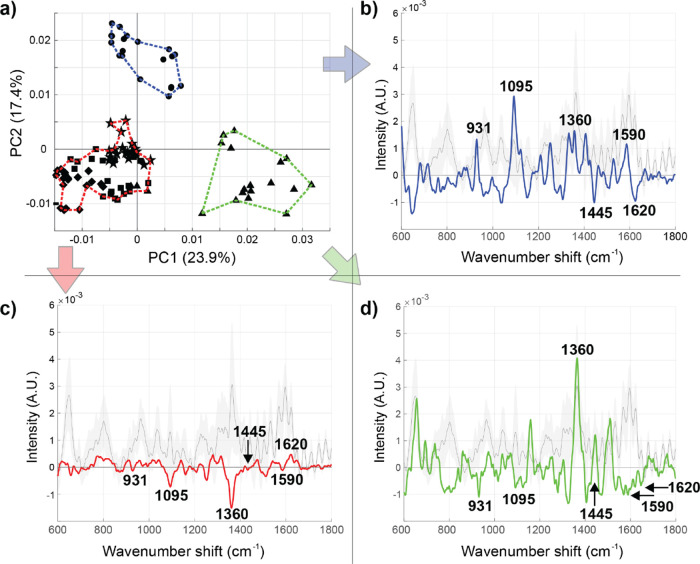
Before analyzing EV samples across a panel of clinical patients, we endeavored to evaluate the inherent heterogeneity within one clinical EV isolate derived from an ovarian cancer patient. Even one patient’s EV sample can reflect various physiological states depending on the individual’s physical activity, nutrition, etc. before the sample was drawn.65,66 Furthermore, even the most subtle sample preparation and transfer practices can alter their internal composition and induce measurable variation between the EV subtypes present.67 As expected, EVs exhibited internal variation even among a single analyzed clinical sample, as visualized by the three clearly separate clusters (blue, red, green) in Figure 5a, with hierarchal clustering analyses determined using the first five PCs. In the cluster-specific spectral loadings (Figure 5b–d), a positive peak indicates the existence of a certain chemical feature whereas a negative peak reports the absence of a proposed chemical entity, compared to the global mean (shown in gray behind the cluster average spectra). The conceptual mathematical processing for obtaining these spectra can be found in the Supporting Information section Mathematical Approach for Deriving Cluster Analysis Spectra. We identified six regions of interest: 931 (C–C ring stretching in, e.g., proline),68 1095 (PO2– stretching, C–C stretching, C–O–C stretching, glycosidic link in DNA/RNA),68 1360 (CH2, CH3 wagging in proteins),68 1445 (CH2 and CH3 deformations in proteins and lipids),24 1590 (C–C ring vibration in aromatic groups),69 and 1620 cm–1 (C=C vibration in, e.g., proteins)70 (Table 1). Based on these peak assignments, the measured EVs forming the blue cluster are seemingly enriched in nucleic acid (DNA/RNA)-related vibrations whereas the red cluster represents the majority of the EVs, which are relatively consistent with the overall global average spectrum with the exception of fewer CH2/CH3 wagging vibrations from proteins. On the contrary, the green cluster EVs are enriched in those vibrations. We additionally implemented PCA analysis for the EVs derived from in vitro SKOV-3 cells to discover whether similar signal variation could be observed. As presented in Figure S5, a similar trend is evident also within the in vitro SKOV-3 EVs. These perceptions stress the remarkable internal heterogeneity even within one EV isolate. It is not clear if this clustering represents an inherent chemically defined subpopulation spread of EVs or rather the relative positioning of EVs toward the plasmonically enhanced electromagnetic fields propagated by the AgNPs throughout the substrate. Based on several scanning results as shown by way of example in Figure 2, we cautiously suggest that the inherent heterogeneity of EVs and their binding via cysteamine dominate variation, rather than the substrates themselves.
Figure 5.
Distinguishable heterogeneity within EVs isolated by UC from a single patient diagnosed with ovarian cancer. (a) Two-dimensional PC score plot revealed three distinguishable clusters (blue, red, and green – defined using the first five PCs). Each point represents a single measurement taken within the substrate, with circles, stars, triangles, squares, and diamonds representing groups of repeated measurements sampled throughout the substrate (20 1 s spectra per spot). The chemical heterogeneity (as evaluated by the Euclidean distance in PC space) is more consistent within a sampled region (e.g., triangles) than within spectral samples in different regions (e.g., triangles vs circles). (b–d) The cluster-specific SERS spectra color coded according to the outlined regions in panel (a).
Evaluation of Native Clinical Samples
Eight clinical serum samples were obtained from the UCDCCC Pathology Biorepository resource as “remnants” – samples to be discarded following doctor-ordered CA125 ELISA assays as part of the patients’ standard clinical care. These deidentified samples were annotated with clinical diagnosis of cancer type and staging, allowing us to bin samples accordingly. Two of these patients turned out to not have malignant lesions and thus served as negative controls. The other six samples represented different cancer subtypes. Here, we used this initial data set to explore the utility of the SERS substrate and evaluate the inherent chemical heterogeneity across samples. Figure S6 shows representative SERS spectra of in vitro SKOV-3, endometrial cancer, ovarian cancer, and benign ovarian neoplasm EV samples used for subsequent multivariate analyses.
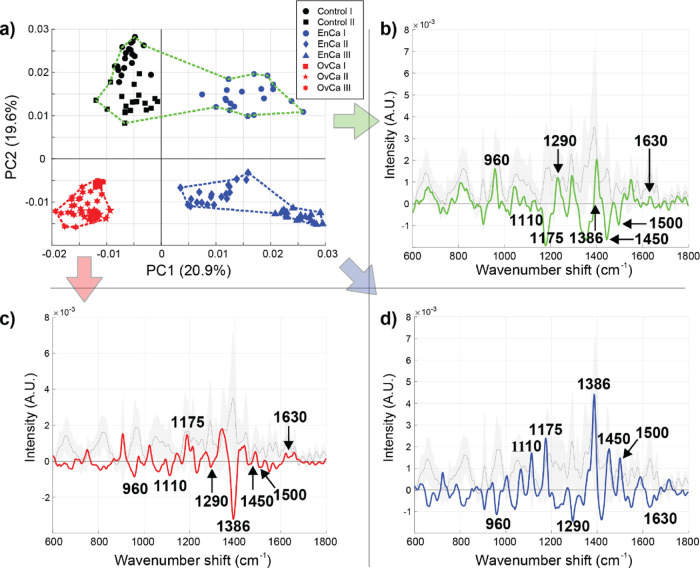
Figure 6 presents the PCA for the acquired SERS spectral data from EVs of eight different individuals. Figure 6a displays the two-dimensional PC score plot (PC1 and PC2 capture ∼40.5% of the total variation within this data set), while Figure 6b–d shows the cluster-specific SERS spectra (blue, red, green). Three of the samples represent different types and stages of endometrioid malignancies (blue), three different types and stages of ovarian malignancies (red), and two controls that were not staged (black). A total of eight spectral regions were selected for the subsequent analysis in order to infer the pertinent chemical differences between these clinical test samples: 960 (protein vibrational modes, e.g., C=C deformation or C–N stretching in amino acids),71,72 1110 (Cα–N, Cα–C, C–N stretching in the protein backbone, C–C stretching in acyl chains of lipids),25,68 1175 (nucleic acid vibrations in DNA/RNA, phenylalanine, or tyrosine vibrations in proteins),25,68 1290 (CH2 deformation in acyl chains of lipids),25,26,68 1386 (symmetrical CH3 deformation in DNA/RNA, proteins, or lipids),70,72 1450 (CH2 and CH3 deformations in proteins and lipids),24 1500 (conjugated −C=C– vibrations in nucleic acids),25,70,73 and 1630 cm–1 (amide I C=O stretching vibrations in proteins)25,74 (Table 1). In addition to PCA and hierarchical clustering analysis, we also performed PCA followed by linear discriminant analysis (LDA), as shown in Figure S7. The control and cancerous samples can be distinctly classified, which is consistent with the obtained results using PCA-hierarchical clustering. An accuracy, sensitivity, and specificity of 99.4, 100, and 99.2% were calculated, respectively. Despite the model misclassifying only a very few spectra, these numbers must be interpreted with caution given the limited sample size, inevitably yielding biased results and necessitating larger clinical cohorts in future studies. PCA-hierarchical clustering is largely used for our analyses since it enables determination of cluster-specific spectra that forecast the chemical differences more explicitly between the investigated sample groups.
Figure 6.
SERS analysis of native EVs isolated from endometrial (EnCa) and ovarian cancer (OvCa) clinical samples. (a) The PCA score plot and (b–d) three cluster-specific spectra derived from hierarchical cluster analysis (blue, red, and green dotted lines – defined using the first five PCs). Given the separation of EVs isolated from clinical samples, it appears that PC1 reports the cancer type while PC2 informs the extent of cancer burden (the EnCa I patient, blue circles, was lower grade than the rest of the EnCa/OvCa samples).
By interpreting the cluster-specific spectra in Figure 6 on the basis of the eight assigned spectral regions, we can deduce tentative chemical contributors responsible for the observed differences. First, the EVs in the blue cluster exhibit distinctly enriched contents of proteins, lipids, and nucleic acids in comparison with the EVs in red and green clusters (positive bands on the blue cluster-specific spectrum at 1110, 1175, 1386, 1450, and 1500 cm–1). Second, EVs from ovarian malignancies (OvCa I-III) have a pronouncedly different lipid composition from the EVs in blue and green clusters (negative bands on the red cluster-specific spectrum at 1110 and 1386 cm–1). Third, the EVs in the green cluster demonstrate certain protein and lipid components being profoundly more represented than the same components in EVs forming the blue and red clusters (positive bands on the green cluster-specific spectrum at 960 and 1290 cm–1). However, simultaneously, many of the nucleic acid- and protein-related vibrations are nearly absent in the green cluster (negative or negligible bands on the green cluster-specific spectrum at 1175, 1450, 1500, and 1630 cm–1). PC1 clearly separated EVs from endometrioid type malignancies (EnCa I-III) from ovarian type malignancies (OvCa I-III). Also, EVs from ovarian malignancies cluster discernibly (red cluster and the associated cluster-specific spectrum). Last, the green cluster is formed by EVs from grade I endometrioid adenocarcinoma (blue markers, EnCa I) and two control samples (black markers): a benign ovarian neoplasm (Control I) and an unspecified gynecologic neoplasm (Control II). Interestingly, while the unspecified control was marked as low CA125 in the clinical testing, it was not able to be explicitly graded. Our analysis may provide a unique angle to better grade such samples where histological analysis is unclear. This green cluster resides on the positive side of the PC2 axis, and the red and blue clusters are located on the negative side along the PC2 axis. Taking all these considerations into account, we posit that PC1 fundamentally reports the disease type while PC2 informs the disease state. To further investigate, whether the hypothesis regarding PC2 interpretation (i.e., containing information about the disease state) was plausible, we cultured an EnCa in vitro cell line and isolated the EVs. We then used the in vitro EnCa and OvCa EVs for validation representing cancerous chemical features, the clinical control samples (Control I and II) were harnessed as representatives of noncancerous characteristics, and the clinical early-stage and late-stage EnCa and OvCa EVs were subjected to the analysis. The results are shown Figure S8. Along the lines with the results displayed in Figure 6, this initial validation emphasizes that cautious interpretation of the disease state can potentially be made, especially for EnCa samples. However, we acknowledge that the sample size in the current study is limited, that the explicit chemical differences remain to be resolved, and the main emphasis of this study was to discern different cancer types from each other using a novel SERS approach. On this note, however, this demonstrated separation capability might suffice as a practical prediagnostic SERS application in a clinical setting complementing conventional screening methods.
Trypsinization of Clinical EVs Greatly Reduces Diagnostic Specificity
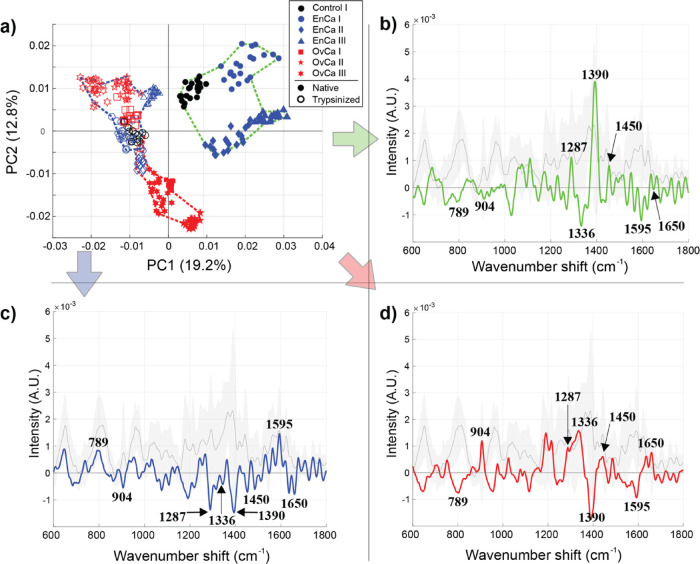
In the final stage of this study, our intention was to subject the analyzed clinical EV samples to trypsin treatment. Such enzymatic treatment would result in (i) cleavage of nonspecific glycocalyx/corona components (wholly or partially) to expose EV inner core structures for plasmonic amplification and (ii) reduction of the inherent chemical heterogeneity. The native EV samples not subjected to trypsin treatment are shown in the Figure 7a PCA score plot as filled markers, and the trypsin-treated samples are represented as empty markers. Trypsinized EVs are analyzed alone in Figure S9. In Figure 7b–d, eight spectral regions were pinpointed in the three cluster-specific spectra (blue, red, green) for further analyses: 789 (vibrations in nucleic acids),23,25,74 904 (carbohydrate-originating vibrations),63 1287 (CH2, CH3 deformation/C–N stretching + N–H deformation; amide III in proteins),25,68 1336 (backbone deformation Cα–H/Cα–C stretching/CH2, CH3 twisting or wagging in proteins),25,68 1390 (CH3 deformation in nucleic acids, proteins, or lipids),68 1450 (CH2 and CH3 deformations in proteins and lipids),24 1595 (vibrations in nucleic acids),25,68,71 and 1650 cm–1 (amide I vibrations in proteins or C=C stretching in lipids)24,25 (Table 1). The experiments and analysis were carried out using the same clinical EV samples with the exception of excluding the unspecified gynecologic neoplasm group (Control II) due to the lack of an adequate amount of sample material. Notwithstanding, the sample groups and experimental parameters were kept consistent.
Figure 7.
SERS of EVs isolated from seven clinical samples without and with trypsin treatment. (a) The PCA score plot and (b–d) the three cluster-specific SERS spectra (blue, red, green – defined using the first five PCs). The native EV samples are shown as filled markers, while the trypsin-treated measurements are shown as empty markers. As visible by their tighter spacing in this PC space, the trypsinized samples were markedly reduced in overall chemical content, indicating that the glycocalyx/corona may also indicate disease-relevant chemical information.
Intriguingly, the band regions at around 789 and 1595 cm–1 (arising from vibrations in nucleic acids) are distinctly positive for the majority of trypsin-treated EVs (blue cluster-specific spectrum) while the same region is clearly negative for nontreated EVs (red and green cluster-specific spectra) implying that trypsinization indeed facilitates exposing more of the intraluminal EV contents (e.g., DNA/RNA) to SERS amplification. Concurrently, a large part of the other bands describing mainly protein and carbohydrate features of the EVs are negative for the trypsin-treated EVs compared to the nontrypsinized EVs further indicating that at least partially these constituents may have been cleaved off by trypsin. The red cluster containing all the nontrypsinized EVs from ovarian malignancy type patients (OvCa I-III) and the trypsinized EVs from a serous endometrial cancer patient (EnCa II, empty blue diamond markers) is highly positive for bands at 904, 1287, 1336, and 1450 cm–1 compared to the global average. This observation may indicate a specific protein and carbohydrate composition in these EVs. Even though some trypsinized serous EnCa EVs (from EnCa II) cluster with the untreated OvCa samples, they are closer in PC space to the trypsin-treated EVs (blue cluster), and therefore, explicit conclusions about their chemical composition following trypsin treatment remain unclear. Similarly, the green cluster comprises untreated EVs isolated from endometrioid malignancy type patients (EnCa I–III, filled markers) and the benign ovarian neoplasm control sample (Control I, filled black circle markers). These samples potentially have a unique – highly likely protein- and/or lipid-related – chemical fingerprint as the band at around 1390 cm–1 discernibly stands out compared to the EVs in blue and red clusters. Importantly, as evidenced by these results, our initial hypothesis of losing some of the diagnostic relevance through the trypsin treatment of EVs is evident. Although we were potentially able to better expose the intraluminal components of EVs for SERS amplification, the separability between samples decreased (Figure 7, nontrypsinized vs trypsinized EVs). LDA analysis of the native EVs and trypsin-treated EVs allow for computing a confusion matrix (Figure S7b,d). We utilized the data from the confusion matrix to assess the accuracy, sensitivity, and specificity of detecting cancer using our SERS substrate and clinical EVs. For native EVs, our sensitivity and specificity are 100 and 99.2%, respectively, and the accuracy is 99.4%. However, after trypsinization of those samples, the sensitivity drops to 45%, specificity to 99.1%, and accuracy to 86.4%.
These results strongly imply that the extraluminal domain, including membrane proteins and glycosylated moieties of lipids and membrane proteins, is critical to indicating the cancer presence. While we acknowledge that the sample size of this clinical data set is small, this is a key finding of this work that warrants further investigation. It is known that variations in cell surface glycoproteins significantly impact the progression of cancer, including the patients’ prognosis.75 Functional and analytical studies will be needed to elucidate the particular glycoproteins that are involved in distinguishing clinical samples from one another and to discern their potential role in EV signaling in cancer.
Conclusions
This report outlined the preparation of a new type of porous, nanoplasmonic substrate for EV analysis. We detailed a methodology for tackling the inherent heterogeneous structure of the scaffolds and rigorous multivariate data analysis steps in order to reproducibly reveal the cancerous SERS spectra features in the measured data sets. As the spectral analyses demonstrate, we have successfully investigated and characterized EVs from in vitro cell cultures and clinical samples with an estimated LOD of ∼600 EVs/mL with low laser powers. We demonstrate both a chemical treatment using cysteamine to nonspecifically bind EVs and also the large effects of extraluminal cleavage to provide complementary chemical information using a SERS approach. Both chemical treatments are generalizable to SERS analysis platforms but are especially useful for our substrate, which is easily washed due to its porous structure. Of note, we report that enzymatic cleavage of the EVs’ extraluminal domain resulted in loss of sensitivity to detect clinical patient samples of endometrial and ovarian cancer, indicating that these components may be of clinical significance. To implement a liquid biopsy methodology based on this platform to the clinic, standardization/automation of isolation and sample handling techniques, evaluation of reproducibility and cost-effectiveness, and validation by clinical trials are required. We envision that this work can act as a step toward a modern, minimally invasive plasmonic liquid biopsy platform with adequate sensitivity, specificity, and economical aspects for future implementation.
Acknowledgments
R.P.C. was supported by a Research Scholar Grant, RSG-19-116-01-CDD, from the American Cancer Society. T.R. gratefully acknowledges The Sigrid Juselius Foundation, Helsinki, Finland. This work was supported by funds from the UC Davis Comprehensive Cancer Center, the Ovarian Cancer Education and Research Network, Inc. (OCERN), and the NIH (1R01CA241666). S.W.H. and J.L. would like to acknowledge financial support from the Natural Sciences and Engineering Research Council of Canada (NSERC), Discovery Grant RGPIN-2018-05675. Special thanks to Maria Navas-Moreno (Lever Photonics LLC) for designing and building the custom Raman confocal microscope.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.0c00953.
Author Contributions
§ T.R. and H.J.K. contributed equally.
The authors declare no competing financial interest.
Notes
As collected data for the main and supporting figures, including raw SEM images, NTA videos, and SERS spectra are deposited at https://doi.org/10.5281/zenodo.3994784.
Supplementary Material
References
- Ferlay J.; Soerjomataram I.; Dikshit R.; Eser S.; Mathers C.; Rebelo M.; Parkin D. M.; Forman D.; Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Ferlay J.; Colombet M.; Soerjomataram I.; Mathers C.; Parkin D. M.; Piñeros M.; Znaor A.; Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- Robertson E. G.; Baxter G. Tumour seeding following percutaneous needle biopsy: the real story!. Clin. Radiol. 2011, 66, 1007–1014. 10.1016/j.crad.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Gerlinger M.; Rowan A. J.; Horswell S.; Math M.; Larkin J.; Endesfelder D.; Math D.; Gronroos E.; Martinez P.; Matthews N.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabières C.; Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- Crowley E.; Di Nicolantonio F.; Loupakis F.; Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- Speicher M. R.; Pantel K. Tumor signatures in the blood. Nat. Biotechnol. 2014, 32, 441–443. 10.1038/nbt.2897. [DOI] [PubMed] [Google Scholar]
- De Rubis G.; Rajeev Krishnan S.; Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Heitzer E.; Perakis S.; Geigl J. B.; Speicher M. R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017, 1, 36. 10.1038/s41698-017-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G.; Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Mó M.; Siljander R. P.-M.; Andreu Z.; Zavec A. B.; Borràs F. E.; Buzas E. I.; Buzas K.; Casal E.; Cappello F.; Carvalho J.; Colás E.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S.; O’Driscoll L.; Borras F. E.; Buzas E.; Camussi G.; Cappello F.; Carvalho J.; Cordeiro da Silva A.; Del Portillo H.; El Andaloussi S.; Trček T. F.; Furlan R.; Hendrix A.; Gursel I.; Kralj-Iglic V.; Kaeffer B.; Kosanovic M.; Lekka M. E.; Lipps G.; Logozzi M.; Marcilla A.; Sammar M.; Llorente A.; Nazarenko I.; Oliveira C.; Pocsfalvi G.; Rajendran L.; Raposo G.; Rohde E.; Siljander P.; van Niel G.; Vasconcelos M. H.; Yáñez-Mó M.; Yliperttula M. L.; Zarovni N.; Zavec A. B.; Giebel B. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano 2016, 10, 3886–3899. 10.1021/acsnano.5b08015. [DOI] [PubMed] [Google Scholar]
- El Andaloussi S.; Mäger I.; Breakefield X. O.; Wood M. J. A. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Zhao H.; Achreja A.; Iessi E.; Logozzi M.; Mizzoni D.; Di Raimo R.; Nagrath D.; Fais S. The key role of extracellular vesicles in the metastatic process. BBA – Rev. Cancer 2008, 1869, 64–77. 10.1016/j.bbcan.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A.; Costa-Silva B.; Shen T.-L.; Rodrigues G.; Hashimoto A.; Mark M. T.; Molina H.; Kohsaka S.; Di Giannnatale A.; Cederm S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-de-Souza P.; Gori V.; Bambi F.; Chiarugi P. Tumor microenvironment: Bone marrow-mesenchymal stem cells as key players. BBA – Rev. Cancer 2013, 1836, 321–335. 10.1016/j.bbcan.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Becker A.; Thakur B. K.; Weiss J. M.; Kim H. S.; Peinado H.; Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Wang X. The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol. Cancer 2017, 16, 92. 10.1186/s12943-017-0659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney R. P.; Hazari S.; Rojalin T.; Knudson A.; Gao T.; Tang Y.; Liu R.; Viitala T.; Yliperttula M.; Lam K. S. Targeting Tumor-Associated Exosomes with Integrin-Binding Peptides. Adv. Biosyst. 2017, 1, 1600038. 10.1002/adbi.201600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M.; Crow J.; Roth M.; Zeng Y.; Godwin A. K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773–3780. 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatischeff I.; Larquet E.; Falcón-Pérez J. M.; Turpin P.-Y.; Kruglik S. G. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J. Extracell. Vesicles 2012, 1, 19179. 10.3402/jev.v1i0.19179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft C.; Wilhelm K.; Eremin A.; Nestel S.; von Bubnoff N.; Schultze-Seemann W.; Popp J.; Nazarenko I. A specific spectral signature of serum and plasma-derived extracellular vesicles for cancer screening. Nanomedicine 2017, 13, 835–841. 10.1016/j.nano.2016.11.016. [DOI] [PubMed] [Google Scholar]
- Gualerzi A.; Niadaia S.; Giannasi C.; Piccolini S.; Morasso C.; Vanna R.; Rosella V.; Masserini M.; Bedoni M.; Ciceri F.; et al. Raman spectroscopy uncovers biochemical tissue-related features of extracellular vesicles from mesenchymal stromal cells. Sci. Rep. 2017, 7, 9820. 10.1038/s41598-017-10448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. J.; Lee C.; Rojalin T.; Carney R. P.; Hazari S.; Knudon A.; Lam K.; Saari H.; Lazaro I. E.; Viitala T.; et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles 2015, 4, 28533. 10.3402/jev.v4.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglik S. G.; Royo F.; Guigner J.-M.; Palomo L.; Seksek O.; Turpin P.-Y.; Tatischeff I.; Falcón-Pérez J. M. Raman tweezers microspectroscopy of circa 100 nm extracellular vesicles. Nanoscale 2019, 11, 1661–1679. 10.1039/c8nr04677h. [DOI] [PubMed] [Google Scholar]
- Lee W.; Nanou A.; Rikkert L.; Coumans F. A. W.; Otto C.; Terstappen L. W. M. M.; Offerhaus H. L. Label-Free Prostate Cancer Detection by Characterization of Extracellular Vesicles Using Raman Spectroscopy. Anal. Chem. 2018, 90, 11290–11296. 10.1021/acs.analchem.8b01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer J.; Jimenez de Aberasturi J.; Aizpurua J.; Alvarez-Puebla R. A.; Auguié B.; Baumberg J. J.; Bazan G. C.; Bell S. E. J.; Boisen A.; Brolo A. G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J. A.; Rahim Ferhan A.; Cho N.-J. Nanoplasmonic sensors for biointerfacial science. Chem. Soc. Rev. 2017, 46, 3615–3660. 10.1039/C6CS00494F. [DOI] [PubMed] [Google Scholar]
- Ferhan A. R.; Jackman J. A.; Park J. H.; Cho N.-J.; Kim D.-H. Nanoplasmonic sensors for detecting circulating cancer biomarkers. Adv. Drug Delivery Rev. 2018, 125, 48–77. 10.1016/j.addr.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Rojalin T.; Phong B.; Koster H. J.; Carney R. P. Nanoplasmonic Approaches for Sensitive Detection and Molecular Characterization of Extracellular Vesicles. Front. Chem. 2019, 7, 279. 10.3389/fchem.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini L.; Alvarez-Puebla R. A. Surface-Enhanced Raman Spectroscopy in Cancer Diagnosis, Prognosis and Monitoring. Cancers 2019, 11, 748. 10.3390/cancers11060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Mi X.; Tan X.; Xiang R. Recent Progress on Liquid Biopsy Analysis using Surface-Enhanced Raman Spectroscopy. Theranostics 2019, 9, 491–525. 10.7150/thno.29875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko M. Y.; Lekchnov E. A.; Vlassov A. V.; Laktionov P. P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed. Res. Int. 2018, 8545347. 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber J.; Clayton A. How pure are your vesicles?. J. Extracell. Vesicles 2013, 2, 19861. 10.3402/jev.v2i0.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz A.; Kenton M.; Aksin G.; Kahraman M.; Wachsmann-Hogiu S. Inexpensive and Flexible SERS Substrates on Adhesive Tape Based on Biosilica Plasmonic Nanocomposites. ACS Appl. Nano Mater. 2018, 1, 5316–5326. 10.1021/acsanm.8b01336. [DOI] [Google Scholar]
- Lee P. C.; Meisel D. Adsorption and Surface-Enhanced Raman of Dyes on Silver and Gold Sols. J. Phys. Chem. 1982, 86, 3391–3395. 10.1021/j100214a025. [DOI] [Google Scholar]
- Polavarapu L.; Liz-Marzán L. M. Towards low-cost flexible substrates for nanoplasmonic sensing. Phys. Chem. Chem. Phys. 2013, 15, 5288–5300. 10.1039/C2CP43642F. [DOI] [PubMed] [Google Scholar]
- John S. Strong localization of photons in certain disordered dielectric superlattices. Phys. Rev. Lett. 1987, 58, 2486–2489. 10.1103/PhysRevLett.58.2486. [DOI] [PubMed] [Google Scholar]
- Yablonovitch E. Inhibited Spontaneous Emission in Solid-State Physics and Electronics. Phys. Rev. Lett. 1987, 58, 2059–2062. 10.1103/PhysRevLett.58.2059. [DOI] [PubMed] [Google Scholar]
- Liu C.; Wang Z.; Li E.; Liang Z.; Chakravarty S.; Xu X.; Wang A. X.; Chen R. T.; Fan D. Electrokinetic Manipulation Integrated Plasmonic-Photonic Hybrid Raman Nanosensors with Dually Enhanced Sensitivity. ACS Sens. 2017, 2, 346–353. 10.1021/acssensors.6b00586. [DOI] [PubMed] [Google Scholar]
- Xu X.; Hasan D.; Wang L.; Chakravarty S.; Chen R. T.; Fan D. L.; Wang A. X. Guided-mode-resonance-coupled plasmonic-active SiO(2) nanotubes for surface enhanced Raman spectroscopy. Appl. Phys. Lett. 2012, 100, 191114–1911145. 10.1063/1.4714710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X.; Squire K.; Li E.; LeDuff P.; Rorrer G. L.; Tang S.; Chen B.; McKay C. P.; Navarro-Gonzalez R.; Wang A. X. Chemical and Biological Sensing Using Diatom Photonic Crystal Biosilica With In-Situ Growth Plasmonic Nanoparticles. IEEE T. NanoBiosci. 2016, 15, 828–834. 10.1109/TNB.2016.2636869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X.; Xi Y.; Le Duff P.; Chong X.; Li E.; Ren F.; Rorrer G. L.; Wang A. X. Detecting explosive molecules from nanoliter solution: A new paradigm of SERS sensing on hydrophilic photonic crystal biosilica. Biosens. Bioelectron. 2017, 88, 63–70. 10.1016/j.bios.2016.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F.; Campbell J.; Rorrer G. L.; Wang A. X. Surface-Enhanced Raman Spectroscopy Sensors From Nanobiosilica With Self-Assembled Plasmonic Nanoparticles. IEEE J. Sel. Top. Quantum Electron. 2014, 20, 127–132. 10.1109/JSTQE.2014.2301016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H.; Shao H.; Park Y. I.; Peterson V. M.; Castro C. M.; Weissleder R.; Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C.; Witwer K. W.; Aikawa E.; Alcaraz M. J.; Anderson J. D.; Andriantsitohaina R.; Antoniou A.; Arab T.; Archer F.; Atkin-Smith G. K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Jiang L.; Mei Y.; Song R.; Tian D.; Huang H. Colorimetric sensing strategy for mercury(ii) and melamine utilizing cysteamine-modified gold nanoparticles. Analyst 2013, 138, 5338–5343. 10.1039/C3AN00690E. [DOI] [PubMed] [Google Scholar]
- Kudelski A.; Hill W. Raman Study on the Structure of Cysteamine Monolayers on Silver. Langmuir 1999, 15, 3162–3168. 10.1021/la9811463. [DOI] [Google Scholar]
- Jiang X.; Yang M.; Meng Y.; Jiang W.; Zhan J. Cysteamine-Modified Silver Nanoparticle Aggregates for Quantitative SERS Sensing of Pentachlorophenol with a Portable Raman Spectrometer. ACS Appl. Mater. Interfaces 2013, 5, 6902–6908. 10.1021/am401718p. [DOI] [PubMed] [Google Scholar]
- Mitchell J. P.; Court J.; Mason M. D.; Tabi Z.; Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J. Immunol. Methods 2008, 335, 98–105. 10.1016/j.jim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Mitra R.; O’Neil G. L.; Harding I. C.; Cheng M. J.; Mensah S. A.; Ebong E. E. Glycocalyx in Atherosclerosis-Relevant Endothelium Function and as a Therapeutic Target. Curr. Atheroscler. Rep. 2017, 19, 63. 10.1007/s11883-017-0691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z.; Fehér B.; Kitka D.; Wacha A.; Bóta A.; Berényi S.; Pipich V.; Fraikin J.-L. Size Measurement of Extracellular Vesicles and Synthetic Liposomes: The Impact of the Hydration Shell and the Protein Corona. Colloids Surf., B 2020, 192, 111053. 10.1016/j.colsurfb.2020.111053. [DOI] [PubMed] [Google Scholar]
- Kennedy B. J.; Spaeth S.; Dickey M.; Carron K. T. Determination of the Distance Dependence and Experimental Effects for Modified SERS Substrates Based on Self-Assembled Monolayers Formed Using Alkanethiols. J. Phys. Chem. B 1999, 103, 3640–3646. 10.1021/jp984454i. [DOI] [Google Scholar]
- Stiles P. L.; Dieringer J. A.; Shah N. C.; van Duyne R. P. Surface-enhanced Raman spectroscopy. Annu. Rev. Anal. Chem. 2008, 1, 601–626. 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- Shurer C. R.; Kuo J. C.-H.; Roberts L. M.; Gandhi J. G.; Colville M. J.; Enoki T.; Pan H.; Su J.; Noble J. M.; Hollander M. J.; et al. Physical Principles of Membrane Shape Regulation by the Glycocalyx. Cell 2019, 177, 1757–1770.e21. 10.1016/j.cell.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo F.; Cossío U.; Ruiz de Angulo A.; Llop J.; Falcon-Perez J. M. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale 2019, 11, 1531–1537. 10.1039/C8NR03900C. [DOI] [PubMed] [Google Scholar]
- Paolini L.; Orizio F.; Busatto S.; Radeghieri A.; Bresciani R.; Bergese P.; Monti E. Exosomes Secreted by HeLa Cells Shuttle on Their Surface Plasma Membrane-Associated Sialidase NEU3. Biochemistry 2017, 56, 6401–6408. 10.1021/acs.biochem.7b00665. [DOI] [PubMed] [Google Scholar]
- Muralidharan-Chari V.; Clancy J. W.; Sedgwick A.; D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci. 2010, 123, 1603–1611. 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley E. A.; Wood D. K.; McCarthy J. B. Carcinoma Cell Hyaluronan as a “Portable” Cancerized Prometastatic Microenvironment. Cancer Res. 2016, 76, 2507–2512. 10.1158/0008-5472.CAN-15-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan P. Structural studies of silicate glasses and melts – applications and limitations of Raman spectroscopy. Am. Mineral. 1984, 69, 622–644. [Google Scholar]
- McMillan P. A Raman spectroscopic study of glasses in the system CaO-MgO-SiO2. Am. Mineral. 1984, 69, 645–659. [Google Scholar]
- Witkowska E.; Korsak D.; Kowalska A.; Janeczek A.; Kamińska A. Strain-level typing and identification of bacteria – a novel approach for SERS active plasmonic nanostructures. Anal. Bioanal. Chem. 2018, 410, 5019–5031. 10.1007/s00216-018-1153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrozek M. F.; Weaver M. J. Detection and Identification of Aqueous Saccharides by Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2002, 74, 4069–4075. 10.1021/ac020115g. [DOI] [PubMed] [Google Scholar]
- Zong S.; Wang L.; Chen C.; Lu J.; Zhu D.; Zhang Y.; Wang Z.; Cui Y. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal. Methods 2016, 8, 5001–5008. 10.1039/C6AY00406G. [DOI] [Google Scholar]
- Frühbeis C.; Helmig S.; Tug S.; Simon P.; Krämer-Albers E.-M. Physical exercise induces rapid release of small extracellular vesicles into the circulation. J. Extracell. Vesicles 2015, 4, 28239. 10.3402/jev.v4.28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovato E.; Di Felice V.; Barone R. Extracellular Vesicles: Delivery Vehicles of Myokines. Front. Physiol. 2019, 10, 522. 10.3389/fphys.2019.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares R.; Tan S.; Gounou C.; Arraud N.; Brisson A. R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29509. 10.3402/jev.v4.29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willets K. A. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal. Bioanal. Chem. 2009, 394, 85–94. 10.1007/s00216-009-2682-3. [DOI] [PubMed] [Google Scholar]
- Seo M. J.; Kim G. W.; Tsalu P. V.; Moon S. W.; Ha J. W. Role of chemical interface damping for tuning chemical enhancement in resonance surface-enhanced Raman scattering of plasmonic gold nanorods. Nanoscale Horiz. 2020, 5, 345–349. 10.1039/C9NH00524B. [DOI] [Google Scholar]
- Stremersch S.; Marro M.; Pinchasik B.-E.; Baatsen P.; Hendrix A.; De Smedt S. C.; Loza-Alvarez P.; Skirtach A. G.; Raemdonck K.; Braeckmans K. Identification of Individual Exosome-Like Vesicles by Surface Enhanced Raman Spectroscopy. Small 2016, 12, 3292–3301. 10.1002/smll.201600393. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Wang L.; Fang Y.; Zheng D.; Lin T.; Wang H. Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis. Molecules 2019, 24, 2947. 10.3390/molecules24162947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.; Jeong H.; Park J.; Hong S.; Choi Y. Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 2018, 3, 2637–2643. 10.1021/acssensors.8b01047. [DOI] [PubMed] [Google Scholar]
- Sivashanmugan K.; Huang W.-L.; Lin C.-H.; Liao J.-D.; Lin C.-C.; Su W.-C.; Wen T.-C. Bimetallic nanoplasmonic gap-mode SERS substrate for lung normal and cancer-derived exosomes detection. J. Taiwan Inst. Chem. E. 2017, 80, 149–155. 10.1016/j.jtice.2017.09.026. [DOI] [Google Scholar]
- Park J.; Hwang M.; Choi B.; Jeong H.; Jung J.-h.; Kim H. K.; Hong S.; Park J.-H.; Choi Y. Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis. Anal. Chem. 2017, 89, 6695–6701. 10.1021/acs.analchem.7b00911. [DOI] [PubMed] [Google Scholar]
- Pinho S. S.; Reis C. A. Glycosylation in cancer: mechanisms and clinical implication. Nat. Rev. Cancer 2015, 15, 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.