Abstract
Purpose
Increasing evidence has shown that the transcription factor SOX4 is closely associated with the development and progression of many malignant tumors. However, the effect of SOX4 on breast cancer is unclear. In this study, we purposed to investigate the role of SOX4 in the growth and metastasis in breast cancer and the underlying mechanism. Moreover, the effect of SOX4 on cancer cell resistance to chemotherapeutic agents was also evaluated in vitro and in vivo.
Methods
We used lentivirus technique to ectopically express SOX4 in MDA-MB-231 and SUM149 cells or knockdown SOX4 in BT474 cells, and examined the effect of these changes on various cellular functions. MTT assay was used to determine the cell viability as well as resistance to chemotherapeutic agents. The regulation of SOX4 on epithelial-mesenchymal transition (EMT)-related genes was analyzed using qRT-PCR. The binding of SOX4 to the CXCR7 gene was demonstrated using chromatin immunoprecipitation assay and dual-luciferase reporter activity assay. The effect of SOX4/CXCR7 axis on metastasis was examined using Transwell migration and Matrigel invasion assays. The expression of SOX4/CXCR7 in primary tumors and metastatic foci in lymph nodes was assessed using immunohistochemistry. Cellular morphology was investigated under phase contrast microscope and transmission electron microscopy. Moreover, the effect of SOX4 on tumor growth, metastasis, and resistance to chemotherapy was also studied in vivo by using bioluminescent imaging.
Results
SOX4 increased breast cancer cell viability, migration, and invasion in vitro and enhanced tumor growth and metastasis in vivo. It regulated EMT-related genes and bound to CXCR7 promoter to upregulate CXCR7 transcription. Both SOX4 and CXCR7 were highly expressed in human primary tumors and metastatic foci in lymph nodes. Treatment of breast cancer cells with the CXCR7 inhibitor CCX771 reversed the SOX4 effect on cell migration and invasion. Ectopic expression of SOX4 increased the susceptibility of cells to paclitaxel.
Conclusions
SOX4 plays an important role in the growth and metastasis of breast cancer. SOX4/CXCR7 may serve as potential therapeutic targets for the treatment. Paclitaxel may be a good therapeutic option if the expression level of SOX4 is high.
Keywords: Breast cancer, Growth, Metastasis, SOX4, CXCR7
Introduction
Breast cancer is the most common cancer and the leading cause of cancer-related death in women [1, 2], accounting for 25% of their cancers and 15% of cancer-related death [2, 3]. The cause of death by breast cancer is often not due to the primary tumor, but due to the metastasis of cancer cells to other tissues and organs. Distant metastasis usually causes the failure of clinical therapy [4]. Therefore, deep understanding of the mechanism underlying metastasis would be crucial to identify novel therapeutic targets and for the treatment of breast cancer.
SOX4 is a member of the SOX (SRY-related HMG box) gene family which consists of transcription factors involved in the development and differentiation of cells and organs, as well as the initiation and development of cancers [5, 6]. The gene of SOX4 is highly conserved in vertebrates and encodes a 47 KD protein containing 474 amino acid residues. SOX4 plays important roles in the development of bone, islet and heart, and is related to osteoporosis, cancers, and other diseases [7–12]. Previous studies have reported that SOX4 is highly expressed in over 20 malignant tumors and promotes malignant phenotypes [13, 14]. It was shown to activate CXCL12 promoter in hepatocellular carcinoma cells to modulate endothelial cell migration and angiogenesis in vivo [15]. Silencing SOX4 could inhibit the growth and metastasis of melanoma and some other tumors [7, 16–18]. These findings indicate that SOX4 functions as an oncogene. Recently, SOX4 was reported to promote epithelial-mesenchymal transition (EMT) in breast cancer [13, 19, 20]. However, the directly SOX4-regulated genes involved in the EMT process are still unknown.
Chemokines are polypeptides that regulate cell migration and serve as key regulators of metastasis. The levels of multiple chemokines are upregulated in the cell matrix of primary or metastatic tumors, compared to normal tissues [21–24]. In breast cancer cells, binding of chemokines to their receptors can activate the transcription of a series of downstream effector genes, resulting in the proliferation and metastasis of tumor cells. CXCR7, also known as RDC1, is one of the CXCR subtype cytokines. The activation of CXCR7 by its ligand CXCL12 could promote cell migration, angiogenesis, tumorigenesis, invasion, metastasis and anti-apoptosis [25]. CXCR7 is considered to be an EMT-related gene and is highly expressed in various cancer cells [26–31]. It was shown to be involved in the regulation of several pathways in cancer cells such as TGF-β1/Smad2/3 [30], PI3K/Akt [32] and β-arrestin [33] signaling pathways. However, the effect of CXCR7 on distant metastasis of breast cancer remains unclear.
In this study, we demonstrated that SOX4 promotes the growth and metastasis of breast cancer both in vitro and in vivo. SOX4 binds to the promoter of CXCR7 and upregulates its transcription. Our results indicated that SOX4/CXCR7 axis plays important roles in breast cancer and may serve as potential novel therapeutic targets in the treatment of this disease.
Materials and methods
Cell lines and cell culture
MDA-MB-231, BT474, SUM149, and 293T cells were purchased from American Type Culture Collection (ATCC). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM, 12800-017; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, 10099-141; Life Technologies, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin in a humidified 5% CO2 atmosphere at 37 °C.
Plasmid construction and generation of stable cell lines
The lentivirus-based vector pLOX/EW-IRES-EGFP [34] was used for ectopic SOX4-expression. SOX4 cDNA was ligated to the Sma I site of the vector and the construct was confirmed by sequencing. The SOX4-expressing vector or the control vector was co-transfected into the 293T packing cells with pAX2 and pMD2G and the lentivirus was collected for subsequent infection into MDA-MB-231 and SUM149 cells. Stably SOX4-expressing cells (MDA-MB-231-SOX4 and SUM149-SOX4) and corresponding control cells (MDA-MB-231-VECT, SUM149-VECT) were sorted using flow cytometry by BD FACS Aria (Becton Dickinson, Franklin Lakes, NJ, USA). For SOX4 knockdown, lentivirus-based pLKO.1-TRC-puro constructs containing a SOX4 short hairpin RNA (shRNA NT or #13-#17) were purchased from Open Biosystems (Huntsville, AL, USA). The lentivirus was prepared as above and used to infect BT474 cells. Stably SOX4-knockdown cells (BT474-14, BT474-16) and control cells (BT474-NT) were yielded after treatment with puromycin (Sigma, San Francisco, CA, USA) for 14 days. To introduce luciferase into tumor cells for bioluminescent imaging in vivo, pMig-Luc-mCherry, pUMVC and pMD2G were co-transfected into the 293T packing cells and the retrovirus was collected to infect tumor cells for xerograph experiments.
Western blot
The protein lysate was resolved by SDS-PAGE and transferred to PVDF membrane. After blocking with 5% bovine serum albumin (BSA), the membrane was incubated with the primary antibody overnight at 4 ºC, followed by incubation with the secondary antibody at room temperature for 1 h. The antibodies used in this study included anti-SOX4 (Cat#H00006659-A01, Abnova, Taipei), anti-GAPDH (Cat#MA5-15738, Invitrogen, Grand Island, NY, USA), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H&L) (Cat#G21040, Invitrogen), and HRP-conjugated goat anti-rabbit IgG (H&L) (Cat#31460, Invitrogen).
MTT assay
Cells were seeded in 96-well plates at a density of 6 × 106 cells per well and incubated at 37 ºC in humidified 5% CO2 for 24 h. The cells were stained with MTT (Cat#CT02, Sigma) at each time point for 1 h at 37 °C. The absorbance was measured at 490 nm using a spectrophotometer (Bio-Rad, Hercules, CA, USA). For the analysis of resistance to chemotherapeutic agents, carboplatin, adriamycin, 5-fluorouracil, or paclitaxel at different concentrations was added at the beginning of the cell culture which was carried out for 48 h.
Transwell assay
Migration was measured using Matrigel-free Transwell plates (Corning, Midland, MI, USA) with an 8 μm porous membrane. For invasion assay, membrane of the upper chambers was coated with Matrigel (BD Biosciences, Sparks, MD, USA) before use. In total, 1 × 105 cells were plated in the upper chambers of the Transwells. After a 24-h incubation, migrating or invading cells were stained with 0.5% crystal violet, then photographed at 200× and counted in five random fields. CXCR7 inhibitor CCX771 was a courtesy from ChemoCentryx (Mountain View, CA, USA).
RT-qPCR
Total RNA was prepared with the Trizol reagent according to the manufacturer’s instructions (Invitrogen). cDNA was synthesized using SuperScript Kit (Invitrogen). Real-time PCR was performed using SYBRH Select Master Mix for CFX (Invitrogen). Relative quantification was achieved by normalization to the amount of GAPDH control RNA. The primers used are shown in Table 1.
Table 1.
Primers for qPCR
| SOX4-F | 5′-GACATGCACAACGCCGAGATCT-3′ |
|---|---|
| SOX4-R | 5′-GTAGTCAGCCATGTGCTTGAGG-3′ |
| CXCR7-F | 5′-CCAAGACCACAGGCTATGACAC-3′ |
| CXCR7-R | 5′-TGGTTGTGCTGCACGAGACTGA-3′ |
| Vimentin-F | 5′-CACAAGCAGAGTGCTGAAGGTG-3′ |
| Vimentin-R | 5′-ATCTGGCGTTCCAGGGACTCAT-3′ |
| N-Cadherin -F | 5′-AGACGCTAGTGGAGGAGTGCAA-3′ |
| N-Cadherin -R | 5′-GTAGGATCTCCGCCACTGATTC-3′ |
| E-Cadherin-F | 5′-GCCTCCTGAAAAGAGAGTGGAAG-3′ |
| E-Cadherin-R | 5′-GGAGATACCAGTTCCACAGGTC-3′ |
| Fibronectin-F | 5′-ACAACACCGAGGTGACTGAGAC-3′ |
| Fibronectin-R | 5′-GGACACAACGATGCTTCCTGAG-3′ |
F forward, R reverse
Chromatin immunoprecipitation assay
Cultured MDA-MB-231-VECT and MDA-MB-231-SOX4 cells were treated with 1% formaldehyde at 37 °C for 15 min, and lysed with nuclei-swelling buffer. After sonication and centrifugation, cytoplasmic extract was incubated at 4 °C overnight with 5 mg of anti-SOX4 (Cat#ab86809, Abcam, Cambridge, UK) or nonspecific IgG control antibody. Cytoplasmic extract not incubated with antibody was saved as an input sample. The antibody-bound complex was precipitated by protein A-Sepharose beads (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). The beads were washed and the protein-DNA complex was eluted from the beads with 250 ml of elution buffer (1% SDS and 0.1 M NaHCO3) at 37 °C for 15 min. The DNA in the immunoprecipitated complex and the DNA in the previously saved input fraction were released by incubation at 65 ℃ for 2 h with 200 nM NaCl and 20 mg of proteinase K. DNA was amplified by PCR under the following conditions: 96 °C for 15 s, 54 °C for 30 s and 72 °C for 30 s, and 40 cycles. The primers used are showed in Table 2.
Table 2.
Primers used in chromatin immunoprecipitation assay
| CXCR7-F | 5′-GAAGAGGCATTCACAGGAGC-3′ |
|---|---|
| CXCR7-R | 5′-CAGAAAGGAGCCTCTAGC-3′ |
F forward, R reverse
Dual-luciferase reporter activity assay
The plasmid pGL3-CXCR7 containing the CXCR7 promoter upstream of the firefly luciferase reporter gene and the corresponding empty vector were purchased from HIBIO (Hangzhou, China). MDA-MB-231-VECT and MDA-MB-231-SOX4 cells were transfected with pGL3-CXCR7 and empty vector, respectively, when the cells grew to 60%-80% confluence. Next day, the cells were lysed and the luciferase activity was tested using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The activity was calibrated to Renilla luciferase activity. Each treatment was carried out in triplicate.
Immunohistochemistry (IHC)
The tissue chip slide (ZL-BRM961, SUPERBIOTEK, Shanghai, China), which contained tissues of primary breast carcinomas and corresponding lymph node metastases (n = 36), was deparaffinized in xylene and rehydrated in graded alcohol solution, followed by treatment with 3% H2O2 to quench the endogenous peroxidase for 15 min and 1% BSA for 1 h at room temperature to block the potential nonspecific binding. The slides were then incubated with primary monoclonal antibody against SOX4 (Cat#ab237903, Abcam) or CXCR7 (Cat#ab38089, Abcam) overnight at 4 °C prior to incubation with biotinylated secondary antibody for 20 min at room temperature. Finally, the slides were treated with avidin–biotin complex reagent and stained with 3, 3 -diaminobenzidine according to manufacturer's protocol. Staining results were analyzed for the staining intensity as well as the percentage of positive cells. The staining intensity was scored 0 point for no obvious coloring, 1 point for mild, 2 points for moderate, or 3 points for strong. The percentage of positive cells for each slide was calculated from 3 different microscopic fields (200x) and was scored as follows: 0 to 5%, 0 point; 6% to 25%, 1 point; 26% to 50%, 2 points; 51% to 75%, 3 points; and > 75%, 4 points. The overall score was the sum of the staining intensity and the percentage of positive cells: 0, negative; 2 to 3, weakly positive; 4 to 5, moderate; and 6 to 7, strongly positive.
Tumor xenograft
Six-week-old male mice with severe combined immunodeficiency (SCID) were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Cells (5 × 106) expressing luciferase were injected into the left mammary fat pad of each mouse. The tumors were examined weekly using bioluminescent imaging after intraperitoneal injection of 150 mg/kg D-luciferin (Caliper LifeSciences, Hopkinton, MA, USA). The length (a) and width (b) of the tumors were also recorded every week. The tumor volume was calculated by the formula of V = ab2/2 mm3. In the study of metastasis, tumors were removed when their sizes exceeded 2 cm × 2 cm. Metastasis was followed by bioluminescent imaging. By the end of the 16th week, mice were euthanized, dissected and examined for organ metastases. In the study of resistance to paclitaxel, 3 weeks after cell injection, i.e., day 0 of treatment, the tumor-bearing mice were administered with paclitaxel at a dose of 10 mg/kg or the same volume of phosphate buffered saline (PBS). The treatment was repeated at day 7 and the tumors were examined using bioluminescent imaging at day 14.
Results
SOX4 promotes breast cancer cell proliferation both in vitro and in vivo
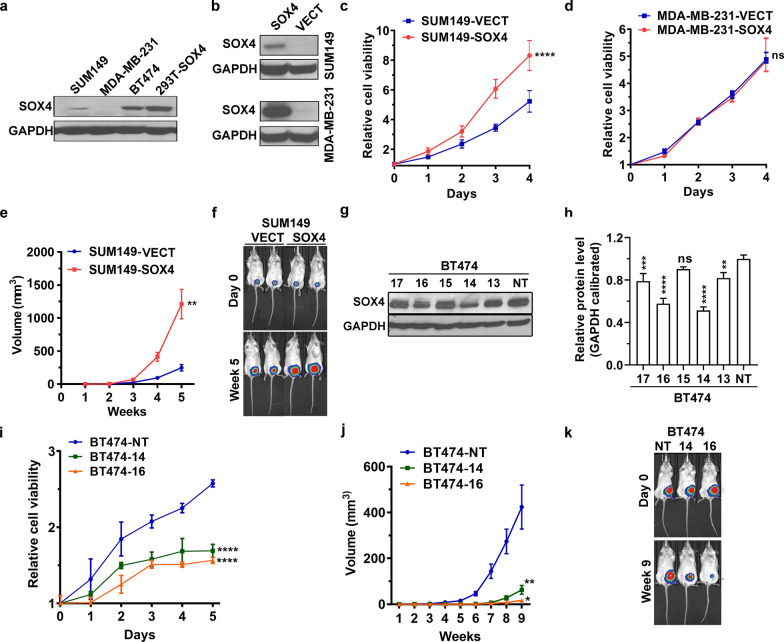
The protein expression levels of SOX4 in breast cancer cell lines SUM149, MDA-MB-231, and BT474 were variable, low in SUM149 and MDA-MB-231 and high in BT474 by Western blot (Fig. 1a). Thus, we chose SUM149 and MDA-MB-231 for ectopic SOX4 expression which was successfully achieved using lentivirus carrying the SOX4 gene (Fig. 1b), with the SOX4 expression levels significantly higher in MDA-MB-231-SOX4 cells than those in SUM149-SOX4 cells.
Fig. 1.
SOX4 promotes breast cancer cell proliferation both in vitro and in vivo. a Western blot analysis of SOX4 expression in three human breast cancer cell lines. Embryonic kidney cell line 293 T with ectopic SOX4 expression (293 T-SOX4) was used as positive control. GAPDH was used as loading control. b Western blot showing SOX4 protein levels in MDA-MB-231-SOX4 cells and SUM149-SOX4 cells as well as the control cells. GAPDH was used as loading control. c and d MTT assay showing cell viability in SOX4-expressing cell lines compared with control. The data are presented as the mean ± s.d. of sextuplicate wells. Unpaired two-tailed t-test: ***p < 0.001. e In vivo tumor growth from SOX4-expressing cells and control cells. 5 × 106 cells were injected into the mammary fat pad of SCID mice. The length a and width b of tumors were recorded weekly. The tumor volume was calculated by the formula of V = ab2/2 (mm3). The data are presented as the mean ± s.e.m. of 6 mice. Unpaired two-tailed t-test: **p < 0.01. f Bioluminescent imaging illustrating tumor growth in vivo. g Western blot showing the SOX4 expression in BT474 cells with individual shRNAs. h Semi-quantitative analysis of western blotting. Unpaired two-tailed t-test: **p < 0.01, ***p < 0.001, ****p < 0.0001. i MTT assay showing decreased cell viability in SOX4-knockdown cells (BT474-14, BT474-16) compared to the control (BT474-NT). j Slower in vivo tumor growth from SOX4 knockdown cells compared to the control cells at different time points. The data are presented as the mean ± s.e.m. of 5 mice. k Bioluminescent imaging showing slower tumor growth with SOX4 knockdown
MTT assay showed that SUM149 cells with ectopic SOX4 expression had higher viability than the control, though no difference in viability was observed between the MDA-MB-231-SOX4 cells and their control cells (Fig. 1c and 1d). To confirm the in vivo findings, we injected the SUM149-SOX4 and SUM149-VECT cells into the mammary fat pad of SCID mice. As shown in Fig. 1e, the tumor volume in the SUM149-SOX4 group was bigger than that in the SUM149-VECT group at all the post-injection time points, which was consistent with the bioluminescent imaging results (Fig. 1f).
On the other hand, we knocked down SOX4 using lentivirus-mediated SOX4 shRNA in BT474 cells whose SOX4 expression was the highest among the cell lines tested. Stable SOX4 knockdown was achieved with shRNA-14 and shRNA-16 (Fig. 1g and h). MTT assay reveled that SOX4 knockdown reduced the viability of BT474 cells (Fig. 1i). Likewise, our in vivo experiments also showed slower growth in the xenografts with SOX4 knockdown (Fig. 1j and k).
SOX4 enhances cancer cell metastasis both in vitro and in vivo
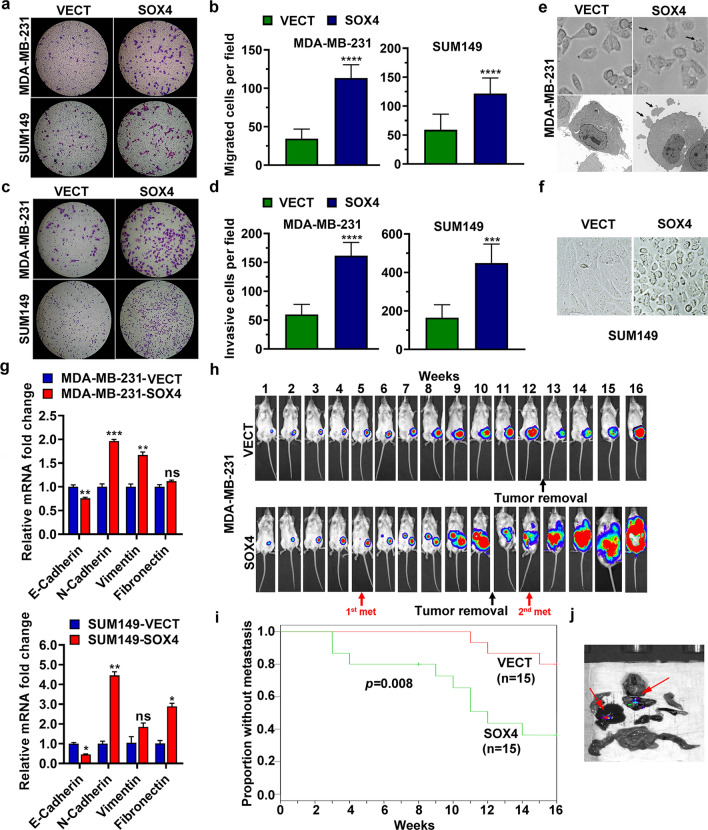
We studied the effect of SOX4 on migration and invasion of breast cancer cells using Transwell migration assay and Matrigel invasion assay, respectively. Ectopic SOX4 expression in SUM149 and MDA-MB-231 cells significantly increased cell migration and invasion (Fig. 2a to d). Interestingly, we observed morphological changes in SOX4-expressing MDA-MB-231 cells which showed more pseudopodia than did the control cells (Fig. 2e). Moreover, there were more intercellular spaces in cultured SUM149 cells with SOX4 expression than in the control cells (Fig. 2f). Since EMT is considered to be an essential change during metastasis, we then evaluated the expression of EMT-related genes E-cadherin, N-cadherin, Vimentin, and Fibronectin, using qRT-PCR [35–38]. Our results showed that, in both MDA-MB-231 cells and SUM149 cells, SOX4 expression reduced the mRNA level of E-cadherin, an epithelial marker, but increased the levels of the three mesenchymal markers, Vimentin, N-cadherin, and Fibronectin (Fig. 2g).
Fig. 2.
SOX4 enhances the cancer cell metastasis both in vitro and in vivo. a Photographs showing Transwell migration assay. b Histograms of the Transwell migration assay results. The data are presented as the mean ± s.d. of triplicate wells. Unpaired two-tailed t-test: *p < 0.05. c Photographs showing Matrigel invasion assay. d Histograms of the Matrigel invasion assay results. The data are presented as the mean ± s.d. of triplicate wells. Unpaired two-tailed t-test: *p < 0.05, ***p < 0.001. e Phase contrast (upper) and electronic microscopic (lower) photographs showing pseudopodia (arrows) in MDA-MB-231 cells. f Phase contrast photographs showing intercellular space in day-4 culture of SUM149 cells. g qRT-PCR analysis of EMT-related genes in MDA-MB-231 (upper panel) and SUM149 (lower panel) cells. The data are presented as the mean ± s.d. of triplicate wells. Unpaired two-tailed t-test: *p < 0.05, **p < 0.01, ***p < 0.001. h Bioluminescent imaging showing tumor growth and metastasis in vivo. 5 × 106 cells (MDA-MB-231-VECT or MDA-MB-231-SOX4) were injected into the mammary fat pad of SCID mice. The primary tumors were removed when their size exceeded 2 cm × 2 cm. i Kaplan and Meier analysis of tumor metastasis for 15 pairs of mice. j Bioluminescent image for metastasis in various organs of a mouse with SOX4-expressing tumor
We further investigated the effect of SOX4 on metastasis in vivo by examining metastasis in tumor bearing mice. We chose MDA-MB-231 cells to study the effect of SOX4 on metastasis in vivo with the view that the observed effect on metastasis would not have been due to an advantage in cell viability since there was no significant increase in cell viability of these cells compared to that of the control cells (Fig. 1d). A representative pair of mice was shown in Fig. 2h; bioluminescent imaging first detected metastasis at the 5th week in the mouse with a tumor derived from the MDA-MB-231-SOX4 cells. After the primary tumor was removed due to oversize, a second metastasis was detected at the 12th week. In contrast, no metastasis was seen in the control mouse with an MDA-MB-231-VECT tumor during the whole period of this experiment. A total of 15 pairs of mice was tested and the time to metastasis was much shorter in the SOX4-expression group than in the control group (p = 0.008, log-rank test, Fig. 2i). Finally, mice were euthanized at the end of 16th week and metastases in various organs were examined by bioluminescent imaging (Fig. 2j). Lung metastases was seen in 87% (13/15) of mice in the group with ectopic SOX4-expression, but only in 47% (7/15) of mice in the control group (p = 0.05, Fisher’s exact test). Liver metastasis was found in 20% (3/15) of mice in the SOX4-expressing group, whereas no liver metastases was observed in the control group.
SOX4 binds to the promoter of the CXCR7 gene and activates its transcription
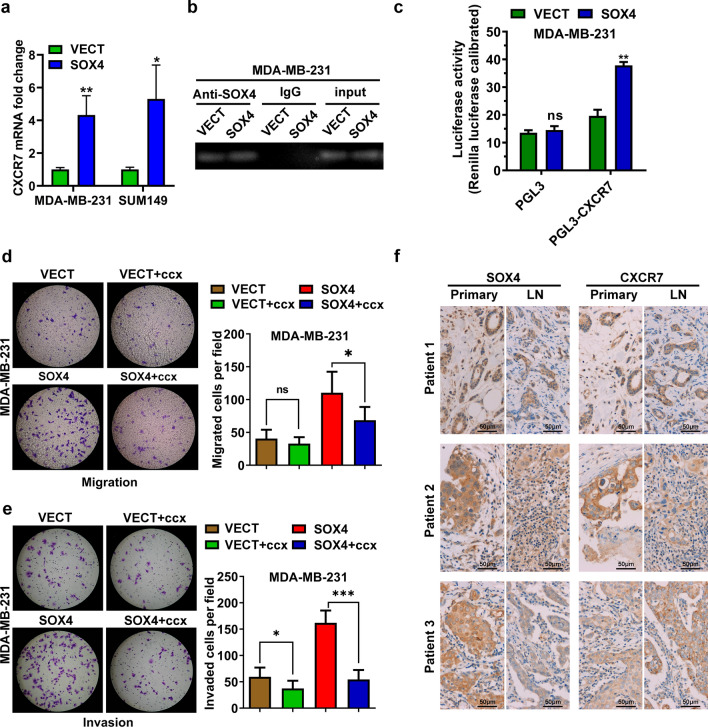
By using gene expression microarray, we found that the expression of CXCR7 was significantly upregulated in SOX4-expressing cells, which was confirmed by qRT-PCR (Fig. 3a). Examination of the CXCR7 gene promoter revealed two DNA sequences, AACAAAG and TACAAAG, which are consistent with the consensus SOX4 binding motif A/T A/T CAA A/T G [39]. We then used chromatin immunoprecipitation assay and amplified, with primers for CXCR7, a 384 bp fragment from MDA-MB-231 cell DNA co-precipitated with the SOX4 antibody (Fig. 3b). No amplified fragment was obtained when non-specific IgG was used as the capture antibody. Moreover, the dual-luciferase reporter assay showed that ectopic SOX4 expression significantly increased the activity of CXCR7 promoter (Fig. 3c). We further found that CXCR7 inhibitor CCX771 was able to reverse the enhanced cell migration and invasion due to ectopic SOX4 expression (Fig. 3d and e). These results indicated that SOX4 directly binds to the promoter of the CXCR7 gene and upregulates its transcription, which might result in an increased metastatic capacity.
Fig. 3.
SOX4 binds to the promoter of the CXCR7 gene and activates its transcription. a Increased amount of CXCR7 mRNA in SOX4-expressing MDA-MB-231 and SUM149 cells. The data are presented as the mean ± s.d. of triple wells. Unpaired two-tailed t-test: *p < 0.05, **p < 0.01. b CXCR7 gene fragments amplified from pull-down samples of the chromatin immunoprecipitation assay using the SOX4 antibody and non-specific IgG, and the input samples. c Dual-luciferase reporter assay showing increased CXCR7 promoter-specific reporter activity associated with SOX4 overexpression. d and e CXCR7 inhibitor CCX771 (ccx) reversed the enhancing effect of SOX4 on migration and invasion. The data are presented as the mean ± s.d. of triplicate wells. Unpaired two-tailed t-test: *p < 0.05, ***p < 0.001. f IHC analysis of SOX4 and CXCR7 expression in human breast primary tumors (Primary) and lymph node metastatic foci (LN)
We performed IHC and detected the SOX4 and CXCR7 expression in all the 36 pairs of human primary breast carcinomas and their corresponding lymph node metastases in a tissue chip slide (Fig. 3f). The expression of SOX4 and CXCR7 was strongly positive in 28/36 and 23/36 of the primary tumors and in 27/36 and 23/36 of the metastatic lymph nodes, respectively, although the IHC staining intensity was not significantly different between the primary tumors and lymph node metastases..
SOX4 increases the susceptibility of breast cancer cells to paclitaxel
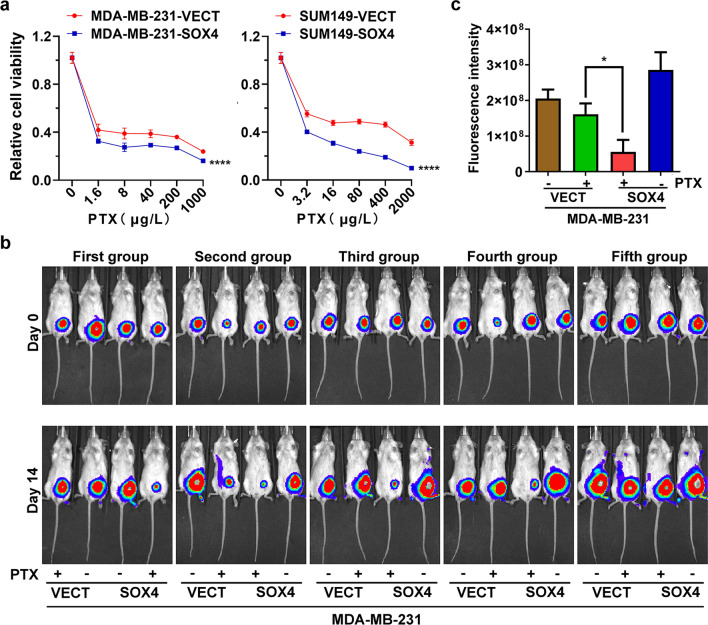
To explore the potential role of SOX4 in breast cancer treatment, we investigated its effect on cancer cell resistance to several commonly used chemotherapeutic agents including ecarboplatin, adriamycin, 5-fluorouracil, and paclitaxel. There was no significant difference in response to ecarboplatin, adriamycin, or 5-fluorouracil between SOX4-expressing cells and the control cells in both MDA-MB-231 and SUM149 (data not shown). However, we observed that SOX4 overexpression significantly increased the susceptibility of MDA-MB-231 and SUM149 cells to paclitaxel (Fig. 4a). On the contrary, SOX4 knockdown increased the IC50 (Additional file 1: Figure S1). In addition, bioluminescent imaging at day 14 showed that treatment with paclitaxel suppressed the tumor growth in vivo (Fig. 4b). Moreover, the fluorescence intensity reduced greater in the SOX4-expression group than the control group (Fig. 4c), indicating that SOX4 increased the susceptibility of MDA-MB-231 cells to paclitaxel.
Fig. 4.
SOX4 increases the susceptibility of breast cancer cells to paclitaxel. a MTT assay showing cell viability in response to paclitaxel (PTX) after 48 h treatment. The data are presented as the mean ± s.d. of sextuple wells. Unpaired two-tailed t-test: ***p < 0.001. b Bioluminescent imaging of tumor growth 14 days after treatment with Paclitaxel. 5 × 106 cells (MDA-MB-231-VECT or MDA-MB-231-SOX4) were injected into the mammary fat pad of SCID mice in 5 groups. Three weeks later (day 0), the tumor-bearing mice were treated with paclitaxel (PTX) or PBS. The treatment was repeated at day 7 and the tumors were examined using bioluminescent imaging at day 14. c Statistical analysis of results in b. The data are presented as the mean ± s.d. of 5 mice. Unpaired two-tailed t-test: *p < 0.05
Discussion
Breast cancer is the most prevalent cancer and the leading cause of cancer-related death in women [1, 40]. Distant metastasis usually leads to the failure of clinical therapy. In this study, we found that transcription factor SOX4 promotes tumor growth and metastasis, likely through binding to the promoter of CXCR7 gene and activating its transcription. CXCR7 was reported to enhance the growth of MDA-MB-435 breast cancer cells, and the proliferation as well as metastasis of lung cancer cells [26]. It was also found to facilitate breast cancer cells to cross the blood–brain barrier, leading to brain metastasis [41]. Our results suggested that CXCR7 is a downstream gene of SOX4. Importantly, as shown by our IHC studies, both SOX4 and CXCR7 are strongly expressed in human primary breast cancer cells and the metastatic foci in lymph nodes. Moreover, treatment of MDA-MB-231 cells with CCX771, a small molecule inhibitor of CXCR7, could effectively reverse the enhanced migration and invasion due to ectopic expression of SOX4. These findings suggested that inhibition of SOX4/CXCR7 pathway may reduce distant metastasis in the treatment for human breast cancer.
Interestingly, cells with ectopic SOX4 expression have more pseudopodia and maintain more intercellular space in culture, changes that are suggestive of EMT. To date, it is well accepted that EMT phenomenon is the favored explanation of distant metastasis of breast cancer [42]. EMT enhances cell motility and helps the release of breast cancer cells from the primary tumor and the establishment of metastatic colonies at distant sites [43]. Previous studies have reported that CXCR7 promotes EMT by upregulating TGF-β1 [29–31]. We determined the expression levels of EMT-related genes in SOX4-expressing cells and found that the expression of three mesenchyme markers Vimentin, N-cadherin, and Fibronectin were upregulated, and that of the epithelial marker E-cadherin was downregulated. These findings indicated that SOX4/CXCR7 axis may promote EMT and play an important role in the metastasis of breast cancer.
One of the important findings in this study is that ectopic expression of SOX4 increases the susceptibility of breast cancer cells to paclitaxel, suggestive of the potential use of paclitaxel in breast cancer treatment if the expression level of SOX4 in the tumor is high. Of note, the MDA-MB-231 and SUM149 cells we tested are triple negative breast cancer cells. Currently, compared to hormonal receptor positive and HER2 positive breast cancers, triple negative cancers lack effective treatment [44]. Our finding may provide a new therapeutic strategy for triple negative breast cancer based on the expression of SOX4. Nevertheless, further studies are required to investigate the status of SOX4 expression in triple negative tumors and to directly test the efficacy of Paclitaxel for their treatment.
In conclusion, our study revealed that SOX4 promotes the growth and metastasis of breast cancer. SOX4/CXCR7 may serve as potential therapeutic targets for the treatment of breast cancer. Paclitaxel may be a good therapeutic option if the expression level of SOX4 in the tumor is high.
Supplementary information
Additional file 1: Figure S1. SOX4 knockdown increases IC50 of paclitaxel in BT474.
Acknowledgements
The authors gratefully acknowledge financial supports from the National Natural Science Foundation of China to JZ (Grant No. 81301809), Zhejiang Provincial Natural Science Foundation to ZL (Grant No. LY15H160012), Zhejiang Provincial Medical and Health Major Science and Technology Plan Project to LT (Grant No. 2017209495), and Science and Technology Major Projects of Ningbo to WW (Grant No. 2015C50003).
Authors’ contributions
JZ, CX, ZF, YG, BS, ZL, YL, XF, and WW carried out the part of experiments and performed the statistical analysis; JZ collected the data and drafted the manuscript; XS and LT conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China to JZ (Grant No. 81301809), Zhejiang Provincial Natural Science Foundation to ZL (Grant No. LY15H160012), Zhejiang Provincial Medical and Health Major Science and Technology Plan Project to LT (Grant No. 2017209495), and Science and Technology Major Projects of Ningbo to WW (Grant No. 2015C50003).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
All animal experiments were approved by Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaoping Sun, Email: xsun@mdanderson.org.
Lisong Teng, Email: lsteng@zju.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-020-01568-2.
References
- 1.Pinheiro CPO, Silva RMD, Brasil CCP, Bezerra IC, Cavalcante ANM, Alexandre AV, Cruz CA. Procrastination in the early detection of breast cancer. Revista brasileira de enfermagem. 2019;72(suppl 3):227–234. doi: 10.1590/0034-7167-2018-0547. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Wu W, Qian L, Chen X, Ding B. Prognostic significance of CXCL12, CXCR4, and CXCR7 in patients with breast cancer. Int J Clin Exp Pathol. 2015;8(10):13217–13224. [PMC free article] [PubMed] [Google Scholar]
- 4.Li CH, Karantza V, Aktan G, Lala M. Current treatment landscape for patients with locally recurrent inoperable or metastatic triple-negative breast cancer: a systematic literature review. Breast Cancer Res. 2019;21(1):143. doi: 10.1186/s13058-019-1210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32(29):3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 6.She ZY, Yang WX. SOX family transcription factors involved in diverse cellular events during development. Eur J Cell Biol. 2015;94(12):547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Can Res. 2009;69(23):9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maschhoff KL, Anziano PQ, Ward P, Baldwin HS. Conservation of Sox4 gene structure and expression during chicken embryogenesis. Gene. 2003;320:23–30. doi: 10.1016/j.gene.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380(6576):711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 11.Mallampati S, Sun B, Lu Y, Ma H, Gong Y, Wang D, Lee JS, Lin K, Sun X. Integrated genetic approaches identify the molecular mechanisms of Sox4 in early B-cell development: intricate roles for RAG1/2 and CK1epsilon. Blood. 2014;123(26):4064–4076. doi: 10.1182/blood-2013-12-543801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun B, Mallampati S, Gong Y, Wang D, Lefebvre V, Sun X. Sox4 is required for the survival of pro-B cells. J Immunol. 2013;190(5):2080–2089. doi: 10.4049/jimmunol.1202736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno CS: SOX4: The unappreciated oncogene. Seminars in cancer biology 2019. [DOI] [PMC free article] [PubMed]
- 14.Ma H, Mallampati S, Lu Y, Sun B, Wang E, Leng X, Gong Y, Shen H, Yin CC, Jones D, et al. The Sox4/Tcf7l1 axis promotes progression of BCR-ABL-positive acute lymphoblastic leukemia. Haematologica. 2014;99(10):1591–1598. doi: 10.3324/haematol.2014.104695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai CN, Yu SC, Lee CW, Pang JS, Wu CH, Lin SE, Chung YH, Tsai CL, Hsieh SY, Yu MC. SOX4 activates CXCL12 in hepatocellular carcinoma cells to modulate endothelial cell migration and angiogenesis in vivo. Oncogene. 2020;39(24):4695–4710. doi: 10.1038/s41388-020-1319-z. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Zhao H, Lu J, Yin J, Zang L, Song N, Dong R, Wu T, Du X. Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma. Diagn Pathol. 2012;7:41. doi: 10.1186/1746-1596-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Zhang J, Yang X, Chang YW, Qi M, Zhou Z, Zhang J, Han B. SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro. Prostate Cancer Prostatic Dis. 2013;16(4):301–307. doi: 10.1038/pcan.2013.25. [DOI] [PubMed] [Google Scholar]
- 18.Bilir B, Osunkoya AO. Wiles WGt, Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD, Moreno CS: SOX4 Is Essential for Prostate Tumorigenesis Initiated by PTEN Ablation. Cancer Res. 2016;76(5):1112–1121. doi: 10.1158/0008-5472.CAN-15-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julian LM, McDonald AC, Stanford WL. Direct reprogramming with SOX factors: masters of cell fate. Curr Opin Genet Dev. 2017;46:24–36. doi: 10.1016/j.gde.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72(17):4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 21.Ali S, Lazennec G. Chemokines: novel targets for breast cancer metastasis. Cancer Metastasis Rev. 2007;26(3–4):401–420. doi: 10.1007/s10555-007-9073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshie O. Chemokine receptors as therapeutic targets. Nihon Rinsho Men'eki Gakkai kaishi. 2013;36(4):189–196. doi: 10.2177/jsci.36.189. [DOI] [PubMed] [Google Scholar]
- 23.Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J. 2018;285(16):2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Do HTT, Lee CH, Cho J. Chemokines and their receptors: multifaceted roles in cancer progression and potential value as cancer prognostic markers. Cancers. 2020;12:2. doi: 10.3390/cancers12020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin Q, Zhang N, Yu HB, Zhang Q, Cui YF, Zhang CS, Ma Z, Yang Y, Liu W. CXCR7/CXCL12 axis is involved in lymph node and liver metastasis of gastric carcinoma. World J Gastroenterol. 2017;23(17):3053–3065. doi: 10.3748/wjg.v23.i17.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao Z, Luker KE, Summers BC, Berahovich R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A, Luker GD, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci USA. 2007;104(40):15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maishi N, Ohga N, Hida Y, Akiyama K, Kitayama K, Osawa T, Onodera Y, Shinohara N, Nonomura K, Shindoh M, et al. CXCR7: a novel tumor endothelial marker in renal cell carcinoma. Pathol Int. 2012;62(5):309–317. doi: 10.1111/j.1440-1827.2012.02792.x. [DOI] [PubMed] [Google Scholar]
- 28.Gentilini A, Caligiuri A, Raggi C, Rombouts K, Pinzani M, Lori G, Correnti M, Invernizzi P, Rovida E, Navari N, et al. CXCR7 contributes to the aggressive phenotype of cholangiocarcinoma cells. Biochim Biophys Acta. 2019;1865(9):2246–2256. doi: 10.1016/j.bbadis.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Guan S, Zhou J. CXCR7 attenuates the TGF-β-induced endothelial-to-mesenchymal transition and pulmonary fibrosis. Mol Biosyst. 2017;13(10):2116–2124. doi: 10.1039/C7MB00247E. [DOI] [PubMed] [Google Scholar]
- 30.Kim N, Ryu H, Kim S, Joo M, Jeon HJ, Lee MW, Song IC, Kim MN, Kim JM, Lee HJ. CXCR7 promotes migration and invasion in head and neck squamous cell carcinoma by upregulating TGF-β1/Smad2/3 signaling. Sci Rep. 2019;9(1):18100. doi: 10.1038/s41598-019-54705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu YC, Tang SJ, Sun GH, Sun KH. CXCR7 mediates TGFβ1-promoted EMT and tumor-initiating features in lung cancer. Oncogene. 2016;35(16):2123–2132. doi: 10.1038/onc.2015.274. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Yang CQ, Gao Y, Wang C, Zhang CL, Zhou XH. Knockdown of CXCR7 inhibits proliferation and invasion of osteosarcoma cells through inhibition of the PI3K/Akt and β-arrestin pathways. Oncol Rep. 2014;32(3):965–972. doi: 10.3892/or.2014.3290. [DOI] [PubMed] [Google Scholar]
- 33.Li XX, Zheng HT, Huang LY, Shi DB, Peng JJ, Liang L, Cai SJ. Silencing of CXCR7 gene represses growth and invasion and induces apoptosis in colorectal cancer through ERK and β-arrestin pathways. Int J Oncol. 2014;45(4):1649–1657. doi: 10.3892/ijo.2014.2547. [DOI] [PubMed] [Google Scholar]
- 34.Ling X, Ma G, Sun T, Liu J, Arlinghaus RB. Bcr and Abl interaction: oncogenic activation of c-Abl by sequestering Bcr. Cancer Res. 2003;63(2):298–303. [PubMed] [Google Scholar]
- 35.Wang W, Yi M, Zhang R, Li J, Chen S, Cai J, Zeng Z, Li X, Xiong W, Wang L, et al. Vimentin is a crucial target for anti-metastasis therapy of nasopharyngeal carcinoma. Mol Cell Biochem. 2018;438(1–2):47–57. doi: 10.1007/s11010-017-3112-z. [DOI] [PubMed] [Google Scholar]
- 36.Canel M. Serrels A Fau - Frame MC, Frame Mc Fau - Brunton VG, Brunton VG: E-cadherin-integrin crosstalk in cancer invasion and metastasis. Cancer Res. 2013;126(2):393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- 37.Mrozik KM, Blaschuk OW, Cheong CM, Zannettino ACW. Vandyke KA-OhooX: N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer. 2018;18(1):939. doi: 10.1186/s12885-018-4845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li CL, Yang D, Cao X, Wang F, Hong DY, Wang J, Shen XC, Chen Y. Fibronectin induces epithelial-mesenchymal transition in human breast cancer MCF-7 cells via activation of calpain. Oncol Lett. 2017;13(5):3889–3895. doi: 10.3892/ol.2017.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27(42):5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 40.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 41.Salmaggi A, Maderna E, Calatozzolo C, Gaviani P, Canazza A, Milanesi I, Silvani A, DiMeco F, Carbone A, Pollo B. CXCL12, CXCR4 and CXCR7 expression in brain metastases. Cancer Biol Ther. 2009;8(17):1608–1614. doi: 10.4161/cbt.8.17.9202. [DOI] [PubMed] [Google Scholar]
- 42.Kotiyal S, Bhattacharya S. Breast cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res Commun. 2014;453(1):112–116. doi: 10.1016/j.bbrc.2014.09.069. [DOI] [PubMed] [Google Scholar]
- 43.Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11(1):28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23(29):7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. SOX4 knockdown increases IC50 of paclitaxel in BT474.
Data Availability Statement
Not applicable.