Abstract
There is an urgent need to understand the behavior of the novel coronavirus (SARS-COV-2), which is the causative agent of COVID-19, and to develop point-of-care diagnostics. Here, a glyconanoparticle platform is used to discover that N-acetyl neuraminic acid has affinity toward the SARS-COV-2 spike glycoprotein, demonstrating its glycan-binding function. Optimization of the particle size and coating enabled detection of the spike glycoprotein in lateral flow and showed selectivity over the SARS-COV-1 spike protein. Using a virus-like particle and a pseudotyped lentivirus model, paper-based lateral flow detection was demonstrated in under 30 min, showing the potential of this system as a low-cost detection platform.
Short abstract
The spike-protein from SARS-COV-2 is shown to bind sialic acids, which is exploited to assemble a lateral flow diagnostic tool, using glycans rather than antibodies, as the recognition unit.
In December 2019, a novel zoonotic coronavirus (SARS-COV-2), reported in Wuhan (China), led to a pandemic of the respiratory disease COVID-19.1 There are currently few therapeutics and no vaccine. Diagnostics, surveillance, and case isolation are the primary tools for controlling the spread of the virus and driving down the basic reproduction (R0) value. Following the successful genome sequencing of SARS-COV-2, RT-PCR-based (reverse transcription polymerase chain reaction) diagnostics were rapidly developed. These require dedicated laboratory facilities, trained personnel, and do not provide an instant output, and while highly specific, false-negative results are reported at 3% versus chest CT scans.2 There are also reports of conflicting RT-PCR results in samples from the same patient.3,4 The results can depend on the sampling location (i.e., throat versus lower respiratory tract),5 and the false-negative rate is highest during the early stages of infection.6
An alternative detection platform to RT-PCR is the lateral flow device (LFD), such as the home-pregnancy test,7 which typically uses antibodies as the detection units immobilized to both the stationary phase (e.g., nitrocellulose paper) and the mobile phase (e.g., gold nanoparticle), forming a “sandwich” with the analyte. Test lines show a positive (red line) response by eye. Such LFDs require little or no clinical infrastructure or training, and they can be used in the patient’s home. The cost-effectiveness of these inexpensive devices has been demonstrated by various studies of malaria rapid diagnostic tests8,9 and were found to compare well to the more expensive RT-PCR for Ebola diagnosis.10 In addition to antibodies, other biological recognition units such as nucleic acids,11 glycans, and lectins12 could be used. Glycans have not been widely applied in lateral flow13 but offer new opportunities and advantages compared with antibody-based systems. Glycans have reduced cold chain needs compared with proteins14 and are thus suited to low resource, triage, or emergency settings. A further benefit of glycans, as the capture unit, would be the detection of intact viruses. For SARS-COV-2, viral RNA (e.g., from a positive RT-PCR result) is detected past the point where patients are no longer infectious, resulting in extended hospital stays.15
Glycans (carbohydrates) direct myriad binding and recognition events in biology from cell–cell communication to being markers of disease. Analysis of the 2009 swine (zoonotic) influenza pandemic showed that porcine viral hemagglutinins, which normally bind α2,3′-linked sialic acids, switched to binding α2,6′-linked sialic acids found in human respiratory tracts.16,17 This demonstrates the importance of glycan-binding during infection. This switch in glycan specificity has enabled the establishment of glycan-functional biosensors to rapidly identify strains without using nucleic acid based detection methods.18,19 All coronaviruses display homotrimers of spike glycoproteins on their surface. Sialic acid binding by the S1 spike protein subunits is crucial for coronavirus to engage host cells, while the S2 domain initiates viral fusion.20 Tortorici et al. showed the structural basis for 9-O-acetylated sialic acid binding to a human coronavirus (strain OC43) by cryo-EM, and affinity to this ligand to the HKU1-HE strain, has also been reported.21,22 MERS S1 preferentially binds α2,3′-linked over α2,6′-linked sialic acids but any acetylation of the sialic acids decreases affinity,23 which is distinct from OC43. This evidence shows that sialic acid binding is crucial in coronavirus infection and potentially in its zoonosis to human hosts,23,24 but that the exact glycan partner can vary between strains. It has also emerged that sulfated glycosaminoglycans (including heparin sulfates) bind SARS-COV-2 spike protein, and can inhibit viral entry.25−27 The above examples demonstrate that glycan “anchoring” of coronaviruses may offer opportunities for detection using capture techniques such as LFD.
Individual glycans display low affinity to their protein targets, but this is overcome in nature by multivalent display. Due to the cluster glycoside effect,28 displaying multiple copies of glycans can result in several orders of magnitude enhancement in the observed affinity. This has been widely exploited in materials chemistry29,30 using dendrimers,31,32 peptides/proteins,33 polymers,34,35 and nanoparticles19,36 to generate high avidity.
Here we report the synthesis of polymer-stabilized, multivalent gold nanoparticles bearing sialic acid derivatives and their interaction with the spike glycoprotein from SARS-COV-2. We find that α,N-acetyl neuraminic acid binds the spike glycoprotein and subsequently exploit this interaction as the detection unit in a prototype lateral flow rapid diagnostic, which requires no centralized infrastructure.
Figure 1A shows the sequence alignments between the S1 domains of coronavirus spike proteins from MERS and SARS-COV-2. There is some conservation of the sialic acid binding site, notably residues His69 and Phe79. Figure 1B,C shows models constructed from the Cryo-EM structure of SARS-COV,37 with missing loops and the α2,3′-sialyllactose modeled into it using the Cryo-EM structure of MERS.38 The modeling data suggests that Arg21 and Leu24 coordinates the glycan, while Gln23 may also have a role in binding. Adjacent to the hairpin containing His69 and Phe79, it seems likely that Arg246 also has a role in coordination. The limited conservation of the sialic acid binding groove sequence (S21, Supporting Information (SI)) is in marked contrast to the entirety of the spike S protein, which is often highly conserved between coronaviruses.39 In the MERS sialic acid binding site in complex with α2,3′-sialyllactose, only the neuraminic acid unit, not the lactose, is engaged. This is in contrast to influenza hemagglutinins, which contact the galactose residues.40 This evidence suggests that N-acetyl neuraminic acid is a reasonable target for SARS-COV-2 binding (also hypothesized here41) and hence a potential capture ligand for a new “glyco-LFD” diagnostic device.
Figure 1.
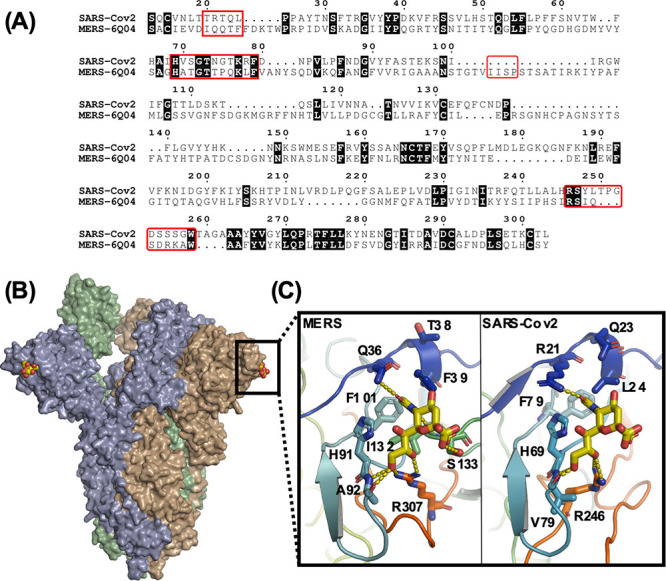
(A) Sequence alignment between the S1 domains of the SARS-COV-2 and MERS spike proteins. Regions important for sialic acid binding are highlighted by red boxes; (B) Model showing the hypothesized sialic acid binding sites (yellow CPK coloring) for the SARS-COV-2 spike protein trimer; (C) A comparison between the sialic acid binding sites from MERS (PDB entry 6Q04) and the SARS-COV-2 model (PDB entry 6VSB) in complex with a2,3′-sialyllactose.
Figure 2A shows a design schematic for a glyco-LFD. Typical LFDs use antibodies, but here the glycan is immobilized (as a BSA-glycoconjugate) on the test strip and also in the mobile phase onboard gold nanoparticles, providing multivalency (and hence affinity), for dissecting SARS-COV-2 binding and for the LFD. Our nanoparticle design concept uses telechelic polymer tethers which conjugate the glycans, by displacement of an ω-terminal pentafluorophenyl (PFP) group, and immobilization onto gold particles via the α-terminal thiol. Poly(N-hydroxyethyl acrylamide), PHEA, was chosen as the polymer to give colloidally stable particles and as an acrylamide, it is not easily hydrolyzable unlike acrylates for example, Figure 2B and Table 1.36,42 PHEA was synthesized using RAFT (reversible addition–fragmentation chain transfer) polymerization resulting in dispersities below 1.3. The PHEA lengths were selected on the basis of performance in initial lateral flow screening assays (data not shown) and from reports of their colloidal stability.36,42 Amino-glycans were synthesized by reduction of anomeric azides and subsequently conjugated to the PHEAs by displacement of the PFP group, which was confirmed by 19F NMR. Polymers were then assembled onto citrate-stabilized gold nanoparticles and excess ligand removed by centrifugation/resuspension cycles. The nanoparticles were characterized by UV–vis, dynamic light scattering (DLS), transmission electron microscopy (TEM), and XPS (x-ray photoelectron spectroscopy) to confirm surface coating (Table 2 and SI). Following observations via DLS and UV–vis that 16 nm sialyllactose particles were less stable than 35 nm particles, the latter were selected for initial glycan-binding assays.
Figure 2.
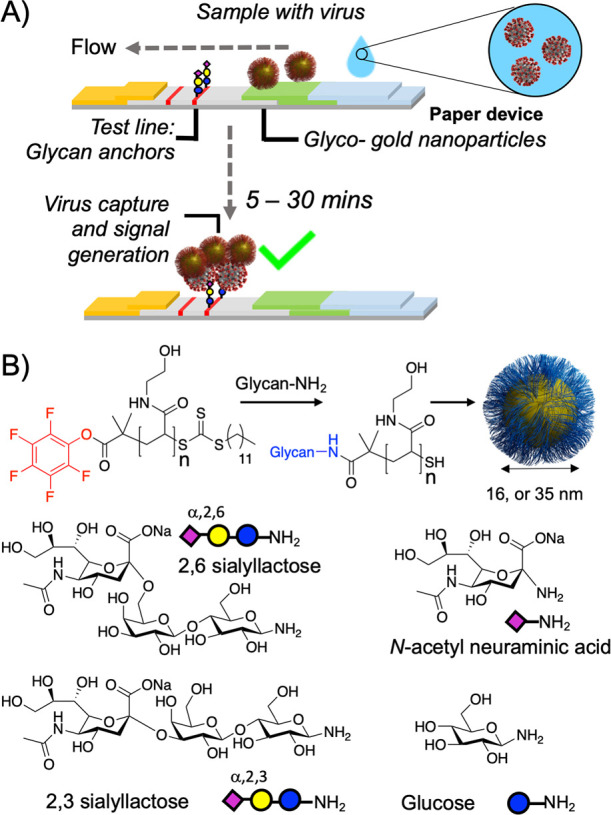
Design concept for glyco-lateral flow devices. (A) Lateral flow assay for virus, using glycan capture units; (B) Synthetic procedure for glyconanoparticles.
Table 1. Polymer Characterization.
| code | M:CTA (−) | MN(theo)a (g mol–1) | MN(SEC)b (g mol–1) | MN(NMR)c (g mol–1) | Đb (−) |
|---|---|---|---|---|---|
| PHEA40 | 20 | 2800 | 5100 | 5000 | 1.19 |
| PHEA50 | 25 | 3400 | 6400 | 5500 | 1.27 |
| PHEA58 | 30 | 4000 | 7200 | 6700 | 1.26 |
Estimated from [M]:[CTA].
From DMF SEC versus PMMA standards.
1H NMR end-group analysis.
Table 2. Nanoparticle Characterization.
| code | UVmaxa (nm) | ASPR/A450b (−) | Dh(UV)c (nm) | Dh (DLS)d (nm) | D(TEM) (nm) |
|---|---|---|---|---|---|
| AuNP16 | 519 | 1.64 | 16 | 20.7 ± 0.8 | 14 ± 2 |
| NeuNAc-PHEA40AuNP16 | 527 | 1.66 | 16 | 40.9 ± 0.5 | (−) |
| NeuNAc-PHEA50AuNP16 | 526 | 1.68 | 18 | 44.2 ± 0.8 | (−) |
| AuNP35 | 526 | 1.91 | 35 | 34.5 ± 0.5 | 35 ± 3 |
| NeuNAc-PHEA40AuNP35 | 531 | 1.98 | 45 | 46.2 ± 0.7 | (−) |
| NeuNAc-PHEA50AuNP35 | 531 | 1.99 | 45 | 55.3 ± 0.8 | (−) |
SPR absorption maximum.
Absorbance ratio of SPR to 450 nm.
Estimated from UV–vis;43
From dynamic light scattering.
From TEM, from an average of >100 particles, showing ± SD.
With the glyconanoparticles on hand, recombinant S1 subunit (SARS-COV-2,S1) spike protein was immobilized onto biolayer interferometry (BLI) sensors19 to replicate a lateral flow situation, which is the primary aim of this work. Since there are 22 N-linked glycans per protein which are not present in bacteria-expressed protein,44 we used protein expressed in mammalian cells (HEK) to ensure glycosylation. Figure 3A shows α,NeuNAc-AuNPs bind to a greater extent compared to both sialyllactose isomers (α2,3′/α2,6′) and the monosaccharide control (glucose). X-ray photoelectron spectroscopy analysis (SI) of these particles revealed that the NeuNAc/Glc monosaccharide-terminated polymers had a higher grafting density than the sialyllactose trisaccharides by a ratio of 2:1 (35 nm)/ 3:1 (16 nm) because of the difference in glycan size. It is therefore important to note that this data does not rule out sialyllactose binding (and indeed, in LFD, we do see binding), but that in this system, NeuNAc gave the strongest response and consequently was taken forward. Thermal shift assays further confirmed that NeuNAc bound selectively over galactose and glucose, and preliminary STD (saturation transfer difference) NMR spectroscopy showed NeuNAc binding also (both in SI). While outside the scope of this work, the identification of the sialic acid binding function of the spike protein may provide fundamental guidance as to how the virus engages host cells, or is processed within them, and this (to the best of our knowledge) is the first report of this matter.
Figure 3.
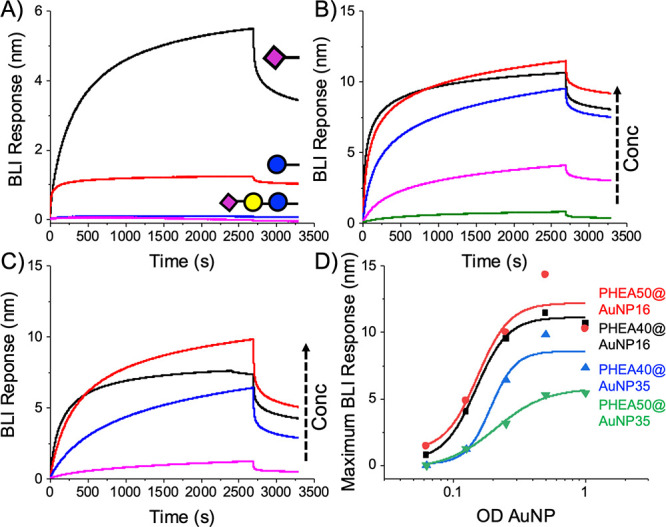
Biolayer interferometry analysis of SARS-COV-2 spike protein with glyconanoparticles. (A) Screening using PHEA50@AuNP35 at OD = 1; Dose-dependent binding of NeuNAc-PHEA50 using (B) @AuNP16 and (C) @AuNP35. OD = 1 (−), 0.5(-), 0.25(-), 0.125(-); (D) Binding curves.
The next step was to evaluate the impact of particle size on binding. Both 16 and 35 nm gold (relevant diameters for LFDs) NeuNAc particles were used to interrogate SARS-COV-2,S1, Figures 3B,C. End-point dose dependency (Figure 3D) showed similar binding trends for both particles, with an apparent Kd ∼ 1 nM, noting that for multivalent systems exact Kd’s cannot be extracted. The plots are shown in terms of OD (SPR absorption maximum) as this is standard for AuNP concentration.
With the successful identification of NeuNAc as a target ligand, its application as the capture unit in lateral flow was examined. The performance of an LFD depends on not only the affinity of the capture ligand (NeuNAc) but also the flow of the particles. “Half” lateral flow assays (Figure 4A) were established to optimize the particles. The negative test line was (commercial) 2,3′-sialyllactose-BSA, which the glyco-nanoparticle should not bind to (to avoid false positives in “full” lateral flow where it would serve to capture viral antigen). The positive control was immobilized SARS-COV-2,S1 (which mimics capture in full lateral flow) and the nanoparticles were flowed over them (original lateral flow strips are in SI). Pleasingly, all particles bound SARS-COV-2,S1 showing this detection method is valid. NeuNAcPHEA50@AuNP35 gave the strongest signal-to-noise compared to other particles, owing to their lower background compared to 16 nm particles (noting that the 16 nm did give a strong signal too). The 16 nm also showed some binding to the 2,3′-sialyllactose-BSA control however. Hence, NeuNAcPHEA50@AuNP35 particles were used from this point onward, Figure 4B/C. Both α2,3′- and α2,6′-sialiyllactosamine particles were also tested, and on larger AuNPs (55–70 nm) and longer polymers (PHEA72) too, but gave no improvement over NeuNAcPHEA50@AuNP35. Blocking of the NeuNAc particles with BSA before running was also explored in an attempt to further reduce the background, as is common in LFDs. BSA blocking did not improve the performance of the NeuNAc systems, but it did reduce off-target binding in the α2,3′-sialyllactosamine systems tested. Encouraged by these results, the specificity and function of the NeuNAcPHEA50@AuNP35 particles were tested against a panel of test-line immobilized lectins [1 mg·mL–1]. Total signal intensity is plotted in Figure 4D, confirming that NeuNAc-AuNPs have no nonspecific binding. The only lectin that bound was RCA120, which is known to have some affinity toward sialic acids.45 WGA and SNA also have some affinity to sialic acids but did not show signal here, highlighting an advantage (and challenge) of LFD, that glycan presentation is a strong determinant of the signal generation in addition to binding affinity. To test binding specificity in a more challenging scenario, the particles were screened against the spike protein, SARS-COV-2,S1, (the desired target) and also against the S1 spike domain of a previous zoonotic coronavirus SARS-COV-1,24 responsible for the 2003 “SARS” outbreak. As can be seen in Figure 4E, the NeuNAc particle system has a clear preference for SARS-COV-2, demonstrating selectivity in this glyconanoparticle system. While this does not rule out binding, it does show the particles/glycan do not generate sufficient signal against SARS-COV-1. This data further supports the notion that the terminal NeuNAc is the key binding motif.
Figure 4.
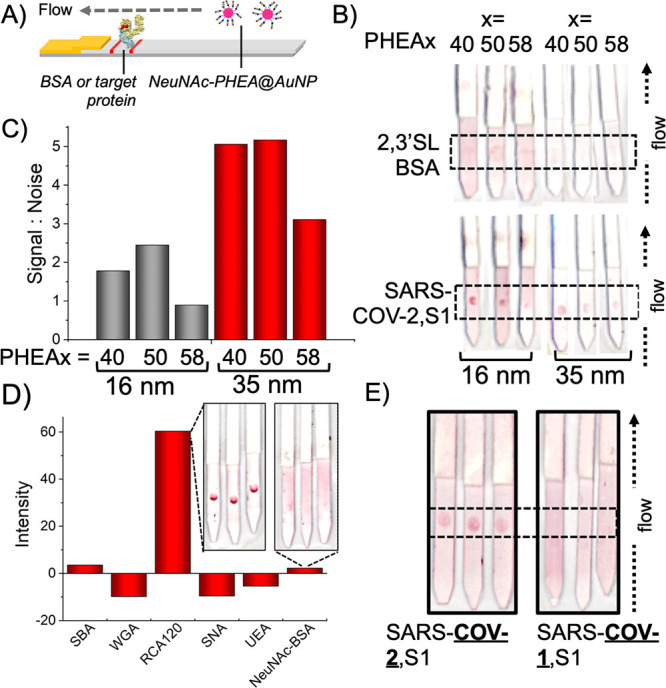
Half lateral flow analysis of NeuNAcPHEAx@AuNPy particles. (A) Half lateral flow assay setup with target protein immobilized on the test line; (B) Effect of polymer chain length and particle size on lateral flow binding; (C) Signal:noise analysis; (D) Selectivity of NeuNAcPHEA50@AuNP35 against a panel of lectins (inset example LFD strips). (E) Selectivity of NeuNAcPHEA50@AuNP35 against S1 protein from different coronavirus strains. Data is the mean from 3 repeats. Original LFD strips are in the SI. Test lines are within the dashed-line box. 2,3′SL-BSA = 2,3′-sialyllactose-functionalized BSA.
To explore the detection limits and specificity of this system, NeuNAc (positive) and galactose (negative) nanoparticles were screened against a dilution series of SARS-COV-2,S1 (Figure 5A,B). At the highest concentration (0.5 mg·mL–1) galactose particles showed weak binding to SARS-COV-2,S1. NeuNAc particles showed significantly stronger binding, with an apparent limit of detection below 8 μg·mL–1 or 8 nM. Encouraged by successful binding, a dipstick sandwich assay was established where the analyte was added to the gold particle solution, rather than dried onto the nitrocellulose paper. The test line was NeuNAc-BSA (validated to capture the particles by BLI, SI) and RCA120 as a control line, which is essential in lateral flow devices to ensure each device is functional. To mimic the virus in a model system without cell debris, which may complicate initial LFD development, the spike glycoprotein was immobilized onto 100 nm polystyrene nanoparticles which match the diameter of the coronavirus. Figure 5C shows the results of testing this system (original lateral flow strips with no image enhancement are in the SI). In this system, 2,3′-sialyllactose-BSA is the test (capture) line as the data in Figure 4 confirmed no nonspecific binding by the nanoparticles to this line. The lateral flow devices could clearly detect the virus-like particles at a concentration of just 5 μg·mL–1 (5 nM) protein, which is in line with the detection limits from Figure 5B. Controls using naked polystyrene colloids showed no binding to the test line, ruling out nonspecific interactions, and a control (with no polystyrene analyte) only showed control line binding. The resolution of the test spots could be further enhanced using a silver-staining protocol,46 which improves the “by eye” detection (Figure 5C). An additional control of two influenza strains (which bind sialyllactoses) were shown to have little off-target binding as influenza hemagglutinins require the galactose linker in addition to the sialic acid, for strong binding (SI).40 As a final proof of concept, SARS-COV-2 spike protein pseudotyped lentivirus were tested in half-lateral flow. Pseudovirus was applied to the test line and ran in a lateral flow cassette (SI for full photos). Using NeuNAc particles, detection at 1.5 × 104 transduction units/mL was achieved. Galactose-functional particles failed to detect the virus, confirming the role of sialic acid binding. Direct comparison of transduction units/mL to viral load (typical copies/mL) is not possible, but values as high as 108 copies·mL–1 are reported from COVID positive swabs,15 suggesting this method may have relevant detection limits. A hybrid LFD using antibody capture in one component may also be possible to further improve this.13
Figure 5.
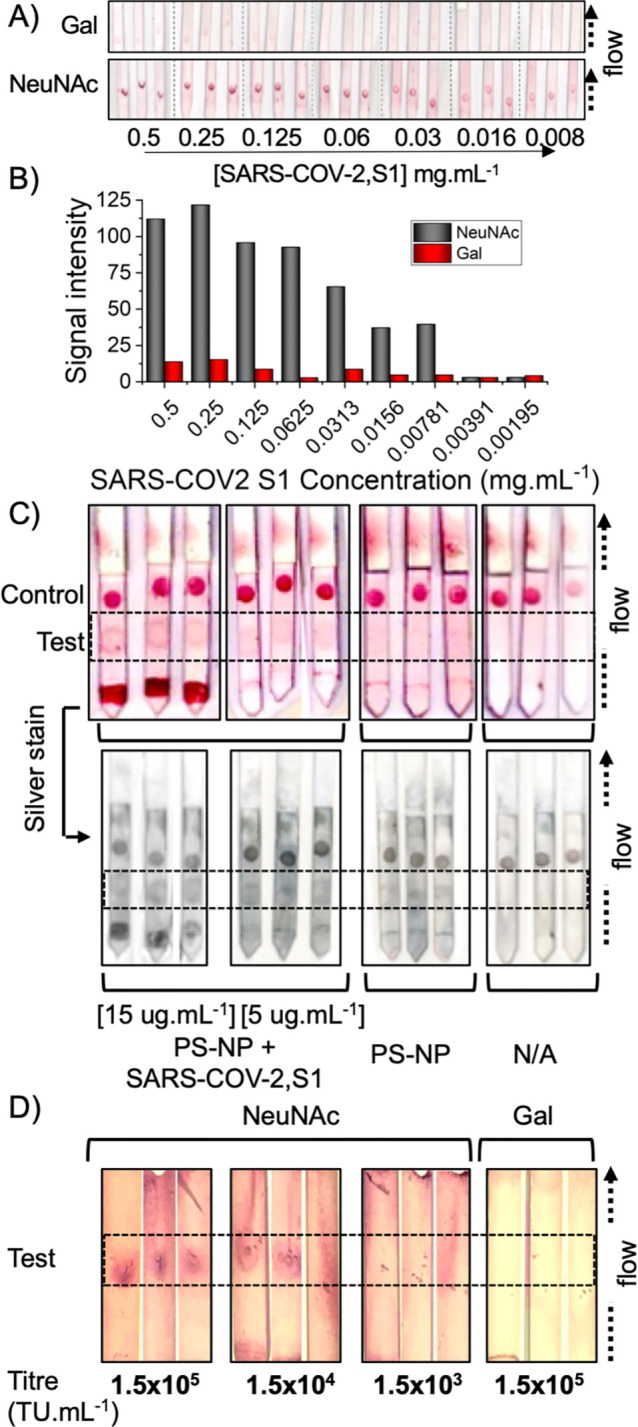
(A) Detection limit analysis of galactose or NeuNAc functionalized AuNPs against immobilized SARS-COV-2,S1 using “half LFD” assays; (B) Signal intensity analysis; (C) Dipstick lateral flow tests using NeuNAcPHEA50@AuNP35 and BSA-NeuNAc as the test line and RCA120 as the control line. PS-NP = 100 nm polystyrene colloid, or + SARS-COV-2,S1. N/A is with no polystyrene analyte. (D) Half lateral flow analysis of SARS-COV-2 Spike pseudotyped lentivirus against NeuNAcPHEA50@AuNP35 or GalPHEA50@AuNP35 nanoparticles. In each image, the test line region is indicated by the dashed box. Complete original images are in Supporting Information.
In conclusion, we have demonstrated a glycan-based lateral flow detection system that can detect the spike glycoprotein from the SARS-COV-2 virus in under 30 min. Guided by sequence alignment against other coronavirus spike proteins, it was hypothesized that sialic acids may bind this protein, to enable capture/detection. Using a nanoparticle-based biolayer interferometry platform, we demonstrated that α,N-acetyl neuraminic acid is a ligand for the spike glycoprotein. The gold nanoparticles and polymer tethers (for glycan capture) were optimized, and it was found in “half sandwich” lateral flow assays that α,N-acetyl neuraminic acid particles have selectivity toward the SARS-COV-2 spike protein, including specificity over SARS-COV-1 and a panel of lectins. Guided by this, we successfully detected a virus mimic particle bearing SARS-COV-2,S1 in under 30 min, with a detection limit of the spike protein around 5 μg·mL–1. Furthermore, a SARS-COV-2 spike protein-presenting pseudotyped lentivirus was successfully detected in a robust proof-of-concept. This work provides proof that glycan binding can be exploited to create rapid point-of-care diagnostics in a format which requires no infrastructure and limited training and, to the best of our knowledge, is the first reported all-glycan lateral flow system. This approach may find application for disease surveillance or mass testing at transport/work hubs or even for self/home testing. Finally, the observation that SARS-COV-2 can engage sialic acids found on human respiratory cells may provide insight into its zoonosis and infection pathways to help guide new interventions.
Acknowledgments
MIG is supported by the ERC (638661) and Royal Society (Industry Fellowship 191037). The BBSRC MIBTP program (BB/M01116X/1) and Iceni Diagnostics Ltd are thanked for support to A.N.B. UoW, EPSRC (EP/R511808/1), and BBSRC (BB/S506783/1) impact acceleration accounts are thanked. A.J.Z. is funded by the MRC DTP (MR/N014294/1). A.S. is a Wellcome Trust Investigator (200870/Z/16/Z). The Leverhulme Trust are thanked for support (RPG-2019-087). P.J.S. thanks Wellcome (208361/Z/17/Z), the MRC (MR/S009213/1), and BBSRC (BB/P01948X/1, BB/R002517/1, and BB/S003339/1). The Warwick Polymer and Electron Microscopy Research Technology Platforms (Y. Han) are acknowledged for the SEC/EM analysis. Dr C. Biggs is thanked for lateral flow device preparation. We sincerely thank the technical and administrative staff of the UoW who enabled our laboratory to remain open during the COVID-19 pandemic. Prof. Peter Scott is thanked for critically reading this manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c00855.
Full experimental details, characterization, BLI curves, unenhanced lateral flow device photos and additional control experiments (PDF)
Author Contributions
¶ (A.N.B., S.-J.R.) These authors contributed equally. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): M.I.G., A.N.B., and S.J.R. are named inventors of a patent relating to this work. R.F. is a shareholder in Iceni diagnostics who part-funded this work.
Notes
Background data is available from wrap.warwick.ac.uk
Supplementary Material
References
- Zhou P.; Yang X.-L.; Wang X.-G.; Hu B.; Zhang L.; Zhang W.; Si H.-R.; Zhu Y.; Li B.; Huang C.-L.; Chen H.-D.; Chen J.; Luo Y.; Guo H.; Jiang R.-D.; Liu M.-Q.; Chen Y.; Shen X.-R.; Wang X.; Zheng X.-S.; Zhao K.; Chen Q.-J.; Deng F.; Liu L.-L.; Yan B.; Zhan F.-X.; Wang Y.-Y.; Xiao G.-F.; Shi Z.-L. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579 (7798), 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X.; Zhong Z.; Zhao W.; Zheng C.; Wang F.; Liu J. Ches CT for Typical 2019-NCoV Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 2020, 296, E41. 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Yao L.; Li J.; Chen L.; Song Y.; Cai Z.; Yang C. Stability Issues of RT-PCR Testing of SARS-CoV-2 for Hospitalized Patients Clinically Diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903. 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.; Liu T.; Huang L.; Liu H.; Lei M.; Xu W.; Hu X.; Chen J.; Liu B. Use of Chest CT in Combination with Negative RT-PCR Assay for the 2019 Novel Coronavirus but High Clinical Suspicion. Radiology 2020, 295, 22–23. 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase R.; Kurita T.; Muranaka E.; Sasazawa H.; Mito H.; Yano Y. A Case of Imported COVID-19 Diagnosed by PCR-Positive Lower Respiratory Specimen but with PCR-Negative Throat Swabs. Infect. Dis. (Auckl). 2020, 52 (6), 423–426. 10.1080/23744235.2020.1744711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucirka L. M.; Lauer S. A.; Laeyendecker O.; Boon D.; Lessler J. Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Ann. Intern. Med. 2020, 173, 262. 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane M. M.; Organon M. V.. Diagnostic Test Device. Patent US3579306A, January 1969.
- Ezennia I. J.; Nduka S. O.; Ekwunife O. I. Cost Benefit Analysis of Malaria Rapid Diagnostic Test: The Perspective of Nigerian Community Pharmacists. Malar. J. 2017, 16 (1), 7–16. 10.1186/s12936-016-1648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawiah T.; Hansen K. S.; Baiden F.; Bruce J.; Tivura M.; Delimini R.; Amengo-Etego S.; Chandramohan D.; Owusu-Agyei S.; Webster J. Cost-Effectiveness Analysis of Test-Based versus Presumptive Treatment of Uncomplicated Malaria in Children under Five Years in an Area of High Transmission in Central Ghana. PLoS One 2016, 11 (10), e0164055. 10.1371/journal.pone.0164055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J. C.; Pettitt J.; George J. S.; Fakoli L. S.; Taweh F. M.; Bateman S. L.; Bennett R. S.; Norris S. L.; Spinnler D. A.; Pimentel G.; Sahr P. K.; Bolay F. K.; Schoepp R. J. Lateral Flow Immunoassays for Ebola Virus Disease Detection in Liberia. J. Infect. Dis. 2016, 214 (suppl 3), S222–S228. 10.1093/infdis/jiw251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X.; Ma Y.; Zhang A.; Zhang L.; Zeng L.; Liu G. Disposable Nucleic Acid Biosensors Based on Gold Nanoparticle Probes and Lateral Flow Strip. Anal. Chem. 2009, 81 (4), 1660–1668. 10.1021/ac8024653. [DOI] [PubMed] [Google Scholar]
- Damborský P.; Koczula K. M.; Gallotta A.; Katrlík J. Lectin-Based Lateral Flow Assay: Proof-of-Concept. Analyst 2016, 141 (23), 6444–6448. 10.1039/C6AN01746K. [DOI] [PubMed] [Google Scholar]
- Ishii J.; Toyoshima M.; Chikae M.; Takamura Y.; Miura Y. Preparation of Glycopolymer-Modified Gold Nanoparticles and a New Approach for a Lateral Flow Assay. Bull. Chem. Soc. Jpn. 2011, 84 (5), 466–470. 10.1246/bcsj.20100303. [DOI] [Google Scholar]
- Jorgensen P.; Chanthap L.; Rebueno A.; Tsuyuoka R.; Bell D. Malaria Rapid Diagnostic Tests in Tropical Climates: The Need for a Cool Chain. Am. J. Trop. Med. Hyg. 2006, 74 (5), 750–754. 10.4269/ajtmh.2006.74.750. [DOI] [PubMed] [Google Scholar]
- Wölfel R.; Corman V. M.; Guggemos W.; Seilmaier M.; Zange S.; Müller M. A.; Niemeyer D.; Jones T. C.; Vollmar P.; Rothe C.; Hoelscher M.; Bleicker T.; Brünink S.; Schneider J.; Ehmann R.; Zwirglmaier K.; Drosten C.; Wendtner C. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581 (7809), 465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Connor R. J.; Kawaoka Y.; Webster R. G.; Paulson J. C. Receptor Specificity in Human, Avian, and Equine H2 and H3 Influenza Virus Isolates. Virology 1994, 205 (1), 17–23. 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Childs R. A.; Palma A. S.; Wharton S.; Matrosovich T.; Liu Y.; Chai W.; Campanero-Rhodes M. A.; Zhang Y.; Eickmann M.; Kiso M.; Hay A.; Matrosovich M.; Feizi T. Receptor-Binding Specificity of Pandemic Influenza A (H1N1) 2009 Virus Determined by Carbohydrate Microarray. Nat. Biotechnol. 2009, 27 (9), 797–799. 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M. J.; Rashid A.; Rejzek M.; Fairhurst S. A.; Wharton S. A.; Martin S. R.; McCauley J. W.; Wileman T.; Field R. A.; Russell D. A. Glyconanoparticles for the Plasmonic Detection and Discrimination between Human and Avian Influenza Virus. Org. Biomol. Chem. 2013, 11 (41), 7101. 10.1039/c3ob41703d. [DOI] [PubMed] [Google Scholar]
- Richards S.-J.; Baker A. N.; Walker M.; Gibson M. I. Polymer-Stabilized Sialylated Nanoparticles: Synthesis, Optimization, and Differential Binding to Influenza Hemagglutinins. Biomacromolecules 2020, 21 (4), 1604–1612. 10.1021/acs.biomac.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E.; Hantak M.; Perlman S.; Gallagher T. Distinct Roles for Sialoside and Protein Receptors in Coronavirus Infection. mBio 2020, 11 (1), e02764-19. 10.1128/mBio.02764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R. J. G.; Lang Y.; Bakkers M. J. G.; Li W.; Li Z.; Schouten A.; Ophorst B.; Van Kuppeveld F. J. M.; Boons G. J.; Bosch B. J.; Huizinga E. G.; De Groot R. J. Human Coronaviruses OC43 and HKU1 Bind to 9-O-Acetylated Sialic Acids via a Conserved Receptor-Binding Site in Spike Protein Domain A. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (7), 2681–2690. 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.; Dong W.; Milewska A.; Golda A.; Qi Y.; Zhu Q. K.; Marasco W. A.; Baric R. S.; Sims A. C.; Pyrc K.; Li W.; Sui J. Human Coronavirus HKU1 Spike Protein Uses O -Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89 (14), 7202–7213. 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Hulswit R. J. G.; Widjaja I.; Raj V. S.; McBride R.; Peng W.; Widagdo W.; Tortorici M. A.; Van Dieren B.; Lang Y.; Van Lent J. W. M.; Paulson J. C.; De Haan C. A. M.; De Groot R. J.; Van Kuppeveld F. J. M.; Haagmans B. L.; Bosch B. J. Identification of Sialic Acid-Binding Function for the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (40), E8508–E8517. 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M.; Donaldson E.; Baric R. SARS-CoV and Emergent Coronaviruses: Viral Determinants of Interspecies Transmission. Curr. Opin. Virol. 2011, 1 (6), 624–634. 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon P. S.; Oh H.; Kwon S. J.; Jin W.; Zhang F.; Fraser K.; Hong J. J.; Linhardt R. J.; Dordick J. S. Sulfated Polysaccharides Effectively Inhibit SARS-CoV-2 in Vitro. Cell Discovery 2020, 6, 50. 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. M.; Sandoval D. R.; Spliid C. B.; Pihl J.; Painter C. D.; Thacker B. E.; Glass C. A.; Narayanan A.; Majowicz S. A.; Zhang Y.; Torres J. L.; Golden G. J.; Porell R.; Garretson A. F.; Laubach L.; Feldman J.; Yin X.; Pu Y.; Hauser B.; Caradonna T. M.; Kellman B. P.; Martino C.; Gordts P. L. S. M.; Leibel S. L.; Chanda S. K.; Schmidt A. G.; Godula K.; Jose J.; Corbett K. D.; Ward A. B.; Carlin A. F.; Esko J. D.. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. bioRxiv, July 14, 2020, 10.1101/2020.07.14.201616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West C. J.; Su D.; Pagani I.; Rudd T. R.; Elli S.; Guimond S. E.; Miller G.; Meneghetti M. C. Z.; Nader H. B.; Li Y.; Nunes Q. M.; Procter P.; Mancini N.; Clementi M.; Bisio A.; Forsyth N. R.; Turnbull J. E.; Guerrini M.; Fernig D. G.; Vicenzi E.; Yates E. A.; Lima M. A.; Skidmore M. A.. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Surface Protein (Spike) S1 Receptor Binding Domain with Heparin. bioRxiv, April 28, 2020, 10.1101/2020.04.28.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist J. J.; Toone E. J. The Cluster Glycoside Effect. Chem. Rev. 2002, 102 (2), 555–578. 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- Reichardt N. C.; Martín-Lomas M.; Penadés S. Glyconanotechnology. Chem. Soc. Rev. 2013, 42 (10), 4358–4376. 10.1039/c2cs35427f. [DOI] [PubMed] [Google Scholar]
- Branson T. R.; Turnbull W. B. Bacterial Toxin Inhibitors Based on Multivalent Scaffolds. Chem. Soc. Rev. 2013, 42 (11), 4613–4622. 10.1039/C2CS35430F. [DOI] [PubMed] [Google Scholar]
- Kiessling L. L.; Gestwicki J. E.; Strong L. E. Synthetic Multivalent Ligands as Probes of Signal Transduction. Angew. Chem., Int. Ed. 2006, 45 (15), 2348–2368. 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Moussodia R.-O.; Vértesy S.; André S.; Klein M. L.; Gabius H.-J.; Percec V. Unraveling Functional Significance of Natural Variations of a Human Galectin by Glycodendrimersomes with Programmable Glycan Surface. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (18), 5585–5590. 10.1073/pnas.1506220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branson T. R.; McAllister T. E.; Garcia-Hartjes J.; Fascione M. A.; Ross J. F.; Warriner S. L.; Wennekes T.; Zuilhof H.; Turnbull W. B. A Protein-Based Pentavalent Inhibitor of the Cholera Toxin B-Subunit. Angew. Chem., Int. Ed. 2014, 53 (32), 8323–8327. 10.1002/anie.201404397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S.-J.; Jones M. W.; Hunaban M.; Haddleton D. M.; Gibson M. I. Probing Bacterial-Toxin Inhibition with Synthetic Glycopolymers Prepared by Tandem Post-Polymerization Modification: Role of Linker Length and Carbohydrate Density. Angew. Chem., Int. Ed. 2012, 51 (31), 7812–7816. 10.1002/anie.201202945. [DOI] [PubMed] [Google Scholar]
- Becer C. R. R.; Gibson M. I. M. I.; Geng J.; Ilyas R.; Wallis R.; Mitchell D. A. D. A.; Haddleton D. M. D. M. High-Affinity Glycopolymer Binding to Human DC-SIGN and Disruption of DC-SIGN Interactions with HIV Envelope Glycoprotein. J. Am. Chem. Soc. 2010, 132 (43), 15130–15132. 10.1021/ja1056714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S.-J.; Gibson M. I. Optimization of the Polymer Coating for Glycosylated Gold Nanoparticle Biosensors to Ensure Stability and Rapid Optical Readouts. ACS Macro Lett. 2014, 3 (10), 1004–1008. 10.1021/mz5004882. [DOI] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367 (6483), 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. J.; Walls A. C.; Wang Z.; Sauer M. M.; Li W.; Tortorici M. A.; Bosch B. J.; DiMaio F.; Veesler D. Structures of MERS-CoV Spike Glycoprotein in Complex with Sialoside Attachment Receptors. Nat. Struct. Mol. Biol. 2019, 26 (12), 1151–1157. 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelli A.; Monti M.; Milanetti E.; Ponti R. D.; Tartaglia G. G.. Structural Analysis of SARS-CoV-2 and Prediction of the Human Interactome. bioRxiv, July 24, 2020, 10.1101/2020.03.28.013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W.; Brown J. H.; Cusack S.; Paulson J. C.; Skehel J. J.; Wiley D. C. Structure of the Influenza Virus Haemagglutinin Complexed with Its Receptor, Sialic Acid. Nature 1988, 333 (6172), 426–431. 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Milanetti E.; Miotto M.; Di Rienzo L.; Monti M.; Gosti G.; Ruocco G.. In-Silico Evidence for Two Receptors Based Strategy of SARS-CoV-2. bioRxiv, April 6, 2020, 10.1101/2020.03.24.006197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou P. G.; Baker A. N.; Richards S. J.; Laezza A.; Walker M.; Gibson M. I. Tuning Aggregative versus Non-Aggregative Lectin Binding with Glycosylated Nanoparticles by the Nature of the Polymer Ligand. J. Mater. Chem. B 2020, 8 (1), 136–145. 10.1039/C9TB02004G. [DOI] [PubMed] [Google Scholar]
- Haiss W.; Thanh N. T. K.; Aveyard J.; Fernig D. G. Determination of Size and Concentration of Gold Nanoparticles from UV - Vis Spectra. Anal. Chem. 2007, 79 (11), 4215–4221. 10.1021/ac0702084. [DOI] [PubMed] [Google Scholar]
- Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-Specific Glycan Analysis of the SARS-CoV-2 Spike. Science 2020, eabb9983. 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X.; Yu H.; Chen X.; Lasanajak Y.; Tappert M. M.; Air G. M.; Tiwari V. K.; Cao H.; Chokhawala H. A.; Zheng H.; Cummings R. D.; Smith D. F. A Sialylated Glycan Microarray Reveals Novel Interactions of Modified Sialic Acids with Proteins and Viruses. J. Biol. Chem. 2011, 286 (36), 31610–31622. 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu E.; Liang T.; Houghtaling J.; Ramachandran S.; Ramsey S. A.; Lutz B.; Yager P. Enhanced Sensitivity of Lateral Flow Tests Using a Two-Dimensional Paper Network Format. Anal. Chem. 2011, 83 (20), 7941–7946. 10.1021/ac201950g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


