Abstract
SARS-CoV-2 is causing a pandemic with currently > 29 million confirmed cases and > 900,000 deaths worldwide. The locations and mechanisms of virus entry into the human respiratory tract are incompletely characterized. We analyzed publicly available RNA microarray datasets for SARS-CoV-2 entry receptors and cofactors ACE2, TMPRSS2, BSG (CD147) and FURIN. We found that ACE2 and TMPRSS2 are upregulated in the airways of smokers. In asthmatics, ACE2 tended to be downregulated in nasal epithelium, and TMPRSS2 was upregulated in the bronchi. Furthermore, respiratory epithelia were negative for ACE-2 and TMPRSS2 protein expression while positive for BSG and furin, suggesting a possible alternative entry route for SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, ACE-2, TMPRSS2, Basigin, Furin, Smoking, Asthma, Respiratory epithelium, Coronavirus
Results & discussion
The current pandemic with the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes coronavirus disease 2019 (COVID-19) is spreading globally with more than 29 million cases and 900,000 deaths worldwide [1, 2]. In a significant fraction of patients, SARS-CoV-2 infection can take a severe course. Especially in the elderly and in those with pre-existing conditions including chronic lung diseases, severe pneumonia and even life-threatening diffuse alveolar damage requiring intensive care and ventilation can occur [3, 4].
The primary infection site for SARS-CoV-2 is the upper respiratory/digestive tract and conjunctival mucosa. The expression and distribution of SARS-CoV-2 entry receptors and cofactors in the human respiratory tract, and how their expression is altered in disease or by environmental and behavioral factors such as air pollution and smoking, is therefore of great interest. This will lead to a better understanding of SARS-CoV-2 biology, the susceptibility of certain populations to COVID-19, and potentially help to develop future therapies.
Angiotensin I converting enzyme 2 (ACE-2) and transmembrane serine protease 2 (TMPRSS2) have been described as the main receptor and cofactor for SARS-CoV-2 cellular entry [5–8]. In addition, emerging reports point towards a role for basigin (BSG / CD147) as receptor [9], and furin as a cofactor [10], in the pathogenicity and virulence of SARS-CoV-2. Here, we examined RNA and protein expression of ACE-2, TMPRSS2, basigin and furin in the human respiratory tract in healthy non-smokers, healthy smokers and asthma patients.
Six RNA microarray datasets of airway epithelial cell brushings, all generated with the Affymetrix Human Genome U133 Plus 2.0 Array, were downloaded from the Gene Expression Omnibus [11–16]. Affymetrix data files (Supplemental Information) were processed and normalized using the robust multiarray average expression measure method using affy and limma packages in R [17, 18]. For genes represented by multiple probes, the probe with the maximum average expression values in all samples was selected to represent that gene’s expression. First, by plotting the first 2 principal components computed on ACE2, TMPRSS2, BSG and FURIN expression across smokers’ and asthmatics’ datasets, we verified that there were no detectable batch effects within each of the six microarray datasets we sought to analyze (Figs. S1A-D and S2A-D). Next, differences in gene expression in smokers vs. non-smokers and asthmatics vs. healthy individuals were modeled using linear regression, including proband age, sex, sample type and dataset as covariates in the model. When modeling the age effect with linear regression, the age of probands for whom age information was not available was set to the average age of all other probands (separately for smokers’ datasets GSE63127 and asthmatics’ dataset GSE4302; for age distribution, see Figs. S1E and S2E). P values were next corrected for multiple hypothesis testing using the Benjamini-Hochberg correction. In post hoc analysis, log2 transformed data not corrected for age and sex were plotted with ggpubr [19], and two sided Mann-Whitney U tests were performed using the Wilcox.test function in R.
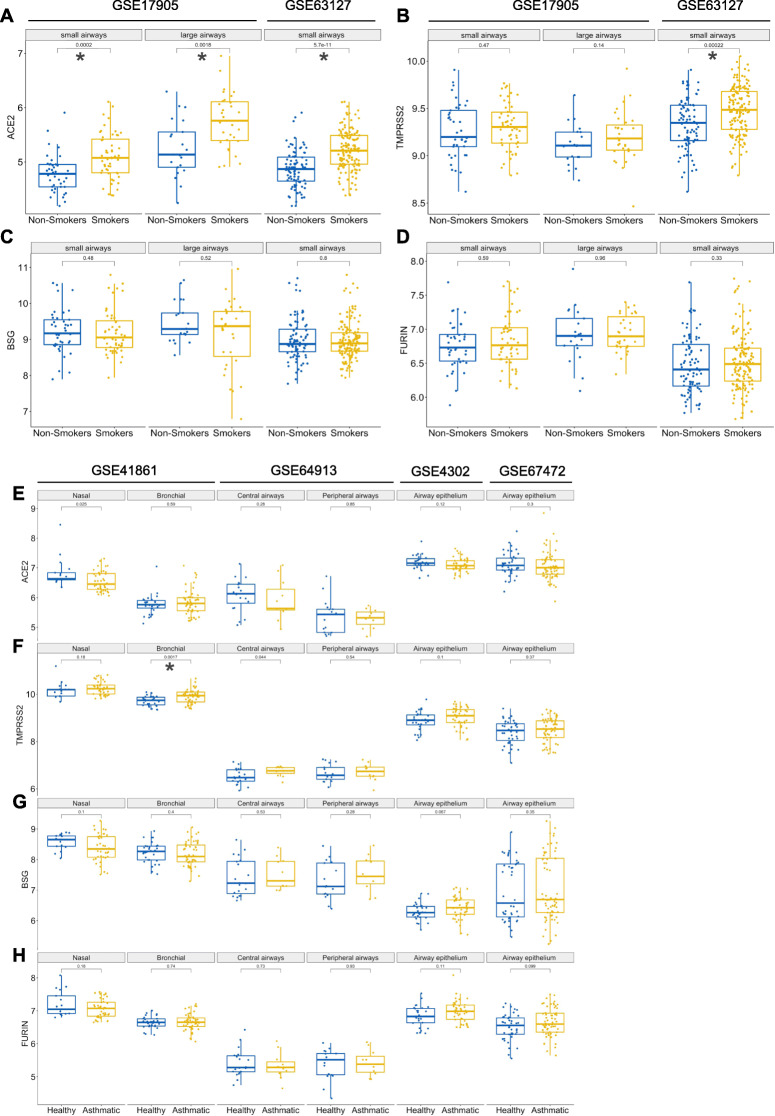
Older age is an important risk factor for adverse COVID-19 outcomes [20]. Another risk factor is male sex [21]. Since there is a very high smoking rate in Chinese males compared to females—66.1% vs. 3.2% according to Ma et al. [22]—it has been suggested that smoking could be a risk factor for the more severe COVID-19 disease course observed in males [20, 23]; however, this topic is controversially discussed [24–26]. In our analysis, we therefore removed possible confounding effects of age and sex on the status of receptor mRNA expression by regressing the linear effects of age and sex and testing our hypothesis on model residuals (Table 1). For samples for which sex information was not available, sex was predicted based on the expression of both X inactive specific transcript (XIST; high expression in females) and ribosomal protein S4 Y-linked 1 (RPS4Y1; high expression in males) simultaneously. In post hoc analysis, we also plotted log2 transformed expression values not corrected for age and sex (Fig. 1). Consistent with previous reports [7, 27], we found significantly higher ACE2 expression in airway epithelia from healthy smokers vs. healthy non-smokers (Fig. 1a, Table 1). Similarly, we found significantly higher TMPRSS2 expression in smokers in one out of two datasets analyzed (Fig. 1b, Table 1), whereas BSG and FURIN expression did not significantly differ between smokers and non-smokers (Fig. 1c-d, Table 1). Taken together, these results indicate that, independently of sex and age, ACE2 and TMPRSS2 are upregulated in the airway epithelia of smokers. In addition, our analyses establish that BSG and FURIN, two alternate potential SARS-CoV-2 receptors, are expressed in the human respiratory tract.
Table 1.
Modeling differences in ACE2, TMPRSS2, BSG and FURIN expression accounting for proband age and sex
| gene | Adjusted p-value smokers vs. non-smokers | β smokers vs. non-smokers | Adjusted p-value asthmatics vs. healthy | β asthmatics vs. healthy |
|---|---|---|---|---|
| ACE2 | 6.80E-16 | 0.36 | 0.186 | −0.074 |
| TMPRSS2 | 0.00044 | 0.1 | 0.0156 | 0.13 |
| BSG | 0.31 | −0.065 | 0.59 | 0.038 |
| FURIN | 0.31 | 0.047 | 0.29 | 0.048 |
Adjusted p-values and linear regression model coefficients (β), testing hypotheses whether expression of ACE2, TMPRSS2, BSG and FURIN differ between smokers vs. non-smokers (columns 1,2) and asthmatics vs. healthy individuals (columns 3,4). The linear regression models were controlled for the linear effect of age, sex, sample type, and dataset
Fig. 1.
Expression of ACE2, TMPRSS2, BSG and FURIN in respiratory epithelium of smokers, asthmatics and healthy individuals. a-d Microarray datasets of bronchial brushings from healthy non-smokers and smokers. GSE17905: non-smokers small airways (n = 41), large airways (n = 21); smokers small airways (n = 52), large airways (n = 31). GSE63127: non-smokers (n = 87); smokers (n = 143). Gene expression for a ACE2, b TMPRSS2, c BSG, and d FURIN. e-h Microarray datasets of airway epithelial brushings from healthy controls (GSE41861, bronchial n = 30 and nasal n = 17; GSE64913, central airway n = 20 and peripheral airway n = 17; GSE4302, n = 28; GSE67472, n = 43) and asthma patients (GSE41861, bronchial n = 51 and nasal n = 40; GSE64913, central airway n = 11 and peripheral airway n = 11; GSE4302, n = 42; GSE67472, n = 62). Gene expression for e ACE2, f TMPRSS2, g BSG, and h FURIN. Data are shown as log2 transformed expression values not corrected for proband age and sex. Multiple comparison significance levels: *p < 0.002
We next examined four RNA microarray datasets for ACE2, TMPRSS2, BSG and FURIN expression in airway epithelia from patients with a common respiratory disease, asthma. Patients with chronic respiratory disorders including asthma are considered a COVID-19 high-risk category [3]. Interestingly, we found that ACE2 expression tended to be downregulated in nasal epithelium, whereas TMPRSS2 was significantly upregulated in bronchi and central airways of asthmatics (Fig. 1e-f, Table 1). ACE2 was proposed to be an interferon-stimulated gene [28]; therefore, a potential explanation for ACE2 downregulation in asthmatics could be corticosteroid use. However, more recent data indicate that a novel, primate-specific ACE2 isoform exists that is incapable of binding SARS-CoV-2, and that this isoform is interferon-stimulated, whereas the canonical ACE2 is not [29, 30]. Further research is needed to address these highly interesting developments in more detail.
Additionally, we did not find any difference in BSG or FURIN expression between healthy and asthmatic individuals (Fig. 1g-h, Table 1). These findings point towards a possible differential regulation of ACE2 and TMPRSS2 expression in airway epithelia and warrant further investigation into the underlying mechanism.
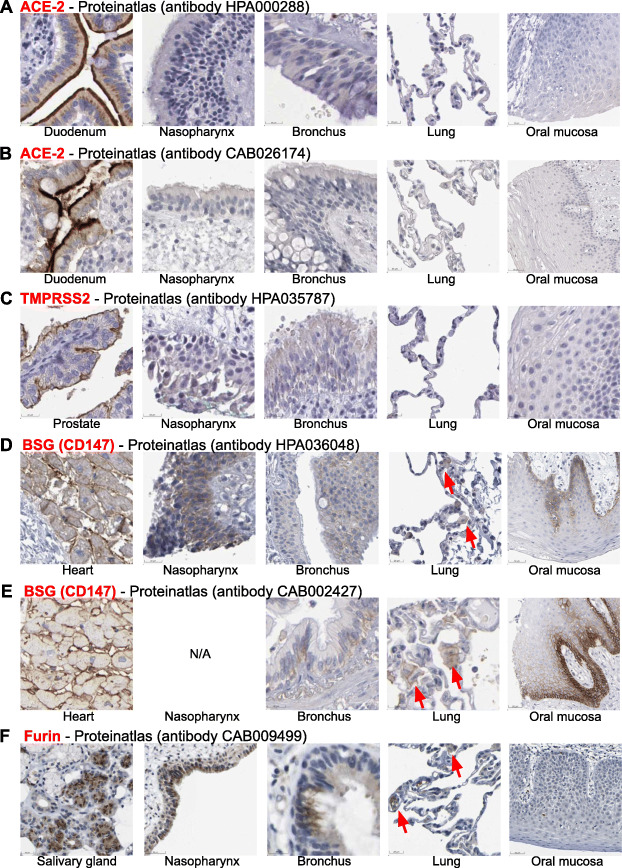
We next aimed to compare RNA expression of these receptors and cofactors to protein expression. Therefore, we examined immunohistochemistry (IHC) images from respiratory and other tissues on The Human Protein Atlas [31]. ACE-2 IHC staining was strong in epithelial cells of the duodenum and was found in other organs including testis and kidney. In contrast, human respiratory epithelial cells in samples from the nasopharynx, bronchi and lungs, as well as squamous epithelial cells from the oral mucosa, were completely negative for ACE-2 staining by IHC with two different antibodies (Fig. 2a-b). Similar negative IHC staining results were also observed for TMPRSS2 protein (Fig. 2c). In contrast, basigin protein was widely expressed in human tissues including heart muscle, brain, liver and kidney, and, importantly, was positive in respiratory epithelial cells from the nasopharynx and bronchi (Fig. 2d-e). Similar to ACE-2 and TMPRSS2, basigin was negative in alveolar epithelial cells but showed multifocal positivity in cells morphologically consistent with alveolar macrophages (Fig. 2d-e). Furthermore, basigin was strongly expressed in the oral mucosa, with a gradient from basal to apical epithelial cells (Fig. 2d-e). Finally, furin IHC stainings were positive in nasopharynx and bronchial epithelial cells, and negative in alveolar epithelial cells, with focal positivity in alveolar macrophages (Fig. 2f). In summary, basigin and furin protein were expressed in airway epithelia of nasopharynx and bronchi, whereas ACE-2 and TMPRSS2 protein stainings were negative.
Fig. 2.
Expression of SARS-CoV-2 receptor and cofactor proteins in respiratory epithelium as analyzed by immunohistochemistry. Representative screenshots of immunohistochemistry (IHC) images from The Human Protein Atlas (www.proteinatlas.org). a-b ACE-2 IHC images from the nasopharynx (n = 4), bronchi (n = 3), lungs (n = 6), and oral mucosa (n = 4). Positive control: duodenum tissue (n = 6). c TMPRSS2 IHC images from the nasopharynx (n = 3), bronchi (n = 2), lungs (n = 3), and oral mucosa (n = 3). Positive control: prostate tissue (n = 3). d-e BSG/CD147 IHC images from the nasopharynx (n = 2), bronchi (n = 3), lungs (n = 6), and oral mucosa (n = 5). Positive control: heart tissue (n = 3). f Furin IHC images from the nasopharynx (n = 3), bronchi (n = 3), lungs (n = 3), and oral mucosa (n = 3). Positive control: salivary gland tissue (n = 3). Red arrows indicate IHC positive cells morphologically consistent with alveolar macrophages. Scale bars, lower left of respective images. Images were analyzed by a board-certified surgical pathologist (C.M.S.)
Our findings are in line with and extend recent studies addressing SARS-CoV-2 receptor and cofactor expression in the respiratory tract [32, 33], but are in stark contrast to a 2004 study by Hamming et al. [34], who found strong and widespread ACE-2 expression in alveolar epithelial cells and basal epithelial cells of the nasopharynx and oral mucosa. Our study highlights the discrepancies between RNA and protein expression of these receptors and cofactors, and points towards potential issues with IHC staining reproducibility and antibody specificity, important factors that need to be addressed in future investigations. One limitation of our study was that in The Human Protein Atlas, only small numbers of IHC stained samples for each tissue and molecule analyzed were available. Therefore, further studies exploring the protein expression and cellular localization of SARS-CoV-2 receptors and cofactors in each of these tissue types, ideally using tissue microarrays with large numbers of samples from multiple donor cohorts, and using multiple different antibodies, are warranted. In addition, these studies should use the recently developed multiplexed microscopy technologies [35] to address protein co-expression patterns and better delineate the cellular subsets expressing these SARS-CoV-2 entry receptors and cofactors.
Supplementary information
Acknowledgements
Not applicable.
Abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- COVID-19
Coronavirus disease 2019
- ACE-2
Angiotensin I converting enzyme 2
- TMPRSS2
Transmembrane serine protease 2
- BSG
Basigin
- XIST
X inactive specific transcript
- RPS4Y1
Ribosomal protein S4 Y-linked 1
Authors’ contributions
C.M.S. conceived of the study. M.M. and C.M.S. analyzed and interpreted data, created the figures, and wrote the manuscript. The authors read and approved the final manuscript.
Funding
Swiss National Science Foundation (P400PM_183915 to C.M.S.).
Availability of data and materials
All data presented are from publicly available datasets, as detailed in the manuscript and supplemental information.
Datasets for smokers vs. non-smokers:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17905
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63127
Datasets for asthmatics vs. healthy individuals:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41861
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64913
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4302
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67472
Datasets for receptor protein immunohistochemistry:
https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue
https://www.proteinatlas.org/ENSG00000184012-TMPRSS2/tissue
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12931-020-01521-x.
References
- 1.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University. Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) [Internet]. 2020 [cited 2020 Jun 19]. Available from: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html?myid=3TGDV68B-3T9VW03K-SMHVE&utm_source=newsletter&utm_medium=email&utm_campaign=kk-2020-corona&utm_content=de#/bda7594740fd40299423467b48e9ecf6.
- 3.Chow N, Fleming-Dutra K, Gierke R, Hall A, Hughes M, Pilishvili T, et al. Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019 — United States, February 12–march 28, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menter T, Haslbauer JD, Nienhold R, Savic S, Hopfer H, Deigendesch N, et al. Post-mortem examination of COVID19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology. 2020;77(2):198–209. [DOI] [PMC free article] [PubMed]
- 5.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–80. [DOI] [PMC free article] [PubMed]
- 6.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: current state of the science. Immunity. 2020:S1074761320301837. [DOI] [PMC free article] [PubMed]
- 7.Leung JM, Yang CX, Tam A, Shaipanich T, Hackett T-L, Singhera GK, et al. ACE-2 Expression in the Small Airway Epithelia of Smokers and COPD Patients: Implications for COVID-19. Eur Respir J. 2020;55:2000688. [DOI] [PMC free article] [PubMed]
- 8.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. 2020;2020(03):14.988345. [Google Scholar]
- 10.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G, Wang R, Ferris B, Salit J, Strulovici-Barel Y, Hackett NR, et al. Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium. Respir Res. 2010;11:150. doi: 10.1186/1465-9921-11-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng D, Staudt M, Tribouley C, Bitter H, Ravindran P, He C, et al. Upper airway gene expression is an effective surrogate biomarker for Th2-driven inflammation in the lower airway. Gene Expression Omnibus 2015. Available from: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41861.
- 14.Christenson SA, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, Postma DS, et al. Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;191:758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilley AE, Staudt MR, Salit J, Van de Graaf B, Strulovici-Barel Y, Kaner RJ, et al. Cigarette smoking induces changes in airway epithelial expression of genes associated with monogenic lung disorders. Am J Respir Crit Care Med. 2016;193:215–217. doi: 10.1164/rccm.201412-2290LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singhania A, Rupani H, Jayasekera N, Lumb S, Hales P, Gozzard N, et al. Altered Epithelial Gene Expression in Peripheral Airways of Severe Asthma. PLoS One. 2017;12:e0168680. doi: 10.1371/journal.pone.0168680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. Oxford Acad. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kassambara A. ggpubr: ‘ggplot2’ Based Publication Ready Plots [Internet]. 2016. Available from: https://rpkgs.datanovia.com/ggpubr/ [cited 2020 12 Apr].
- 20.Jordan RE, Adab P, Cheng KK. Covid-19: risk factors for severe disease and death. BMJ. 2020:368 Available from: https://www.bmj.com/content/368/bmj.m1198 [cited 2020 19 Jun]. [DOI] [PubMed]
- 21.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis [Internet]. 2020; Available from: http://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa270/5805508 [cited 2020 9 Apr].
- 22.Ma Y, Wen L, Cui W, Yuan W, Yang Z, Jiang K, et al. Prevalence of Cigarette Smoking and Nicotine Dependence in Men and Women Residing in Two Provinces in China. Front Psychiatr. 2017;8:254. [DOI] [PMC free article] [PubMed]
- 23.Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. 2020;8:e20. doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippi G, Henry BM. Active smoking is not associated with severity of coronavirus disease 2019 (COVID-19). European Journal of Internal Medicine [Internet]. Elsevier; 2020;0. Available from: https://www.ejinme.com/article/S0953-6205(20)30110-2/abstract [cited 2020 9 Apr]. [DOI] [PMC free article] [PubMed]
- 25.Vardavas CI, Nikitara K. COVID-19 and smoking: A systematic review of the evidence. Tob Induc Dis. 2020;18(March):20. [DOI] [PMC free article] [PubMed]
- 26.Rossato M, Russo L, Mazzocut S, Vincenzo AD, Fioretto P, Vettor R. Current Smoking is Not Associated with COVID-19. Eur Respir J. 2020;55:2001290. [DOI] [PMC free article] [PubMed]
- 27.Cai G. Bulk and single-cell transcriptomics identify tobacco-use disparity in lung gene expression of ACE2, the receptor of 2019-nCov. medRxiv. 2020;2020.02.05.20020107.
- 28.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onabajo OO, Banday AR, Yan W, Obajemu A, Stanifer ML, Santer DM, et al. Interferons and viruses induce a novel primate-specific isoform dACE2 and not the SARS-CoV-2 receptor ACE2. bioRxiv; 2020;2020.07.19.210955. [DOI] [PMC free article] [PubMed]
- 30.Ng K, Attig J, Bolland W, Young GR, Major J, Wack A, et al. Tissue-specific and interferon-inducible expression of non-functional ACE2 through endogenous retrovirus co-option. bioRxiv; 2020;2020.07.24.219139. [DOI] [PMC free article] [PubMed]
- 31.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based human protein atlas. Nat Biotechnol Nat Publishing Group. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 32.Hikmet F, Mear L, Uhlen M, Lindskog C. The protein expression profile of ACE2 in human tissues. bioRxiv; 2020;2020.03.31.016048. [DOI] [PMC free article] [PubMed]
- 33.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schürch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, et al. Coordinated cellular neighborhoods orchestrate Antitumoral immunity at the colorectal Cancer invasive front. Cell. 2020;S0092867420308709. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented are from publicly available datasets, as detailed in the manuscript and supplemental information.
Datasets for smokers vs. non-smokers:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17905
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63127
Datasets for asthmatics vs. healthy individuals:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41861
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64913
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4302
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67472
Datasets for receptor protein immunohistochemistry:
https://www.proteinatlas.org/ENSG00000130234-ACE2/tissue
https://www.proteinatlas.org/ENSG00000184012-TMPRSS2/tissue