Abstract
Hepatic arterial infusion (HAI) of chemotherapy is an experimental treatment option for patients with colorectal cancer liver metastases (CRCLM). The current study aimed to investigate the predictive and prognostic value of cell free DNA (cfDNA) in patients with CRCLM receiving HAI with oxaliplatin and systemic capecitabine. Plasma samples from 62 patients were investigated who were included into a single arm phase II study investigating HAI treatment for patients with CRCLM. The clinical outcome of the trial has been presented previously. In brief, treatment consisted of intrahepatic infusion of oxaliplatin 100 mg/m2 every second week with concomitant oral capecitabine 3,500 mg/m2 every second week for up to 12 cycles. Blood samples were drawn at baseline and follow-up and plasma was analyzed for cell free DNA using a direct fluorescent assay. The baseline level of plasma cfDNA was 0.92 ng/µl (95% CI 0.84-1.00). Patients with a baseline value of cfDNA above the 75th quartile had a median overall survival of 2.4 years (95% CI 0.7-2.8), compared with 3.9 years (95% CI 2.8-5.9) for patients below the 75th quartile (P=0.02). The baseline level of cfDNA was significantly lower (0.91 ng/µl, 95% CI 0.76-0.98) in patients who achieved an objective response compared to non-responders (1.79 ng/µl; 95% CI 0.99-2.57; P=0.02). The current study demonstrated a possible prognostic and predictive value of cfDNA for patients with CRCLM undergoing HAI with oxaliplatin and concomitant capecitabine.
Keywords: intrahepatic chemotherapy, oxaliplatin, liver metastases, circulating free DNA, fluorescence
Introduction
Colorectal cancer (CRC) is the third most common malignancy worldwide (1) being a frequent cause of cancer related death as close to 50% of patients diagnosed with CRC eventually are diagnosed with liver metastases (CRCLM) (2). Appropriately selected patients with CRCLM can be treated in a potential curative setting including both surgery and non-operable metastasis directed therapies with reported 5 years survival rates of around 40% (3-5). Hepatic arterial infusion (HAI) of chemotherapy for CRCLM has been explored since the 1970s (6), yet, unable to be an established standard of care in current guidelines (7). The theoretical advantage of HAI is the direct delivery of cytotoxic agents into the metastases, with oxaliplatin showing a favorable pharmacokinetic profile for this administration (8). As only 10-15% of CRCLM are upfront resectable, there is an unmet need for non-operative therapies and corresponding prognostic and/or predictive biomarkers to individualize treatment.
Cell free DNA (cfDNA) is present in the blood stream as a mixture of healthy and mutated tumor specific DNA (9). Although known for decades (10), cfDNA has in recent years attracted increasing attention as a strong prognostic marker for patients with metastatic colorectal cancer (mCRC) (11). The methodology for cfDNA analysis is complex and heterogeneous, thus we have optimized and validated a direct fluorescent assay (DFA) for the total cfDNA analysis. This allows for a rapid quantification of cfDNA using only low volume (40 µl) of plasma and no DNA preparation.
Cell free DNA can emerge as an essential future biomarker, with possible applications in both systemic and localized cancer treatment. Tumor specific cfDNA might display unique mutational status regarding RAS, BRAF, hypermethylation and microsatellite instability, while the total level of cfDNA can serve as a more universal biomarker (11,12). Here, we investigate the level of cfDNA, assessed by DFA, as a prognostic and predictive marker for patients with CRCLM undergoing HAI with oxaliplatin together with oral capecitabine.
Materials and methods
Patients and materials
Patients were treated according to a single arm phase II study including patients with liver limited mCRC from November 2004 to May 2010, who were not eligible for any other standard local treatment. Therapy comprised intrahepatic infusion of oxaliplatin 100 mg/m2 every second week with concomitant oral capecitabine 3,500 mg/m2 every second week for up to 12 cycles. The clinical data and outcome of the study has been presented separately (13), thus only translational aspects are addressed here. For comparison, we had blood samples available from 94 healthy individuals from a Danish biobank, as previously described (14).
After HAI treatment patients underwent standard of care surveillance with regular Computed Tomography (CT) scans. The first occurrence of radiologically progression according to the RESICT criteria was defined as the first progression.
Laboratory investigations
Blood samples for translational use were drawn at baseline prior to the administration of the first intrahepatic dose of oxaliplatin and at a fixed schedule during follow up. Analysis of cfDNA was done using a direct fluorescent assay for cfDNA analysis, not requiring any prior processing, as preliminary reported by Douvdevani and colleagues (15,16) and further modified by our group, as previously published (14).
In short, 40 µl plasma, not requiring any prior processing, were used and adding SYBR® Gold Nucleic Acid Gel Stain (1:8,000). The quantification of fluorescence was performed with a 96-well fluorometer (Infinite F200 PRO, Tecan) at an emission wavelength of 535 nm and an excitation wavelength of 485 nm in a black 96-well plate (Bio-Plex Pro Flat Bottom Plates, Bio-Rad). Using dilution with a 10% bovine serum albumin (BSA; Sigma® Life Science) solution, DNA standards were prepared from Human Control Genomic DNA (Life Technologies). From a standard curve the concentration of the samples were calculated. For each sample, we determined the concentration of cfDNA by calculating the median value of four measurements, removing outliers following Dixons q test if the standard deviation exceeded 10%. Carcino-Embryonic Antigen (CEA) was measured with routine analysis with an upper normal limit (UNL) of 5 µg/l.
Statistics
The level of plasma cfDNA is expressed as median value with 95% confidence interval (95% CI). Survival was calculated from time of inclusion until death of any cause or censoring at end of follow-up. We analyzed survival by the Kaplan-Meier method and comparison between groups by log rank test. The comparison of cfDNA levels between groups was done by the Mann-Whitney U test and comparison of categorical variables between groups by contingency tables and χ2 test or Fischer exact test when appropriate. In Cox proportional hazard model, we computed Hazard Ratios (HR) for mortality and included age, gender, site of primary tumor, WHO Performance Status (PS), cfDNA, CEA and KRAS mutational in archival tissue in the multivariate model.
Results
Baseline cfDNA and comparison with healthy controls
Baseline blood samples for plasma cfDNA measurement were available for 62 patients who all completed at least one HAI treatment. The gender distribution was 61% male and the median age was 61.3 years (range 40.8-74.8 years). Colon cancer was the site of primary for 68%. The baseline clinical characteristics and corresponding plasma cfDNA levels are presented in Table I. The only significant difference in cfDNA levels among baseline characteristics, were patients with a WHO PS of 1 or 2 having a higher level than patients with a WHO PS of 0 (P<0.001). The baseline median level of CEA was 53 ng/l (95% CI 28-97) (n=60).
Table I.
Patient characteristics and corresponding level of plasma cfDNA.
| Characteristic | Number (%) N=62 | cfDNA, ng/ml (95%CI) | P-value |
|---|---|---|---|
| Sex | 0.3 | ||
| Male | 38(61) | 0.91 (0.75-0.99) | |
| Female | 24(39) | 0.97 (0.86-1.33) | |
| Age | 0.4 | ||
| Median | 61.3 | ||
| Range | 40.8-74.8 | ||
| ≤65 | 0.91 (0.76-0.99) | ||
| >65 | 0.98 (0.77-1.44) | ||
| Performance status | <0.001 | ||
| 0 | 54(87) | 0.90 (0.75-0.94) | |
| 1 | 6(10) | 1.97 (1.07-2.74) | |
| 2 | 2(3) | 3.02 (2.51-3.52) | |
| Site of primary | 0.4 | ||
| Colon | 42(68) | 1.08 (0.91-1.68) | |
| Ascending | 11(18) | ||
| Transverse | 1 (1.5) | ||
| Descending | 3 (4.5) | ||
| Sigmoid | 27(44) | ||
| Rectal | 20(32) | 0.96 (0.86-1.28) | |
| Debut of metastases | 0.6 | ||
| Synchronous | 33(53) | 1.16 (0.98-1.84) | |
| Metachronous | 29(47) | 1.50 (0.94-2.05) | |
| Response | 0.02 | ||
| CR+PR | 56(90) | 0.91 (0.76-0.98) | |
| SD+PD | 4(7) | 1.79 (0.99-2.57) | |
| Not RECIST | 2(3) | ||
| KRAS status | 0.21 | ||
| Wt | 36(58) | 0.87 (0.75-0.98) | |
| mut | 26(42) | 1.01 (0.89-1.29) |
cfDNA, cell free DNA; CR, complete remission; PR, partial remission; SD, stable disease; PD, progressive disease; KRAS, Kirsten RAt Sarcoma oncogene; Wt, wild type; mut, mutation.
The median plasma cfDNA level for healthy controls (n=94) was 0.52 ng/µl (95%CI 0.48-0.57) significantly lower than the HAI cohort (P<0.01). The discriminatory power of cfDNA between healthy individuals and patients with CRCLM receiving HAI was high with a ROC curve (Fig. 1) with an AUC value of 0.86.
Figure 1.
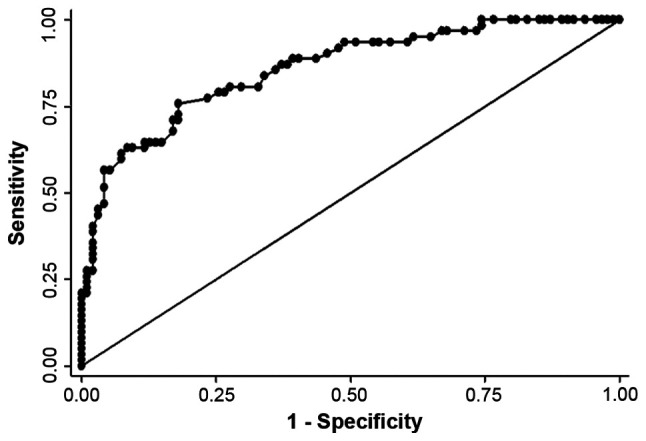
Receiver operating characteristics curve with diagnostic accuracy for cell free DNA in separating healthy individuals from patients with CRCLM receiving intrahepatic chemotherapy with an area under the curve value of 0.86. CRCLM, colorectal cancer liver metastases.
Sequential samples
The median baseline level of plasma cfDNA was 0.92 ng/µl (95%CI 0.84-1.00) (n=62). At the end of HAI treatment blood samples were available for 56 patients with a median plasma cfDNA level of 0.82 ng/µl (95% CI 0.73-0.89) (P=0.06 for comparison with baseline). At the first time of progression, plasma samples available from 32 patients at with a median level of 0.80 ng/µl (95% CI 0.66-0.94) (P=0.01; compared with baseline). The last data point was at 3 years follow-up were only 9 patients contributed with plasma samples with a median cfDNA level of 0.82 µg/µl (95% CI 0.61-0.91) (P=0.5). At 3 years the estimated overall survival was 48% (95% CI 35-59).
During the HAI treatment 20 patients had an increasing level of cfDNA while 32 patients had a decreasing value. The median value of value of change in cfDNA level was a decline of 0.14 ng/µl (95%CI 0.00-0.21).
Clinical correlation of cfDNA
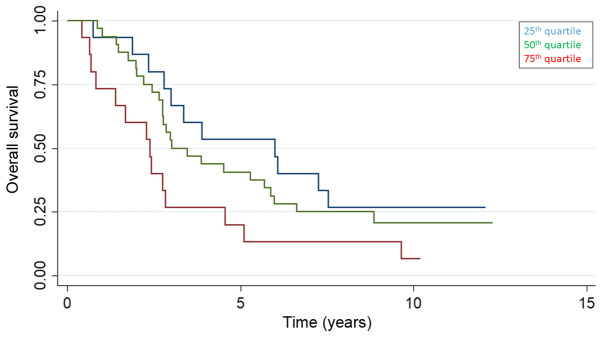
The 25, 50 and 75th quartile of baseline cfDNA were 0.71 (95% CI 0.61-0.81), 0.92 (95% CI 0.84-1.00) and 1.30 ng/µl (1.00-1.67) and the overall survival stratified by baseline cfDNA quartiles is presented in Fig. 2. Patients with a baseline value of cfDNA above the 75th quartile had a median overall survival (OS) of 2.4 years (95% CI 0.7-2.8), compared to 3.9 years (95% CI 2.8-5.9) for patients below the 75th quartile (P=0.02) (Fig. 3). Plasma samples at the end of HAI (n=56) showed a tendency for a longer survival for patients below the 75th quartile with a median survival of 3.5 years (95% CI 2.76-5.69) compared to 2.4 years (95% CI 1.67-4.55) for patients above the 75th quartile (P=0.5).
Figure 2.
Overall survival (%) stratified by the 25, 50 and 75th quartile of baseline cell free DNA.
Figure 3.
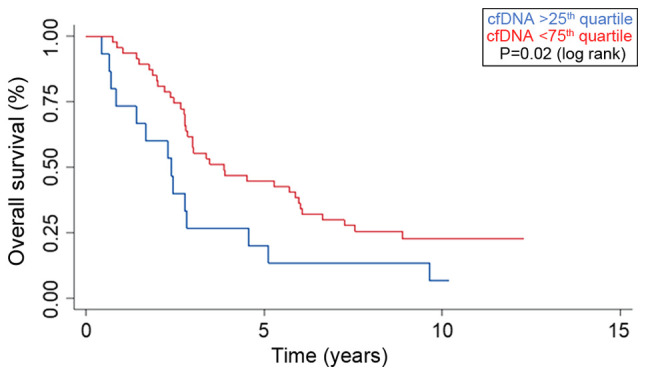
Survival curves displaying the overall survival (%) stratified by the 75th quartile into high (blue) and low (red) baseline level of plasma cell free DNA (P=0.02; log rank). Time from hepatic arterial infusion treatment on x-axis.
Separating the survival of patients by the baseline level of CEA showed no significant differences whether the UNL (P=0.3) or 75th quartile (P=0.18) were applied. Patients who achieved a best objective response of either a complete (CR) or partial response (PR) had a baseline cfDNA concentration of 0.91 ng/µl (95% CI 0.76-0.98) compared to patients who only obtained stable disease (SD) or progression (PD) with a level of 1.79 ng/µl (95% CI 0.99-2.57) (P=0.02). The change in plasma cfDNA during HAI treatment was not correlated to any clinical outcomes in term of survival or response. Selectively looking at patients with the longest survival (above the 75th quartile of survival=7.3 years) (n=13) did not show any trends in cfDNA change.
The diagnostic accuracy of cfDNA is presented in Table II by a contingency table displaying the value of cfDNA above/below the baseline 75q for patients with either response (CR/PR, n=56) versus patient with no response (SD/PR, n=4) (P=0.03 Fisher exact test). The sensitivity, specificity, positive and negative predictive values are 80, 75, 97 and 21%.
Table II.
Diagnostic accuracy of baseline cfDNA (above/below the 75th quartile) in cross tabulation with response (CR/PR) versus not response (SD/PD) (P=0.03; Fisher exact test).
| Response | >75q baseline cfDNA | <75q baseline cfDNA | Total |
|---|---|---|---|
| CR/PR (n=56) | 11 | 45 | 56 |
| SD/PD (n=4) | 3 | 1 | 4 |
| Total | 14 | 46 |
cfDNA, cell free DNA; CR, complete remission; PR, partial remission; SD, stable disease; PD, progressive disease.
Prognostic factors
In both uni- and multivariate model increasing baseline level of cfDNA were associated with increased mortality with HR of 2.39 (95% CI 1.51-3.76, P<0.001) and 1.90 (95% CI 1.07-3.38, P<0.03), respectively. The only other variable associated with a sustained impact on survival in the multivariate model was the mutational status of the KRAS oncogene where patients with a KRAS mutation had a HR for mortality of 2.93 (95% CI 1.66-5.18, P<0.001) and 3.17 (95% CI 1.67-6.03, P<0.001) in the univariate and multivariate analysis, respectively (Table III).
Table III.
Uni and multivariate analysis displaying the HR for mortality.
| Variable | Univariate HR (95% CI) | P-value | Multivariate HR (95%CI) | P-value |
|---|---|---|---|---|
| Sex | ||||
| Male (reference) | ||||
| Female | 0.69 (0.38-1.25) | 0.20 | 0.54 (0.28-1.02) | 0.06 |
| Age | ||||
| Continuous variable | 0.99 (0.96-1.02) | 0.40 | 0.98 (0.94-1.01) | 0.20 |
| Site of primary | ||||
| Colon (reference) | ||||
| Rectum | 0.89 (0.48-1.68) | 0.70 | 0.90 (0.48-1.70) | 0.70 |
| Performance status | ||||
| 0 (reference) | ||||
| 1 | 5.74 (2.24-14.74) | <0.01 | 2.19 (0.74-6.43) | 0.15 |
| 2 | 13.63 (2.84-65.52) | 0.01 | 3.84 (0.51-28.91) | 0.19 |
| cfDNA (baseline) | ||||
| Continous variable | 2.39 (1.51-3.76) | <0.01 | 1.90 (1.07-3.38) | 0.03 |
| CEA (baseline) | ||||
| Continous variable | 1.49 (0.70-3.18) | 0.3 | 1.01 (0.45-2.23) | 0.90 |
| KRAS status | ||||
| Wild-type (reference) | ||||
| Mutated | 2.93 (1.66-5.18) | <0.01 | 3.17 (1.67-6.03) | <0.01 |
cfDNA, cell free DNA; CEA, carcinoembryonic antigen; CI, confidence interval; HR, hazard ratio; KRAS, kirsten RAt sarcoma oncogene.
Discussion
Due to the direct intrahepatic administration of chemotherapy, HAI is an attractive treatment option for patients with CRCLM. Over the past decades, the use of HAI has been subjected to clinical trials both in first line treatment of non-resectable metastases (17) and as an adjuvant treatment subsequent to metastasectomy (18). Alongside chemo-embolization, radiofrequency ablation (RFA), stereotactic body radiotherapy (SBRT) and selective internal radiotherapy (SIRT), HAI constitutes a loco-regional toolbox of treatment option for patients with CRCLM. Seeking to increase the optimal use of these loco-regional modalities, new prognostic and/or predictive circulating biomarkers are needed (19).
Here, we have demonstrated that patients with CRCLM and a high baseline level of cfDNA have an inferior outcome, both in term of objective response and survival. Although the negative prognostic impact of a high cfDNA level is well described in term of survival (11), it is especially interesting to report a possible predictive value of cfDNA, as patients who subsequent developed a partial or complete response had a lower baseline cfDNA compared to patients who only obtained stable disease or progression. For patients with CRCLM not upfront eligible for resection, a predictive marker can have a major impact, as responding patients might be converted to resectability. In contrast, patients not obtaining an objective response might benefit mostly from purely systemic chemotherapy, avoiding the invasive procedure and possible complications from the catheter placement.
We have analyzed the total level of cfDNA using a new fluorescent assay applied directly to the biological sample (plasma). We have refined, optimized and validated this technique since the first reported use in the literature in 2009(14), being an attractive option for cfDNA analysis due to a high degree of laboratory feasibility, no DNA preparation and low financial costs. Considering the total amount of cfDNA, this allows for a detectable biomarker in practically all patients, not restricting the analysis to patients with tumor specific mutations. This is, in turn, limited by the potential pitfalls of this assay being a potential contamination from degenerated lymphocytes and falsely elevated levels due to various medical disorders known to affect the total level of plasma cfDNA (20).
We demonstrate that the total level of cfDNA has a possible prognostic and predictive value, not considering any tumor specific mutations in the blood. As reported from the clinical data of this trial (13), patients with KRAS wild-type status in archival tissue had a significant improved survival, which is maintained in this translational supplement, as increasing level cfDNA and the existence of KRAS mutation are the only two independent variables associated with increased mortality in the multivariate model (Table III). As a limitation, with DFA analysis, we are unable to quantify any RAS mutations in the blood stream, hence to analyze any concordance between archival tissue and the blood stream. Analysis of tumor specific cfDNA alterations could have further applications in translational oncology, both in term of early detection of recurrence or resistance to e.g. anti-EGFR therapy (21).
CEA has been the hallmark of blood borne biomarkers for patients with mCRC for decades, although the routinely use of CEA plays a minor role in current guidelines (7,22,23). For patients with CRCLM, CEA has been widely studied in patients undergoing surgical resection of liver metastases and well established as a prognostic marker for survival (4), yet a predictive value of CEA for any loco-regional treatment has never been established.
In our analysis, cfDNA is a stronger prognostic marker for mortality than CEA, as increasing CEA fails to show independent association with mortality with a HR of 1.01 in multivariate analysis. In contrast, increasing cfDNA levels show sustained prognostic value in both uni- and multivariate model. This is in concordance with a recent comparative study of CEA and cfDNA for patients with mCRC, demonstrating a possible diagnostic superiority of cfDNA compared to CEA (24). The majority of studies on cfDNA and CRC has emphasis on either widespread metastatic disease, in a palliative setting (11) or patients with locally advanced rectal cancer scheduled for chemo-radiation (25,26). Few studies have explored the utility of cfDNA in the clinical setting of CRCLM, where some patients might still be within the range of curability. We have previously, also by DFA quantification, examined the cfDNA levels for 14 patients with CRCLM who received chemo-embolization, reporting a numerical shorter survival for patients with a high baseline level of cfDNA, but unable to display statistical significance with the low sample size (27). The possible efficacy of HAI treatment as a modality cannot be addressed in this single armed study, and the relatively long survival can be a consequence of selection bias.
In conclusion, we have demonstrated an inferior outcome for patients with CRCLM with a high level of cfDNA who undergo intrahepatic infusion with oxaliplatin and systemic capecitabine. Cell free DNA in plasma could hold both prognostic and predictive value for this group of patients emphasizing the need for biobanking biological material for translational analysis.
Acknowledgements
Not applicable.
Funding
This study was supported by the Danish Cancer Society, the Novo Nordisk Foundation and Aase og Ejnar Danielsens Foundation.
Availability of data and materials
All raw data material from this study is available for review upon contact to the corresponding author.
Authors' contributions
AKB, JVS, BVJ, DN, BSS, JSJ and KLGS designed the study, AKB and BSS performed the laboratory analysis, AKB, BSS and KLGS performed the data analysis, AKB drafted the manuscript, AKB, JVS, BVJ, DN, BSS, JSJ and KLGS reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients consented verbal and written to protocol inclusion. The protocol was approved by the regional ethical committee and The Danish Data Protection Agency. The study was conducted according to the principles of the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jamal A. Cancer statistics 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM, McWilliams RR. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27:3677–3683. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: Analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–321. doi: 10.1097/00000658-199909000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boysen AK, Spindler KL, Høyer M, Mortensen FV, Christensen TD, Farkas DK, Ording AG. Metastasis directed therapy for liver and lung metastases from colorectal cancer-a population based study. Int J Cancer. 2018;143:3218–3226. doi: 10.1002/ijc.31626. [DOI] [PubMed] [Google Scholar]
- 6.Ramming KP, Sparks FC, Eilber FR, Holmes EC, Morton DL. Hepatic artery ligation and 5-fluorouracil infusion for metastatic colon carcinoma and primary hepatoma. Am J Surg. 1976;132:236–242. doi: 10.1016/0002-9610(76)90054-4. [DOI] [PubMed] [Google Scholar]
- 7.van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 8.Dzodic R, Gomez-Abuin G, Rougier P, Bonnay M, Ardouin P, Gouyette A, Rixe O, Ducreux M, Munck JN. Pharmacokinetic advantage of intra-arterial hepatic oxaliplatin administration: Comparative results with cisplatin using a rabbit VX2 tumor model. Anticancer Drugs. 2004;15:647–650. doi: 10.1097/01.cad.0000131684.06390.fe. [DOI] [PubMed] [Google Scholar]
- 9.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224ra24) doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 11.Spindler KG, Boysen AK, Pallisgaard N, Johansen JS, Tabernero J, Sørensen MM, Jensen BV, Hansen TF, Sefrioui D, Andersen RF, et al. Cell free DNA in metastatic colorectal cancer: A systematic review and meta analysis. Oncologist. 2017;22:1049–1055. doi: 10.1634/theoncologist.2016-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarkavelis G, Boussios S, Papadaki A, Katsanos KH, Christodoulou DK, Pentheroudakis G. Current and future biomarkers in colorectal cancer. Ann Gastroenterol. 2017;30:613–621. doi: 10.20524/aog.2017.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vittrup BV, Nørgaard H, Bergenfeldt M, Larsen FO, Høgdall E, Johansen JS, Sidenius Johansen JS, Hermann HK, Nielsen DL. Hepatic arterial infusion (HAI) of oxaliplatin with capecitabine in first line treatment of patients (pts) with liver limited metastases from colorectal cancer (LLmCRC) Abstracts Gastrointestinal Tumours, Colorectal. 2018;29: Suppl 8(viii173) [Google Scholar]
- 14.Boysen AK, Sørensen BS, Lefevre AC, Abrantes R, Johansen JS, Jensen BV, Schou JV, Larsen FO, Nielsen D, Taflin H, et al. Methodological development and biological observations of cell free DNA with a simple direct fluorescent assay in colorectal cancer. Clin Chim Acta. 2018;487:107–111. doi: 10.1016/j.cca.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 15.Goldshtein H, Hausmann MJ, Douvdevani A. A rapid direct fluorescent assay for cell-free DNA quantification in biological fluids. Ann ClinBiochem. 2009;46:488–494. doi: 10.1258/acb.2009.009002. [DOI] [PubMed] [Google Scholar]
- 16.Czeiger D, Shaked G, Sebbag G, Vakhrushev A, Flomboym A, Lior Y, Belochitski O, Ariad S, Douvdevani A. Elevated cell-free DNA measured by a simple assay is associated with increased rate of colorectal cancer relapse. Am J Clin Pathol. 2016;145:852–857. doi: 10.1093/ajcp/aqw068. [DOI] [PubMed] [Google Scholar]
- 17.Mocellin S, Pilati P, Lise M, Nitti D. Meta-analysis of hepatic arterial infusion for unresectable liver metastases from colorectal cancer: The end of an era? J Clin Oncol. 2007;25:5649–5654. doi: 10.1200/JCO.2007.12.1764. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Zhong Y, Weixin N, Xinyu Q, Yanhan L, Li R, Jianhua W, Zhiping Y, Jiemin C. Preoperative hepatic and regional arterial chemotherapy in the prevention of liver metastasis after colorectal cancer surgery. Ann Surg. 2007;245:583–590. doi: 10.1097/01.sla.0000250453.34507.d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly CM, Kemeny NE. Liver-directed therapy in metastatic colorectal cancer. Expert Rev Anticancer Ther. 2017;17:745–758. doi: 10.1080/14737140.2017.1345629. [DOI] [PubMed] [Google Scholar]
- 20.Spindler KL, Appelt AL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int J Cancer. 2014;135:2984–2991. doi: 10.1002/ijc.28946. [DOI] [PubMed] [Google Scholar]
- 21.Boussios S, Ozturk MA, Moschetta M, Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, Christodoulou DK, Pavlidis N. The developing story of predictive biomarkers in colorectal cancer. J Pers Med. 2019;9(12) doi: 10.3390/jpm9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim IH, Lee JE, Yang JH, Jeong JW, Ro S, Oh ST, Kim JG, Choi MH, Lee MA. Clinical significance of discordance between carcinoembryonic antigen levels and RECIST in metastatic colorectal cancer. Cancer Res Treat. 2018;50:283–292. doi: 10.4143/crt.2016.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermunen K, Lantto E, Poussa T, Haglund C, Österlund P. Can carcinoembryonic antigen replace computed tomography in response evaluation of metastatic colorectal cancer? Acta Oncol. 2018;57:750–758. doi: 10.1080/0284186X.2018.1431400. [DOI] [PubMed] [Google Scholar]
- 24.Berger AW, Schwerdel D, Welz H, Marienfeld R, Schmidt SA, Kleger A, Ettrich TJ, Seufferlein T. Treatment monitoring in metastatic colorectal cancer patients by quantification and KRAS genotyping of circulating cell-free DNA. PLoS One. 2017;12(e0174308) doi: 10.1371/journal.pone.0174308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boysen AK, Wettergren Y, Sorensen BS, Taflin H, Gustavson B, Spindler KG. Cell-free DNA levels and correlation to stage and outcome following treatment of locally advanced rectal cancer. Tumour Biol. 2017;39(1010428317730976) doi: 10.1177/1010428317730976. [DOI] [PubMed] [Google Scholar]
- 26.Schou JV, Larsen FO, Sørensen BS, Abrantes R, Boysen AK, Johansen JS, Jensen BV, Nielsen DL, Spindler KL. Circulating cell-free DNA as predictor of treatment failure after neoadjuvant chemo-radiotherapy before surgery in patients with locally advanced rectal cancer. Ann Oncol. 2018;29:610–615. doi: 10.1093/annonc/mdx778. [DOI] [PubMed] [Google Scholar]
- 27.Boysen AK, Jensen M, Nielsen DT, Mortensen FV, Sørensen BS, Jensen AR, Spindler KL. Cell-free DNA and chemoembolization in patients with liver metastases from colorectal cancer. Oncol Lett. 2018;16:2654–2660. doi: 10.3892/ol.2018.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw data material from this study is available for review upon contact to the corresponding author.