Abstract
Background
Diffuse large B-cell lymphoma (DLBCL) is a common malignant tumor in the immune system with high mortality. We investigated the functional effects of long non-coding RNA paternally expressed imprinted gene 10 (PEG10) on DLBCL progression.
Material/Methods
Real-time quantitative polymerase chain reaction was used to measure the level of PEG10, kinesin family member 2A (KIF2A) and microRNA-101-3p (miR-101-3p) in DLBCL tissues and cell lines. The relative protein level was detected by western blot analysis. The biological behaviors including cell proliferation, apoptosis, migration, and invasion were determined by MTT assay, flow cytometry analysis, and Transwell assays, respectively. Bioinformatics analysis and dual-luciferase reporter assay were performed to evaluate the interaction among PEG10, miR-101-3p, and KIF2A.
Results
PEG10 and KIF2A level were significantly upregulated, while miR-101-3p was downregulated in DLBCL tissues and cells. PEG10 positively regulated KIF2A level in DLBCL. PEG10, or KIF2A deletion significantly inhibited the proliferative, migratory, and invasive abilities of DLBCL cells and elevated cell apoptosis in DLBCL cells. KIF2A upregulation partially reversed the effects of PEG10 downregulation on cell growth, metastasis, and apoptosis in DLBCL. Moreover, PEG10 negatively regulated miR-101-3p level and miR-101-3p upregulation exerted inhibition effects on the progression of DLBCL. Besides, miR-101-3p was a target of PEG10 and miR-101-3p could directly target KIF2A. PEG10 promoted KIF2A level by sponging miR-101-3p.
Conclusions
Our findings revealed that PEG10 played an oncogenic role in DLBCL progression, which might be a potential target for the treatment of DLBCL.
MeSH Keywords: Cell Proliferation; Lymphoma, B-Cell; MicroRNAs
Background
Diffuse large B-cell lymphoma (DLBCL) is a solid tumor of the immune system with a fast-growing incidence, accounting for 30% to 40% in non-Hodgkin lymphomas [1–3]. Previous studies reported that DLBCL frequently occurs in patients older than 60 and 70 years old [4]. The diagnosis for DLBCL patients is based on the clinical features, including a high degree of proliferation and strong metastasis, which resulted in highly variable treatment outcomes and prognosis for DLBCL patients [5]. The common lesion sites of the solid tumor DLBCL are mainly in the thymus, spleen, lymph nodes, and other lymphoid organs [6]. Genetic alternation, virus infection, and disorders of the immune system exerted crucial effects on the biological behaviors in the initiation and development of DLBCL. Although the diagnosis and treatment methods of DLBCL have achieved rapid development in recent years, there are still about 40% of DLBCL patients at an advanced stage fail due to remission and relapse, leading to the high mortality rate. The diagnosis biomarkers for early DLBCL patients remain lacking. Thus, it is of great importance to find efficient therapeutic targets for DLBCL patients.
Long non-coding RNAs (lncRNAs) with the length >200 nucleotides are a group of non-protein-coding RNAs that act as regulators in the processes of human cancers [7]. LncRNAs are involved in biological processes by interacting with DNA, RNA, and protein and by modulating the transcriptional or post-transcriptional expression level [8,9]. To date, accumulating evidence indicates that aberrantly expressed lncRNAs are closely related to the progression and prognosis of tumors [10]. Multiple research studies reported that dysregulation of lncRNAs was observed in DLBCL [11]. The LncRNA HULC deletion can attenuate cell growth in DLBCL cells by suppressing the level of cyclinD1 [12]. TUG1 has been identified as an oncogene in DLBCL, which could inhibit the degradation of MET and repress DLBCL cell growth and proliferation [13]. A previous study revealed that SNHG16 elevated the progression of DLBCL by boosting cell growth and inhibiting cell apoptosis through targeting miR-497-5p [14]. LncRNA paternally expressed imprinted gene 10 (PEG10) located on the chromosome 7q21 was first reported in 2001 [15]. PEG10 was confirmed to contribute to multiple functions including cell growth, differentiation, and apoptosis [16,17]. Additionally, PEG10 was involved in various malignancies, including DLBCL [18]. However, the molecular mechanism of PEG10 in DLBCL is still largely unknown.
PEG10 has been proven to function as competing endogenous RNA to sponge miRNAs and exert its functional effects. For example, PEG10 directly targeted miR-134 to regulate cell proliferation and metastasis in bladder cancer and affect the proliferative ability and apoptotic rate of HCT-116 cells via sponging miR-491 [19,20]. MicroRNA-101-3p (miR-101-3p) acted as a suppressor in bladder and gastric cancer [21,22]. However, the functional role of miR-101-3p in DLBCL is unclear. In the current research, starBase v2.0 predicted that kinesin family member 2A (KIF2A) contained the putative binding site of miR-101-3p. Thus, we determined PEG10, miR-101-3p, and KIF2A levels in DLBCL tissues, as well as in C1R-neo, SU-DHL-8, and OCI-LY-8 cells. Moreover, we aimed to explore the functional effects of PEG10 on DLBCL. Besides, the interaction among PEG10, miR-101-3p, and KIF2A in DLBCL was investigated. We attempted to find a potential therapeutic approach for DLBCL.
Material and Methods
Tissues samples
The 25 individuals with normal lymph nodes were used as controls and the 25 patients diagnosed with DLBCL participated in this study and signed informed consents. The normal individuals and the DLBCL patients from the Affiliated Shanxi Tumor Hospital of Shanxi Medical University had no prior local or systemic treatment. These experiments were approved by the Human Research Ethics Committee of Affiliated Shanxi Tumor Hospital of Shanxi Medical University.
Cell lines
BLBCL cell lines (SU-DHL-8 and OCI-LY-8) and normal lymphoblast cell line (C1R-neo) were used in this study. C1R-neo and SU-DHL-8 cells were obtained from American Type Culture Collection (Manassas, VA, USA) and OCI-LY-8 cells were bought from Shanghai Zishi Biological Technology Co., Ltd. (Shanghai, China). Streptomycin/penicillin (100 U/mL; Invitrogen, Carlsbad, CA, USA) and 10% fetal bovine serum (FBS; Invitrogen) were added into Roswell Park Memorial Institute 1640 (RPMI-1640) medium (Gibco, Carlsbad, CA, USA), which was used to culture cells at a humidified chamber at 37°C supplemented with 5% CO2.
Cell transfection
SiRNA against PEG10 (si-PEG10) or KIF2A (si-KIF2A) and the negative controls (si-NC), KIF2A overexpression vector (KIF2A) and the control (pcDNA), PEG10 overexpression vector (PEG10) and the blank control (pcDNA) were purchased from Genepharma (Shanghai, China). MiR-101-3p mimic (miR-101-3p) and control (miR-NC), miR-101-3p inhibitor (anti-miR-101-3p) and control inhibitor (anti-miR-NC) were bought from Ribobio (Guangzhou, China). All these vectors were transfected into DLBCL cells using Lipofectamine 2000 Reagent (Invitrogen).
Real-time quantitative polymerase chain reaction (RT-qPCR)
The isolation of total RNA from DLBCL tumor tissues and cells was carried out using TRIzol reagent (Invitrogen). The level of PEG10, miR-101-3p, and KIF2A was analyzed by RT-qPCR analysis using an ABI 7500 Real-time PCR system (Bio-Rad Laboratories Inc., Hercules, CA, USA). The levels of miR-101-3p, PEG10, and KIF2A were normalized by U6 and glyceraldehyde 3-phosphate dehydrogenase (DAPDH), respectively. The specific primers were as follows:
-
PEG10: CATCCTTCCTGTCTTCGC (sense) and
CCCTCTTCCACTCCTTCTTT (antisense),
-
GAPDH: TGGTATCGTGGAAGGACTCA (sense) and
CCAGTAGAGGCAGGGATGAT (antisense),
-
miR-101-3p: GCGCGCATACAGTACTGTGATA (sense) and
CGGCCCAGTGTTCAGACTAC (antisense),
-
KIF2A: GCCTTTGATGACTCAGCTCC (sense) and
TTCCTGAAAAGTCACCACCC (antisense),
-
U6: TCCGATCGTGAAGCGTTC (sense) and
GTGCAGGGTCCGAGGT (antisense).
The 2−ΔΔCt method was used to calculate the level of PEG10, miR-101-3p,and KIF2A.
Western blot analysis
Total protein was extracted using RIPA lysis buffer (Beyotime, Shanghai, China). The isolated total protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The primary antibodies including anti-KIF2A (1: 500, ab197988, Abcam, Cambridge, UK), anti-Bax (1: 1000, ab32503, Abcam), anti-caspase-3 (1: 500, ab4051, Abcam), anti-Bcl-2 (1: 1000, ab59348, Abcam), anti-cyclinD1 (1: 200, ab16663, Abcam), and GAPDH (1: 1000, ab8245, Abcam) were used in this study. Secondary antibodies included goat-anti-rabbit IgG H&L (1: 2000, ab6721, Abcam) and goat anti-mouse IgG H&L (1: 2000, ab205719, Abcam). Finally, the protein signals were measured by the enhanced chemiluminescence kit.
Cell proliferation assay
DLBCL cells were transfected for 48 hours. Then, cells were seeded in 96-well plates and cultured at the humidified chamber for 24, 48, and 72 hours. Then 20 μL MTT reagent (5 mg/mL) was added into the medium for 4 hours. Subsequently, the blue crystal in cells was dissolved by dimethyl sulfoxide (150 μL per well). Finally, cell viability was measured using a microplate reader at 490 nm.
Cell apoptosis assay
After transfection, the binding buffer was used to resuspend the DLBCL cells (1 × 106 cells/mL). Then, SU-DHL-8 and OCI-LY-8 cells were double stained with 5 μL Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (BestBio, Shanghai, China). Cell apoptosis was measured by a FACScan flow cytometer (Becton Dickinson, Mountain View, CA, USA).
Transwell assay
For examination of the migrated and invasive cells, SU-DHL-8 and OCI-LY-8 cells mixed with the serum-free medium were seeded on the upper chamber (Corning Incorporated, Big Flats, NY, USA) pre-coated with Matrigel for the invasion assay, while the chambers were non-coated with Matrigel in the Transwell migration assay. DMEM with 10% FBS was provided for the lower chamber, regarding as a chemoattractant. The not-migrated or not-invaded cells on the top surface were wiped off using a cotton swab, whereas the migrated or invaded cells on the lower chamber were fixed, stained, and then counted using a microscope.
Dual-luciferase reporter assay
The 3′UTR fragment of KIF2A (KIF2A-3′UTR-WT) and the wide-type PEG10 sequence (WT-PEG10) containing the predictive binding sites of miR-101-3p were inserted into pGL3 promoter vectors (Invitrogen). The mutant sequences of PEG10 and KIF2A were used to establish PEG10-mutant (MUT-PEG10) and KIF2A-mutant (KIF2A-3′UTR-MUT) reporter vectors. Then, SU-DHL-8 and OCI-LY-8 cells were co-transfected with miR-NC or miR-101-3p mimics and MUT-PEG10, WT-PEG10, KIF2A-3′UTR-WT or KIF2A-3′UTR-MUT. Finally, the dual-luciferase reporter assay system (Promega, Madison, WI, USA) was carried out to determine the luciferase activity in SU-DHL-8 and OCI-LY-8 cells.
Statistical analysis
The data in the present research were displayed as the means±the standard deviations (SD). Student’s t-test for comparison between 2 groups and one-way analysis of variance (ANOVA) for differences among more than 2 groups followed by Turkey’s test were used. The interaction between variables was analyzed by Pearson correlation analysis. The value of P<0.05 means a statistically significant difference.
Results
PEG10 and KIF2A were upregulated in DLBCL tissues and cells
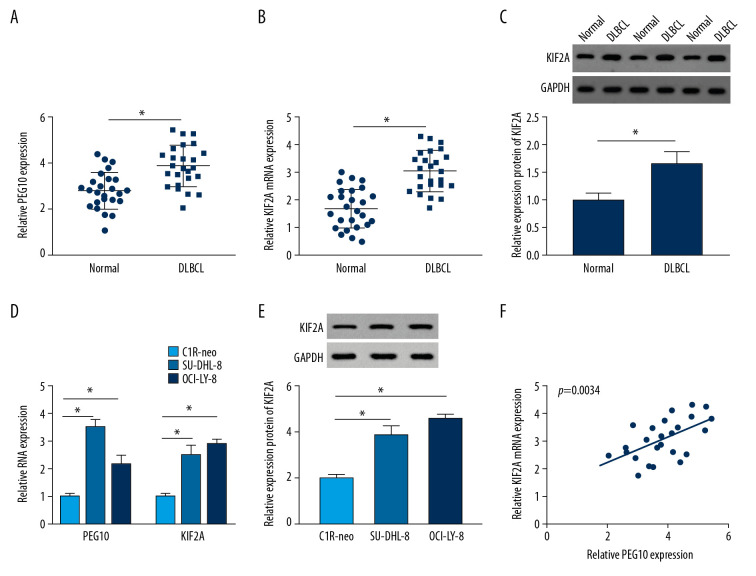
RT-qPCR indicated that the level of PEG10 and KIF2A was boosted in DLBCL tissues (Figure 1A, 1B). As shown in Figure 1C, KIF2A was highly expressed in DLBCL tissues relative to that in normal tissues. Moreover, we also performed RT-qPCR and western blot assay to determine the level of PEG10 and KIF2A in C1R-neo, SU-DHL-8, and OCI-LY-8 cells. PEG10 and KIF2A levels were significantly increased in SU-DHL-8 and OCI-LY-8 cells compared with C1R-neo cells (Figure 1D). Similarly, the protein level of KIF2A was markedly enhanced in DLBCL cells (Figure 1E). Besides, we found that there was a positive relationship between PEG10 level and KIF2A level (Figure 1F). Taken together, these data indicated that PEG10 and KIF2A played vital roles in DLBCL.
Figure 1.
PEG10 and KIF2A are upregulated in DLBCL tissues and cells. (A, B) Detection of PEG10 and KIF2A level by RT-qPCR. (C) The protein level of KIF2A was measured by western blot. (D) RT-qPCR was performed to examine the level of PEG10 and KIF2A in C1R-neo, SU-DHL-8, and OCI-LY-8 cells. (E) The protein level of KIF2A in C1R-neo, SU-DHL-8, and OCI-LY-8 cells was measured by western blot. (F) Pearson correlation analysis revealed a positive relationship between PEG10 and KIF2A (R2=0.3173, P=0.0034). * P<0.05. PEG10 – paternally expressed imprinted gene 10; KIF2A – kinesin family member 2A; DLBCL – diffuse large B-cell lymphoma; RT-qPCR – real-time quantitative polymerase chain reaction.
PEG10 deletion inhibited cell growth and metastasis and enhanced cell apoptosis in DLBCL
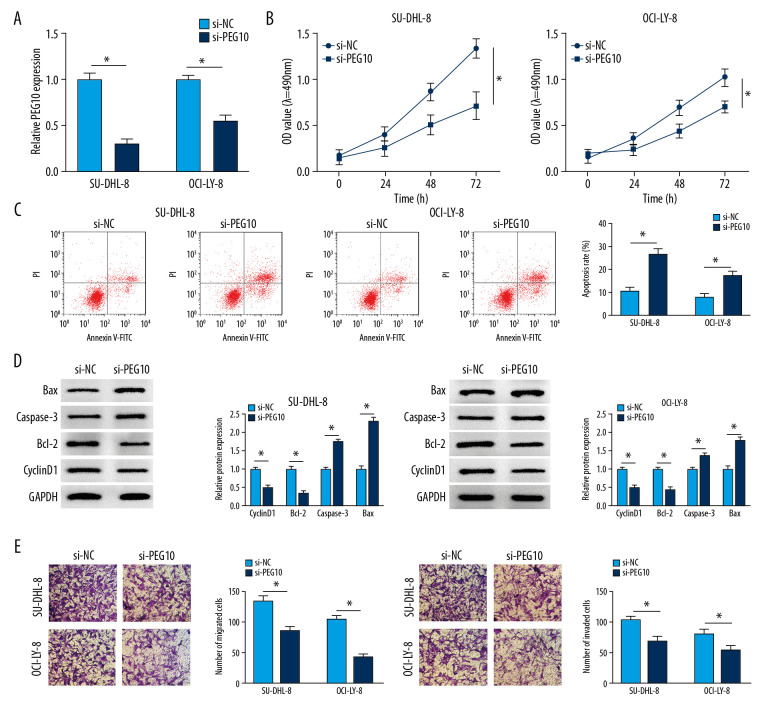
We explored the functional effects of PEG10 on DLBCL. SU-DHL-8, and OCI-LY-8 cells were transfected with si-NC or si-PEG10. The data showed that PEG10 deletion significantly suppressed the level of PEG10 in both SU-DHL-8 and OCI-LY-8 cells (Figure 2A). Knockdown of PEG10 markedly inhibited cell proliferation in both SU-DHL-8 and OCI-LY-8 cells (Figure 2B). Flow cytometry analysis indicated that cell apoptosis was significantly enhanced by downregulating PEG10 in DLBCL cells (Figure 2C). As described in Figure 2D, cyclinD1 and Bcl-2 were decreased, while the level of caspase-3 and Bax was significantly increased in both SU-DHL-8 and OCI-LY-8 cells transfected with si-PEG10. The results suggested that knockdown of PEG10 could significantly suppress cell metastasis in DLBCL cells (Figure 2E). All these results demonstrated that PEG10 promoted the progression of DLBCL.
Figure 2.
PEG10 deletion inhibits cell growth, metastasis and enhances cell apoptosis in DLBCL. (A) The knockdown efficiency of PEG10 in DLBCL cells. (B, C) Detection of cell proliferation and apoptosis in SU-DHL-8 and OCI-LY-8 cells using MTT assay and low cytometry analysis, respectively. (D) The proliferation-related protein cyclinD1 and the apoptosis-related proteins Bcl-2, caspase-3 and Bax level in DLBCL cells were detected by western blot. (E) Transwell assays were used to evaluate the migratory and invasive abilities of DLBCL cells. * P<0.05. PEG10 – paternally expressed imprinted gene 10; DLBCL – diffuse large B-cell lymphoma.
KIF2A deletion suppressed cell proliferation, migration, invasion and elevated cell apoptosis in DLBCL
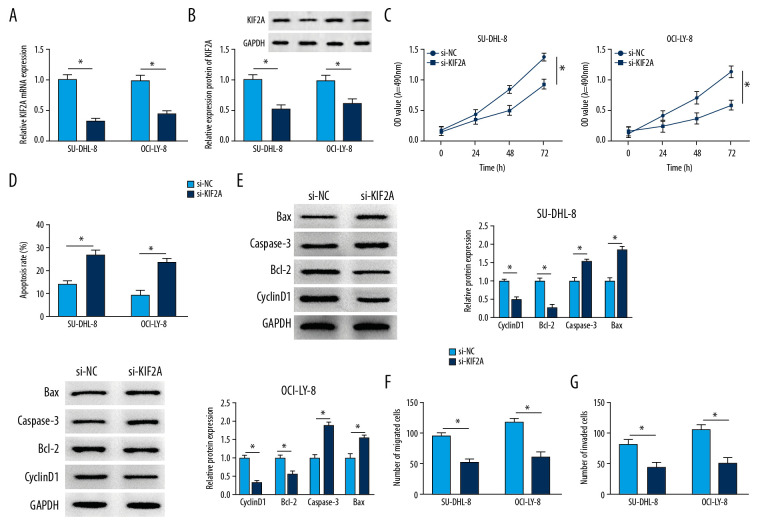
Firstly, we measured the knockdown efficiency of si-KIF2A in DLBCL. The data indicated that the mRNA level of KIF2A was markedly alleviated in SU-DHL-8 and OCI-LY-8 cells after transfection of si-KIF2A (Figure 3A). Similarly, we discovered that knockdown of KIF2A could repress the protein level of KIF2A (Figure 3B). MTT assay indicated that KIF2A deletion significantly inhibited cell proliferation in both SU-DHL-8 and OCI-LY-8 cells (Figure 3C). Cell apoptosis was significantly promoted by downregulating KIF2A in DLBCL cells (Figure 3D). Moreover, the proliferation and apoptosis related proteins were measured by western blot analysis. The apoptosis-related protein Bcl-2 and the proliferation-related protein cyclinD1 were repressed by KIF2A knockdown, whereas caspase-3 and Bax were highly expressed in DLBCL cells transfected with si-KIF2A (Figure 3E). Besides, we observed that the migratory and invasive abilities were significantly attenuated in SU-DHL-8 and OCI-LY-8 cells transfected with si-KIF2A (Figure 3F, 3G). All these data suggested that KIF2A deletion suppressed the progression of DLBCL.
Figure 3.
KIF2A deletion suppresses cell growth, metastasis and elevates cell apoptosis in DLBCL. (A) KIF2A level was detected by RT-qPCR. (B) The protein level of KIF2A was measured by western blot. (C) MTT assay for cell proliferation. (D) Flow cytometry for cell apoptosis. (E) Western blot analysis for the determination of cyclinD1, Bcl-2, caspase-3 and Bax level. (F, G) Transwell assays for cell migration and invasion. * P<0.05. KIF2A – kinesin family member 2A; DLBCL – diffuse large B-cell lymphoma; RT-qPCR – real-time quantitative polymerase chain reaction.
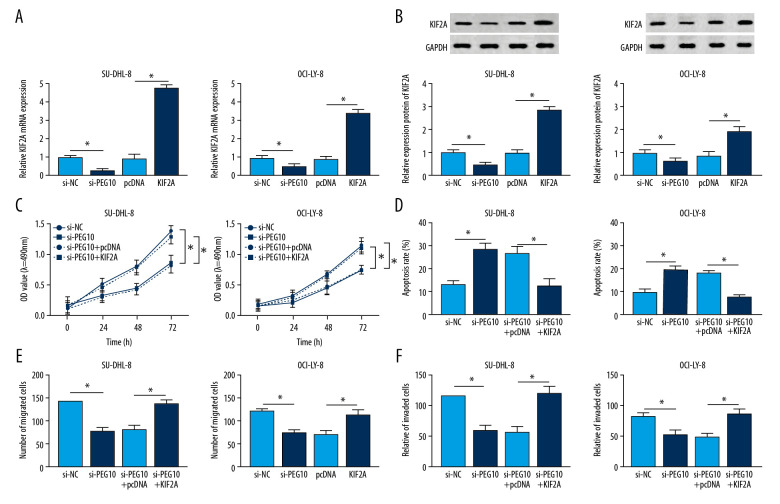
KIF2A overexpression reversed the effects of PEG10 knockdown on DLBCL progression
Using RT-qPCR, we confirmed that PEG10 deletion decreased the level of KIF2A, while KIF2A was highly expressed in both SU-DHL-8 and OCI-LY-8 cells transfected with KIF2A overexpression vector (Figure 4A). Similarly, the protein level of KIF2A was suppressed by PEG10 downregulation and enhanced by KIF2A overexpression in DLBCL cells (Figure 4B). KIF2A overexpression reversed the inhibitory effect of PEG10 deletion on cell proliferation in both SU-DHL-8 and OCI-LY-8 cells (Figure 4C). Our data also noted that PEG10 deletion induced cell apoptosis was markedly attenuated by overexpression of KIF2A (Figure 4D). In Transwell assay, we discovered that the inhibitory effects of PEG10 deletion on cell migration and invasion were reversed by upregulating KIF2A (Figure 4E, 4F). Taken together, upregulation of KIF2A could reverse the effects of PEG10 knockdown on cell growth, apoptosis and metastasis in DLBCL.
Figure 4.
KIF2A overexpression reverses the effects of PEG10 knockdown on the progression of DLBCL. (A, B) The mRNA and protein level of KIF2A in SU-DHL-8 and OCI-LY-8 cells. (C, D) Measurement of cell proliferation and apoptosis by MTT and flow cytometry, respectively. (E, F) Transwell assays used for the measurement of the migratory and invasive abilities of SU-DHL-8 and OCI-LY-8 cells. * P<0.05. KIF2A – kinesin family member 2A; DLBCL,– diffuse large B-cell lymphoma; RT-qPCR – real-time quantitative polymerase chain reaction.
MiR-101-3p overexpression inhibited cell growth, metastasis and enhanced cell apoptosis in DLBCL
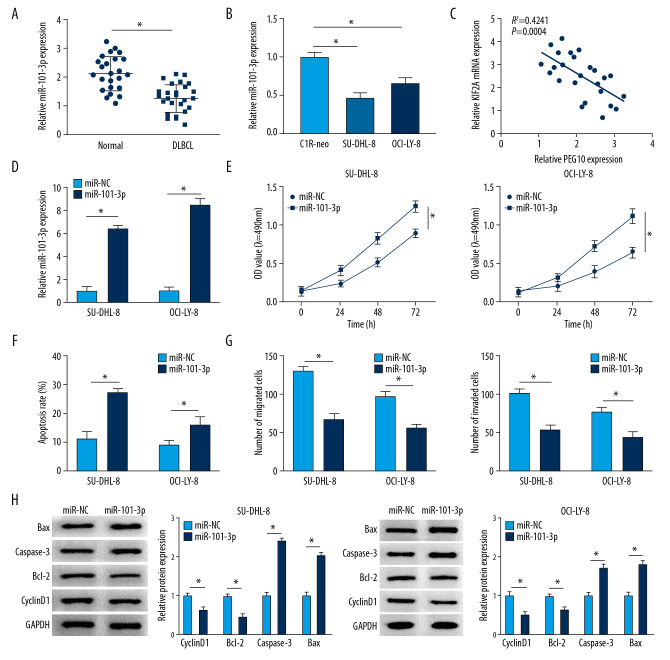
As shown in Figure 5A and 5B, miR-101-3p level was significantly repressed in DLBCL tissues and cells compared with the level in normal tissues and cells. Moreover, we discovered a negative relationship between PEG10 level and miR-101-3p level (Figure 5C). The level of miR-101-3p was boosted in DLBCL cells transfected with miR-101-3p mimics (Figure 5D). MiR-101-3p overexpression significantly inhibited the proliferative ability of SU-DHL-8 and OCI-LY-8 cells (Figure 5E). However, the apoptosis rate of DLBCL cells was markedly elevated by upregulating miR-101-3p (Figure 5F). Increased level of miR-101-3p suppressed cell migration and invasion in both SU-DHL-8 and OCI-LY-8 cells (Figure 5G). We also found that miR-101-3p upregulation decreased the level of cyclinD1 and Bcl-2 and boosted the level of Bax and caspase-3 in DLBCL cells (Figure 5H). Overall, miR-101-3p might play a suppressor role in DLBCL.
Figure 5.
MiR-101-3p overexpression inhibits cell growth, metastasis and improves cell apoptosis in DLBCL. (A, B) MiR-101-3p level in DLBCL tissues and cells. (C) A negative relationship between PEG10 level and miR-101-3p level (R2=0.4241, P=0.0004). (D) The transfection efficiency of miR-101-3p in DLBCL cells. (E, F) Determination of cell growth and apoptosis in DLBCL cells. (G) Cell migration and invasion of DLBCL cells were measured by Transwell assays. (H) Western blot was used to detect the level of cyclinD1, Bcl-2, caspase-3, and Bax in SU-DHL-8 and OCI-LY-8 cells using western blot. * P<0.05. DLBCL – diffuse large B-cell lymphoma; PEG10 – paternally expressed imprinted gene 10.
PEG10 regulated KIF2A level by sponging miR-101-3p
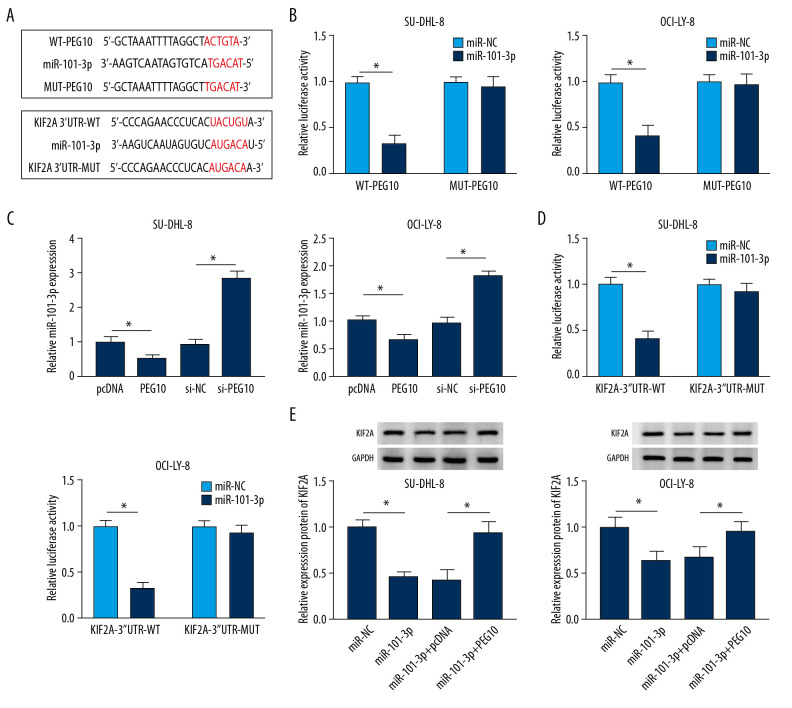
MiRcode predicted that there were potential binding sites between PEG10 and miR-101-3p, while starBase v2.0 showed that KIF2A might be a target of miR-101-3p (Figure 6A). The results noted that the luciferase activity was significantly decreased in DLBCL cells co-transfected with miR-101-3p mimic and WT-PEG10 (Figure 6B). Moreover, miR-101-3p was inversely regulated by PEG10 in both SU-DHL-8 and OCI-LY-8 cells (Figure 6C). We also discovered that miR-101-3p mimic repressed the luciferase activity in DLBCL cells transfected with KIF2A-3′UTR-WT. However, for cells transfected with KIF2A-3′UTR-MUT, the luciferase activity was not changed in the miR-101-3p group in comparison to the control group (Figure 6D). MiR-101-3p overexpression inhibited KIF2A protein level, while PEG10 upregulation reversed the inhibitory effect of miR-101-3p overexpression on KIF2A level in both SU-DHL-8 and OCI-LY-8 cells (Figure 6E). All these data demonstrated that PEG10 regulated the protein level of KIF2A by targeting miR-101-3p in DLBCL.
Figure 6.
PEG10 regulates KIF2A level by sponging miR-101-3p. (A) MiRcode showed the putative binding sites between PEG10 and miR-101-3p, while starBase2.0 predicted that the KIF2A mRNA 3′UTR sequence contained the binding sites of miR-101-3p. (B) The luciferase activity in DLBCL cells co-transfected with miR-NC, miR-101-3p, WT-PEG10, and MUT-PEG10 was detected by dual-luciferase reporter assay. (C) RT-qPCR was used to examine miR-101-3p level in DLBCL cells transfected with pcDNA, PEG10, si-NC, and si-PEG10. (D) Detection of the luciferase activity in DLBCL cells co-transfected with miR-NC, miR-101-3p, KIF2A-3′UTR-WT, and KIF2A-3′UTR-MUT by dual-luciferase reporter assay. (E) The protein KIF2A level was detected by western blot analysis in DLBCL cells transfected with miR-NC, miR-101-3p, miR-101-3p+pcDNA or miR-101-3p+PEG10. * P<0.05. PEG10 – paternally expressed imprinted gene 10; KIF2A – kinesin family member 2A; DLBCL – diffuse large B-cell lymphoma; RT-qPCR – real-time quantitative polymerase chain reaction.
PEG10 regulated cell proliferation and apoptosis via miR-101-3p/KIF2A axis in DLBCL
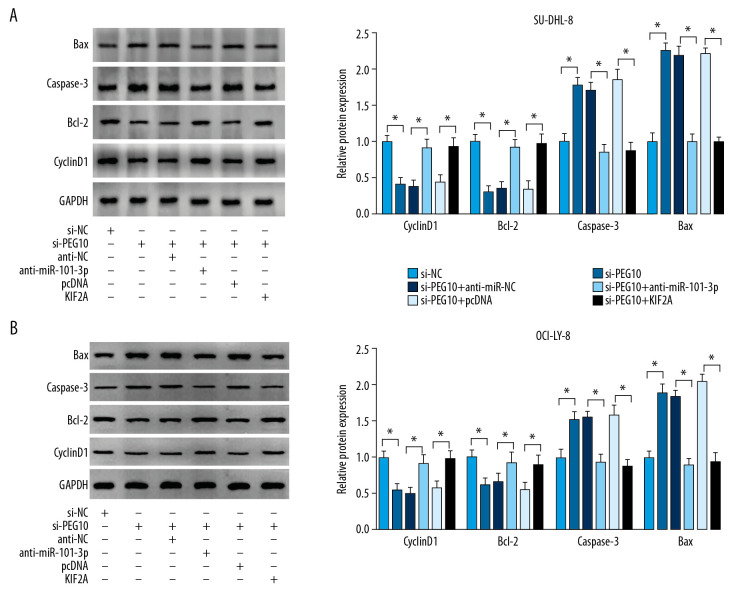
PEG10 knockdown significantly decreased cyclinD1 and Bcl-2 level and facilitated caspase-3 and Bax level in DLBCL cells, which were eliminated after the transfection of miR-101-3p inhibitor or overexpression of KIF2A (Figure 7A, 7B). These data suggested that PEG10 regulated KIF2A to further affect the levels of proliferation and apoptosis related proteins, which played crucial roles in DLBCL cell growth and apoptosis.
Figure 7.
PEG10 regulates cell growth and apoptosis via miR-101-3p/KIF2A axis in DLBCL. (A, B) cyclinD1, Bcl-2, caspase-3 and Bax level in SU-DHL-8 and OCI-LY-8 cells were detected by western blot. * P<0.05. PEG10 – paternally expressed imprinted gene 10; DLBCL – diffuse large B-cell lymphoma.
Discussion
Previous researches demonstrated that lncRNAs participated in DLBCL development [12,23]. Moreover, PEG10 was demonstrated to play a vital role in various cancers. For instance, PEG10 level was markedly upregulated in bladder cancer and pancreatic cancer and enhanced the abilities of cell growth, as well as cell metastasis in bladder cancer cells and pancreatic cancer cells in vitro, which might be a prognosis biomarker [24,25]. The similar phenomenon was discovered in breast cancer, the proliferative ability of breast cancer cells was significantly enhanced by PEG10 [26]. Zang et al. indicated that PEG10 made a contribution to cell invasion and proliferation in esophageal cancer [27]. However, the level of PEG10 in DLBCL and its functional role remain largely uninvestigated. In this research, we discovered the high level of PEG10 in DLBCL tumor tissues. Similarly, upregulated level of PEG10 was discovered in DLBCL cells in comparison to normal B lymphocytes. Moreover, PEG10 deletion suppressed cell proliferation and promoted the apoptotic rate in DLBCL cells. Overall, knockdown of PEG10 exerted a suppressive role in the biological processes of DLBCL. Subsequently, we explored the underlying mechanism of PEG10 in the progression of DLBCL.
MicroRNAs (miRNAs) could regulate the translation of mRNAs through binding to mRNAs 3′-UTR [28]. Accumulating evidence demonstrated that miRNAs dysregulation were closely correlated with various cancers initiation and development [29]. It has been reported that the prognosis of DLBCL patients might be associated with the specific miRNAs [30]. For example, miR-155 might be a potential prognosis biomarker in DLBCL [31]. He et al. reported that miR-195 upregulation could alleviate the immune escape of DLBCL and regulate the release of interferon gamma (IFN-γ), interleukin-10 (IL-10), tumor necrosis factor alpha (TNF-α) in PD-1T cells [32]. Besides, the aberrant levels of miR-22, miR-23a, and miR-4638-5p were also observed in DLBCL [33–35]. We observed that miR-101-3p was upregulated in DLBCL tumor tissues and cells. Moreover, we discovered a negative relationship between miR-101-3p level and PEG10 level. MiR-101-3p acted the inhibitory effects on cell proliferation and metastasis and increased the apoptotic rate of SU-DHL-8 and OCI-LY-8 cells. Besides, miR-101-3p was testified as a target of PEG10 in DLBCL.
KIF2A is a member of kinesin superfamily proteins with the oncogenic function involving in human cancers [36]. The regulatory role of KIF2A in carcinogenesis of various tumors has received extensive attention. Zhao et al. reported that KIF2A deletion exerted cell growth and invasion repression effects in MKN-45 and SGC-7901 cells through repressing the level of MTI-MMP [37]. Furthermore, KIF2A was increased in lung adenocarcinoma and the high level of KIF2A played a promotion role in cell growth and metastasis of lung adenocarcinoma cells, which might serve as an effective prognosis biomarker for lung adenocarcinoma [38]. Zhang et al. noted that enhanced level KIF2A was discovered in DLBCL that related to the prognosis of DLBCL [39]. Our data suggested that KIF2A was upregulated in DLBCL. The effects of KIF2A knockdown on DLBCL cells were similar to PEG10 deletion. KIF2A deletion inhibited cell growth and metastasis and induced cell apoptosis in SU-DHL-8 and OCI-LY-8 cells. Besides, we observed that PEG10 could regulate KIF2A level by sponging miR-101-3p in DLBCL cells. PEG10 deletion could suppress cell growth, metastasis, and promoted cell apoptosis through miR-101-3p/KIF2A axis in DLBCL.
Conclusions
In conclusion, we discovered that PEG10 and KIF2A level were markedly increased, while miR-101-3p was decreased in DLBCL tumor tissues and cells. Functionally, PEG10 deletion significantly suppressed cell proliferation and metastasis, and enhanced cell apoptosis in DLBCL cells. Moreover, our findings demonstrated that PEG10 directly targeted miR-101-3p and that miR-101-3p was a target of KIF2A. Besides, we found that PEG10 regulated cell proliferation, apoptosis, and metastasis through the miR-101-3p/KIF2A axis, providing a new therapeutic target for the diagnosis and treatment of DLBCL.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Ren W, Ye X, Su H, et al. Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood. 2018;131(24):2670–81. doi: 10.1182/blood-2017-11-817601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oarbeascoa G, Rodriguez-Macias G, Guzman-de-Villoria JA, et al. Methotrexate-induced subacute neurotoxicity surrounding an ommaya reservoir in a patient with lymphoma. Am J Case Rep. 2019;20:1002–5. doi: 10.12659/AJCR.915632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokushima M, Katsuki NE, Tago M, et al. Intravascular large B-cell lymphoma presenting with hypoxemia without any abnormalities on standard imaging studies. 2019;20:1199–204. doi: 10.12659/AJCR.916877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaito S, Muto H, Miyawaki S, et al. Timely diagnosis and successful treatment of intravascular large B-cell lymphoma in an elderly patient with poor performance status. Am J Case Rep. 2019;20:780–84. doi: 10.12659/AJCR.915189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 6.Dogan A, Bagdi E, Munson P, et al. CD10 and BCL-6 expression in paraffin sections of normal lymphoid tissue and B-cell lymphomas. Am J Surg Pathol. 2000;24(6):846–52. doi: 10.1097/00000478-200006000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan H, Bu P. Non-coding RNAs in cancer stem cells. Cancer Lett. 2018;421:121–26. doi: 10.1016/j.canlet.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham SW, Gruhl F, Mattick JS, et al. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–25. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dousti F, Shahrisa A, Ansari H, et al. Long non-coding RNAs expression levels in diffuse large B-cell lymphoma: An in silico analysis. Pathol Res Pract. 2018;214(9):1462–66. doi: 10.1016/j.prp.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Peng W, Wu J, Feng J. Long noncoding RNA HULC predicts poor clinical outcome and represents pro-oncogenic activity in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;79:188–93. doi: 10.1016/j.biopha.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H, Yan Z, Wang X, et al. Downregulation of long non-coding RNA TUG1 suppresses tumor growth by promoting ubiquitination of MET in diffuse large B-cell lymphoma. Mol Cell Biochem. 2019;461(1–2):47–56. doi: 10.1007/s11010-019-03588-7. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Q, Li Y, Guo Y, et al. Long non-coding RNA SNHG16 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med. 2019;23(11):7395–405. doi: 10.1111/jcmm.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono R, Kobayashi S, Wagatsuma H, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73(2):232–37. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- 16.Li CM, Margolin AA, Salas M, et al. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006;66(2):665–72. doi: 10.1158/0008-5472.CAN-05-1553. [DOI] [PubMed] [Google Scholar]
- 17.Hishida T, Naito K, Osada S, et al. Peg10, an imprinted gene, plays a crucial role in adipocyte differentiation. FEBS Lett. 2007;581(22):4272–78. doi: 10.1016/j.febslet.2007.07.074. [DOI] [PubMed] [Google Scholar]
- 18.Peng W, Fan H, Wu G, et al. Upregulation of long noncoding RNA PEG10 associates with poor prognosis in diffuse large B cell lymphoma with facilitating tumorigenicity. Clin Exp Med. 2016;16(2):177–82. doi: 10.1007/s10238-015-0350-9. [DOI] [PubMed] [Google Scholar]
- 19.Jiang F, Qi W, Wang Y, et al. LncRNA PEG10 promotes cell survival, invasion and migration by sponging miR-134 in human bladder cancer. Biomed Pharmacother. 2019;114:108814. doi: 10.1016/j.biopha.2019.108814. [DOI] [PubMed] [Google Scholar]
- 20.Li B, Shi C, Li B, et al. The effects of Curcumin on HCT-116 cells proliferation and apoptosis via the miR-491/PEG10 pathway. J Cell Biochem. 2018;119(4):3091–98. doi: 10.1002/jcb.26449. [DOI] [PubMed] [Google Scholar]
- 21.Cao C, Xu Y, Du K, et al. LINC01303 functions as a competing endogenous RNA to regulate EZH2 expression by sponging miR-101-3p in gastric cancer. J Cell Mol Med. 2019;23(11):7342–48. doi: 10.1111/jcmm.14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D, Li Y, Luo G, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–91. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M, Zhao H, Xu W, et al. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16(1):16. doi: 10.1186/s12943-017-0580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng YP, Zhu Y, Yin LD, et al. PEG10 overexpression induced by E2F-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J Exp Clin Cancer Res. 2017;36(1):30. doi: 10.1186/s13046-017-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang T, Wang Y, Wang Y, et al. Long noncoding RNA PEG10 facilitates bladder cancer cells proliferation, migration, and invasion via repressing microRNA-29b. J Cell Physiol. 2019;234(11):19740–49. doi: 10.1002/jcp.28574. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Xiao R, Tembo K, et al. PEG10 promotes human breast cancer cell proliferation, migration and invasion. Int J Oncol. 2016;48(5):1933–42. doi: 10.3892/ijo.2016.3406. [DOI] [PubMed] [Google Scholar]
- 27.Zang W, Wang T, Huang J, et al. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther. 2015;22(3):138–44. doi: 10.1038/cgt.2014.77. [DOI] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.D’Angelo B, Benedetti E, Cimini A, et al. MicroRNAs: A puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36(11):5571–75. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 30.Roehle A, Hoefig KP, Repsilber D, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142(5):732–44. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 31.Ahmadvand M, Eskandari M, Pashaiefar H, et al. Over expression of circulating miR-155 predicts prognosis in diffuse large B-cell lymphoma. Leuk Res. 2018;70:45–48. doi: 10.1016/j.leukres.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 32.He B, Yan F, Wu C. Overexpressed miR-195 attenuated immune escape of diffuse large B-cell lymphoma by targeting PD-L1. Biomed Pharmacother. 2018;98:95–101. doi: 10.1016/j.biopha.2017.11.146. [DOI] [PubMed] [Google Scholar]
- 33.Marchesi F, Regazzo G, Palombi F, et al. Serum miR-22 as potential non-invasive predictor of poor clinical outcome in newly diagnosed, uniformly treated patients with diffuse large B-cell lymphoma: An explorative pilot study. J Exp Clin Cancer Res. 2018;37(1):95. doi: 10.1186/s13046-018-0768-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Wang L, Cheng L. Aberrant ERG expression associates with downregulation of miR-4638-5p and selected genomic alterations in a subset of diffuse large B-cell lymphoma. Mol Carcinog. 2019;58(10):1846–54. doi: 10.1002/mc.23074. [DOI] [PubMed] [Google Scholar]
- 35.Xu M, Xu T. Expression and clinical significance of miR-23a and MTSS1 in diffuse large B-cell lymphoma. Oncol Lett. 2018;16(1):371–77. doi: 10.3892/ol.2018.8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe T, Kakeno M, Matsui T, et al. TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J Cell Biol. 2015;210(5):737–51. doi: 10.1083/jcb.201412075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao P, Lan F, Zhang H, et al. Down-regulation of KIF2A inhibits gastric cancer cell invasion via suppressing MT1-MMP. Clin Exp Pharmacol Physiol. 2018;45(10):1010–18. doi: 10.1111/1440-1681.12974. [DOI] [PubMed] [Google Scholar]
- 38.Xie T, Li X, Ye F, et al. High KIF2A expression promotes proliferation, migration and predicts poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2018;497(1):65–72. doi: 10.1016/j.bbrc.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, You X, Liu H, et al. High KIF2A expression predicts unfavorable prognosis in diffuse large B cell lymphoma. Ann Hematol. 2017;96(9):1485–91. doi: 10.1007/s00277-017-3047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]